Abstract
A large body of data have accumulated that characterize the gene regulatory network of stem cells. Yet, a comprehensive and integrative understanding of this complex network is lacking. Network reverse engineering methods that use transcriptome data to derive these networks may help to uncover the topology in an unbiased way. Many methods exist that use co-expression to reconstruct networks. However, it remains unclear how these methods perform in the context of stem cell differentiation, as most systematic assessments have been made for regulatory networks of unicellular organisms. Here, we report a systematic benchmark of different reverse engineering methods against functional data. We show that network pruning is critical for reconstruction performance. We also find that performance is similar for algorithms that use different co-expression measures, i.e. mutual information or correlation. In addition, different methods yield very different network topologies, highlighting the challenge of interpreting these resulting networks as a whole.
This article is part of the theme issue ‘Designer human tissue: coming to a lab near you’.
Keywords: network reconstruction, transcriptional network, network inference
1. Background
Despite huge efforts to map out gene regulatory interactions between genes in many different cell types using ChIP-seq and similar methods, a comprehensive understanding of the gene regulatory network governing cell state transitions is still lacking [1]. In addition to functional annotations of the genome, a large body of gene expression data has been collected during the past decades. We therefore decided to investigate if such data could be leveraged to derive the topology of the gene regulatory network governing the complex transition from a stem cell to a differentiated cell. A number of approaches that address the problem of inferring gene regulatory networks from high-throughput data have been developed in the past 20 years. Most network reconstruction methods that use expression data make the assumption that the rate of change of a gene's mRNA concentration is a function of the mRNA concentration of all other genes [2]. Such an approach assumes that the protein concentration of each gene is determined purely by its mRNA concentration, and it ignores the influence of post-translational modifications.
Many network reconstruction approaches have been systematically benchmarked at the DREAM project [3]. However, these benchmarks have mainly been done on data from microorganisms such as E. coli and Saccharomyces cerevisiae. Also using data from Escherichia coli, Allen et al. [4] studied network reconstruction using data from 500 microarrays, and examined the influence of the number of samples on reconstruction quality. As the complexity of mammalian regulatory networks is much larger than that of yeast or bacteria, the results of those benchmarks may not be transferable to reconstructing more complex networks. Only very few studies have benchmarked reconstruction algorithms on mammalian datasets, largely because of missing gold standards. One study compared different methods on multiple datasets [5], showing that the ARACNE algorithm, which is based on information theory, performs best on the B-cell data, where there is a good gold standard [6]. Another study used mammalian datasets but used only GO enrichment, an indirect measure, as a performance metric, limiting the interpretation [7].
Two recent publications attempted to reconstruct mammalian embryonic stem cell networks. Cegli et al. [8] used ARACNE on 171 microarrays to reconstruct a gene regulatory network in mouse embryonic stem cells (mESC). Reconstruction quality was assessed using Reactome and transcription factor perturbation data. Cahan et al. [9] used approximately 4000 samples from different mouse tissues to train a gene regulatory network using the CLR [10] algorithm. They used the area under the precision-recall-curve (AUPR) to estimate the performance, using gold standards from ChIP-Chip data in the Escape database [11], a dataset measuring differential expression after overexpression, and the Encode ChIP-seq data. However, neither of these works included any comparison of reconstruction methods, making the improvement over random values hard to interpret.
Other attempts to reconstruct the gene regulatory network in mESC [10,12] used ChIP-seq data additionally or exclusively for training and are thus not directly comparable to approaches using only transcriptome data. Finally, some reconstruction efforts aim at only reconstructing a core pluripotency network containing a couple of nodes, using time-resolved data [13] or culture condition-dependent expression [14].
In this paper, we will use and compare different state-of-the-art network reconstruction methods to reconstruct the topology of the gene regulatory network governing the differentiation process of mESCs from a large body of transcriptome data. We evaluate these reconstructed networks using functional data on regulatory interactions, such as ChIP-seq data or transcription factor perturbation experiments.
2. Material and methods
(a). Algorithms and packages used
All calculations were performed in R. We compared different similarity measures (Pearson and Spearman correlation, mututal information, ARACNE [15], CLR [16], MRNET [17], partial correlation [18]), as implemented in base R, in the parmigene R-package, or in the parcor package. Most algorithms did not require a parameter, except for ARACNE (τ), partial correlation (parameter k, which we set to 3) and mutual information (estimation related parameter k, which was set to 9). All topological properties were computed using the R package igraph.
(b). Gold standards
A list of transcription factors in mouse ‘Gene list of TFs’ was obtained from the Animal Transcription Factor Database (ATFDB) [19], located at http://www.bioguo.org/AnimalTFDB/, which was used to limit the genes analysed to transcription factors. To define the ChIP-seq gold standard, we downloaded the Mouse ES Cell ChIP-Seq Compendium maintained by the Bioinformatics Core at Wellcome Trust—MRC Stem Cell Institute, available at http://lila.results.cscr.cam.ac.uk/ES_Cell_ChIP-seq_compendium_UPDATED.html. We computed the transcription factor association score for each gene in each dataset, as defined in [20], with characteristic length scale at 1000 bp. For the overexpression gold standard, we downloaded the GEO-datasets GSE31381 and GSE14559 using the R package GEOquery. Because these series contain a mix of already logarithmized expression values and linear expression values, all samples with median greater than 10 were logarithmized. To rank genes, we calculated empirical z-values. We first calculated the mean expression and the standard deviation for each gene using the four samples labelled as ‘Control’, and we then defined the standard deviation by a Loess fit on the standard deviation versus the mean with a span of 3. We then used a z-score threshold of 2.9 and a fold change of more than 2, which results in an estimated false discovery rate of less than 0.001. The loss of function (LoF) benchmark data were obtained from the Escape database [11]. The list of differential genes for each transcription factor assayed was downloaded from http://www.maayanlab.net/ESCAPE/download/logof.txt.zip. From these data, we kept only experiments marked as LoF. Gene symbols were translated to Ensembl gene ids using the symbolToGene function of the R package annmap (Ensembl v. 74). The transcription factor knockdown data were obtained from publication [21]. We downloaded the list of differential genes for each transcription factor assayed from http://www.nature.com/srep/2013/130306/srep01390/extref/srep01390-s1.xls. Gene symbols were translated to Ensembl gene ids using the symbolToGene function of the R package annmap (Ensembl v. 74). The literature-based PluriNetWork described in [22] was downloaded from WikiPathways (http://wikipathways.org/index.php/Pathway:WP1763) on 15 April 2014 using Cytoscape 3.1.0 and exported as plain text file. It was subsequently imported into R and restricted to node pairs that could be mapped to Ensembl gene ids using Ensembl v. 79. We furthermore retained only interactions where the first node was a transcription factor, yielding 362 interactions.
(c). Transcriptome data
Microarray samples were obtained from the GEO database automatically using the R packages GEOquery for data retrieval and GEOmetadb for data selection. Using GEOmetadb (time stamp 11/2013), the GEO database was searched for entries corresponding to the Affymetrix Mouse Gene St platform (GPL6246). These entries were filtered by the following criteria. First, the raw CEL data had to be present. Second, the description had to contain at least one of the following keywords: mESC, stem cell, stem cells, Oct4, Sox2, Nanog, Pou5f1, embryonic. The corresponding 1194 samples were then downloaded. These samples were normalized together using the rma function from the R package oligo. Probesets were annotated with Ensembl gene ids using the R package mogene10sttranscriptcluster.db. Probesets that were associated with more than one gene were omitted. The final expression matrix contained 19 615 genes measured in 1194 samples.
(d). Differentiated tissue annotations
We downloaded raw data for the microarray samples referenced in the manually curated CellNet tissue atlas [9] and normalized samples using RMA implemented in the R package oligo. Probesets were annotated with ensembl gene ids using the R package mogene10sttranscriptcluster.db. To compare the expression values of the CellNet samples with the keyword-based samples, both were re-normalized together using quantile normalization.
(e). Calculation of area under the precision-recall
We ranked all predictions based on the score, and calculated the precision-recall curve using the ROCR package [23] together with the interpolation algorithm of Boyd et al. [24], available at https://github.com/kboyd/raucpr [25]. The obtained AUPRs were used to calculate the improvement of the predictions over random.
(f). Size-dependent performance
From the expression dataset of 1194 samples, five random samples were drawn with the sizes 16, 32, 64, 128, 256, 512 and 1024. The reconstruction algorithms were then applied on the same random samples.
3. Results
Our approach to benchmark network reconstruction algorithms was as follows: we compiled a large collection of 1194 publicly available transcriptomes of mESC by querying the GEO database with keywords. By co-normalization, these data then represented a large data matrix of expression of genes in mESCs under different conditions and with different differentiation status. We subsequently applied a set of co-expression metrics and published state-of-the-art network reconstruction algorithms (figure 1a) to these data that generated ranked lists of potential transcription factor target gene pairs. We then compared the predictions of these algorithms with different sets of gold standards that link transcription factors and their target genes. To this end, we used ChIP-seq data, data on differential expression after transcription factor perturbation and a manually curated literature-based network. Using these gold standards, we could then rank individual algorithms by their average performance using the area under the precision-recall curve.
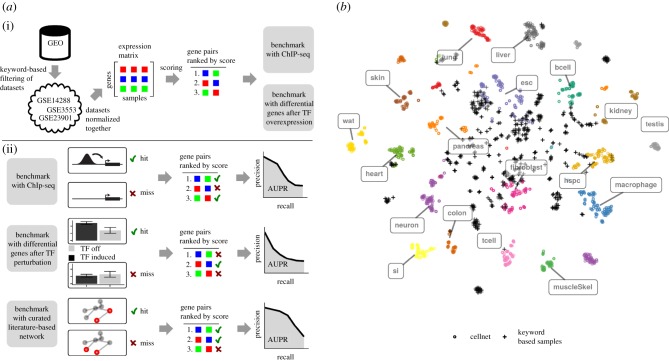
Figure 1.
(a) Pipeline for the benchmarking of network prediction algorithms. (i) Microarray samples from a fixed Affymetrix mouse array type that are identified by keywords related to embryonic stem cells are downloaded from the GEO database. All samples are normalized together, yielding the gene expression matrix. Different algorithms that predict interactions based on co-expression are then applied to the expression matrix to obtain a list of gene pairs ranked by their interaction score. As we investigate gene regulation by TFs, only TF-gene pairs are retained. The quality of predictions is evaluated using two kinds of evidence for direct interaction, ChIP-seq and differential expression after TF perturbation. (ii) TF-gene pairs are classified as hits and misses according to ChIP-seq and TF perturbation evidence. For the ChIP-seq gold standard, a TF-gene pair is classified as hit when the TF binds close enough to the promoter of the gene. For the perturbation gold standard, genes with a significant fold change upon TF perturbation are classified as targets of the respective TF. In addtion, a literature-based network is used for benchmarking. From the rank of the hit and miss TF-gene pairs in the list ranked by interaction score, the precision-recall curve is calculated. The area under the precision-recall curve (AUPR) serves as quality benchmark of the network predictions. (b) Visualization of keyword-based samples relative to annotated differentiated tissue and ESC samples using tSNE.
(a). A large dataset of gene expression in mESCs
To aggregate a large mESC transcriptome compendium, we used the GEO database and searched for the following keywords in the abstracts: mESC, stem cell, stem cells, Oct4, Sox2, Nanog, Pou5f1 and embryonic. We then downloaded all datasets as raw data that were from one array platform (Affymetrix Mouse Gene 1.0 ST Array (Gene St) platform). This yielded 1194 transcriptomes. These data were then normalized together to obtain an expression matrix (figure 1a). When we visualized the samples using t-distributed stochastic neighbour embedding (t-SNE) together with samples from annotated tissues (CellNet dataset, figure 1b), we noted that our selected samples form a heterogeneous group where most samples cluster with annotated stem cells, many are not assigned to specific tissues and some co-cluster with tissues. Such heterogeneity is potentially important to reconstruct the network, since then the data contain sufficient variance in the expression of pluripotency-associated TFs, which in turn allows to identify genes that covary with these TFs.
(b). Selection of methods for network reverse engineering
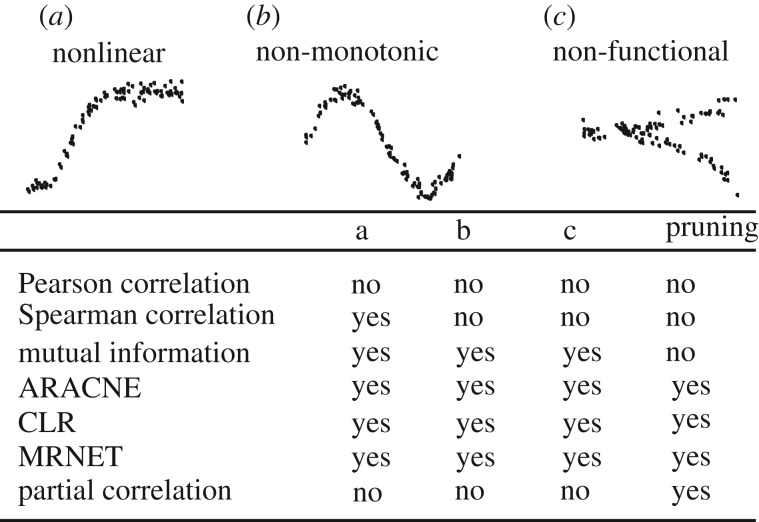
Most network reconstruction methods use a measure for statistical association of two variables [26] to predict links. In this work, we decided to benchmark similarity measures that score non-functional (mutual information), functional monotonic nonlinear (Spearman correlation) and linear (Pearson correlation) relations [27–29] (figure 2). In addition, we added algorithms that are based on these measures, but in addition prune these network either on mutual information (ARACNE [15], MRNET [17], CLR [16]), or Pearson correlation (partial correlation).
Figure 2.
Classification of scores with the help of the scheme proposed in [27]. Scores are classified by their ability to detect co-expression relationships with the indicated properties, nonlinear, non-monotonic and non-functional. The pruning column indicates whether the algorithm tries to remove indirect links.
All algorithms take a n × m expression matrix as input, where n is the number of genes and m is the number of (microarray) samples. The output is an n × n matrix that contains an association score for each gene pair. The ARACNE algorithm is the only one that needs an input parameter. This parameter τ determines how aggressively the algorithm tries to prune indirect links. The value τ = 1 corresponds to no pruning, while for τ = 0, the weakest link between three mutually connected nodes will always be deleted. In this work, we employed τ = 0.15 (denoted by aracne_15) and τ = 0.5 (denoted by aracne_50) corresponding to the preconfigured standard value and very weak pruning, respectively. To benchmark partial correlation, we chose different implementations, namely a sparse (pcor_lasso) and a non-sparse method (pcor_pls, partial least squares) for estimating partial correlation [18].
(c). Gold standards for determining direct TF-gene interactions
We compiled different complementary gold standards to benchmark the network reconstructions, covering TF binding and regulation by a TF [30]. First, we used a collection of ChIP-seq datasets (Mouse ES Cell ChIP-Seq Compendium [31]) that provides information about TF binding. Second, we used three collections of perturbation experiments, where transcriptomes are measured after TF knockout [11], TF knockdown (Kd) [21] and TF overexpression [32,33] (see §2). These gold standards emphasize different aspects. While TF overexpression will unveil the targets of TFs that are not active in mESCs, TF knockdown and knockout will only unveil targets of TFs that are already expressed in mESCs. Note also that the number and identity of the TFs are different between different gold standards. Additionally, we used the manually curated PluriNetWork [22], which concentrates on the core pluripotency factors. The overlap in interactions between these gold standards is shown in electronic supplementary material, figure S1.
Each gold standard was then filtered to those genes that are annotated as transcription factors using transcription factor annotation from the animal transcription factor database (ATFDB [19]). For details, consult §2.
(d). Benchmarking network prediction algorithms: pruning is important
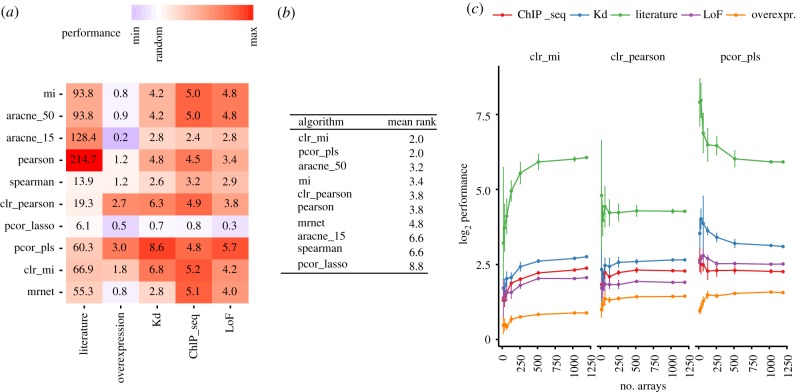
For benchmarking the algorithms, we decided first to concentrate on the top predictions, as one of the main aims of network reconstruction is to generate a list of interesting candidates for further testing. We thus profiled the algorithms based on the area under the precision-recall curve for the top 5% recall (AUPR0.05; see full precision-recall curves in electronic supplementary material, figure S2). For each algorithm, TF–target interactions were ranked based on the absolute value of the corresponding score. As the pure AUPR0.05 score is hard to interpret, we decided to divide it by the AUPR0.05 score for random predictions. These resulting scores quantify the improvement over random predictions for individual algorithms with respect to the different gold standards and are shown in figure 3a. The ChIP-seq and LoF gold standards are less sensitive to differences between the algorithms, as can be seen from the relatively small performance differences of the top-performing algorithms. By contrast, in the case of the overexpression and Kd benchmark, two and three algorithms, respectively, clearly outperform the rest.
Figure 3.
(a) Improvement of the AUPR0.05 over random predictions (performance) for networks predicted by individual scores. Performance is shown for gold standards based on ChIP-seq measurements (ChIP-seq), differential gene expression after TF knockdown (Kd), TF loss of function experiments (LoF), differential gene expression upon TF overexpression (overexpression) and a curated literature-based network (literature). Algorithms used for the comparison are Spearman correlation (spearman), Pearson correlation (pearson), mutual information (mi), ARACNE (with cut-off parameter 0.15 and 0.5, respectively), CLR, MRNET and partial correlation in the pls (pcor_pls), lasso (pcor_lasso) implementation. (b) Mean rank of the algorithm for all gold standards. (c) Size dependence of the log2 performance (AUPR0.05 over random). Points and errorbars indicate the median and standard deviation of the performance on five randomly sampled groups of arrays (same samples were taken for different scores) of the indicated size.
When comparing individual algorithms, partial correlation (pls implementation) and clr_mi attain the highest average rank across gold standards of all algorithms (figure 3b). Except for the ChIP-seq and literature-based gold standards, pls-based partial correlation ranks top for each gold standard. The two algorithms clr_mi and ARACNE with cut-off parameter τ = 0.5, both based on mutual information but using additional strategies for eliminating indirect links, perform similarly to pcor_pls. Therefore, all three top-performing algorithms employ a strategy for pruning indirect links. On the other hand, being able to detect only linear relationships does not impact performance strongly. This is indicated by the fact that partial correlation is among the best-performing algorithms and that Pearson correlation and mutual information attain a similar average rank. We concluded that reconstruction success of an algorithm is determined more by the pruning strategy used than by which measure is used to score co-expression patterns.
Somewhat surprisingly, the two implementations for partial correlation performed very differently, with the pls implementation ranking top, although this implementation has been shown to have the highest error for estimating the partial correlation matrix on synthetic data [18].
As experimental data are costly, it is also of interest how well an algorithm performs with respect to the amount of available data. Therefore, we also benchmarked the two top-performing algorithms clr_mi and pcor_pls on random subsamples of the entire expression dataset (figure 3c). The two algorithms differ markedly with respect to their size-dependent performance. The algorithm pcor_pls shows top performance even at the smallest subsamples (16 arrays) while performance saturates at approximately 500 arrays for clr_mi. To understand why clr_mi needs large amounts of data for top performance, we additionally computed the size dependence of the CLR algorithm based on Pearson correlation, CLR_Pearson. For this algorithm, performance quickly saturates similar to pcor_pls. This suggests that the slowly saturating behaviour of clr_mi is due to the relatively large amount of data needed to obtain good estimates for mutual information.
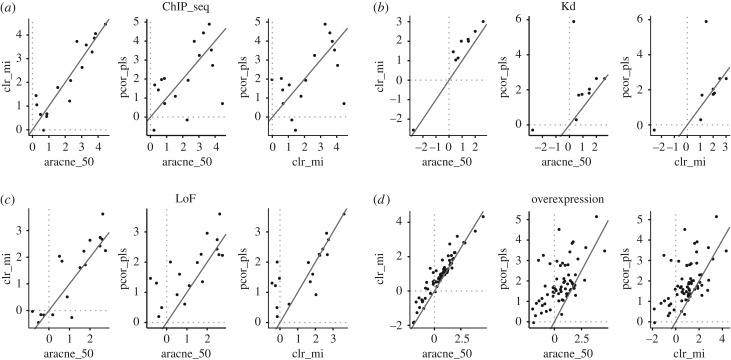
We were next interested to see whether some TF targets can be predicted better then others. We therefore assessed the performance of different algorithms on predicting targets for individual TFs present in the different gold standards, and compared the AUPR0.05. The resulting improvements over random predictions are shown in figure 4. We found that the performance varies considerably among TFs, although some of this variation could be due to different numbers of true targets for different TFs (in the Kd, LoF and overexpression gold standards). However, even for the ChIP-seq gold standard, where we had a fixed number of targets, there is huge variation. For example, the log2 improvement over random of the clr_mi algorithm ranges from approximately 1 to more than 4.
Figure 4.
(a–d) Comparison of the performance improvement over random log2(AUPR0.05/random AUPR0.05) for individual TFs for aracne_50 clr_mi and pcor_pls. Scatter plot of the improvement of the AUPR0.05 over random predictions for the clr_mi and pcor_pls algorithms for the different gold standards as indicated.
When we clustered the different algorithms by using the correlations in their performance for the TFs (electronic supplementary material, figure S3), we noted that the pcor_pls score is the only score that is both not correlated to the mutual information-derived scores and performs well with respect to the overall improvement over random. That makes this score an interesting candidate for providing TF targets that complement the predictions of other well-performing scores. This is especially obvious when comparing the performance of pcor_pls and the clr_mi algorithm for individual TFs determined on the overexpression gold standard (figure 4).
(e). Predicted topologies of the TF–TF network differ strongly
We next used the top-performing algorithms to predict the topology of the gene regulatory network in differentiating mESC, and asked if there is a clear hierarchy between core pluripotency factors and ancillary pluripotency factors [14]. Another interesting aspect that could be inferred from the topology is how pluripotency factors are coupled to lineage-specific factors in order to regulate the transition from the maintenance of pluripotency to the commitment to certain lineages [34].
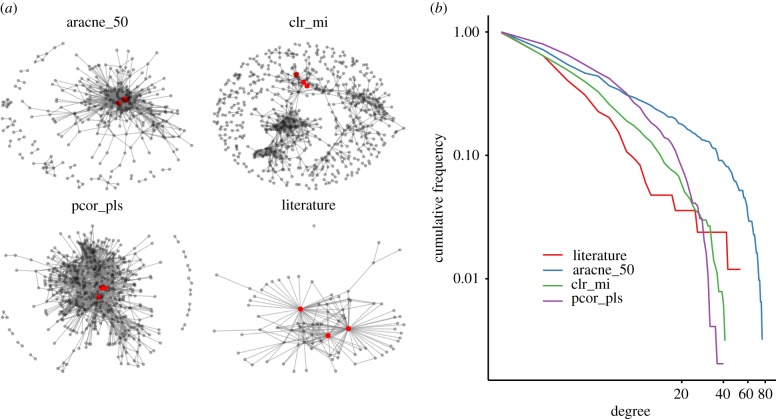
To derive the network topology, we applied the three top-performing algorithms and took the top 0.1% of interactions across the predicted network. The predicted networks are shown together with the literature-based network in figure 5a. These visualizations unveil that the networks exhibit very different topologies. We next compared the topological properties of the literature network and the predicted networks by quantifying some standard measures (table 1). The degree distribution (figure 5b) of the literature network shows an approximately linear decrease in the log–log-plot. Contrary to this, the predicted networks show a marked deviation from a linear degree distribution for the highest degrees. The linear degree distribution of the literature network up to the highest degrees is reflected by the fact that the most important nodes, Nanog and Pou5f1, are involved in 50% of all interactions. Their targets typically have no interactions among themselves, leading to a star-like structure centered on Nanog and Pou5f1 (figure 5). This fact is reflected in the low transitivity of the literature network when compared with the predicted networks. A related quantity, the degree correlation coefficient indicates whether nodes of high degree are typically connected to other nodes of high degree (positive degree correlation) or to nodes of low degree (negative degree correlation). The degree correlation coefficient of −0.47 confirms the star-like structure.
Figure 5.
(a) Comparison of the topologies of the predicted TF–TF networks with the literature-based network for the top-ranking algorithms. The transcription factors Pou5f1, Sox2 and Nanog are indicated by magenta dots. (b) Degree distribution for the TF–TF gene regulatory networks predicted by the indicated algorithms, with aracne_15 and aracne_50 denoting the ARACNE algorithm with 0.15 and 0.5 as cut-off parameter and pcor_pls denoting partial correlation in the pls implementation.
Table 1.
Quantification of topological properties of the predicted networks. The networks predicted by the algorithms clr_mi, partial correlation in the pls implementation and ARACNE with 0.50 as cut-off parameters are compared with the literature network. Mean rank OSN (Oct4, Sox2, Nanog): denotes the mean degree rank of the core triad, with low values indicating high degrees; % in largest component: the fraction of vertices contained in the largest connected component; no connected components: the number of connected components; modularity edge betweenness: the modularity of the graph according to the edge betweenness community measure; modularity fastgreedy: the modularity of the graph according to the fastgreedy community measure; diameter, the graph diameter; transitivity, the graph transitivity.
| clr | pcor_pls | aracne_50 | literature | |
|---|---|---|---|---|
| mean rank OSN | 172.33 | 66 | 7 | 2 |
| % in largest component | 0.69 | 0.95 | 0.78 | 0.99 |
| no connected components | 61 | 12 | 22 | 2 |
| modularity edge betweenness | 0.72 | 0.56 | 0.21 | 0.12 |
| modularity fastgreedy | 0.72 | 0.54 | 0.29 | 0.34 |
| diameter | 22 | 10 | 9 | 5 |
| transitivity | 0.48 | 0.26 | 0.53 | 0.16 |
| degree correlation | 0.63 | −0.01 | 0.12 | −0.47 |
In the network reconstructed by ARACNE, interactions are focused on a small fraction of all nodes, which form the centre of the largest connected component. The centre of this connected component is formed by Oct4, Sox2 and Nanog (OSN), among others. This is reflected in the very low mean rank of the degrees of this core triad. Similar to the literature network, the ARACNE network has a low modularity and also a relatively low diameter. It differs from the literature network in its degree correlation, which is positive. This fact points towards a hierarchy of nodes, with the highest degree nodes connected to the next highest degree nodes, and so on.
The network predicted by partial correlation has the highest fraction of nodes with small degree among the predicted networks. Also, the degree correlation is near zero, indicating that there is little substructure in the largest connected component. There are no communities that are only connected to other communities via gateway nodes, resulting in a comparably low diameter and the largest connected component of all predicted networks. The mean rank of the degree of the core triad is higher than for ARACNE, reflecting a less central place of these TFs in the network.
Finally, the network predicted by the clr_mi algorithm shows the most pronounced modular structure with groups of nodes that have high connectivity among themselves but low connectivity with outside nodes. The modular structure is reflected in the highest modularity index of all predicted networks as well as the highest transitivity and degree correlation. Though the core triad is located in a community of nodes with high connectivity, the smallness of this community leads to the highest mean rank of the degree of OSN among the predicted networks. The large diameter of the clr_mi network is also a consequence of the modular structure. Nodes from different communities can only be connected by paths traversing the few gateway nodes that connect communities.
It is known that reconstruction algorithms, in general, tend to enrich different types of motifs [26], affecting the topology of the predicted network. Some of the different topological features of the predicted networks can be explicitly traced back to the way the employed algorithm works. Networks induced by correlation are more transitive than random networks, whereas those induced by partial correlation are less transitive than random networks [35]. The ARACNE algorithm also directly influences transitivity by cutting all weakest links in triangles unless their strength is above the tolerance parameter. Here, ARACNE shows rather high transitivity because the cutoff parameter τ was set to the large value of 0.5. The clr_mi algorithm does not influence the topology in an explicit way, but induces a network that exhibits a unique modular structure among the predicted networks. In comparison with these reconstructed networks, the literature network is strongly shaped by the prominent roles of Pou5f1 and Nanog and to a lesser degree Sox2. Thus, its structure may to some degree also be a consequence of a publication bias. Taking into account the bias introduced by the different algorithms and the extreme focus of the literature network on the core triad, it seems that each algorithm emphasizes different aspects about the topology of the TF–TF network in mESC from those unbiased network reconstruction attempts.
4. Discussion and conclusion
Understanding how development and differentiation are controlled at the molecular level depends largely on understanding the underlying gene regulatory networks [36]. Both for human and mouse ESC, a large body of published transcriptome data has accumulated from which regulatory interactions can be deduced. Network reconstruction based on co-expression measures seems to be a promising approach for deducing interactions from this data.
Here, we analysed how useful different reconstruction algorithms are for inferring the gene regulatory network underlying differentiation of embryonic stem cells. We focus on mouse, as for mESCs there are several datasets that can serve as a gold standard. We benchmarked the predictions generated by different algorithms using orthogonal high-throughput experiments. We could show that the best-performing algorithms use pruning schemes that remove indirect links. One of the top-performing algorithms, partial correlation, was particularly interesting because its predictions are not strongly correlated with those of the other well-performing algorithms. A topological analysis of the TF–TF regulatory network predicted by the three top-performing algorithms highlighted markedly different topologies whose features can be traced back to the design of the algorithm. Thus, while evidence from high-throughput data supports the usefulness of the predictions based on co-expression, the observed network structures are strongly influenced by the employed algorithm. This is one of the aspects currently limiting the usefulness of network reconstruction algorithms.
Network reconstruction has been approached with differing scope and multiple tools. Reconstruction of small scale networks is often used to integrate information about multiple perturbation experiments into a single model. This model can then be used to predict untested perturbations without tedious or costly experiments. This approach to molecular networks has proved to be fruitful in devising ways to manipulate cellular signal transduction [37]. In the field of stem cells, it can be used to understand how the wiring between signalling pathways and pluripotency-associated factors determines maintenance of the pluripotent state under different culture conditions [14,38].
Network reconstruction on a larger scale was often done with the goal of generating lists of candidate genes for follow-up experiments [8,12]. Another aspect that has been studied with large-scale networks is the overall structure of the network and its relation to selective pressures that gave rise to the observed network structure [39]. Important concepts in this context are overrepresented motifs and the frequency with which feedback is encountered in biological networks [40].
Current limits in the quality of network reconstructions, as also observed in this work, show that the goal of obtaining a comprehensive reliable representation of the regulatory network is still distant. How can progress in this direction be made? New data may significantly enhance our ability to infer networks. In particular, single cell data may open up the possibility of observing distinct states of a network that get blurred in bulk data [41–43]. This may help to resolve the temporal sequence of gene silencing and activation. Perturbations in conjunction with well-resolved time series may also yield important insights as one can separate early, presumably direct, from late, presumably indirect effects. The currently available transcription factor perturbation data are usually optimized for detecting even weakly deregulated genes [32]. As this is achieved by assessing the transcriptome days after the start of the perturbation, a large amount of differential expression is caused by indirect effects.
How can these approaches help experimental researchers interested in studying differentiation? First, reverse engineering methods are good enough to leverage the large body of available transcriptomes to identify likely TF–target interactions that can then be validated experimentally. Second, our results also show that different methodologies unveil different sets of TF–target interactions, thus it is valuable to use different algorithms. However, it is clear that the networks reconstruction algorithms are clearly not reliable enough (yet) that their results can be taken for granted, and can only be taken as starting points as candidates for further experimental validation.
Supplementary Material
Data accessibility
R-scripts for the benchmark are available at https://github.com/johannesmg/benchmark_gene_regulation_mesc.git and normalized transcriptome data are available at http://sysbio.net/download/expression_set_gpl_6246_full.RData.
Author's contributions
N.B. and J.M. conceived the study and wrote the manuscript. J.M. carried out the computational analysis. Both authors approved the submission.
Competing interests
We have no competing interests.
Funding
This study was funded by the Federal Ministry of Education and Research of Germany (BMBF), grant SysDT-trans (031L0117D).
References
- 1.EP Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. ( 10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margolin AA, Califano A. 2007. Theory and limitations of genetic network inference from microarray data. Ann. N. Y. Acad. Sci. 1115, 51–72. ( 10.1196/annals.1407.019) [DOI] [PubMed] [Google Scholar]
- 3.Stolovitzky G, Monroe D, Califano A. 2007. Dialogue on reverse-engineering assessment and methods. Ann. N. Y. Acad. Sci. 1115, 1–22. ( 10.1196/annals.1407.021) [DOI] [PubMed] [Google Scholar]
- 4.Allen JD, Xie Y, Chen M, Girard L, Xiao G. 2012. Comparing statistical methods for constructing large scale gene networks. PLoS ONE 7, e29348 ( 10.1371/journal.pone.0029348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. 2007. How to infer gene networks from expression profiles. Mol. Syst. Biol. 3, 78 ( 10.1038/msb4100120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. 2005. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390. ( 10.1038/ng1532) [DOI] [PubMed] [Google Scholar]
- 7.Song L, Langfelder P, Horvath S. 2012. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinf. 13, 328 ( 10.1186/1471-2105-13-328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cegli RD. et al. 2013. Reverse engineering a mouse embryonic stem cell-specific transcriptional network reveals a new modulator of neuronal differentiation. Nucleic Acids Res. 41, 711–726. ( 10.1093/nar/gks1136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahan P, Li H, Morris S, Lummertz da Rocha E, Daley G, Collins J. 2014. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915. ( 10.1016/j.cell.2014.07.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061. ( 10.1016/j.cell.2008.02.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, Baroukh C, Dannenfelser R, Chen EY, Tan CM, Kou Y, Kim YE, Lemischka IR, Ma'ayan A. 2013. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database 2013, bat045 ( 10.1093/database/bat045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowell KG, Simons AK, Wang ZZ, Yun K, Hibbs MA. 2013. Cell-type-specific predictive network yields novel insights into mouse embryonic stem cell self-renewal and cell fate. PLoS ONE 8, e56810 ( 10.1371/journal.pone.0056810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anchang B, Sadeh MJ, Jacob J, Tresch A, Vlad MO, Oefner PJ, Spang R. 2009. Modeling the temporal interplay of molecular signaling and gene expression by using dynamic nested effects models. Proc. Natl Acad. Sci. USA 106, 6447–6452. ( 10.1073/pnas.0809822106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. 2014. Defining an essential transcription factor program for naïve pluripotency. Science 344, 1156–1160. ( 10.1126/science.1248882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Favera RD, Califano A. 2006. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinf. 7, 1–15. ( 10.1186/1471-2105-7-S1-S7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol 5, e8 ( 10.1371/journal.pbio.0050008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer PE, Kontos K, Lafitte F, Bontempi G. 2007. Information-theoretic inference of large transcriptional regulatory networks. EURASIP J. Bioinf. Syst. Biol. 2007, 79879 ( 10.1155/2007/79879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krämer N, Schäfer J, Boulesteix AL. 2009. Regularized estimation of large-scale gene association networks using graphical Gaussian models. BMC Bioinf. 10, 384 ( 10.1186/1471-2105-10-384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang H, Guo A-Y. 2011. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40, D144–D149. ( 10.1093/nar/gkr965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouyang Z, Zhou Q, Wong WH. 2009. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl Acad. Sci. USA 106, 21 521–21 526. ( 10.1073/pnas.0904863106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama A. et al. 2013. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci. Rep. 3, srep01390 ( 10.1038/srep01390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Som A, Harder C, Greber B, Siatkowski M, Paudel Y, Warsow G, Cap C, Schöler H, Fuellen G. 2010. The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS ONE 5, e15165 ( 10.1371/journal.pone.0015165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sing T, Sander O, Beerenwinkel N, Lengauer T. 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941. ( 10.1093/bioinformatics/bti623) [DOI] [PubMed] [Google Scholar]
- 24.Boyd K, Eng KH, Page CD. 2013. Area under the precision-recall curve: point estimates and confidence intervals. In Machine learning and knowledge discovery in databases (eds Blockeel H, Kersting K, Nijssen S, Železný F). Lecture Notes in Computer Science, no. 8190, pp. 451–466. Berlin, Germany: Springer. [Google Scholar]
- 25.Davis J, Goadrich M. 2006. The relationship between precision-recall and ROC curves. In Proc. of the 23rd Int. Conf. on Machine Learning. ICML '06, Pittsburgh, PA, USA, 25–29 June 2006, pp. 233–240. New York, NY, USA: ACM. [Google Scholar]
- 26.Marbach D. et al. 2012. Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804. ( 10.1038/nmeth.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos SdS, Takahashi DY, Nakata A, Fujita A. 2013. A comparative study of statistical methods used to identify dependencies between gene expression signals. Brief. Bioinf. 15, 906–918. ( 10.1093/bib/bbt051) [DOI] [PubMed] [Google Scholar]
- 28.de Jong H. 2002. Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103. ( 10.1089/10665270252833208) [DOI] [PubMed] [Google Scholar]
- 29.He F, Balling R, Zeng AP. 2009. Reverse engineering and verification of gene networks: principles, assumptions, and limitations of present methods and future perspectives. J. Biotechnol. 144, 190–203. ( 10.1016/j.jbiotec.2009.07.013) [DOI] [PubMed] [Google Scholar]
- 30.Cusanovich DA, Pavlovic B, Pritchard JK, Gilad Y. 2014. The functional consequences of variation in transcription factor binding. PLoS Genet. 10, e1004226 ( 10.1371/journal.pgen.1004226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Berthold Göttgens B, Niwa H, Smith A. 2012. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491–504. ( 10.1016/j.stem.2012.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiyama A. et al. 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell. Stem. Cell. 5, 420–433. ( 10.1016/j.stem.2009.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correa-Cerro LS. et al. 2011. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Sci. Rep. 1, 167 ( 10.1038/srep00167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalkan T, Smith A. 2014. Mapping the route from naive pluripotency to lineage specification. Phil. Trans. R. Soc. B 369, 20130540 ( 10.1098/rstb.2013.0540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zalesky A, Fornito A, Bullmore E. 2012. On the use of correlation as a measure of network connectivity. NeuroImage 60, 2096–2106. ( 10.1016/j.neuroimage.2012.02.001) [DOI] [PubMed] [Google Scholar]
- 36.Peter IS, Davidson EH. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985. ( 10.1016/j.cell.2011.02.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinger B. et al. 2013. Network quantification of EGFR signaling unveils potential for targeted combination therapy. Mol. Syst. Biol. 9, 673 ( 10.1038/msb.2013.29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Ang YS, Sevilla A, Lemischka IR, Ma'ayan A. 2014. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Comput. Biol. 10, e1003777 ( 10.1371/journal.pcbi.1003777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson G, Niehrs C. 2010. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell. Biol. 20, 453–460. ( 10.1016/j.tcb.2010.05.002) [DOI] [PubMed] [Google Scholar]
- 40.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298, 824–827. ( 10.1126/science.298.5594.824) [DOI] [PubMed] [Google Scholar]
- 41.Buganim Y, Faddah D, Cheng A, Itskovich E, Markoulaki S, Ganz K, Klemm S, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209–1222. ( 10.1016/j.cell.2012.08.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trott J, Hayashi K, Surani A, Babu MM, Martinez-Arias A. 2012. Dissecting ensemble networks in ES cell populations reveals micro-heterogeneity underlying pluripotency. Mol. Biosyst. 8, 744–752. ( 10.1039/c1mb05398a) [DOI] [PubMed] [Google Scholar]
- 43.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 18, 675–685. ( 10.1016/j.devcel.2010.02.012) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R-scripts for the benchmark are available at https://github.com/johannesmg/benchmark_gene_regulation_mesc.git and normalized transcriptome data are available at http://sysbio.net/download/expression_set_gpl_6246_full.RData.