ABSTRACT
Multidrug-resistant (MDR) tuberculosis, defined as tuberculosis resistant to the two first-line drugs isoniazid and rifampin, poses a serious problem for global tuberculosis control strategies. Lack of a safe and convenient model organism hampers progress in combating the spread of MDR strains of Mycobacterium tuberculosis. We reasoned that auxotrophic MDR mutants of M. tuberculosis would provide a safe means for studying MDR M. tuberculosis without the need for a biosafety level 3 (BSL3) laboratory. Two different sets of triple auxotrophic mutants of M. tuberculosis were generated, which were auxotrophic for the nutrients leucine, pantothenate, and arginine or for leucine, pantothenate, and methionine. These triple auxotrophic strains retained their acid-fastness, their ability to generate both a drug persistence phenotype and drug-resistant mutants, and their susceptibility to plaque-forming mycobacterial phages. MDR triple auxotrophic mutants were obtained in a two-step fashion, selecting first for solely isoniazid-resistant or rifampin-resistant mutants. Interestingly, selection for isoniazid-resistant mutants of the methionine auxotroph generated isolates with single point mutations in katG, which encodes an isoniazid-activating enzyme, whereas similar selection using the arginine auxotroph yielded isoniazid-resistant mutants with large deletions in the chromosomal region containing katG. These M. tuberculosis MDR strains were readily sterilized by second-line tuberculosis drugs and failed to kill immunocompromised mice. These strains provide attractive candidates for M. tuberculosis biology studies and drug screening outside the BSL3 facility.
KEYWORDS: Mycobacterium tuberculosis, arginine, auxotrophy, methionine, multidrug resistance
IMPORTANCE
Elimination of Mycobacterium tuberculosis, the bacterium causing tuberculosis, requires enhanced understanding of its biology in order to identify new drugs against drug-susceptible and drug-resistant M. tuberculosis as well as uncovering novel pathways that lead to M. tuberculosis death. To circumvent the need for a biosafety level 3 (BSL3) laboratory when conducting research on M. tuberculosis, we have generated drug-susceptible and drug-resistant triple auxotrophic strains of M. tuberculosis suitable for use in a BSL2 laboratory. These strains originate from a double auxotrophic M. tuberculosis strain, H37Rv ΔpanCD ΔleuCD, which was reclassified as a BSL2 strain based on its lack of lethality in immunocompromised and immunocompetent mice. A third auxotrophy (methionine or arginine) was introduced via deletion of metA or argB, respectively, since M. tuberculosis ΔmetA and M. tuberculosis ΔargB are unable to survive amino acid auxotrophy and infect their host. The resulting triple auxotrophic M. tuberculosis strains retained characteristics of M. tuberculosis relevant for most types of investigations.
INTRODUCTION
The World Health Organization (WHO) reported that half a million new tuberculosis (TB) cases in 2017 were multidrug resistant (MDR) (1). Strains of Mycobacterium tuberculosis, the causative agent of TB, are defined as MDR when they are resistant to the two most potent first-line TB drugs, isoniazid (INH) and rifampin (RIF). MDR-TB treatment requires 2 years with second-line TB drugs that have very significant side effects. The TB community is actively pursuing the development of new vaccines and new drugs to reach the WHO goals of eradicating TB by 2050.
Screening for new drugs active against MDR M. tuberculosis strains can pose serious issues of worker safety because of the potential aerosolization of these virulent and hard-to-treat bacteria, which require biosafety level 3 (BSL3) containment. These containment requirements limit the number of laboratories that can evaluate new TB drugs or screen existing libraries in a high-throughput manner. Even when BSL3 facilities are available, many institutions do not allow investigators to study MDR M. tuberculosis strains for safety reasons. To meet this unmet research need, we have constructed M. tuberculosis strains that have been reclassified as BSL2 strains based on attenuation of the virulence achieved by specific deletions of genes involved in de novo biosyntheses of various amino acids and vitamins. We and others have demonstrated that M. tuberculosis strains that are auxotrophic for amino acids or vitamins failed to grow in mice (2–5). Whereas previous studies showed that the pantothenate auxotroph M. tuberculosis H37Rv ΔpanCD can replicate at low levels in mice (5), the leucine auxotroph M. tuberculosis H37Rv ΔleuCD does not (2). The double pantothenate-leucine auxotrophic M. tuberculosis strain mc26206 (H37Rv ΔpanCD ΔleuCD) was shown to be safer than Mycobacterium bovis BCG in immunodeficient mice; mc26206 was therefore approved to be reclassified as a BSL2 strain by the Albert Einstein College of Medicine Institutional Biosafety Committee (6) and many other institutions (personal communications). We reasoned that the addition of two independent, nonreversible auxotrophic mutations would further improve the safety of mc26206. We separately deleted the genes encoding the homoserine O-acetyltransferase MetA and the acetylglutamate kinase ArgB in mc26206, leading to methionine and arginine auxotrophs, respectively. Methionine and arginine starvation are bactericidal events in M. tuberculosis (7, 8). M. tuberculosis H37Rv ΔmetA and M. tuberculosis H37Rv ΔargB failed to grow in immunocompetent and immunodeficient mice (7; S. Tiwari, A. V. Tonder, C. Vilchèze, B. Weinrick, V. Mendes, S. E. Thomas, A. Malek, B. Chen, M. Chen, J. Kim, M. Berney, T. L. Blundell, J. Parkhill, and W. R. Jacobs, Jr., submitted for publication). We anticipate that these triple auxotrophic strains could be useful progenitors to construct BSL2-safe, MDR M. tuberculosis mutants for safely studying MDR strains and for screening for new TB drugs against MDR M. tuberculosis. Interestingly, screening for INH resistance in the arginine triple auxotrophic mutant yielded INH-resistant isolates with large genomic deletions that removed the katG gene, involved in INH resistance, and over 44 kb of additional DNA. In this work, we describe the successful construction of BSL2-approved triple auxotrophic antibiotic-sensitive and MDR strains of M. tuberculosis and characterize their genetic and phenotypic features in vitro and in vivo.
RESULTS
Construction of BSL2 triple auxotrophic strains.
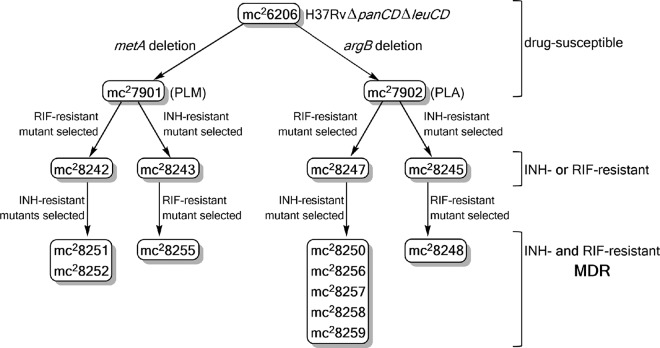
The leucine and pantothenate auxotrophic strain mc26206 (Table 1), an M. tuberculosis H37Rv strain reclassified as a BSL2 strain (6), was the basis for the construction of triple auxotrophs (Fig. 1). Using a specialized transduction system (6), methionine or arginine auxotrophy was introduced in mc26206 by deleting metA (Rv3341) or argB (Rv1654), respectively, The hygromycin cassette selectable marker for the specialized transduction was removed from the knockout strains using γδ resolvase (6), yielding the unmarked pantothenate-leucine-methionine (PLM) auxotroph mc27901 and the unmarked pantothenate-leucine-arginine (PLA) auxotroph mc27902 (Table 1). Whole-genome sequencing of mc27901 and mc27902 confirmed the expected deletions of metA and argB from mc26206; other mutations were identified by genomic comparison with the laboratory strain M. tuberculosis H37Rv. Two mutations were found in both strains: a stop codon to an arginine codon in Rv0401, a gene of unknown function, and an L262R mutation in Rv1272c, a gene encoding an ABC transporter. These mutations were also present in the parental strain mc26206. Additionally, mc27901 carried a mutation in Rv2941 (R562G), a gene involved in phthiocerol dimycocerosate biosynthesis, whereas mc27902 had mutations in Rv2566 (L864P), a gene of unknown function, and the sigma factor A gene Rv2703 (Q425H).
TABLE 1 .
Bacterial strains
| Strain | Genotype | Drug resistance | Reference or source |
|---|---|---|---|
| mc26206 | ΔpanCD ΔleuCD | 6 | |
| mc27271 | ΔpanCD ΔleuCD ΔmetA–hyg-sacB | Hyga | This work |
| mc27272 | ΔpanCD ΔleuCD ΔargB–hyg-sacB | Hyg | This work |
| mc27901 | ΔpanCD ΔleuCD ΔmetA | This work | |
| mc27902 | ΔpanCD ΔleuCD ΔargB | This work | |
| mc28242 | ΔpanCD ΔleuCD ΔmetA rpoB (H445Y) | RIF | This work |
| mc28243 | ΔpanCD ΔleuCD ΔmetA katG (W728stop codon) | INH | This work |
| mc28245 | ΔpanCD ΔleuCD ΔargB Δ2116169–2162530 | INH | This work |
| mc28247 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) | RIF | This work |
| mc28248 | ΔpanCD ΔleuCD ΔargB Δ2116169–2162530 rpoB (S450L) | INH, RIF | This work |
| mc28250 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) Δ2122397–2170320 | INH, RIF | This work |
| mc28251 | ΔpanCD ΔleuCD ΔmetA rpoB (H445Y) katG (S315N) | INH, RIF | This work |
| mc28252 | ΔpanCD ΔleuCD ΔmetA rpoB (H445Y) katG (S315N) | INH, RIF | This work |
| mc28255 | ΔpanCD ΔleuCD ΔmetA katG (W728stop codon) rpoB (S450L) | INH, RIF | This work |
| mc28256 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) katG (Δ305–312 bp, frameshift/stop codon) | INH, RIF | This work |
| mc28257 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) katG (V1A) | INH, RIF | This work |
| mc28258 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) katG (W438R) | INH, RIF | This work |
| mc28259 | ΔpanCD ΔleuCD ΔargB rpoB (H445Y) katG (W198stop codon) | INH, RIF | This work |
Hyg, hygromycin.
FIG 1 .
Schematic construction of drug-susceptible and drug-resistant M. tuberculosis triple auxotrophs.
Characterization of the drug-susceptible, triple auxotrophic strains.
To test the suitability of these triple auxotrophic strains as surrogates for virulent M. tuberculosis, in vitro growth, acid-fast staining, phage infectibility, and drug susceptibility were assessed. In the experiments described below, a mixture of pantothenate, leucine, arginine, and methionine (PLAM) was added to the growth medium of the triple auxotrophic strains unless otherwise stated.
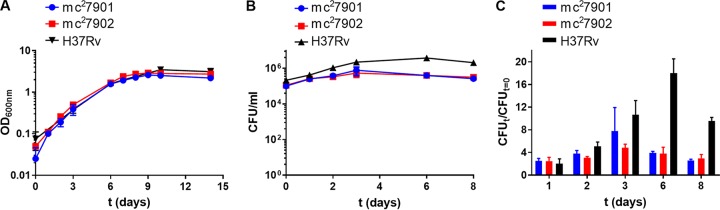
Cultures of mc27901 and mc27902 were first tested in Middlebrook 7H9 medium. mc27901 and mc27902 grew similarly to H37Rv under broth conditions (Fig. 2A). Intracellular growth was evaluated in RAW 264.7 macrophages (Fig. 2B). The triple auxotrophs followed a similar growth pattern as that of H37Rv for the first 2 days of infection. PLA-mc27902 grew more slowly than PLM-mc27901 and H37Rv, but both triple auxotrophs exhibited a 4- to 8-fold increase in CFU (Fig. 2C). Between days 3 and 6 postinfection, the triple auxotrophs stopped growing, and CFU decreased: this phenotype was observed in H37Rv only after day 6 postinfection.
FIG 2 .
mc27901 and mc27902 grow similarly to virulent M. tuberculosis in vitro. (A) Log-phase cultures of mc27901 and mc27902 grown in Middlebrook 7H9-OADC-glycerol-tyloxapol-PLAM were diluted 1/100, and growth was followed by recording optical density at 600 nm (OD600) over time. Mean with standard deviation is plotted (n = 2). (B) RAW 264.7 macrophages were infected at an MOI of 1 with mc27901, mc27902, or H37Rv. At the indicated time points, macrophages were lysed, and bacterial titers were determined by plating for CFU on Middlebrook 7H10-OADC-glycerol-PLAM plates. PLAM was added to the macrophage growth medium, and the medium was changed at each time point. (C) Growth of mc27901 and mc27902 in RAW 264.7 macrophages relative to the inocula (same experiment as in panel B). Mean with standard deviation is plotted (n = 2).
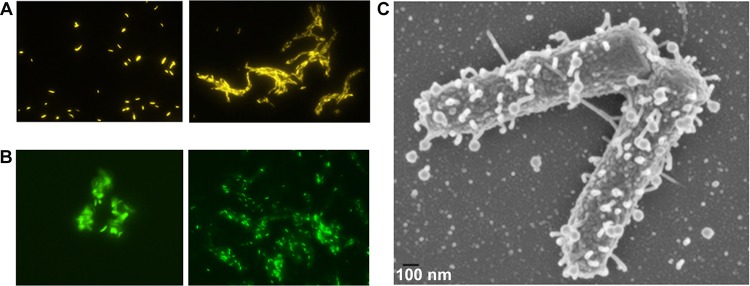
Acid-fast staining has been the basis of clinical diagnosis of TB for over a century. Loss of acid-fastness in M. tuberculosis is linked to genetic mutations, alteration in cell wall-associated lipids, lipid accumulation, and persistence (9–11). mc27901 and mc27902 were acid-fast positive (Fig. 3A), indicating that they could be used to study acid-fastness in a BSL2 environment.
FIG 3 .
mc27901 and mc27902 retain acid-fastness and susceptibility to phage infection. (A) Acid-fast staining of mc27901 (left) and mc27902 (right) using an auramine kit. Magnification, ×60. (B) Infection of mc27901 (left) and mc27902 (right) cells with phAE912. Magnification, ×60. (C) Scanning electron microscopy of mc27901 (~106 CFU) infected with phAE732 (Φ2DRM9, 3 × 107 PFU).
Mycobacteriophages (MP) are important tools for mycobacterial genetic studies (12) and TB drug susceptibility testing (13, 14). Through research programs aimed at introducing young students to scientific research (PHIRE and HHMI SEA-PHAGES), thousands of novel mycobacteriophages were discovered using Mycobacterium smegmatis, a nonpathogenic fast-growing mycobacterial species, as the host strain (12). However, many mycobacteriophages that infect M. smegmatis may not infect M. tuberculosis (15). The ability of the BSL2-safe M. tuberculosis strains mc27901 and mc27902 to serve as mycobacteriophage hosts was therefore examined. The mycobacteriophages phAE912 (16), a DS6A phage restricted to the M. tuberculosis complex and expressing mVenus, and Φ2DRM9 (17), which infects both M. smegmatis and M. tuberculosis, infected mc27901 and mc27902 (Fig. 3B and C). Thus, mc27901 and mc27902 are not restricted for phage infection and phage DNA delivery, demonstrating their suitability as mycobacteriophage hosts and substrates for specialized transduction.
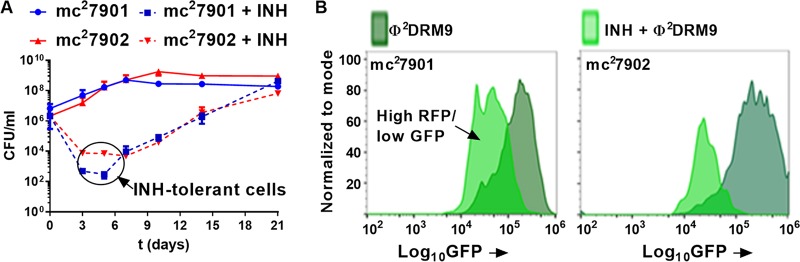
The availability of safe M. tuberculosis strains for testing novel therapeutics would circumvent the need for a BSL3 laboratory and allow for high-throughput screening in a BSL2 environment. To test whether triple auxotrophy altered drug susceptibility, the MICs of various first-line and second-line TB drugs were measured (Table 2); mc27901, mc27902, and H37Rv were found equally sensitive to all these drugs. A detailed kinetic analysis of the susceptibility of these triple auxotroph mutants to INH was examined. INH was chosen because it has a very distinctive killing pattern for H37Rv, with rapid death of the bacteria (2- to 3-log decrease in CFU within 4 days). After 4 days of INH treatment, the remaining INH-sensitive bacterial population consists of INH-tolerant cells from which INH-resistant mutants emerge (17). In the presence of INH, mc27901 and mc27902 followed the same death kinetics described above for H37Rv. A rapid decrease in CFU followed by a stabilization of the bacterial population, now consisting of INH-tolerant bacteria, was observed before INH-resistant mutants emerged (Fig. 4A). The presence of an INH-tolerant population was confirmed with the dual green fluorescent protein reporter (GFP)/red fluorescent protein (RFP) reporter phage Φ2DRM9 (Fig. 4B) (17). Flow cytometry analysis of INH-treated mc27901 and mc27902 cells infected with Φ2DRM9 revealed the presence of a high-RFP/low-GFP population, characteristic of INH persisters (17). Their triple auxotrophy did not impair the ability of the strains to be killed by TB drugs or to produce an INH-tolerant population.
TABLE 2 .
MICs of first-line and second-line TB drugs against the triple auxotrophic strainsa
| Strain | MIC (mg/liter) of TB drug |
|||||||
|---|---|---|---|---|---|---|---|---|
| First line |
Second line |
|||||||
| INH | RIF | OF | Km | Moxi | ETH | CFZ | Ami | |
| mc27901 | 0.06 | 0.06 | 0.5 | 2 | 0.125 | 0.625 | 0.5 | 0.5 |
| mc28251 | 1 | >4 | 0.5 | 4 | 0.125 | 0.625 | 0.5 | 0.5 |
| mc28255 | 1 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc27902 | 0.06 | 0.06 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28248 | >4 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28250 | >4 | >4 | 0.5 | 4 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28256 | >4 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28257 | >4 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28258 | >4 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
| mc28259 | >4 | >4 | 0.5 | 2 | 0.125 | 0.625 | 0.25 | 0.5 |
Abbreviations: Ami, amikacin; CFZ, clofazimine; ETH, ethionamide; INH, isoniazid; Km, kanamycin; Moxi, moxifloxacin; OF, ofloxacin; RIF, rifampin.
FIG 4 .
mc27901 and mc27902 generate INH persisters in culture. (A) Log-phase cultures of mc27901 and mc27902 grown in Middlebrook 7H9-OADC-glycerol-tyloxapol-PLAM were diluted 1/100 and treated with INH (1 mg/liter). Samples were taken at the indicated time points, diluted, and plated for CFU. Mean with standard deviation is plotted (n = 2). (B) mc27901 and mc27902 cultures treated or not with INH (1 mg/liter) for 2 days were infected with the phage Φ2DRM9 and analyzed by flow cytometry. Phage Φ2DRM9 contains both the L5 promoter driving GFP (mVenus) expression and the INH persister-specific dnaK promoter fused to the red fluorescent protein gene (RFP, tdTomato). The panels show the high-RFP population back-gated for GFP expression, representing the persister population (low GFP/high RFP).
Next, we examined whether there is an alteration in the mutation rate in the triple auxotrophs. Following exposure to INH or RIF, mc27901 and mc27902 yielded INH-resistant and RIF-resistant mutants, respectively, at frequencies similar to those found with H37Rv. INH-resistant mutants in mc27901 and mc27902 were isolated at frequencies of 2 × 10−5 and 9 × 10−6, respectively, compared to 6 × 10−6 in H37Rv. RIF-resistant mutants occurred less often than INH-resistant mutants, with frequencies of 4 × 10−8, 2 × 10−7, and 4 × 10−8 for mc27901, mc27902, and H37Rv, respectively. The primary means of resistance to INH and RIF in M. tuberculosis are mutations in katG (Rv1908c), encoding the activator of INH (18), and in rpoB (Rv0667), encoding an RNA polymerase (19), respectively. Sequence analysis of the rpoB and katG genes from one drug-resistant isolate from each selection confirmed the presence of an rpoB mutation (H445Y) in mc28242 and mc28247, the RIF-resistant mutants of mc27901 and mc27902, and a katG mutation in the mc27901-derived, INH-resistant mutant mc28243 (Table 1; Fig. 1). Surprisingly, we could not amplify katG from the mc27902-derived, INH-resistant strain mc28245. To characterize mc28245, whole-genome sequencing was performed and revealed a large genomic deletion of 46.3 kbp encompassing katG and 50 other genes (Table 3). After the isolation of mc27901 and mc27902 mutants with either INH or RIF monoresistance, we next explored the development of BSL2-safe MDR mutants.
TABLE 3 .
Genes deleted from mc28245 and mc28250
| Gene | Coordinates | Cla | Product |
|---|---|---|---|
| Rv1867 | 2115764–2117248 | 1 | Conserved hypothetical protein |
| Rv1868 | 2117347–2119446 | 10 | Conserved hypothetical protein |
| Rv1869c | 2119460–2120695 | 7 | Probable reductase |
| Rv1870c | 2120795–2121430 | 10 | Conserved hypothetical protein |
| Rv1871c | 2121495–2121884 | 10 | Conserved hypothetical protein |
| lldD2 | 2121907–2123151 | 7 | Possible l-lactate dehydrogenase (cytochrome) |
| Rv1873 | 2123174–2123611 | 10 | Conserved hypothetical protein |
| Rv1874 | 2123684–2124370 | 10 | Hypothetical protein |
| Rv1875 | 2124381–2124824 | 10 | Conserved hypothetical protein |
| bfrA | 2125340–2125819 | 7 | Probable bacterioferritin |
| Rv1877 | 2125904–2127967 | 3 | Probable conserved integral membrane protein |
| glnA3 | 2128022–2129374 | 7 | Probable glutamine synthetase |
| Rv1879 | 2129377–2130513 | 10 | Conserved hypothetical protein |
| cyp140 | 2130541–2131857 | 7 | Probable cytochrome P450 140 |
| lppE | 2131907–2132329 | 3 | Possible conserved lipoprotein |
| Rv1882c | 2132370–2133203 | 7 | Probable short-chain-type dehydrogenase |
| Rv1883c | 2133231–2133692 | 10 | Conserved hypothetical protein |
| rpfC | 2133731–2134261 | 3 | Probable resuscitation-promoting factor |
| Rv1885c | 2134273–2134872 | 7 | Conserved hypothetical protein |
| fbpB | 2134890–2135867 | 1 | Secreted antigen 85-B |
| Rv1887 | 2136258–2137400 | 10 | Hypothetical protein |
| Rv1888c | 2137519–2138079 | 10 | Possible transmembrane protein |
| Rv1888A | 2138444–2138617 | 3 | Conserved hypothetical protein |
| Rv1889c | 2138661–2139017 | 10 | Conserved hypothetical protein |
| Rv1890c | 2139076–2139687 | 10 | Hypothetical protein |
| Rv1891 | 2139741–2140148 | 10 | Conserved hypothetical protein |
| Rv1892 | 2140165–2140476 | 3 | Probable membrane proteins |
| Rv1893 | 2140486–2140704 | 10 | Conserved hypothetical protein |
| Rv1894c | 2140739–2141869 | 10 | Conserved hypothetical protein |
| Rv1895 | 2142521–2143675 | 7 | Possible dehydrogenase |
| Rv1896c | 2143535–2144446 | 10 | Conserved hypothetical protein |
| Rv1897c | 2144451–2144882 | 10 | Conserved hypothetical protein |
| Rv1898 | 2144940–2145248 | 10 | Conserved hypothetical protein |
| lppD | 2145214–2146245 | 3 | Possible lipoprotein |
| lipJ | 2146245–2147633 | 7 | Probable lignin peroxidase |
| cinA | 2147662–2148954 | 0 | Probable CinA-like protein |
| nanT | 2149006–2150274 | 3 | Probable sialic acid transport membrane proteins |
| Rv1903 | 2150364–2150768 | 3 | Probable conserved membrane proteins |
| Rv1904 | 2150954–2151385 | 10 | Conserved hypothetical protein |
| aao | 2151433–2152395 | 7 | Probable d-amino acid oxidase |
| Rv1906c | 2152425–2152895 | 10 | Conserved hypothetical protein |
| Rv1907c | 2153235–2153882 | 10 | Hypothetical protein |
| katG | 2153889–2156111 | 0 | Catalase-peroxidase-peroxynitritase T |
| furA | 2156149–2156601 | 9 | Ferric uptake regulation protein |
| Rv1910c | 2156706–2157299 | 3 | Probable exported protein |
| lppC | 2157382–2157987 | 3 | Probable lipoprotein |
| fadB5 | 2158087–2159091 | 1 | Possible oxidoreductase |
| Rv1913 | 2159191–2159943 | 10 | Conserved hypothetical protein |
| Rv1914c | 2159921–2160328 | 10 | Hypothetical protein |
| aceAa | 2160463–2161566 | 7 | Probable isocitrate lyase (first part) |
| aceAb | 2161566–2162762 | 7 | Probable isocitrate lyase (second part) |
| PPE34 | 2162932–2167311 | 6 | PPE family protein |
| PPE35 | 2167649–2170612 | 6 | PPE family protein |
Cl, classification, based on the TubercuList website (http://genolist.pasteur.fr/TubercuList/), with the following categories: 0, virulence, detoxification, adaptation; 1, lipid metabolism; 3, cell wall and cell processes; 6, PE/PPE; 7, intermediary metabolism and respiration; 9, regulatory proteins; 10, conserved hypothetical proteins.
Construction of BSL2 triple auxotrophic MDR strains.
The mono-INH- and mono-RIF-resistant mutants mc27901 and mc27902 were used to isolate MDR strains (Fig. 1). INH-resistant mutants were isolated at a frequency of 3 × 10−7 from mc28242 (PLM-mc27901 derived, RIF resistant) and 7 × 10−7 from mc28247 (PLA-mc27902 derived, RIF resistant), while RIF-resistant mutants were isolated at a frequency of 2 × 10−8 from mc28243 (mc27901 derived, INH resistant) and 1 × 10−8 from mc28245 (mc27902 derived, INH resistant). The resulting MDR strains were found to be highly resistant to INH and RIF (Table 2).
While rpoB and katG mutations were identified in most of the MDR mutants (Table 1), selection for INH resistance in mc28247, the RIF-resistant PLA-mc27902 mutant, afforded one MDR mutant, mc28250, from which katG could not be amplified by PCR. Whole-genome sequencing of mc28250 identified another large genomic deletion (47.9 kbp) encompassing the katG gene (Table 3). The genome of mc28245, the INH-resistant PLA-mc27902 mutant, had a deletion from position 2116169 to 2162530 (Rv1867 to Rv1916), whereas mc28250, the INH- and RIF-resistant PLA-mc27902 mutant, had a deletion from position 2122397 to 2170320 (Rv1872c to Rv1918c). These large deletions contain up to 50 nonessential genes, 37 of them of unknown function (Table 3). In contrast, selecting for INH-resistant mutants in mc28242, the RIF-resistant PLM-mc27901 mutant, led to the isolation of MDR clones having the same mutations in katG (S315N) (Table 1).
Testing of the BSL2 MDR strains in vitro.
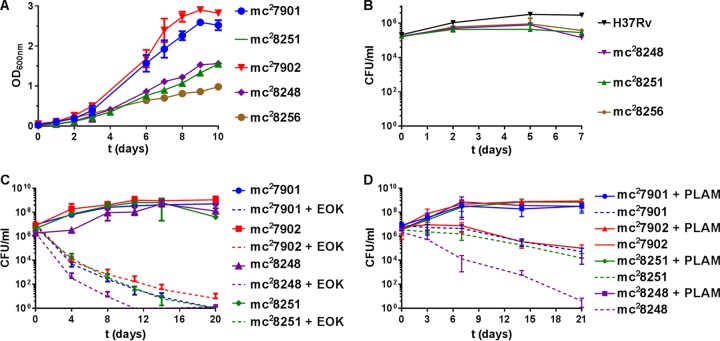
To ensure that these MDR strains could be used to study drug resistance or for drug screening, we examined their in vitro growth patterns, drug susceptibility, and safety. In liquid cultures (Fig. 5A) and in macrophages (Fig. 5B), the MDR strains grew slower than their parental strains or virulent M. tuberculosis in fully supplemented medium.
FIG 5 .
mc27901- and mc27902-derived MDR strains grow slower than parental strains in vitro and are killed by second-line TB drugs or nutrient starvation. (A) Log-phase cultures were diluted 1/100, and growth was followed by recording optical density at 600 nm. Mean with standard deviation is plotted (n = 2). (B) RAW 264.7 macrophages were infected at an MOI of 1. At the indicated time points, macrophages were lysed to determine bacterial titers. PLAM was added to the macrophage growth medium, and the medium was changed at each time point. (C) PLAM-supplemented log-phase cultures of mc27901, mc27902, mc28248, and mc28251 were treated with EOK (ethionamide [25 mg/liter], ofloxacin [5 mg/liter], kanamycin [20 mg/liter]). (D) Log-phase cultures of mc27901, mc27902, mc28248, and mc28251 were washed five times in PBS and resuspended in Middlebrook 7H9-OADC-glycerol-tyloxapol containing PLAM (dilution factor 1/100) or not. In experiments in panels B, C, and D, the strains were initially grown in Middlebrook 7H9-OADC-glycerol-tyloxapol-PLAM. Samples were taken at the indicated time points, diluted, and plated for CFU on Middlebrook 7H10-OADC-glycerol-PLAM plates. Means with standard deviations are plotted (n = 3).
Susceptibility testing with second-line TB drugs indicated that the MDR strains were as susceptible as H37Rv to these drugs (Table 2). Next, killing of the MDR strains and that of their parental strains by second-line TB drugs were compared. The triple auxotrophic drug-susceptible parental strains and MDR strains were simultaneously treated with three drugs representing a typical treatment combination for MDR-TB, i.e., one fluoroquinolone (ofloxacin), one injectable drug (kanamycin), and one conventional second-line drug (ethionamide). This combination was efficient in sterilizing the BSL2 drug-susceptible and MDR strains. The MDR PLA-mc28248 strain with a 46-kb genomic deletion was the most susceptible to the treatment, reaching a 6-log decrease in CFU within 10 days (Fig. 5C).
We also examined the susceptibility of these triple auxotrophic strains to nutrient starvation. Starvation of the drug-susceptible triple auxotrophs mc27901 and mc27902 for pantothenate, leucine, arginine, and methionine led to a 2-log reduction in CFU (Fig. 5D). The same result was observed for the PLM-mc27901-derived MDR strain mc28251, but not for the PLA-mc27902-derived mc28248, which had a 6-log reduction in CFU during nutrient starvation (Fig. 5D). These experiments demonstrate that the BSL2 MDR strains lose viability when grown without nutrient supplements or when treated with a second-line TB drug combination.
Testing the BSL2 MDR strains in mice.
Auxotrophic mutants of M. tuberculosis have been shown to be avirulent and highly attenuated for growth in immunocompromised SCID mice (20). The safety of these triple auxotrophic MDR strains was assessed in SCID mice. The mice were infected via tail vein injection with a high dose (5 × 105 CFU) of H37Rv, mc28248 (PLA-MDR), and mc28251 (PLM-MDR). Mice infected with H37Rv died by 14 days postinfection. Conversely, mice infected with the MDR strains were still alive 5 months postinfection (Fig. 6A).
FIG 6 .
mc27901- and mc27902-derived MDR strains are attenuated in mice and protect against virulent M. tuberculosis. (A) Survival of immunocompromised SCID mice (7 mice per group) infected with H37Rv and two MDR strains, mc28248 and mc28251, via tail vein injection at a dose of 5 × 105 CFU. (B and C) Immunocompetent C57BL/6 mice were immunized with mc28248, mc28250, or BCG and challenged with H37Rv (low-dose aerosol infection) 6 weeks later. Mice were euthanized 3 and 15 weeks postchallenge to determine lung (B) and spleen (C) bacterial burden.
Vaccination with single or double auxotrophic M. tuberculosis strains offers protection against virulent M. tuberculosis challenge as effective as, although not better than, the vaccine strain M. bovis BCG (20). We were curious whether the large deletions (46 to 48 kbp) from the genomes of the MDR strains mc28248 and mc28250 would remove overall immunogenicity and the ability to protect against M. tuberculosis. To examine this, immunocompetent (C57BL/6) mice were immunized by subcutaneous injection with 5 × 105 CFU of mc28248, mc28250, or BCG followed by an immunization boost 3 weeks later. Six weeks after the primary immunization, the mice were challenged with a low dose (48 CFU) of virulent H37Rv via the aerosol route. The mice were euthanized at 3 and 15 weeks postchallenge to estimate lung and spleen bacterial burdens. Against H37Rv challenge, mc28248 and mc28250 protected the mice as well as BCG did but not better (Fig. 6B and C). Furthermore, none of the colonies isolated from the lung and spleen homogenates at 3 and 15 weeks postchallenge were auxotrophs for PLA or PLM, suggesting that mc28248 and mc28250 had been cleared by the host.
DISCUSSION
Studies on the physiology, biochemistry, cell structure, and mycobacteriophage infection of M. tuberculosis are hampered by the need for BSL3 containment. Analogous studies on MDR M. tuberculosis are even more hazardous due to the difficulty of treating infections with these strains, so we sought to make safe variants of MDR M. tuberculosis. The generation of auxotrophic mutants can attenuate virulent isolates of M. tuberculosis in defined ways (2–5). The most recognized attenuated mutant of the M. tuberculosis complex is BCG, which was first isolated from M. bovis in 1904 and has been used to vaccinate children since 1921 (21). Although BCG can be used in BSL2 laboratories, there are great variations in the many BCG isolates passaged over the years in different laboratories (22–24).
We reasoned that the introduction of a mutation inducing a third auxotrophy in the leucine and pantothenate auxotroph mc26206 would provide an ideal level of safety. First, both the leucine and pantothenate auxotrophies rendered significant attenuation in immunocompromised mice (6), and the deletion of two individual genes from the leucine or pantothenate pathway prevented the reversion or suppression of an auxotrophic phenotype to a prototrophic phenotype. Methionine auxotrophy conferred by metA deletion and arginine auxotrophy mediated by argB deletion are nonreversible, nonsuppressible, and bactericidal under starvation for their respective auxotrophy, and the strains are fully safe in immunodeficient mice (7; Tiwari et al., submitted). Thus, we reasoned that three independent auxotrophies resulting from well-separated genomic deletions would create an insurmountable barrier to restoration of virulence by reversion, suppression, or complementation with genes obtained from other bacteria. Importantly, the two resulting triple auxotrophic strains mc27901 and mc27902 retained signature M. tuberculosis properties such as acid-fast staining, INH tolerance, and susceptibility to diverse mycobacteriophages. Interestingly, both triple auxotrophs can be supplemented for growth in macrophage cultures for two to four generations. These properties could be useful in developing high-throughput screens for defective growth in macrophages as well as in assessing M. tuberculosis killing by new drugs.
The generation of MDR versions of mc27901 and mc27902 did not greatly alter in vitro growth properties. However, these strains were safe as they failed to kill immunocompromised mice after 5 months. Interestingly, a vaccination experiment showed that they protected as well as BCG against virulent M. tuberculosis. Thus, we propose that the INH- and/or RIF-resistant version of these triple auxotrophic strains could be used for diverse physiological studies, including assaying the bactericidal properties of novel TB drugs. The discovery and development of new drugs targeting M. tuberculosis and particularly drug-resistant M. tuberculosis strains are difficult processes encumbered by BSL3 laboratory requirements. However, our triple auxotrophic MDR strains can be used for large chemical library screenings without the need for BSL3 containment, which will reduce the cost of drug screening and encourage other laboratories to reproduce findings.
Surprisingly, two independent, large (46- to 48-kbp) deletions in the PLA auxotrophs were isolated while selecting for INH-resistant mutants using two different parental strains (mc27902 and mc28245). It is worth noting that these deletions were obtained only in strains carrying the argB deletion. The INH-resistant mutants isolated from the PLM auxotroph mc27901 carried point mutations in katG. Deletions in katG (either partial or total) are found in INH-resistant clinical isolates (25), and large genomic deletions (from 2 to 34 kbp, containing katG and/or the furA gene) were identified in a study of six INH-resistant clinical isolates from Japanese patients (26). The largest deletion identified in the Japanese study extended from genomic position 2130514 to 2164879, a region that overlaps the two deletions in mc28248 and mc28250. We hypothesize that the argB deletion or arginine supplementation might favor the formation of these large deletions. De novo arginine biosynthesis involves the acetylglutamate kinase ArgB and several other genes, but no genes of the arginine biosynthetic pathway were part of the 46- to 48-kbp deletions. This selectivity for deletion suggests that INH killing and arginine auxotrophy have some heretofore undiscovered commonality.
Arginine or methionine starvation of M. tuberculosis lacking argB or metA, respectively, is a bactericidal event leading to a 3- to 4-log reduction in CFU within 10 days (7; Tiwari et al., submitted), while starving M. tuberculosis ΔleuCD or M. tuberculosis ΔpanCD for leucine or pantothenate, respectively, for 10 days resulted in no loss of viability (7). Nutrient starvation of the PLM strain mc27901 and the PLA strain mc27902 was static for the first 10 days before becoming bactericidal, suggesting that the static phenotype of leucine or pantothenate starvation is dominant compared to the bactericidal effect of arginine or methionine starvation. On the other hand, nutrient starvation of mc28248, the PLA-MDR strain with the 46-kbp deletion, resulted in sterilization of the strain with similar killing kinetics as those found with M. tuberculosis ΔmetA and M. tuberculosis ΔargB. It is possible that among the 50 deleted genes, the majority of unknown function, one or more genes exist that would antagonize the static effect of the leucine and pantothenate starvation. It is unlikely, though, that this sterilization upon nutrient starvation is due to the katG or rpoB mutation, since this nutrient starvation sterilization was not observed in mc28251, the PLM-MDR strain (Fig. 5D).
In conclusion, these triple auxotrophic strains are not only valuable to research laboratories but could also be important tools for clinical and microbiology laboratories as training tools since they retain most of the properties of the tubercle bacillus except virulence. We anticipate that these strains will reveal new discoveries about the biology of M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains and reagents.
The M. tuberculosis strains mc26206 (H37Rv ΔpanCD ΔleuCD) and H37Rv and the M. bovis BCG Danish strain were obtained from laboratory stocks. The strains were grown in Middlebrook 7H9 (Difco, Sparks, MD) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; Difco), 0.2% (vol/vol) glycerol, and 0.05% (vol/vol) tyloxapol at 37°C with shaking. Middlebrook 7H10 (Difco) supplemented with 10% (vol/vol) OADC and 0.2% (vol/vol) glycerol was used as solid medium. PLAM nutrient supplements were used at the following concentrations: l-pantothenate, 24 mg/liter; l-leucine, 50 mg/liter; l-arginine, 200 mg/liter; and l-methionine, 50 mg/liter. Plates were incubated at 37°C for 4 to 8 weeks. The plasmid pYUB1471 (6), shuttle phasmid phAE159 (27), and phage phAE280 (6) were obtained from laboratory stocks. Hygromycin (Gold Biotechnology, St. Louis, MO) was used at concentrations of 50 mg/liter for mycobacteria and 150 mg/liter for Escherichia coli. Phosphate-buffered saline (PBS) was obtained from Corning Cellgro (Manassas, VA). All other chemicals were obtained from Sigma-Aldrich or Thermo (Fisher) Scientific.
Construction and unmarking of mc27901 and mc27902.
Deletion of metA or argB in mc26206 was carried out by specialized transduction (6). The transductants were selected on plates containing hygromycin as the selective marker and either methionine or arginine. The hygromycin cassette was excised from the knockout strains using the phage phAE280 and sucrose selection (6). The deletion and unmarked strains were confirmed by PCR and by whole-genome sequencing performed on a MiSeq instrument (Illumina, San Diego, CA).
Murine macrophage infection.
RAW 264.7 murine macrophages were obtained from laboratory stocks; subcultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), penicillin (100 units), and streptomycin (100 mg/liter); and grown to confluence. Macrophages were seeded into 24-well tissue culture plates at a concentration of ~5 × 105 cells per well and incubated at 37°C overnight for adherence. The M. tuberculosis strains were grown to an optical density at 600 nm (OD600) of ~1, washed twice in PBS, sonicated twice for 10 s, and resuspended in DMEM. The macrophages were infected at a multiplicity of infection (MOI) of 1, and plates were incubated at 37°C in 5% CO2 for 3 h to allow for bacterial uptake. Medium was then removed, and the wells were washed twice with warm PBS and made replete with 500 µl/well of DMEM containing 10% FBS and PLAM. At specific time points, medium was removed, the wells were washed once with PBS, and the macrophages were lysed using 0.0625% aqueous sodium dodecyl sulfate. The lysates were serially diluted and plated to determine CFU.
Phage infection.
M. tuberculosis strains were grown at 37°C to an OD600 of approximately 0.7 to 1.0 in fully supplemented Middlebrook 7H9. The cultures (1 ml) were washed three times with mycobacteriophage (MP) buffer (50 mM Tris, 150 mM NaCl, 10 mM MgCl2, 2 mM CaCl2) and resuspended in 0.1 ml MP buffer. For visualization using a Nikon Eclipse Ti microscope, mycobacterial phages (109 PFU/ml; 5 or 25 µl) were added to a bacterial suspension (0.1 ml), and the suspension was incubated at 37°C overnight. The cells were spun down, washed with PBS, resuspended in MP buffer (0.01 ml), and spread on a microscope slide. For visualization by scanning electron microscopy, the bacterial suspension (0.01 ml) was mixed with phage lysate (109 PFU/ml; 0.03 ml) for 5 min prior to fixation (see below).
Scanning electron microscopy.
The bacterium-phage lysate mix was diluted 1/1 with a fixative solution (2.5% glutaraldehyde, 0.1 M sodium cacodylate, 0.2 M sucrose, 5 mM MgCl2, pH 7.4). The samples were plated onto poly-l-lysine-coated coverslips, dehydrated through a graded series of ethanol, critical point dried using liquid carbon dioxide in a Tousimis Samdri 795 critical point dryer (Rockville, MD), and sputter coated with chromium in a Quorum EMS 150T ES (Quorum Technologies Ltd., United Kingdom). The samples were examined in a Zeiss Supra field emission scanning electron microscope (Carl Zeiss Microscopy, LLC, North America) using an accelerating voltage of 2 kV.
Detection of the M. tuberculosis persister population using Φ2DRM9.
M. tuberculosis strains were grown as described for phage infection (see above), washed twice with MP buffer, and diluted to an OD600 of ≈0.1 in 7H9 medium without tyloxapol in 96-well plates. Each well containing 0.1 ml of culture was treated with or without INH (1 mg/liter) and incubated at 37°C for 2 days. Samples and untreated controls were infected with 0.1 ml of the phage Φ2DRM9 at an MOI of 10 for 16 h at 37°C. For each sample and control, 10,000 events were acquired on an S3e cell sorter (Bio-Rad, CA) after gating the singlets on forward scatter (FSC) A and side scatter (SSC) A on a log10 scale. The data were analyzed using the FlowJo software package (version 10.0.7; Tree Star, Inc., Ashland, OR) by gating for GFP+ and GFP+ tdTomato+ cells in comparison to uninfected cells. The RFP population was back-gated to determine GFP expression distribution in these cells.
MIC determination.
Serial 2-fold dilutions of each drug were prepared in the inside wells of sterile 96-well plates for a final volume of 0.1 ml. The outside wells of the plates were filled with 0.2 ml PBS. The strains were grown to mid-log phase (OD600 of ≈0.8 to 1) and diluted 1/1,000, and 0.1 ml of diluted culture was then added to each inside well. The plates were incubated at 37°C for 7 days. OD590 was read on a Victor 3V plate reader (PerkinElmer, Waltham, MA), and the MIC was determined as the lowest concentration of drug that prevented growth. MIC data were confirmed by adding an aqueous solution of resazurin (0.03 ml; 0.2 mg/ml) to each well and further incubating the plates for 2 to 3 days. The MIC was determined as the lowest concentration of drug that prevented the conversion of resazurin (blue) to resorufin (pink).
Isolation of INH- and RIF-resistant strains.
Cultures of mc27901 and mc27902 grown to an OD600 of ≈2 were plated on 7H10-OADC-glycerol-PLAM plates containing INH or RIF at a concentration of 1 mg/liter. The plates were incubated at 37°C for up to 8 weeks. INH- and RIF-resistant mutants were patched onto 7H10-OADC-glycerol-PLAM plates containing INH (0 and 1 mg/liter) or RIF (0 and 1 mg/liter), respectively, to confirm drug resistance. Following incubation for 4 weeks, one INH-resistant and one RIF-resistant mutant from mc27901 and mc27902 were then cultured in 7H9-OADC-glycerol-tyloxapol-PLAM to an OD600 of ≈2. The INH-resistant and RIF-resistant mutants were plated on Middlebrook 7H10-OADC-glycerol-PLAM plates containing RIF (1 mg/liter) or INH (1 mg/liter), respectively, and incubated at 37°C for up to 8 weeks. INH-RIF-resistant mutants were patched onto 7H10-OADC-glycerol-PLAM plates containing INH (0 and 1 mg/liter) or RIF (0 and 1 mg/liter) to check for drug resistance. The plates were incubated at 37°C for 4 weeks.
Mouse infection.
C57BL/6 and SCID female mice (6 to 8 weeks old) were obtained from Envigo (Somerset, NJ). The animal protocol “Evaluation of the safety and the efficacy of attenuated mycobacterial vaccine vectors” used in this study was approved by the Einstein Animal Institute, which is accredited by the “American Association for the Use of Laboratory Animals” and accepts as mandatory the NIH “Principles for the Use of Animals.” The mycobacterial strains were grown to mid-log phase (OD600 of ~0.6 to 0.8), centrifuged, and washed twice with PBS containing 0.05% tyloxapol. Cell pellets were resuspended in PBS-tyloxapol, sonicated twice, and diluted in PBS-tyloxapol. SCID mice were infected via tail vein injection with 5 × 105 CFU of each strain. C57BL/6 mice were infected via the subcutaneous route at a dose of 5 × 105 CFU (control mice received 0.1 ml PBS subcutaneously). C57BL/6 mice were boosted 3 weeks later with another dose of 5 × 105 CFU of each strain. Six weeks postimmunization, C57BL/6 mice were infected via low-dose aerosol (28) with H37Rv (48 CFU). For organ bacterial burden determination, mice were euthanized, and lungs and spleens were collected and homogenized in PBS-tyloxapol. The lung and spleen homogenates were plated on Middlebrook 7H10-OADC-glycerol and Middlebrook 7H10-OADC-glycerol-PLAM. CFU were counted after 4 and 8 weeks of incubation at 37°C. Colonies that grew on Middlebrook 7H10-OADC-glycerol-PLAM plates were picked and patched onto Middlebrook 7H10-OADC-glycerol and Middlebrook 7H10-OADC-glycerol-PLAM plates, and the plates were incubated at 37°C for 4 weeks to check for nutrient requirements.
ACKNOWLEDGMENTS
We are grateful to Mei Chen and John Kim for technical assistance with the animal work. We acknowledge the Analytical Imaging Facility from the Albert Einstein College of Medicine.
Research reported in this publication was supported by the Albert Einstein Cancer Center support grant of the National Institutes of Health under award number P30CA013330. P.J. acknowledges the support from the Stony Wold Foundation and the Potts Memorial Foundation. This work was supported by the National Institutes of Health grant AI26170 and U19AI111276 (to W.R.J.).
Footnotes
Citation Vilchèze C, Copeland J, Keiser TL, Weisbrod T, Washington J, Jain P, Malek A, Weinrick B, Jacobs WR, Jr. 2018. Rational design of biosafety level 2-approved, multidrug-resistant strains of Mycobacterium tuberculosis through nutrient auxotrophy. mBio 9:e00938-18. https://doi.org/10.1128/mBio.00938-18.
REFERENCES
- 1.World Health Organization 2017. Multidrug-resistant tuberculosis (MDR-TB). 2017 update. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/challenges/mdr/MDR-RR_TB_factsheet_2017.pdf?ua=1. [Google Scholar]
- 2.Hondalus MK, Bardarov S, Russell R, Chan J, Jacobs WR Jr, Bloom BR. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect Immun 68:2888–2898. doi: 10.1128/IAI.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson M, Phalen SW, Lagranderie M, Ensergueix D, Chavarot P, Marchal G, McMurray DN, Gicquel B, Guilhot C. 1999. Persistence and protective efficacy of a Mycobacterium tuberculosis auxotroph vaccine. Infect Immun 67:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAdam RA, Weisbrod TR, Martin J, Scuderi JD, Brown AM, Cirillo JD, Bloom BR, Jacobs WR Jr.. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun 63:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR Jr.. 2006. Mycobacterium tuberculosis DeltaRD1 DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309–6320. doi: 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- 6.Jain P, Hsu T, Arai M, Biermann K, Thaler DS, Nguyen A, González PA, Tufariello JM, Kriakov J, Chen B, Larsen MH, Jacobs WR Jr.. 2014. Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio 5:e01245-14. doi: 10.1128/mBio.01245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berney M, Berney-Meyer L, Wong KW, Chen B, Chen M, Kim J, Wang J, Harris D, Parkhill J, Chan J, Wang F, Jacobs WR Jr.. 2015. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 112:10008–10013. doi: 10.1073/pnas.1513033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordhan BG, Smith DA, Alderton H, McAdam RA, Bancroft GJ, Mizrahi V. 2002. Construction and phenotypic characterization of an auxotrophic mutant of Mycobacterium tuberculosis defective in l-arginine biosynthesis. Infect Immun 70:3080–3084. doi: 10.1128/IAI.70.6.3080-3084.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deb C, Lee CM, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan GJ, Hoff DR, Driver ER, Voskuil MI, Gonzalez-Juarrero M, Basaraba RJ, Crick DC, Spencer JS, Lenaerts AJ. 2010. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PLoS One 5:e11108. doi: 10.1371/journal.pone.0011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilchèze C, Kremer L. 2017. Acid-fast positive and acid-fast negative Mycobacterium tuberculosis: the Koch paradox. Microbiol Spectr 5. doi: 10.1128/microbiolspec.TBTB2-0003-2015. [DOI] [PubMed] [Google Scholar]
- 12.Hatfull GF. 2014. Mycobacteriophages: windows into tuberculosis. PLoS Pathog 10:e1003953. doi: 10.1371/journal.ppat.1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain P, Hartman TE, Eisenberg N, O’Donnell MR, Kriakov J, Govender K, Makume M, Thaler DS, Hatfull GF, Sturm AW, Larsen MH, Moodley P, Jacobs WR Jr.. 2012. phi(2)GFP10, a high-intensity fluorophage, enables detection and rapid drug susceptibility testing of Mycobacterium tuberculosis directly from sputum samples. J Clin Microbiol 50:1362–1369. doi: 10.1128/JCM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell MR, Pym A, Jain P, Munsamy V, Wolf A, Karim F, Jacobs WR Jr, Larsen MH. 2015. A novel reporter phage to detect tuberculosis and rifampin resistance in a high-HIV-burden population. J Clin Microbiol 53:2188–2194. doi: 10.1128/JCM.03530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing Genomics And Evolutionary Science (SEA-PHAGES) Program, Modlin RL, Hendrix RW, Hatfull GF. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayer O, Jain P, Weisbrod TR, Biro D, Ho L, Jacobs-Sera D, Hatfull GF, Jacobs WR Jr.. 2016. Fluorescent reporter DS6A mycobacteriophages reveal unique variations in infectibility and phage production in mycobacteria. J Bacteriol 198:3220–3232. doi: 10.1128/JB.00592-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain P, Weinrick BC, Kalivoda EJ, Yang H, Munsamy V, Vilcheze C, Weisbrod TR, Larsen MH, O’Donnell MR, Pym A, Jacobs WR Jr.. 2016. Dual-reporter mycobacteriophages (Phi2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 7:e01023-16. doi: 10.1128/mBio.01023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 19.Miller LP, Crawford JT, Shinnick TM. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother 38:805–811. doi: 10.1128/AAC.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng TW, Saavedra-Ávila NA, Kennedy SC, Carreño LJ, Porcelli SA. 2015. Current efforts and future prospects in the development of live mycobacteria as vaccines. Expert Rev Vaccines 14:1493–1507. doi: 10.1586/14760584.2015.1089175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calmette A. 1931. Preventive vaccination against tuberculosis with BCG. Proc R Soc Med 24:1481–1490. [PMC free article] [PubMed] [Google Scholar]
- 22.Behr MA. 2002. BCG—different strains, different vaccines? Lancet Infect Dis 2:86–92. doi: 10.1016/S1473-3099(02)00182-2. [DOI] [PubMed] [Google Scholar]
- 23.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Zhang Y, Zheng H, Pan Y, Liu H, Du P, Wan L, Liu J, Zhu B, Zhao G, Chen C, Wan K. 2013. Genome sequencing and analysis of BCG vaccine strains. PLoS One 8:e71243. doi: 10.1371/journal.pone.0071243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilchèze C, Jacobs WR Jr.. 2014. Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013. doi: 10.1128/microbiolspec.MGM2-0014-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ando H, Kondo Y, Suetake T, Toyota E, Kato S, Mori T, Kirikae T. 2010. Identification of katG mutations associated with high-level isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 54:1793–1799. doi: 10.1128/AAC.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr.. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 28.Chen B, Weisbrod TR, Hsu T, Sambandamurthy V, Vieira-Cruz D, Chibbaro A, Ghidoni D, Kile T, Barkley WE, Vilchèze C, Colon-Berezin C, Thaler DS, Larsen MH, Sturm AW, Jacobs WR Jr.. 2011. Einstein contained aerosol pulmonizer (ECAP): improved biosafety for multi-drug resistant (MDR) and extensively drug resistant (XDR) Mycobacterium tuberculosis aerosol infection studies. Appl Biosaf 16:134–138. doi: 10.1177/153567601101600302. [DOI] [PMC free article] [PubMed] [Google Scholar]