ABSTRACT
Several Old World and New World arenaviruses are responsible for severe endemic and epidemic hemorrhagic fevers, whereas other members of the Arenaviridae family are nonpathogenic. To date, no approved vaccines, antivirals, or specific treatments are available, except for Junín virus. However, protection of nonhuman primates against Lassa fever virus (LASV) is possible through the inoculation of the closely related but nonpathogenic Mopeia virus (MOPV) before challenge with LASV. We reasoned that this virus, modified by using reverse genetics, would represent the basis for the generation of a vaccine platform against LASV and other pathogenic arenaviruses. After showing evidence of exoribonuclease (ExoN) activity in NP of MOPV, we found that this activity was essential for multiplication in antigen-presenting cells. The introduction of multiple mutations in the ExoN site of MOPV NP generated a hyperattenuated strain (MOPVExoN6b) that is (i) genetically stable over passages, (ii) has increased immunogenic properties compared to those of MOPV, and (iii) still promotes a strong type I interferon (IFN) response. MOPVExoN6b was further modified to harbor the envelope glycoproteins of heterologous pathogenic arenaviruses, such as LASV or Lujo, Machupo, Guanarito, Chapare, or Sabia virus in order to broaden specific antigenicity while preserving the hyperattenuated characteristics of the parental strain. Our MOPV-based vaccine candidate for LASV, MOPEVACLASV, was used in a one-shot immunization assay in nonhuman primates and fully protected them from a lethal challenge with LASV. Thus, our hyperattenuated strain of MOPV constitutes a promising new live-attenuated vaccine platform to immunize against several, if not all, pathogenic arenaviruses.
IMPORTANCE Arenaviruses are emerging pathogens transmitted to humans by rodents and responsible for endemic and epidemic hemorrhagic fevers of global concern. Nonspecific symptoms associated with the onset of infection make these viruses difficult to distinguish from other endemic pathogens. Moreover, the unavailability of rapid diagnosis in the field delays the identification of the virus and early care for treatment and favors spreading. The vaccination of exposed populations would be of great help to decrease morbidity and human-to-human transmission. Using reverse genetics, we generated a vaccine platform for pathogenic arenaviruses based on a modified and hyperattenuated strain of the nonpathogenic Mopeia virus and showed that the Lassa virus candidate fully protected nonhuman primates from a lethal challenge. These results showed that a rationally designed recombinant MOPV-based vaccine is safe, immunogenic, and efficacious in nonhuman primates.
KEYWORDS: viral hemorrhagic fevers, arenavirus, innate immunity, Lassa fever, live-vector vaccines
INTRODUCTION
Lassa virus (LASV) is an Old World arenavirus responsible for a hemorrhagic fever that is endemic in West Africa. Epidemiological data are scarce, but up to 300,000 people may be infected annually, with several thousand of them succumbing to the disease (1), while survivors often experience debilitating sequelae, such as irreversible deafness (2). Lassa fever (LF) is the viral hemorrhagic fever (VHF) the most frequently imported to northern countries, with approximately 30 cases during the last 10 years. More worrying, the area of circulation of LASV is expanding, as illustrated by the outbreak of LF that occurred in Benin in 2015, a country previously not affected by this virus, or by the sporadic cases observed in Mali, Burkina Faso, Ghana, and Ivory Coast. Although the mortality rate of LASV is frequently between 5 and 15%, outbreaks associated with a mortality rate of approximately 30 to 40% have been observed recently in Nigeria and Benin, and almost half of imported LF cases have resulted in death.
Arenaviruses are enveloped viruses with a bisegmented single-stranded RNA genome with an ambisense coding strategy. The large (L) segment encodes matrix protein Z and RNA-dependent RNA polymerase L, whereas the small (S) segment encodes the nucleoprotein (NP) and the precursor GPC of the envelope glycoprotein GP1-GP2 (3). Except for pathogenic arenaviruses isolated in humans, most arenaviruses have been isolated from their natural rodent reservoirs and have never been associated with human disease. Among Old World arenaviruses, Mopeia virus (MOPV) has been isolated in Mozambique from the same natural reservoir as LASV, Mastomys natalensis, and is closely related to LASV, sharing approximately 75% amino acid homology (4). MOPV is nonpathogenic in nonhuman primates (NHP) and can even protect them against a lethal challenge with LASV (5, 6). We have previously shown that both viruses share specific tropism for antigen-presenting cells (APC), in particular dendritic cells (DC) and macrophages (MP) (7, 8), but only MOPV (i) activates APC, (ii) induces type I interferon (IFN) production (9), and (iii) promotes a robust activation of T cells harboring functional cytotoxic properties (10). The greater immunogenicity of MOPV than of LASV, together with their close relationship, may explain the fact that MOPV is a “natural live-attenuated” vaccine against LASV. However, the complete absence of pathogenicity in NHP has not been demonstrated (6), and limited virulence has been observed in a mouse model (11). Thus, this natural virus cannot be used in humans as a vaccine.
Reverse genetics greatly improved the range of possibilities to study arenaviruses, which also included the generation of attenuated viruses for vaccine purposes (12, 13). Hence, viruses with a switch of the open reading frame (ORF) or codon deoptimization in the S segment of lymphocytic choriomeningitis virus (LCMV) are viable and attenuated and protected mice from lethal challenge with wild-type (WT) LCMV (14–16). Similar results were obtained with the replacement of the intergenic region of the L segment with that of the S segment of LCMV (17, 18). To combat LF, several vaccine candidates against LASV have already been assayed in NHP models and showed promising results. A vesicular stomatitis virus (VSV)-based candidate expressing LASV GPC fully protected NHP against several LASV strains (19, 20), but the use of the VSV backbone in humans might be hampered because of possible side effects (21). The expression of LASV GPC is sufficient to promote strong CD4+ and CD8+ T-cell responses, but other virus-derived antigens may also confer protection (20). To date, the only arenavirus-based vaccine for LF is the Mopeia/Lassa virus reassortant virus ML29, which harbors the L segment of MOPV and the S segment of LASV. ML29 was shown to protect NHP in a challenge with LASV, but the lack of reverse genetics for ML29 impedes the manipulation and control of its genome, particularly with WT NP of LASV, a known virulence factor that drastically dampens the type I IFN response in APC (22). Taken together, these results suggest that if MOPV can act as a vaccine for LASV, a reverse-genetics system for MOPV used with an attenuated virulence factor(s) and specific LASV antigenic properties would represent a promising new vaccine strategy.
NP of arenaviruses contains a DEDDH exoribonuclease (ExoN) domain that can digest double-stranded RNA (dsRNA), limiting the recognition of pathogen-associated molecular patterns by cell sensors and suppressing the induction of type I IFN responses (23, 24). We first confirmed that NP of MOPV has 3′-to-5′ exoribonuclease activity and used reverse genetics to generate a recombinant MOPV in which this function of NP was stably abrogated. This virus is highly attenuated in APC, more immunogenic than MOPV itself in terms of cell activation, and promotes a type I IFN response comparable to that with WT MOPV (MOPVWT). Further modifications of the genome of this “hyperattenuated” version of MOPV, named MOPEVAC, allowed the introduction of the heterologous GPC ORF to generate MOPV-based strains against pathogenic Old World and New World arenaviruses. The MOPVExoN6b GPC LASV candidate (MOPEVACLAS) was then used to immunize cynomolgus macaques before challenge with LASV. All immunized animal survived challenge, while all terminally ill control animals were euthanized. Our results establish the basis of an efficacious and versatile recombinant MOPV-based arenavirus vaccine platform against pathogenic arenaviruses.
RESULTS
Reverse genetics for generation of an ExoN knockout-based attenuated MOPV.
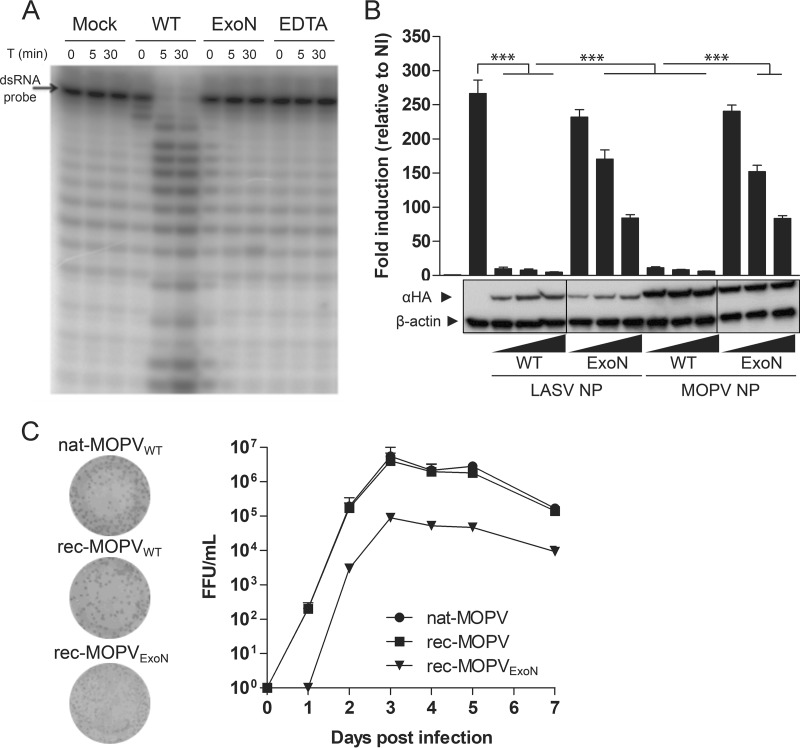
Previous studies have demonstrated the potential of MOPV to provide protection from LF (5, 6, 25). We propose the use of MOPV as a molecular basis for the development of a putative vaccine platform for pathogenic arenaviruses. NP of most arenaviruses is a virulence factor that is able to hinder the recognition of dsRNA intermediates by cellular sensors through their ExoN activity. We assessed the potential ExoN activity of NP of MOPV and its inhibition by synthesizing the C-terminal domains (residues 354 to 569) of WT MOPV and the D390A/G393A (NP ExoN) double mutant and incubating them for up to 30 min with radiolabeled 20-bp-long dsRNA probes. Gel separation of the reaction products showed that the WT domain was able to degrade the probe in a time- and Mn2+-dependent manner, contrary to the D390A/G393A mutant, which was unable to degrade the probe (Fig. 1A). We further assessed the role of ExoN activity in the inhibition of dsRNA sensing by cell host factors. We cotransfected HEK293T cells with a plasmid driving the expression of firefly luciferase (FF-Luc) under the control of an IFN regulatory 3 (IRF3)-dependent promoter along with expression plasmids for hemagglutinin (HA)-tagged versions of NP from the WT or the ExoN mutant (D390A/G393A) of MOPV, before infection with Sendai virus (SeV). The HA-tagged versions of the WT or the ExoN mutant (D389A/G392A) of LASV were used as controls. Western blot analysis of the cellular NP content showed a general decrease in the expression level of LASV NP relative to that of MOPV NP in our assay, regardless of the amount of transfected plasmids used. Both the WT and the ExoN mutant of MOPV were expressed at similar levels. In the absence of NP, SeV infection increased IRF3-dependent FF-Luc activity (mock) by 260-fold. The WT versions of both LASV and MOPV NP significantly inhibited this activity by 95 and 90%, respectively, whereas mutated NP was unable to inhibit FF-Luc activity by more than 10% at the lowest concentration and 60% at the highest (Fig. 1B). Hence, MOPV NP has ExoN activity that can be reduced by the introduction of mutations in the active site of the enzyme.
FIG 1.
MOPV NP has ExoN activity required for efficient multiplication. (A) In vitro degradation of a 5′-γ-32P-labeled dsRNA probe by the recombinant C-terminal domain of WT or D390A/G393A mutant MOPV NP. Purified ExoN domains and substrate probes were incubated with 5 mM MnCl2 for up to 30 min. The WT ExoN domain was incubated with EDTA (10 mM) as a positive control for the abrogation of ExoN activity. T, time. (B) Inhibition of SeV-induced IRF3 activation by ExoN activity of MOPV NP. HEK293T cells were transfected with plasmids encoding WT or ExoN mutant NP of MOPV or LASV along with a plasmid with FF-Luc under the control of an IRF3 response element (p55CIB-Luc) for 24 h prior to infection with SeV. At 24 h postinfection, cells were lysed, and Luc activities were measured by using the Dual-Glo luciferase assay (Promega). FF-Luc activity was normalized to that of renilla Luc. Both levels of luciferase activity are expressed as arbitrary units, and the results are expressed as the means ± standard errors of the means of data from three independent experiments performed in triplicate. ***, P < 0.001. The vertical lines indicate where the original blots were spliced together. NI, noninfected. (C) Reverse genetics for MOPV. Transfection of mouse Pol-I-driven expression plasmids for the S (with the WT or D390A/G393A mutant NP ORF) and WT L segments with plasmids encoding WT NP and L-polymerase (Lpol) ORFs of MOPV allowed the rescue of rec-MOPVWT and rec-MOPVExoN. The replication kinetics of passage 2 recombinant viruses were compared to those of nat-MOPVWT in Vero E6 cells infected at an MOI of 0.001. Supernatants were collected and titrated. Results are expressed as FFU per milliliter. Plaque phenotypes for the three viruses harvested 72 h after infection are shown.
We next investigated the consequence of the introduction of the same mutations in NP of MOPV in an infectious context by setting up a four-plasmid reverse-genetics system for MOPV similar to that of LASV (26). Both WT recombinant MOPV (rec-MOPVWT) and the recombinant ExoN mutant of MOPV (rec-MOPVExoN) were rescued, and the kinetics of their growth were compared to that of the WT natural virus (nat-MOPVWT) in Vero E6 cells infected at a multiplicity of infection (MOI) of 0.001 (Fig. 1C). The growth rates of nat- and rec-MOPVWT rapidly reached a plateau and were indistinguishable from each other. The growth curve of rec-MOPVExoN was parallel to that of the WT counterparts but with a 15-fold-lower titer. Similar results were obtained with Vero E6 cells infected at an MOI of 0.01 (data not shown). Both versions of MOPVWT had similar plaque phenotypes, whereas rec-MOPVExoN had a small-plaque phenotype. The delayed and reduced multiplication of MOPV ExoN in Vero E6 cells suggests that the introduction of these mutations is responsible for the observed attenuation.
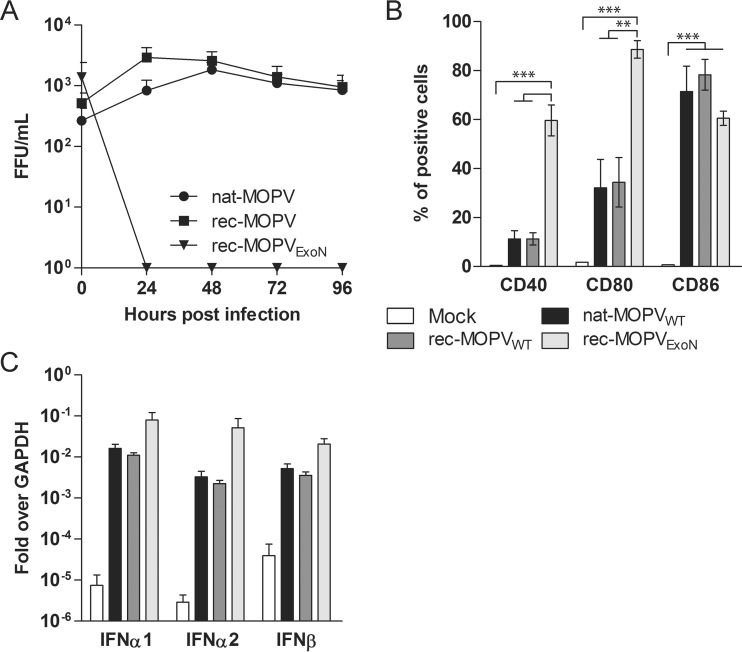
To verify the attenuated profile of our rec-MOPVExoN, we infected monocyte-derived MP along with nat-MOPVWT or rec-MOPVWT. MP were infected with all three viruses at an MOI of 0.1, and the cell culture supernatants were collected for 4 days and titrated in order to compare their capacities to replicate (Fig. 2A). Both natural and recombinant strains of MOPVWT were able to sustain multiplication throughout the experiments. We were unable to detect infectious rec-MOPVExoN in the supernatants of MP. To compare the immunogenicity of rec-MOPVExoN with those of the WT viruses, MP infected with these viruses at an MOI of 1 were monitored for the cell surface expression of activation markers 48 h after infection (Fig. 2B). Infection with nat- or rec-MOPVWT promoted similar increases in the expression levels of CD40 and CD80 in MP compared to those under mock conditions, but this difference was not significant. In contrast, there was a large and significant increase in the percentage of cells that were positive for the surface expression of both molecules following rec-MOPVExoN infection (P < 0.01 and P < 0.001, respectively). All three viruses promoted similar increases in CD86 expression levels in MP compared to mock-infected MP (P < 0.001). We then compared the type I IFN responses to infection with the three viruses by the quantification of mRNA levels of IFN-β, IFN-α1, and IFN-α2 in infected MP. All viruses promoted robust increases in the mRNA levels of all three IFNs compared to mock infection. Our results indicate that the knockdown of ExoN activity in MOPV prevents the replication of the virus and promotes strong cellular activation in MP but has a limited impact on increasing an already strong type I IFN response. We reasoned that the generation of this hyperattenuated MOPV could constitute the starting point for a MOPV-based vaccine platform for pathogenic arenaviruses.
FIG 2.
ExoN mutant MOPV is a potent activator of MP and the type I IFN response. MP were infected with nat-MOPVWT, rec-MOPVWT, or rec-MOPVExoN for 1 h; washed twice; and cultured in complete MP medium. (A) The supernatants of MP infected at an MOI of 0.1 were collected after infection, and every 24 h for 4 days, and titrated. (B) MP infected at an MOI of 1 were detached 48 h after infection; saturated with human IgG; surface stained with antibodies to CD40, CD80, and CD86 before final fixation in PBS–1% PFA; and analyzed by flow cytometry (BD Biosciences). **, P < 0.01; ***, P < 0.001. (C) RT-qPCR analysis of IFN-α1, -α2, and -β; TNF-α; and CXCL10 mRNA levels in mock-infected MP and MP infected at an MOI of 1, 24 h after infection. The expression levels of all genes were normalized to GAPDH mRNA levels. The final results are expressed as fold induction relative to GAPDH. The results represent the means ± standard errors of the means of data gathered from four different donors.
Reinforcement of the mutation at the ExoN site of MOPV NP.
The attenuation of ExoN of MOPV relies on the introduction of two mutations, in the active site of the enzyme, of residues involved in the divalent cation binding site. Since we previously observed reversion to the WT in a single ExoN NP mutant of LASV, we substituted up to five additional residues (mutant 3 [M3] to M7) (Fig. 3A) of the DEDDH domain of MOPV NP that are in direct or close contact with the divalent cation required for ExoN activity to strongly minimize the probability of the reversion of the ExoN-mutated NP ORF to the WT (27). The selected mutations were first introduced into an HA-tagged MOPV NP expression plasmid, and their capacity to block the IRF3-driven response to SeV infection in HEK293T cells was measured. All mutated NP had similar expression profiles, regardless of the number of mutations of the ExoN active site (Fig. 3A). WT MOPV NP inhibited 95% of FF-Luc IRF3-dependent induction, whereas all other mutants had at least a 10-fold decrease in ExoN activity (Fig. 3A). We then introduced these mutations into the NP ORF of the MOPV S segment, and the corresponding viruses were rescued. The ExoN4, ExoN6a, and ExoN7 mutant viruses were discontinued, as they had low titers and delayed kinetics incompatible with further studies. The ExoN3, ExoN5, and ExoN6b variants had titers, growth kinetics, and plaque phenotypes in Vero E6 cells that were similar to those of MOPVExoN (Fig. 3B).
FIG 3.
Reinforcement of the mutated ExoN site of MOPV NP. (A) Schematic representation of the DEDDH domain of MOPV NP and nomenclature of the introduced mutations. The capacities of WT and mutated NP to inhibit the IRF3-dependent expression of FF-Luc activity in HEK293T cells infected by SeV were measured and normalized as described in the legend of Fig. 1B. The results obtained are shown as the means ± standard errors of the means of data from three independent experiments performed in triplicate. (B) Reverse genetics for the NP mutant viruses MOPVExo3, MOPVExo5, and MOPVExoN6b and comparison of growth kinetics with those of MOPVWT and MOPVExoN in Vero E6 cells. Infected-cell supernatants were collected and titrated as described in the legend of Fig. 1D. (C to F) MP were infected with MOPVWT, MOPVExoN, and MOPVExoN6b for 1 h; washed twice; and cultured in complete MP medium. (C and D) The supernatants of MP infected at an MOI of 0.1 were collected and titrated (C), and NP RNA was quantified from total RNA extracted from infected cells by using an in-house RT-qPCR assay (D). (E) MP infected at an MOI of 1 were stained 48 h after infection for cell surface expression of the CD40, CD80, and CD86 activation markers before analysis by flow cytometry. (F) RT-qPCR quantification of IFN-α1, -α2, and -β mRNA levels in mock-infected MP and MP infected at an MOI of 1, 24 h after infection, normalized to GAPDH mRNA levels, as described in the legend of Fig. 2C. The results represent the means ± standard errors of the means of data collected from three different donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The MOPVExoN6b variant was selected for further analysis, as it contained the highest number of mutations. We investigated its capacity to replicate and induce an immune response in APC. MP were infected at an MOI of 0.1, and cell culture supernatants were titrated. Similar to MOPVExoN, MOPVExoNb6 was not released from infected MP, in contrast to MOPVWT (Fig. 3C). Quantification of viral RNA in infected MP showed an increase in viral RNA copy numbers for MOPVWT starting 24 h after infection. In contrast, the viral RNA content in MOPVExoN- and MOPVExoN6b-infected cells decreased throughout the experiment (Fig. 3D). We monitored the cell surface activation markers CD40, CD80, and CD86 after infection at an MOI of 1. Both MOPVExoN and MOPVExoN6b promoted enhanced numbers of CD40- and CD80-positive MP compared to MOPVWT, but only CD80 expression was significantly increased between WT and mutated MOPV (P < 0.01). CD86 was highly upregulated in infected MP relative to mock-infected cells for all three viruses (P < 0.001) (Fig. 3E). The type I IFN response of infected MP was 1,000- to 10,000-fold greater than that in noninfected MP for all three viruses (Fig. 3F). MOPVExoN6b showed the same growth characteristics as those of MOPVExoN, despite the four additional mutations being introduced into the ExoN active site of the NP. Both viruses were unable to replicate in MP but promoted stronger activation than did the WT virus and a comparable type I IFN response. We thus propose the use of hyperattenuated MOPVExoN6b, with the reinforced knockout of ExoN activity, as a platform for the design of vaccine candidates against pathogenic arenaviruses and name it “MOPEVAC.”
Swapping the GPC ORF of LASV and MOPV to generate a vaccine specific for Lassa fever.
In order to use GPC of pathogenic arenaviruses as specific antigens for immune activation in the MOPEVAC backbone, we investigated whether the envelopes of MOPV and LASV might be involved in virulence. We modified the S segments of both viruses and used reverse genetics to generate MOPV expressing GPC of LASV (MOPVGPC LASV) and LASV expressing GPC of MOPV (LASVGPC MOPV) (Fig. 4A). We first analyzed their replication along with those of their WT counterparts in Vero E6 cells and found that all four viruses had similar titers and growth kinetics, reaching a plateau 3 days after infection. Only LASVGPC MOPV showed a slight delay 24 h after infection relative to the other viruses (Fig. 4B). We then used these viruses to infect MP at an MOI of 1 and quantified the type I IFN response by reverse transcription-quantitative PCR (qRT-PCR) 24 h after infection (Fig. 4C). We confirmed that MOPVWT is more immunogenic than LASVWT, as type I IFN mRNA levels induced by MOPV were higher than those induced by LASV. The GPC chimeric viruses induced type I IFN mRNA levels comparable to those of their respective WT counterparts. Our results provide direct evidence that GPC does not play a role in the control of the type I IFN response in MP.
FIG 4.
GPC of LASV and MOPV are not involved in the type I IFN response. (A) Schematic representation of the GPC-deleted version of the LASV or MOPV S segments used to generate the chimeric viruses MOPVGPC LASV and LASVGPC MOPV. (B) Growth kinetics of the recombinant viruses MOPVWT, MOPVGPC LASV, LASVWT, and LASVGPC MOPV in Vero E6 cells infected at an MOI of 0.01. Supernatants of infected cells were titrated as described above. (C) RT-qPCR quantification of IFN-α1, -α2, and -β mRNA levels in mock-infected MP and MP infected at an MOI of 1, 24 h after infection, normalized to GAPDH mRNA levels. Results represent the means ± standard errors of the means of data collected from three different donors.
MOPEVAC as a vaccine platform for pathogenic Old World and New World arenaviruses.
Based on the use of the MOPEVAC platform, we developed a common strategy to produce vaccines against pathogenic arenaviruses. A GPC-deleted version of the MOPVExoN6b S segment was generated for the direct insertion of the GPC ORF of related pathogenic Old World or New World arenaviruses (Fig. 5A). We generated six chimeric MOPVExoN6b expressing GPC from LASV, Lujo virus (LUJV), Machupo virus (MACV), Guanarito virus (GUAV), Chapare virus (CHAV), and Sabia virus (SABV). The GPC chimeras of LASV (MOPEVACLAS), LUJV (MOPEVACLUJ), and MACV (MOPEVACMAC) had plaque phenotypes, titers, and growth rates similar to those of the parental MOPEVAC in Vero E6 cells (Fig. 5A). The GPC chimeras of GUAV (MOPEVACGUA), CHAV (MOPEVACCHA), and SABV (MOPEVACSAB) had 5- to 10-fold-lower titers and smaller-plaque phenotypes (Fig. 5A, right). To assess their stability, MOPEVAC, MOPEVACLAS, MOPEVACLUJ, and MOPEVACMAC were passaged iteratively up to 10 times in Vero E6 cells at an MOI of 0.01. The titers of the mutated viruses were compared with that of MOPVWT obtained under identical conditions (Fig. 5B). We observed a general decrease in the titers of all viruses from passage 2 to passage 5, which was more pronounced for MOPEVAC and MOPEVACMAC than for the others. Except for MOPEVAC, all other viruses recovered titers similar to titers obtained at passage 1 from passages 6 to 10. We then investigated the stability of the genome of MOPVWT, MOPEVACLAS, MOPEVACLUJ, and MOPEVACMAC throughout these passages. Viral RNA was extracted from stocks obtained at passages 1 and 5 and submitted to deep-sequencing analysis. The results showed an almost total coverage of the genome sequence, except for the intergenic region of the S segment, which is notably difficult to sequence because of the repeated sequences and secondary structures (data not shown). At passage 5, sequences were identical to the respective cloned sequences used for rescue, with no mutation in MOPVWT and only the ExoN mutations in the MOPEVAC-based viruses. All heterologous GPC sequences introduced into MOPEVAC were also identical to their respective sequences of reference. We did not detect the emergence of mutations in heterologous GPC or other regions of the MOPEVAC genome. Notably, we noted the presence of WT NP sequences in the first passages that likely originated from the residual presence of the pTM1-NP plasmid transfected at the time of rescue. However, 100% of the sequences were ExoN6b at passage 5. We then evaluated the ability of MOPEVACLAS to activate the immune response in MP and compared it to those of MOPVWT and MOPEVAC. The kinetics of replication in MP after infection at an MOI of 0.1 showed that MOPEVACLAS was unable to proliferate in MP, similarly to MOPEVAC and in contrast to MOPVWT (Fig. 6A). Viral RNA levels in cells infected with the MOPEVAC-based constructs slowly decreased during the course of the infection (Fig. 6B). Forty-eight hours after infection, 80% of MP infected with MOPEVAC and MOPEVACLAS at an MOI of 1 expressed CD40 at their cell surface, a 2-fold increase compared to MOPVWT-infected MP (P < 0.01). An 8- to 10-fold increase in the number of CD80-positive MP was observed for MOPEVAC- and MOPEVACLAS-infected cells compared to MOPVWT-infected cells (P < 0.01 and P < 0.001). On the contrary, CD86 was expressed at the surface of fewer MOPEVAC- and MOPEVACLAS-infected cells than in MOPVWT-infected MP (P < 0.01) (Fig. 6C). The type I IFN response for the MOPEVAC-based constructs was similar to that for MOPVWT, which was at least 3 logs higher than those under mock conditions (Fig. 6D). The quantification of IFN-α2 levels in the supernatants of the corresponding infected MP (MOI of 0.1) confirmed the robust induction observed at the mRNA level. We detected 27- and 108-fold more IFN-α2 in the supernatants of MOPEVAC- and MOPEVACLAS-infected cells, respectively, than in the supernatants of WT MOPV-infected MP (Fig. 6E). Taken together, our results show that MOPEVACLAS induced cellular activation and a strong type I IFN response in infected MP similar to those induced by its parental MOPEVAC. Therefore, the GPC ORF present on the surface of our vaccine candidates does not modify their capacity to activate APC.
FIG 5.
Generation of a vaccine strain platform for arenaviruses based on attenuated recombinant MOPV. (A) Reverse-genetics strategy for the generation of MOPVExoN6b expressing GPC of the pathogenic arenaviruses LASV, LUJV, MACV, GUAV, CHAV, and SABV. Passage 2 viruses were used to infect Vero E6 cells at an MOI of 0.01, and supernatants were collected and titrated. A representative example of the plaque phenotype of passage 2 viruses is shown. (B) Multiple passages of MOPVWT, MOPVExoN6b, MOPVExoN6b GPC LASV, MOPVExoN6b GPC LUJV, and MOPVExoN6b GPC MACV in Vero E6 cells. De novo stocks of all five viruses, from passage 1 to passage 10, were used to infect cells for 4 days at an MOI of 0.01.
FIG 6.
The MOPEVACLAS strain is an effective activator of MP and the type I IFN response. MP were infected with MOPVWT, MOPVExoN6b, or MOPVExoN6b GPC LASV for 1 h, washed twice, and cultured in complete MP medium. (A) The supernatants of MP infected at an MOI of 0.1 were collected after infection for 4 days and titrated as described in Materials and Methods. (B) Total RNA was extracted from the cells, and NP RNA levels were quantified by using an in-house RT-qPCR assay. (C) Cell surface expression of CD40, CD80, and CD86 in mock-infected MP and MP infected at an MOI of 1, 48 h after infection. (D) Total RNA in MP infected at an MOI of 1 were collected 24 h after infection and DNase treated, and mRNA levels of IFN-α1, -α2, and -β were quantified after oligo(dT) cDNA-driven RT and primer/probe PCR quantification (Applied Biosystems). The expression levels of all genes were normalized to GAPDH mRNA levels. The final results are expressed as fold induction relative to GAPDH. (E) Quantification of IFN-α2 levels in supernatants of infected MP (MOI of 0.1) harvested 24 h postinfection by an ELISA. For panels C to E, results represent the means ± standard errors of the means of data collected from three different donors. **, P < 0.01; ***, P < 0.001.
A single shot of MOPEVACLAS protects cynomolgus monkeys against LASV challenge.
To confirm the potential of our MOPEVACLAS candidate as an effective vaccine against LASV, we immunized four cynomolgus monkeys with 6 × 106 focus-forming units (FFU) 37 days before challenge with LASV (Josiah strain). Three animals were treated with an irrelevant vaccine and used as controls. A single dose of either vaccine was injected during the study. Following the subcutaneous injection of MOPEVACLAS, we did not record any sign of clinical symptoms or viremia during the 37 days before challenge, except for low-grade fever observed for all animals on the day of immunization. Blood samples were collected at several time points, and the activation of CD4+ and CD8+ T cells was measured after stimulation with LASV GPC-derived peptides. Our results showed that tumor necrosis factor alpha (TNF-α)-producing CD8+ and CD4+ T cells were induced 14 days after immunization in most immunized animals in response to LASV GPC-derived peptides. In one animal, GPC-specific CD8+ T cells were rather detected 30 days after immunization, while in another one, GPC-specific CD4 T cells circulated for 14 days postimmunization (Table 1). Importantly, neutralizing antibodies specific for MOPEVACLAS were detected in the plasma of all immunized animals 23 days after immunization. Thus, a single shot of MOPEVACLAS was able to induce both cellular and humoral immune responses against LASV.
TABLE 1.
Immunogenicity and efficacy of MOPEVACLAS in LASV NHP challenge
| Animal | Vaccine | MOPEVACLAS-NAb titera | % of CD8+ T cells producing TNF-αb | % of CD4+ T cells producing TNF-αb | Day of death |
|---|---|---|---|---|---|
| 1 | MeV Schwarz | Neg | Neg | Neg | 12 |
| 2 | MeV Schwarz | Neg | Neg | Neg | 14 |
| 3 | MeV Schwarz | Neg | Neg | Neg | 15 |
| 4 | MOPEVACLAS | 1:100 | 0.07c | 0.15 | Survived |
| 5 | MOPEVACLAS | 1:250 | 0.05 | 0.11d | Survived |
| 6 | MOPEVACLAS | 1:100 | 2.62 | 1.12 | Survived |
| 7 | MOPEVACLAS | 1:100 | 0.37 | 0.39 | Survived |
Fifty percent MOPEVACLAS-neutralizing antibody (NAb) titers.
Percentages of LASV GPC-specific CD8+ and CD4+ T cells producing TNF-α at day 21 postimmunization. Neg, negative.
Percentage of LASV GPC-specific CD8+ T cells producing TNF-α at day 30 postimmunization.
Percentage of LASV GPC-specific CD4+ T cells producing TNF-α at day 14 postimmunization.
All animals were then submitted to a challenge with LASV injected subcutaneously (1,500 FFU). The three control animals presented severe clinical signs, including fever, anorexia, prostration, and final hypothermia, and were killed at between days 12 and 15 after reaching the endpoint defined in the protocol. The four animals immunized with MOPEVACLAS survived the challenge. Three animals presented a fever from days 5 to 10, while the temperature of the fourth animal remained within the normal range. No other symptoms were recorded throughout the course of the challenge. Our results showed that MOPEVACLAS fully protects animals against a lethal challenge with LASV. Taken together, these results showed that our hyperattenuated MOPV-based LASV vaccine candidate is effective.
DISCUSSION
The close relatedness of MOPV to LASV makes it a live-attenuated vaccine against LASV (5, 25, 28). However, the lack of a complete understanding of MOPV behavior in NHP and, more importantly, the lack of data concerning MOPV infection in humans impede its use as a vaccine against LASV. The reassorted virus ML29 allows the presentation of LASV GP1 and GP2 to the immune system, both of which are antigens that are known to be sufficient for protection against LASV (19, 28–31). Indeed, ML29 is highly immunogenic in NHP, and a single shot fully protects them against a lethal LASV challenge (31). The viability and attenuation profile of ML29 likely rely on the close relationship between both viruses, but such a strategy to generate vaccines for other pathogenic arenaviruses may not be extendable. Moreover, the prolonged presence of infectious material in blood and/or organs, including brain and lungs, after immunization and also the possibility of shedding through oral, rectal, and vaginal secretions make it difficult to cope with today's standard for human vaccine purposes (31). These results suggest that WT LASV ExoN activity renders ML29 capable of multiplying in monkey cells, thereby explaining the presence of infectious material weeks after immunization in NHP and an even higher level of viremia in immunocompromised NHP. Taken together, these results raise questions regarding the host control of this vaccine strain despite its efficacy (31, 32). Here, we developed a rationally designed vaccine platform that is easily applicable to Old World and New World arenaviruses and quickly adaptable to new emerging viruses. Our approach is based on the modification of MOPV, which is possible by using a reverse-genetics system similar to the one that we developed for LASV (26). The ExoN activity located in the C-terminal domain of NP of LASV digests the dsRNA intermediates generated during viral replication, making its RNA less visible to retinoic acid-inducible gene I (RIG-I) and MDA5, the main cellular sensors involved in the type I IFN response induced by arenaviruses (22, 33). The DEDDH motif characteristics of this ExoN activity is conserved among arenaviruses, even in those not associated with pathogenicity in humans or NHP, including MOPV. We show here that the ExoN activity of MOPV is functional, is capable of digesting dsRNA, and inhibits type I IFN induction. We have not been able to compare the activity of MOPV ExoN with the one of LASV at the enzymatic level, but it seems that the increases of type I IFN mRNA synthesis in infected MP compared to the mock-infected counterparts are equivalent (about 3 logs) after the abrogation of LASV (26) and MOPV ExoN (this study) activities. However, the variations of type I IFN mRNA synthesis and IFN-α2 release between MOPV or LASV and its ExoN-deficient counterpart are more elevated in the case of LASV (26). This result suggests that a more efficacious ExoN activity of LASV, fine-tuning of NP expression, the ability to interact and inhibit the cellular partner's activity, and/or other virulence factors may be responsible for the differences in type I IFN inhibition and pathogenicity between LASV and MOPV. A recent report suggested that the Z protein of LASV, but not that of MOPV, can bind RIG-I and MDA5 and suppress subsequent type I IFN induction (34). Moreover, LASV NP has been shown to bind IκB kinase ε (IKKε) and to inhibit the further phosphorylation of IRF3 (35). The ability of MOPV NP to bind this factor is not known, and no cellular partners have been identified so far. Finally, it was recently demonstrated that the polymerase of MOPV, but not that of LASV, is able to induce the activation of RIG-I and MDA5, probably through the synthesis of dsRNA (36). Altogether, these additional mechanisms may account for the differences in pathogenicity and immunogenicity between LASV and MOPV, thereby explaining the attenuation of ML29.
The ExoN mutation in MOPVExoN induced attenuation in Vero E6 cells, which are unable to produce type I IFN, and completely impeded replication in human MP, in contrast to the WT virus. Furthermore, MOPVExoN was more immunogenic than its WT counterpart in human MP, with a slight increase of type I IFN mRNA synthesis and a strong upregulation of surface activation molecules. The abrogation of the ExoN activity of NP gave rise to a hyperattenuated MOPV, which is immunogenic in APC but unable to replicate in these cells and constitutes the cornerstone for the development of our live-attenuated vaccine against LASV. The risk of reversion toward a WT genotype/phenotype during virus passages was considered, as the abrogation of the ExoN domain of MOPVExoN relied on only two amino acid changes. Additional mutations targeting the residues involved in the catalytic site of the enzyme were introduced to circumvent this issue. We generated MOPVExoN6b, in which the abrogation of ExoN activity relies on six amino acid changes, and found that this virus is stable and retained the same replication and immunogenic properties as those of the initial MOPVExoN. This virus therefore constituted our vaccine backbone and was named MOPEVAC (Fig. 3).
Anti-GP immunity is mandatory for an arenavirus vaccine (19, 25). As this antigen has been shown to be sufficient to induce protective immunity, it would be unreasonable to use WT MOPV GPC as a heterologous antigen considering the wide array of circulating LASV strains (37). Previous studies suggested that GPC of New World arenaviruses may be involved in virulence (38–41). On the contrary, a large body of evidence arises from ML29 studies and argues against a role of LASV GPC in the virulence of LASV compared to MOPV. Indeed, no adverse effect has been observed after the inoculation of ML29, expressing LASV GPC, in guinea pigs or NHP (30, 31). To confirm that LASV GPC will not modify the virulence of MOPV, we assessed whether exchanging GPC of MOPV with that of LASV modifies the profile of the type I IFN response in ExoN mutant and WT viruses. Chimeric LASV and MOPV carrying GPC of the other virus had the same type I IFN response as their WT counterparts (Fig. 4C). These results suggest that MOPEVAC will be a safe platform, regardless of the GPC encoded. The knockdown of the ExoN function abrogates effective replication within human APC, despite infection, thereby preventing any virulence from the expression of the viral envelope. Consequently, it is likely that this vector will not persist in the host and will be rapidly eliminated without spreading. This property is an important feature for an arenavirus-based vaccine, given the ability of some arenaviruses, such as LCMV, to induce persistent infection. We therefore swapped GPC of MOPV with GPC of Old World and New World arenaviruses known to induce VHF, thereby allowing the rescue of six potential vaccine candidates.
To demonstrate that MOPEVAC is a safe and immunogenic vaccine platform that is able to provide protection after a single shot, we performed a proof-of-concept experiment with MOPEVACLAS in the relevant cynomolgus monkey model. Previous results demonstrated that MOPV can induce robust T-cell responses, including cytotoxic T cells, in an in vitro model for the induction of primary T-cell responses (10). We show here that the MOPEVACLAS is able to generate robust humoral and T-cell responses within weeks following immunization. The LASV challenge succeeded with 100% protection in immunized animals, while all mock-treated animals were euthanized due to advanced LF symptoms. Further studies are still required to assess the complete potential of our MOPV-based vaccine platform. First, the safety of the MOPV backbone must be confirmed by investigating the presence or the absence of shedding and of sterilizing immunity in NHP. We also need to understand the molecular mechanisms that drive the antigen presentation of the MOPEVACLAS live-attenuated virus for T- and B-cell activation. We detected the presence of neutralizing antibodies specific for MOPEVACLAS in immunized NHP before challenge. This significant amount of neutralizing antibodies demonstrates that the vaccine has been able to induce a functional humoral response directed against LASV GPC. Also of note is the potency of MOPEVAC in inducing neutralizing antibodies, suggesting that this platform will probably be efficient against New World arenaviruses, given the role of neutralizing antibodies for the control of these viruses (42–45). The induction of humoral and cellular responses against MOPV NP and the possible cross-reactivity against LASV NP, as well as their role in the control of LASV infection in NHP, will be further addressed. Nevertheless, the ability of vaccine candidates based only on LASV GPC to provide full protection against LASV infection was demonstrated previously (19, 25), suggesting that the protective immunity induced by MOPEVACLAS is probably mainly GPC mediated.
The specifications of an “ideal” vaccine against LASV, or other arenaviruses, would be (i) to induce T-cell responses, which are mandatory for protection; (ii) to be effective after a single inoculation, to facilitate vaccination programs in countries of endemicity and because ring vaccination is very useful for the control of outbreaks (46); (iii) to use homologous GPC, as this antigen is necessary and sufficient for protection; (iv) to be rapidly eliminated by the host and not persist; (v) to be genetically stable and safe; and (vi) to induce long-lasting protection. We believe that MOPEVACLAS presents all these characteristics. Outbreaks of VHF due to arenaviruses occur mainly in rural areas with poor health care infrastructures and a limited ability for diagnosis. The human and economic consequences of these outbreaks are a major threat to public health and require an appropriate answer. Given the dynamics of the Arenaviridae family, with reports of newly identified viruses as well as the emergence of known Arenaviridae in areas where they have not been reported previously (exemplified by the recent outbreaks of LF in Benin and Togo), the most efficient means to combat these diseases is to immunize people at risk. One of the advantage of our platform-based vaccine approach is its versatility toward known arenaviruses and their genetic diversity but also toward these new viruses (47, 48). We have demonstrated that the MOPEVACLAS vaccine showed promising results in the NHP model, and further investigations will be performed to confirm the complete efficacy, safety, and immunogenicity of our platform.
MATERIALS AND METHODS
Cells and viruses.
BHKT7/9 cells were used to rescue recombinant viruses and were maintained as described previously (26). Vero E6 cells and HEK293T cells were grown in GlutaMAX Dulbecco modified Eagle's medium (DMEM; Life Technologies) supplemented with 5% fetal calf serum (FCS), 1% HEPES, and 0.5% penicillin-streptomycin. Blood samples were obtained from the Etablissement Français du Sang (EFS) (Lyon, France). Mononuclear cells were purified by Ficoll density gradient centrifugation (GE Healthcare). Monocytes were first separated from peripheral blood mononuclear cells by centrifugation on a cushion of 50% Percoll (GE Healthcare, Velizy, France) in phosphate-buffered saline (PBS) and then purified by using monocyte isolation kit II according to the manufacturer's instructions (Miltenyi Biotec, Paris, France). MP were obtained by incubating monocytes for 6 days in a solution containing RPMI, 10% FCS, and 10% autologous serum supplemented with 50 ng/ml of macrophage colony-stimulating factor (M-CSF). M-CSF was added every 2 days, and 40% of the culture medium was replaced. Strain AN21366 (GenBank accession numbers JN561684 and JN561685) was used for infection and to establish the reverse-genetics system for MOPV. The LASV AV reverse-genetics system was described previously (26). The GPC ORF for LASV (strain Josiah; GenBank accession number J04324), LUJV (GenBank accession number NC_012776), MACV (GenBank accession number AY619643), GUAV (strain INH 95551; GenBank accession number AY129247), CHAV (GenBank accession number NC_010562), and SABV (GenBank accession number NC_006317) were used in this study. The DI-H4 strain of SeV was a gift from D. Garcin (University of Geneva, Switzerland).
Plasmid constructs.
For exonuclease activity experiments, the regions spanning residues 365 to 570 of WT and D390A/G393A ExoN mutant MOPV NP were cloned in frame with the N-terminal cleavable thioredoxin-hexahistidine tag into the pETG20A expression vector (Gateway; Life Technologies, Carlsbad, CA). For the reverse-genetics system of MOPV, a four-plasmid strategy was used for virus rescue. The L and S segments of MOPV were reverse transcribed from viral RNA extracts, and the resulting cDNA was cloned into the pRF108 plasmid between mouse polymerase I (Pol-I) promoter/terminator signals. An extra nontemplated G base was included at the 5′ end of the cloned sequences. The L and NP ORF were cloned into the pTM1 plasmid (T7 promoter/terminator signals) to support both the transcription and replication of the viral segments. To generate the S-segment-derived GPC chimera, GPC ORF-deleted versions of the plasmids encoding the S segments of either MOPV or LASV were generated with BsmBI restriction sites introduced downstream and upstream of the start and stop codons of the deleted ORF, respectively. The GPC ORF of heterologous arenaviruses were then cloned into the modified plasmids. For the SeV-dependent IRF3 activation reporter assay, MOPV NP with a C-terminal HA tag was expressed from the phCMV plasmid. All mutations were introduced by using a site-directed mutagenesis strategy according to the manufacturer's instructions (Agilent). All plasmid constructs were verified by sequencing.
Protein expression and exonuclease activity assay.
Transformed bacteria were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.5. Expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and bacteria were grown at 17°C overnight after supplementation of the medium with ZnCl2 to 100 μM. Cell pellets were frozen, stored at −80°C, and then resuspended in a buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 5 mM imidazole, and 5% glycerol, to which 0.04 mg/ml of DNase and 0.08 mg/ml of lysozyme had been added. The solution was lysed by using a sonicator, and the lysate was clarified by centrifugation at 20,000 rpm for 30 min at 4°C. The protein was first purified by cobalt affinity chromatography. To remove the fusion tag, the elution fraction containing the protein sample was cleaved with tobacco etch virus (TEV) and reapplied onto a second cobalt resin column. A final purification was performed by size exclusion chromatography (Superdex 75 column) in a solution containing 20 mM Tris (pH 7.5), 300 mM NaCl, 2 mM MnCl2, and 5% glycerol. High-performance liquid chromatography (HPLC)-grade synthetic 20-nucleotide (nt)-long RNA probes were purchased from Dharmacon (GE Healthcare) or Biomers (Ulm, Germany) and 5′ labeled with [γ-32P]ATP, using protein nucleotide kinase, according to the manufacturer's instructions (New England BioLabs, Ipswich, MA). In brief, standard reaction mixtures contained 0.25 μM NP C-terminal domains, 5 mM MnCl2, and 1.25 μM radiolabeled dsRNA probe. Reactions were quenched at intervals with buffer containing formamide and EDTA (10 mM), and the samples were resolved by separation on a 20% polyacrylamide gel containing 8 M urea. Gels were exposed to a phosphor screen and visualized by using a FLA-300 phosphorimager (Fujifilm, Saint-Quentin-en-Yvelines, France).
Virus rescue and titration.
The rescue procedure for the recombinant viruses was reported previously (26). Briefly, 1 × 106 BHKT7/9 cells were transfected with plasmids encoding NP and L, as well as S and L segments, by using Fugene HD (Promega, Charbonnières-les-Bains, France). Supernatants of the BHKT7/9 cells constituted the seed stock used to infect Vero E6 cells. Virus from this first passage was titrated and used to infect Vero E6 cells at an MOI of 0.01. Titrated viruses from the second passages in Vero E6 cells provided the viral stocks for all other experiments. The absence of mycoplasma contamination was confirmed for all viral stocks. For neutralization experiments, Vero E6 cells were infected with 50 to 60 FFU/well of MOPEVACLAS incubated previously for 1 h at 37°C with serial dilutions of NHP plasma samples (starting 1/40 to 1/1,500). For viral titration, cells were fixed with formaldehyde (FA) and permeabilized with PBS–0.1% Triton X-100 prior to the revelation of the presence of virus by immunostaining with a polyclonal rabbit antibody that recognizes either the LASV or the MOPV Z protein (Agrobio, France), a phosphatase alkaline-conjugated polyclonal goat anti-rabbit antibody (Sigma), and the 1-Step Nitro Blue Tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate (Thermo Fisher Scientific, Waltham, MA). Results are expressed in FFU per milliliter.
Quantitative RNA analysis.
For RT-qPCR experiments, total RNA was isolated from mock-infected or infected cells by using the RNeasy minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions, and a supplementary DNase step was added using the Turbo DNA-free kit (Ambion; Thermo Fisher Scientific). Synthesis of cDNA was performed by using SuperScript III, and amplification was performed by using the gene expression master mix kit (Applied Biosystems, Thermo Fisher Scientific). For type I IFN, the primer/probe mix was developed in-house (9). Runs of qPCR assays were performed with a LightCycler 480 instrument (Roche Diagnostics, Meylan, France). The expression levels of all genes were standardized to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene and expressed as fold induction relative to GAPDH. For viral RNA quantification, an RNA probe of the region spanning bp 771 to 934 of the NP ORF was cloned into the pGEM vector (Promega) to generate T7 polymerase-driven transcripts. The RNA probe was DNase treated, purified, and quantified (Dropsense96; Trinean, Ghent, Belgium). Quantitative PCR for viral RNA was performed with the EuroBioGreen Lo-ROX qPCR mix (Eurobio, Les Ulis, France), using primers 5′-CTTTCCCCTGGCGTGTCA-3′ and 5′-GAATTTTGAAGGCTGCCTTGA-3′.
Detection of IFN-α2 in supernatants of macrophages.
The IFN-α2 level in the supernatants of macrophages was quantified by using enzyme-linked immunosorbent assay (ELISA) matched-pair antibodies (IFN-α human ELISA kit; Thermo Fisher Scientific) according to the manufacturer's instructions.
Deep-sequencing analysis of viral genomes.
Purified RNAs were treated with 6 U Turbo DNase (Ambion) to digest contaminating DNA. First- and second-strand cDNA syntheses were conducted with random hexamer primers using SuperScript III (Thermo Fisher Scientific) and 5 U Klenow fragment (New England BioLabs), respectively. Finally, double-stranded cDNAs were purified with the MinElute PCR purification kit (Qiagen) according to the manufacturer's instructions. Libraries were prepared by using the NEBNext fast DNA fragmentation and library prep for Ion Torrent. Size selection using AMPure XP beads (Agencourt, Beckman Coulter, Brea, CA) to obtain 200-bp reads was performed, and 14 to 20 cycles of amplification were done. Finally, the enriched libraries were purified with Agencourt AMPure XP beads. The 2100 bioanalyzer (Agilent, Santa Clara, CA) and the high-sensitivity DNA kit (Agilent) were used to determine the quality and concentration of the libraries. Sequencing was performed by using the Personal Genome Machine (PGM) Ion Torrent technology. Emulsion PCR and enrichment steps were carried out by using the Ion PGM Hi-Q OT2 kit. Sequencing was undertaken by using a 318v2 chip and the Ion PGM Hi-Q sequencing kit according to the manufacturer's instructions. Automated read data sets provided by Torrent software suite 5.0 were trimmed according to the quality score (99%) and length (reads of <75 bp were removed). Trimmed fastq files were then mapped onto a reference genome using bowtie2 and filtering to remove PCR duplicates (Picard tools). Finally, variants were called by using SAMtools mpileup and varscan 2.3.6. Tables of nucleotide frequencies generated were then used to call the consensus genome in fasta format with a custom python script.
Flow cytometry for MP activation and LASV-specific T-cell activation.
Mock-infected MP and MP infected at an MOI of 1 were detached 48 h after infection, saturated with human IgG, and surface stained with antibodies to CD40, CD80, and CD86 (BD Biosciences, Le-Pont-de-Claix, France) before final fixation in PBS–1% paraformaldehyde (PFA). During the NHP challenge, LASV antigen-specific T cells from fresh whole blood incubated with a pool of overlapping GPC peptides in the presence of CD28 and CD49d antibodies (2 μg/ml) and brefeldin A (10 μg/ml) for 6 h at 37°C were analyzed. Staphylococcal enterotoxin A (SEA) (1 μg/ml) or PBS was used as the positive or negative control, respectively, for activation. Peptides are 15-mer long (1 μg/ml each), with an overlap of 11 residues, and spanned the complete GPC ORF of the LASV Josiah strain. PBS–20 mM EDTA was added to samples before cell surface staining for CD3, CD4, and CD8 (BD Biosciences). Red blood cells were then lysed by using PharmLyse (BD Biosciences). Cells were then fixed and permeabilized for intracellular staining with antibodies to TNF-α (BioLegend). Cells were analyzed by flow cytometry using an LSR Fortessa cytometer (BD Biosciences). Data were analyzed by using Kaluza software (Beckman Coulter).
Cynomolgus monkey challenge with LASV.
Briefly, a group of 4 male cynomolgus monkeys (Macaca fascicularis; 31 to 38 months of age and 3.2 to 4 kg) were immunized in biosafety level ABSL2 facilities (SILABE, Strasbourg, France) by the intramuscular injection of 6 × 106 FFU of MOPEVACLAS. Another group of 3 monkeys was immunized with measles virus (MeV) vaccine strain Schwarz, as an irrelevant vaccine. Blood draws were performed at 2 weeks postimmunization in order to assess T-cell responses against LASV GP. After 5 weeks, monkeys were transported to biosafety level 4 (BSL4) facilities (Laboratoire P4-INSERM Jean Mérieux) and challenged subcutaneously by using 1,500 FFU of the LASV Josiah strain. Animals were monitored for clinical signs of disease and euthanized according to scoring made based on body temperature, body weight, feeding, hydrating, behavior, and clinical signs. All animals still alive at 29 days postchallenge were euthanized according to validated experimental procedures. This study was approved by the Comité Régional d'Ethique en Matière d'Expérimentation Animale de Strasbourg (APAFIS 6543-20160826144775) and by the Comité Régional d'Ethique pour l'Expérimentation Animale Rhône Alpes (CECCAPP 20161110143954).
Statistical analysis.
Statistical analyses were performed by using SigmaPlot (Systat Software, San Jose, CA, USA) and GraphPad Prism (GraphPad Software, La Jolla, CA, USA). Data were analyzed by one-way analysis of variance (ANOVA) with a Bonferroni posttest.
ACKNOWLEDGMENTS
All experiments using LASV, LASVGPC MOPV, and MOPVGPC LASV were carried out in the Laboratoire Jean Mérieux-INSERM BSL4 facility. We are grateful to the BSL4 team members for support while performing the experiments. We thank R. Flick and M. Bouloy for providing the pRF108 and pTM1 plasmids and BHKT7/9 cells. p55C1B-Luc was kindly provided by T. Fujita (Institute for Virus Research, Kyoto University, Kyoto, Japan) and Luiz Martinez-Sobrído (University of Rochester, Rochester, NY, USA). We also thank D. Garcin (University of Geneva, Switzerland) for providing the DI-H4 strain of Sendai virus. We thank C. Clegg and G. Lloyd (Public Health England, Porton Down, Salisbury, United Kingdom) for the gift of MOPV; S. Becker for providing us with the AV strain of LASV; M. Bouloy for the tools used to develop the LASV and MOPV reverse-genetics systems; T. G. Ksiazek, P. E. Rollin, and P. Jahrling for LASV monoclonal antibodies; and the Etablissement Français du Sang for providing human blood.
This work was supported by the Fondation pour l'Innovation en Infectiologie (FINOVI) (Lyon, France), the Fondation pour la Recherche Médicale (FRM) (France), French National Research Agency grant ANR-11-BSV_019-02, and the Fondation Méditerranée Infection. This work was also supported by Labex Ecofect (grant ANR-11-LABX-0048, Lyon University) within the program Investissements d'Avenir (grant ANR-11-IDEX-0007, French National Research Agency).
REFERENCES
- 1.McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. 1987. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis 155:437–444. doi: 10.1093/infdis/155.3.437. [DOI] [PubMed] [Google Scholar]
- 2.Cummins D, McCormick JB, Bennett D, Samba JA, Farrar B, Machin SJ, Fisher-Hoch SP. 1990. Acute sensorineural deafness in Lassa fever. JAMA 264:2093–2096. doi: 10.1001/jama.1990.03450160063030. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier MJ, de la Torre J-C, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1827. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Wulff H, McIntosh B, Hamner D, Johnson K. 1977. Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ 55:441–444. [PMC free article] [PubMed] [Google Scholar]
- 5.Kiley MP, Lange JV, Johnson KM. 1979. Protection of rhesus monkeys from Lassa virus by immunisation with closely related arenavirus. Lancet ii:738–745. [DOI] [PubMed] [Google Scholar]
- 6.Walker DH, Johnson KM, Lange JV, Gardner JJ, Kiley MP, McCormick JB. 1982. Experimental infection of rhesus monkeys with Lassa virus and a closely related arenavirus, Mozambique virus. J Infect Dis 146:360–368. doi: 10.1093/infdis/146.3.360. [DOI] [PubMed] [Google Scholar]
- 7.Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot MC, Deubel V. 2004. Lassa virus infection of dendritic cells and macrophages is productive but fails to activate cells. J Immunol 172:2861–2869. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 8.Pannetier D, Faure C, Georges-Courbot MC, Deubel V, Baize S. 2004. Human macrophages, but not dendritic cells, are activated and produce type I interferons in response to Mopeia virus infection. J Virol 78:10516–10524. doi: 10.1128/JVI.78.19.10516-10524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baize S, Pannetier D, Faure C, Marianneau P, Marendat I, Georges-Courbot MC, Deubel V. 2006. Role of interferons in the control of Lassa virus replication in human dendritic cells and macrophages. Microbes Infect 8:1193–1202. doi: 10.1016/j.micinf.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Pannetier D, Reynard S, Russier M, Journeaux A, Tordo N, Deubel V, Baize S. 2011. Human dendritic cells infected with the non-pathogenic Mopeia virus induce stronger T-cell responses than with Lassa virus. J Virol 85:8293–8306. doi: 10.1128/JVI.02120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieger T, Merkler D, Günther S. 2013. Infection of type I interferon receptor-deficient mice with various Old World arenaviruses: a model for studying virulence and host species barriers. PLoS One 8:e72290. doi: 10.1371/journal.pone.0072290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergthaler A, Gerber NU, Merkler D, Horvath E, De la Torre J, Pinschewer DD. 2006. Envelope exchange for the generation of live-attenuated arenavirus vaccines. PLoS Pathog 2:e51. doi: 10.1371/journal.ppat.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Sobrido L, de la Torre JC. 2017. Development of recombinant arenavirus-based vaccines, p 133–149. In Ferran MC, Skuse GR (ed), Recombinant virus vaccines: methods and protocols. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 14.Cheng BYH, Ortiz-Riaño E, Nogales A, de la Torre JC, Martínez-Sobrido L. 2015. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J Virol 89:3523–3533. doi: 10.1128/JVI.03401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng BYH, Ortiz-Riaño E, de la Torre JC, Martínez-Sobrido L. 2015. Arenavirus genome rearrangement for the development of live attenuated vaccines. J Virol 89:7373–7384. doi: 10.1128/JVI.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng BYH, Nogales A, de la Torre JC, Martínez-Sobrido L. 2017. Development of live-attenuated arenavirus vaccines based on codon deoptimization of the viral glycoprotein. Virology 501:35–46. doi: 10.1016/j.virol.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasaki M, Ngo N, Cubitt B, Teijaro JR, de la Torre JC. 2015. General molecular strategy for development of arenavirus live-attenuated vaccines. J Virol 89:12166–12177. doi: 10.1128/JVI.02075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki M, Cubitt B, Sullivan BM, de la Torre JC. 2016. The high degree of sequence plasticity of the arenavirus noncoding intergenic region (IGR) enables the use of a nonviral universal synthetic IGR to attenuate arenaviruses. J Virol 90:3187–3197. doi: 10.1128/JVI.03145-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisbert TW, Jones S, Fritz EA, Shurtleff AC, Geisbert JB, Liebscher R, Grolla A, Ströher U, Fernando L, Daddario KM, Guttieri MC, Mothé BR, Larsen T, Hensley LE, Jahrling PB, Feldmann H. 2005. Development of a new vaccine for the prevention of Lassa fever. PLoS Med 2:537–545. doi: 10.1371/journal.pmed.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safronetz D, Mire C, Rosenke K, Feldmann F, Haddock E, Geisbert T, Feldmann H. 2015. A recombinant vesicular stomatitis virus-based Lassa fever vaccine protects guinea pigs and macaques against challenge with geographically and genetically distinct Lassa viruses. PLoS Negl Trop Dis 9:e0003736. doi: 10.1371/journal.pntd.0003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Hu Z, Muñoz P, Moon JE, Ruck RC, Bennett JW, Twomey PS, Gutiérrez RL, Remich SA, Hack HR, Wisniewski ML, Josleyn MD, Kwilas SA, Van Deusen N, Mbaya OT, Zhou Y, Stanley DA, Jing W, Smith KS, Shi M, Ledgerwood JE, Graham BS, Sullivan NJ, Jagodzinski LL, Peel SA, Alimonti JB, Hooper JW, Silvera PM, Martin BK, Monath TP, Ramsey WJ, Link CJ, Lane HC, Michael NL, Davey RT Jr, Thomas SJ. 2017. A recombinant vesicular stomatitis virus Ebola vaccine. N Engl J Med 376:330–341. doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynard S, Russier M, Fizet A, Carnec X, Baize S. 2014. Exonuclease domain of the Lassa virus nucleoprotein is critical to avoid retinoic acid-inducible gene I signaling and to inhibit the innate immune response. J Virol 88:13923–13927. doi: 10.1128/JVI.01923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–785. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher-Hoch SP, Hutwagner L, Brown B, McCormick JB. 2000. Effective vaccine for Lassa fever. J Virol 74:6777–6783. doi: 10.1128/JVI.74.15.6777-6783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carnec X, Baize S, Reynard S, Diancourt L, Caro V, Tordo N, Bouloy M. 2011. Lassa virus nucleoprotein mutants generated by reverse genetics induce robust type I interferon response in human dendritic cells and macrophages. J Virol 85:12093–12097. doi: 10.1128/JVI.00429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yekwa E, Khourieh J, Canard B, Papageorgiou N, Ferron F. 2017. Activity inhibition and crystal polymorphism induced by active-site metal swapping. Acta Crystallogr D Struct Biol 73:641–649. doi: 10.1107/S205979831700866X. [DOI] [PubMed] [Google Scholar]
- 28.Fisher-Hoch SP, McCormick JB, Auperin D, Brown BG, Castor M, Perez G, Ruo S, Conaty A, Brammer L, Bauer S. 1989. Protection of rhesus monkeys from fatal Lassa fever by vaccination with a recombinant vaccinia virus containing the Lassa virus glycoprotein gene. Proc Natl Acad Sci U S A 86:317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukashevich IS. 1992. Generation of reassortants between African arenaviruses. Virology 188:600–605. doi: 10.1016/0042-6822(92)90514-P. [DOI] [PubMed] [Google Scholar]
- 30.Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, Brasky K, Geiger R, Hubbard GB, Bryant J, Salvato MS. 2005. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol 79:13934–13942. doi: 10.1128/JVI.79.22.13934-13942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukashevich IS, Carrion R Jr, Salvato MS, Mansfield K, Brasky K, Zapata J, Cairo C, Goicochea M, Hoosien GE, Ticer A, Bryant J, Davis H, Hammamieh R, Mayda M, Jett M, Patterson J. 2008. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine 26:5246–5254. doi: 10.1016/j.vaccine.2008.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zapata JC, Poonia B, Bryant J, Davis H, Ateh E, George L, Crasta O, Zhang Y, Slezak T, Jaing C, Pauza CD, Goicochea M, Moshkoff D, Lukashevich IS, Salvato MS. 2013. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol J 10:52. doi: 10.1186/1743-422X-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Cerny AM, Zacharia A, Fitzgerald KA, Kurt-Jones EA, Finberg RW. 2010. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J Virol 84:9452–9462. doi: 10.1128/JVI.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing J, Ly H, Liang Y. 2015. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-I-like receptor-dependent interferon production. J Virol 89:2944–2955. doi: 10.1128/JVI.03349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pythoud C, Rodrigo WWSI, Pasqual G, Rothenberger S, Martínez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L-K, Xin Q-L, Zhu S-L, Wan W-W, Wang W, Xiao G. 2016. Activation of the RLR/MAVS signaling pathway by the L protein of Mopeia virus. J Virol 90:10259–10270. doi: 10.1128/JVI.01292-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen KG, Shapiro BJ, Matranga CB, Sealfon R, Lin AE, Moses LM, Folarin OA, Goba A, Odia I, Ehiane PE, Momoh M, England EM, Winnicki S, Branco LM, Gire SK, Phelan E, Tariyal R, Tewhey R, Omoniwa O, Fullah M, Fonnie R, Fonnie M, Kanneh L, Jalloh S, Gbakie M, Saffa S, Karbo K, Gladden AD, Qu J, Stremlau M, Nekoui M, Finucane HK, Tabrizi S, Vitti JJ, Birren B, Fitzgerald M, McCowan C, Ireland A, Berlin AM, Bochicchio J, Tazon-Vega B, Lennon NJ, Ryan EM, Bjornson Z, Milner DA Jr, Lukens AK, Broodie N, Rowland M, Heinrich M, Akdag M, et al. 2015. Clinical sequencing uncovers origins and evolution of Lassa virus. Cell 162:738–750. doi: 10.1016/j.cell.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seregin AV, Yun NE, Miller M, Aronson J, Smith JK, Walker AG, Smith JN, Huang C, Manning JT, de la Torre JC, Paessler S. 2015. The glycoprotein precursor gene of Junin virus determines the virulence of the Romero strain and the attenuation of the Candid #1 strain in a representative animal model of argentine hemorrhagic fever. J Virol 89:5949–5956. doi: 10.1128/JVI.00104-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar N, Wang J, Lan S, Danzy S, McLay Schelde L, Seladi-Schulman J, Ly H, Liang Y. 2012. Characterization of virulence-associated determinants in the envelope glycoprotein of Pichinde virus. Virology 433:97–103. doi: 10.1016/j.virol.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron É, White DM, Nichol ST. 2011. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol 85:10404–10408. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golden JW, Maes P, Kwilas SA, Ballantyne J, Hooper JW. 2016. Glycoprotein-specific antibodies produced by DNA vaccination protect guinea pigs from lethal Argentine and Venezuelan hemorrhagic fever. J Virol 90:3515–3529. doi: 10.1128/JVI.02969-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmutovic S, Clark L, Levis SC, Briggiler AM, Enria DA, Harrison SC, Abraham J. 2015. Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses. Cell Host Microbe 18:705–713. doi: 10.1016/j.chom.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brouillette RB, Phillips EK, Ayithan N, Maury W. 2017. Differences in glycoprotein complex receptor binding site accessibility prompt poor cross-reactivity of neutralizing antibodies between closely related arenaviruses. J Virol 91:e01454-. doi: 10.1128/JVI.01454-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeltina A, Krumm SA, Sahin M, Struwe WB, Harlos K, Nunberg JH, Crispin M, Pinschewer DD, Doores KJ, Bowden TA. 2017. Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization. Proc Natl Acad Sci U S A 114:7031–7036. doi: 10.1073/pnas.1702127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeitlin L, Geisbert JB, Deer DJ, Fenton KA, Bohorov O, Bohorova N, Goodman C, Kim D, Hiatt A, Pauly MH, Velasco J, Whaley KJ, Altmann F, Gruber C, Steinkellner H, Honko AN, Kuehne AI, Aman MJ, Sahandi S, Enterlein S, Zhan X, Enria D, Geisbert TW. 2016. Monoclonal antibody therapy for Junin virus infection. Proc Natl Acad Sci U S A 113:4458–4463. doi: 10.1073/pnas.1600996113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell P-S, Hossmann S, Watle SV, Kondé MK, Kéïta S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Røttingen J-A, Kieny M-P. 2017. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet 389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasdell KR, Duong V, Eloit M, Chretien F, Ly S, Hul V, Deubel V, Morand S, Buchy P. 2016. Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia. Elife 5:e13135. doi: 10.7554/eLife.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gryseels S, Rieger T, Oestereich L, Cuypers B, Borremans B, Makundi R, Leirs H, Günther S, Goüy de Bellocq J. 2015. Gairo virus, a novel arenavirus of the widespread Mastomys natalensis: genetically divergent, but ecologically similar to Lassa and Morogoro viruses. Virology 476:249–256. doi: 10.1016/j.virol.2014.12.011. [DOI] [PubMed] [Google Scholar]