ABSTRACT
Eastern equine encephalitis virus (EEEV) has a high case-fatality rate in horses and humans, and Florida has been hypothesized to be the source of EEEV epidemics for the northeastern United States. To test this hypothesis, we sequenced complete genomes of 433 EEEV strains collected within the United States from 1934 to 2014. Phylogenetic analysis suggested EEEV evolves relatively slowly and that transmission is enzootic in Florida, characterized by higher genetic diversity and long-term local persistence. In contrast, EEEV strains in New York and Massachusetts were characterized by lower genetic diversity, multiple introductions, and shorter local persistence. Our phylogeographic analysis supported a source-sink model in which Florida is the major source of EEEV compared to the other localities sampled. In sum, this study revealed the complex epidemiological dynamics of EEEV in different geographic regions in the United States and provided general insights into the evolution and transmission of other avian mosquito-borne viruses in this region.
IMPORTANCE Eastern equine encephalitis virus (EEEV) infections are severe in horses and humans on the east coast of the United States with a >90% mortality rate in horses, an ∼33% mortality rate in humans, and significant brain damage in most human survivors. However, little is known about the evolutionary characteristics of EEEV due to the lack of genome sequences. By generating large collection of publicly available complete genome sequences, this study comprehensively determined the evolution of the virus, described the epidemiological dynamics of EEEV in different states in the United States, and identified Florida as one of the major sources. These results may have important implications for the control and prevention of other mosquito-borne viruses in the Americas.
KEYWORDS: EEEV, evolution, next-generation sequencing, NGS, phylodynamics, phylogeography, source-sink dynamics
INTRODUCTION
Eastern equine encephalitis virus (EEEV) is a positive-sense, single-stranded RNA arbovirus (arthropod-borne virus) classified in the family Togaviridae (genus Alphavirus). The EEEV genome is approximately 11-kb in length and comprises four nonstructural proteins (nsP1 to nsP4) and five structural proteins (Capsid, E3, E2, 6K/TF, and E1) that are flanked by 5′ and 3′ noncoding sequences (1, 2). Although EEEV rarely infects humans, human cases usually lead to neurological disease that is often fatal; survivors often experience long-term sequelae involving a range of neurological impairments.
The first human cases of EEEV infection were identified in 1938 in Massachusetts following an outbreak in horses in the surrounding area (3–5). Prior to this, EEEV was known to be maintained in an enzootic transmission cycle between birds and mosquitoes that would periodically cause epizootics in horses (5). Previous studies suggested that four subtypes/lineages of EEEV circulated throughout the Americas (1, 2, 6–11). Subtype/lineage I was primarily found in North America east of the Mississippi River and the Caribbean (and denoted as NA EEEV strains), while subtypes/lineages II, III, and IV circulated in Central and South America (SA EEEV strains). Unlike the highly pathogenic and conserved NA EEEV strains, the SA EEEV strains are generally less pathogenic and highly divergent within and among these three subtypes/lineages (9, 12–15). Recently, SA EEEV has been reclassified into a separate species, Madariaga virus, because of its distinct geographic distribution, human pathogenicity, and genetic diversity (9, 16, 17). Accordingly, the EEEV designation now only corresponds to NA EEEV or subtype/lineage I.
Two major EEEV outbreaks in humans occurred within the past 11 years. Between August and September 2005 in Massachusetts and New Hampshire, an outbreak of 11 cases caused four deaths (18). Also, between late July and early September 2012 in Massachusetts, an outbreak infected 7 humans (3 deaths), 6 horses, and 2 alpacas due to 267 EEEV-positive mosquito pools (19). From 2004 to 2016, there were a total of 103 nationwide human cases (outbreak and sporadic), of which approximately 95% were neuroinvasive with a case-fatality rate of over 40%. The highest number of cases (24 cases) occurred in Massachusetts. Florida ranks second in the number (18 cases) of neuroinvasive cases of EEEV infection (20) due to its subtropical climate and year-round mosquito activity, which accounts for circulation of EEEV throughout the year (21). Conversely, a far more seasonal pattern is observed in the temperate northeastern United States with EEEV activity reported from July to October in such states as New York, Massachusetts, Connecticut, Vermont, and New Hampshire (7, 8, 22–24). Despite this seasonality, there is evidence that the virus may have overwintered and persisted for multiple seasons in these localities (7, 8, 25). However, the epidemiological dynamics of EEEV, including its relative persistence in temperate and subtropical regions of the United States, remain poorly understood.
Previous studies showed that EEEV isolates from Florida were related to those from northern states that do not exhibit year-round transmission and hence proposed that Florida may serve as a reservoir that introduced the virus to these states (6–8, 24). However, these studies were based on a limited number of samples and only partial genome sequences and were unable to delineate the direction of virus movement. In addition, due to the limited data (only 16 EEEV strains from 1959 to 2012 sequenced as of November 2016), little is known about the evolutionary characteristics of EEEV at a genomic scale.
To help fill the gaps in our understanding of EEEV evolution and spread in these regions and across the United States, we sequenced and analyzed the complete genomes of 433 strains collected from Florida, New York, Massachusetts, and 15 other states between 1934 and 2014. These data greatly expanded the existing data set of EEEV genome sequences, and allowed us to determine the long-term genomic evolution and molecular epidemiology of EEEV across spatial and temporal scales. Importantly, our data enabled us to establish that Florida is one of the major sources of EEEV epidemics in the northeastern United States.
RESULTS
Characteristics of the EEEV complete genomes.
Next-generation sequencing (NGS) of EEEV resulted in an average of 17,317 reads per sample with an average coverage of 233.85× (minimum coverage of 11.65× and maximum average coverage of 430.96×). Notably, these new complete genome sequences greatly increased the number of EEEV genomic data available in GenBank, from 16 to 437. There was no significant evidence of recombination. Overall, the EEEV genome was found to be highly conserved, with an average nucleotide similarity of 99.17% and an average amino acid similarity 99.76%. Nonstructural protein genes (nsP1 to nsP4) showed similar levels of heterogeneity as structural protein genes (Capsid, E3, E2, 6K/TF, and E1), with an average nucleotide similarity ranging from 98.99% (nsP3) to 99.37% (capsid) and an average amino acid similarity ranging from 99.34% (nsP3) to 99.92% (E3). nsP3 was the most variable gene across the genome as a whole.
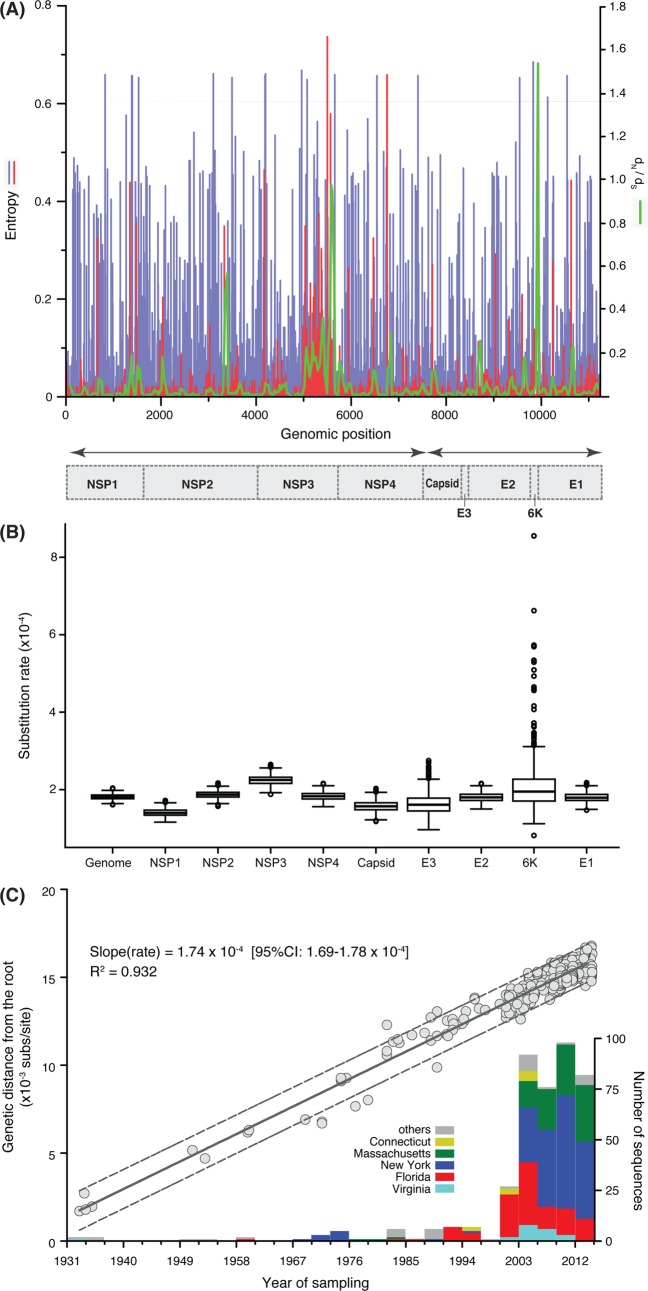
Entropy values greater than 0.8 in nucleotide alignments and greater than 0.6 in amino acid alignments were considered highly variable, based on the level of diversity in our data set and previously determined entropy values (26). Within the nucleotide alignment, different genomic regions showed similar levels of entropy, and there was no highly variable region found with entropy over 0.8, even in the nsP3 gene (Fig. 1A, blue bars). However, entropy analysis in amino acid alignments showed that greater entropy was present in nsP3, and there were two relatively variable regions, one in nsP3 and one in nsP4, with entropy higher than 0.6 (Fig. 1A, red bars). Amino acid site 45 in the structural protein 6K/TF was identified as subject to putative positive selection in all methods (single- likelihood ancestral reconstruction [SLAC], fixed-effects likelihood [FEL], and IFEL [the same as FEL, except that selection is only tested along internal branches of the phylogeny]) at significance level of <0.05. This site is positioned between the two predicted helical transmembrane domains (27) and exposed on the surface of the virion (Fig. 2). Analysis of the substitution patterns in the phylogeny revealed a total of 10 substitutions at this site, all from alanine (Ala) to valine (Val), most (n = 7) of which occurred at the terminal branches (Fig. 3). Finally, the sliding window analysis revealed that the dN/dS values (i.e., the ratios of nonsynonymous versus synonymous substitutions per site) at the 3′ termini of the nsP3 and 6K genes are the highest (0.94 and 1.54, respectively) across the whole genome (Fig. 1A, green line).
FIG 1.
Evolution of EEEV genomes. (A) Entropy analyses of the genomic nucleotide sequences (blue bars) and translated amino acid sequences (red bars) of EEEV isolates (n = 437) from 1934 to 2014. Estimates of dN/dS (green line) of the sliding windows (window size = 30 codons) across the genome. The x axis is the genomic position, while the left y axis is the entropy level, and the right y axis is dN/dS. (B) Evolutionary rates (nucleotide substitutions/site/year) of the complete genome and each individual gene. They were estimated using a Bayesian protocol and displayed in boxplot where the whiskers extend to the 95% highest posterior density (HPD) interval, and the boxes indicate the 25 to 75% interquartile range of the posterior distribution of rate estimates, thus describing its central tendency. (C) Root-to-tip regression analysis for complete genome sequences. Dash lines are the 95% prediction bands of the regression. Frequencies of sequences sampled in different states and time are summarized in the stacked bar chart, with the y axis shown on the right.
FIG 2.
Threading of EEEV structural polyprotein through an endoplasmic reticulum membrane. Structure proteins are colored separately: Capsid in yellow, E3 in green, E2 in blue, 6K in red, and E1 in magenta. The positive selection pressure site, residue 45 in 6K, is indicated by a black arrow.
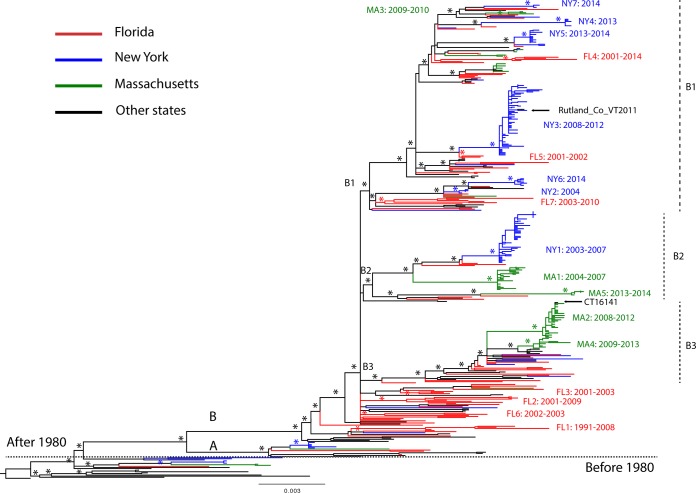
FIG 3.
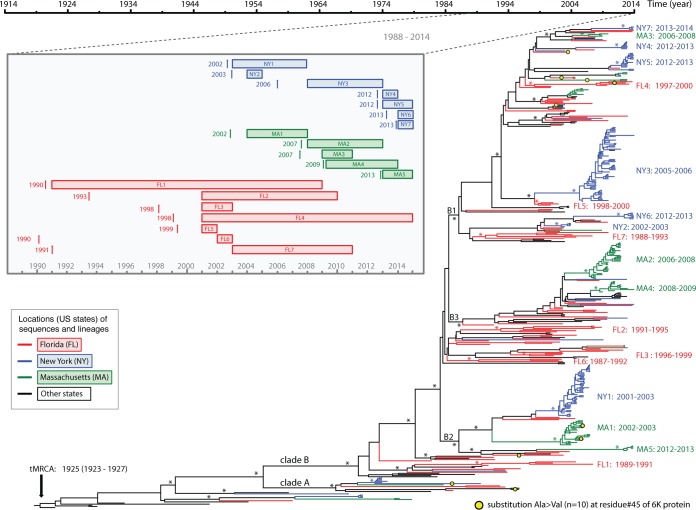
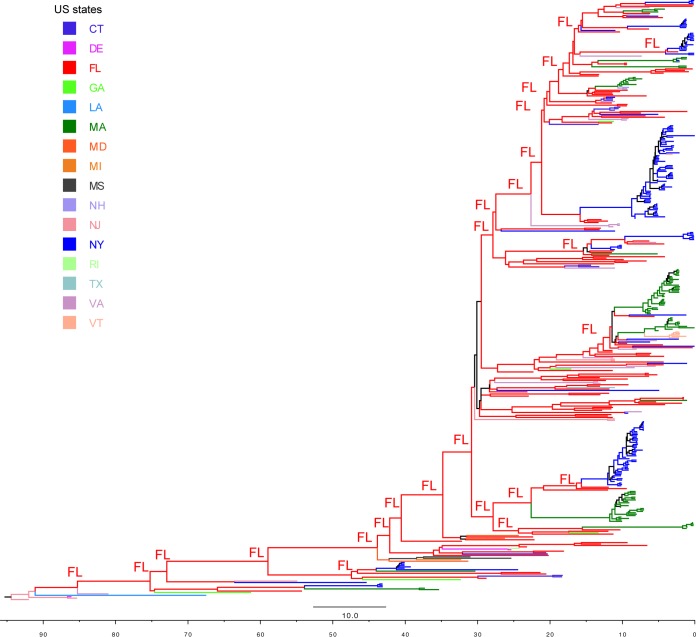
Time-scaled phylogeny of EEEV. A maximum clade credibility (MCC) tree of complete genome sequences of EEEV is shown. Major nodes with posterior probabilities over 0.9 are marked by asterisks. Sequences from Florida, New York, Massachusetts, and other places are colored and described in the legend key. Clades A and B and subclades B1, B2, and B3, as well as the 19 monophyletic groups further defined for Florida, New York, and Massachusetts viruses, are shown. The times of the most recent common ancestors (tMRCAs) of all EEEVs and the monophyletic groups are provided after the colons. Mutations at residue 45 (Ala→Vla) in the 6K protein are mapped on the branches of the tree using yellow circles. The inset diagram on the left shows the time spans of these monophyletic groups, including the range of the sampling time (rectangular boxes) and the mean of the tMRCAs (vertical arrow bars).
Differing epidemiological dynamics of EEEV in Florida, New York, and Massachusetts.
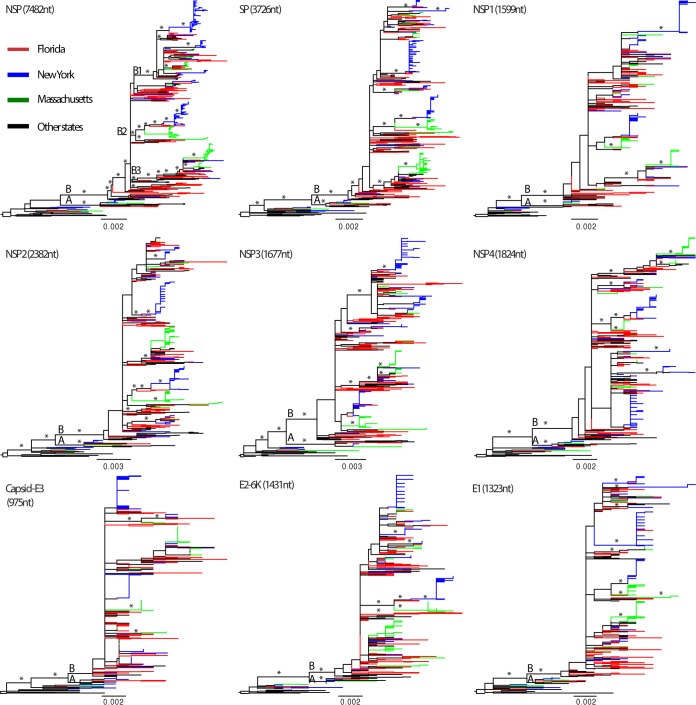
The maximum-likelihood (ML) phylogeny of the 437_genome data set (Fig. 4) shows that those EEEV sequences sampled after 1980 fell into two clades, here defined as clades A and B. Clade A was small (n = 13), containing the sequences sampled from Florida, Massachusetts, New York, Georgia, and Connecticut during 1974 to 1996. Clade B was larger, containing the sequences collected from different states since 1982. In clade B, we observed three large, well-supported subclades, B1, B2, and B3, together with some other smaller, well-supported subclades. Interestingly, the phylogeny suggests that clade B has continuously circulated in the United States since the 1980s. Clade A was not sampled after 1996, even in more heavily sampled states, including Florida, New York, and Massachusetts, suggesting that it may have gone extinct. Gene-specific data sets presented similar topologies as the complete genome data set (Fig. 5); however, most of the individual gene trees were relatively poorly resolved since the bootstrap levels for most nodes were lower than 70%.
FIG 4.
ML phylogenetic tree of complete genome sequences of EEEV. Well-supported nodes by bootstrap values over 70% and posterior probabilities > 0.9 from BMCMC analyses are marked by asterisks. Clades (A and B) and subclades (B1, B2, and B3) are described in the trees. Sequences sampled from Florida, New York, and Massachusetts are shown in red, blue, and green, respectively, and described in the key. The monophyletic groups of Florida, New York, and Massachusetts sequences are indicated in the trees. Phylogenetic trees were rooted by the oldest EEEV strain, Ten Broeck, collected in 1933 in Virginia. Scale bars represent the number of nucleotide substitutions per site.
FIG 5.
Phylogenetic trees of individual genes of EEEV. ML phylogenetic trees inferred for different regions of the EEEV genome are indicated in black. Well-supported nodes by bootstrap values over 70% are marked by asterisks. Sequences from Florida, New York, Massachusetts, and other places are colored and described in the key. Phylogenetic trees were rooted using the oldest EEEV strain, Ten Broeck, collected in 1933, and scale bars represent the number of nucleotide substitutions per site.
Extensive genetic diversity and long-term local persistence of EEEV in Florida.
One of the most notable observations from the phylogenies was the extensive genetic diversity of EEEV in Florida. In particular, the Florida sequences were paraphyletic as viruses from different states were mixed with Florida sequences throughout the tree, a finding suggestive of extensive spatial dispersal of the virus in the United States. Despite the large degree of mixing between sequences from Florida and other states, we identified seven small monophyletic groups dominated by Floridian viruses (FL1, FL2, FL3, FL4, FL5, FL6, and FL7) and (i) defined by containing at least three sequences from Florida and (ii) supported by both high bootstrap values (>70%) in the ML phylogeny and high BPP values (>0.9) in the Bayesian Markov chain Monte Carlo (BMCMC) phylogeny (Fig. 4). Closer examination of these seven monophyletic groups yielded evidence of multiyear local persistence in Florida. These groups were sampled over a range of 8 to 18 years: FL1 from 1991 to 2008, FL2 from 2001 to 2009, FL3 from 2001 to 2003, FL4 from 2001 to 2014, FL5 from 2001 to 2002, FL6 from 2002 to 2003, and FL7 from 2003 to 2010 (Fig. 4).
Multiple viral introductions, short-term local persistence, and strain replacement in New York state and Massachusetts.
The EEEV sequences from New York and Massachusetts sequences also formed distinct, large, and well-supported monophyletic groups with long internal branches. Even though these two states are geographically close and viruses were sampled close in time, the viruses from New York and Massachusetts sequences often did not cluster together. In clade B, we identified seven monophyletic groups of New York sequences and five monophyletic groups of Massachusetts sequences (Fig. 4), with two exceptions, NY3 and MA2. NY3 includes a strain collected in Vermont (Rutland_Co_VT2011_mu), while MA2 includes an isolate originally from Connecticut (CT16141) (Fig. 4). The monophyletic groups also showed evidence of multiyear local persistence of EEEV in New York and Massachusetts, although the duration of each ranged from 1 to 5 years, shorter than that seen in Florida (Fig. 3 and 4).
Significant phylogeographic clustering of EEEV.
To determine the phylogeographic structure of EEEV, we performed phylogeny-trait association (BaTS) tests (Table 1) on the phylogenies of complete genome sequences. This revealed stronger clustering by location than expected by chance alone (P values for the association index [AI] and the parsimony score [PS] < 0.001). Indeed, the maximum clade statistics for the sites with more than five sequences were significant (P < 0.01), suggesting the significant spatial structure and more localized evolution of EEEV in these regions. Notably, differences in the observed and expected maximum clade values also suggested that New York and Massachusetts exhibited the strongest spatial structure (difference of 43.98 and 19.49 separately), while the Florida isolates were relatively less structured, indicative of more geographic mixing (difference of 3.60, Table 1).
TABLE 1.
Results of phylogeny-trait association tests of EEEV
| Statistica | n | Observed mean | Null mean | Significance | Difference |
|---|---|---|---|---|---|
| AI | 7.88 | 32.71 | 0 | ||
| PS | 86.94 | 230.03 | 0 | ||
| MC_state: New York | 170 | 48 | 4.02 | 0.001 | 43.98 |
| MC_state: Florida | 102 | 6.34 | 2.75 | 0.003 | 3.60 |
| MC_state: Massachusetts | 89 | 22 | 2.51 | 0.001 | 19.49 |
| MC_state: Virginia | 20 | 3 | 1.21 | 0.003 | 1.79 |
| MC_state: Connecticut | 10 | 3 | 1.07 | 0.001 | 1.93 |
| MC_state: New Jersey | 7 | 2 | 1.02 | 0.011 | 0.98 |
| MC_state: Vermont | 6 | 5 | 1.02 | 0.001 | 3.98 |
| MC_state: Georgia | 5 | 1 | 1.02 | 1 | −0.02 |
| MC_state: Maryland | 3 | 1.19 | 1.01 | 1 | 0.18 |
| MC_state: New Hampshire | 2 | 1.14 | 1.001 | 1 | 0.14 |
| MC_state: Delaware | 2 | 1 | 1.002 | 1 | −0.002 |
| MC_state: Mississippi | 2 | 1 | 1.004 | 1 | −0.004 |
| MC_state: Michigan | 1 | 1 | 1 | 1 | 0 |
| MC_state: Louisiana | 1 | 1 | 1 | 1 | 0 |
| MC_state: Rhode Island | 1 | 1 | 1 | 1 | 0 |
| MC_state: Texas | 1 | 1 | 1 | 1 | 0 |
AI, association index; PS, parsimony score; MC, maximum clade.
Evolutionary rates and tMRCAs of EEEV in the United States.
The root-to-tip regression analysis revealed that EEEV evolution in the United States was characterized by a very strong temporal structure (R2 = 0.93), indicative of strongly clock-like evolution, with a relatively low evolutionary rate of 1.74 × 10−4 nucleotide substitutions/site/year (95% confidence interval = 1.69 × 10−4 to 1.78 × 10−4; Fig. 1C). A very similar genomic substitution rate was observed using the Bayesian method available in the BEAST package: mean rate of 1.81 × 10−4 subs/site/year (95% highest posterior density [HPD] of 1.69 × 10−4 to 1.94 × 10−4 subs/site/year). The mean substitution rates of the nine individual EEEV genes ranged from 1.4 × 10−4 substitutions (subs)/site/year for nsP1 to 2.24 × 10−4 subs/site/year for nsP3. The rate estimate for the 6K/TF gene had the widest 95% HPD, likely due to the short length of the gene and the lack of genetic information (Fig. 1B). Correspondingly, the most recent common ancestor (tMRCA) of all EEEV strains in the United States was dated to 1923 in a root-to-tip regression analysis. Two independent BEAST runs resulted highly consistent results, with mean tMRCAs dated to 1925 (95% HPD = 1923 to 1927) in the genomic analysis and 1925 (95% HPD = 1923 to 1927) in the gene-partitioned analysis (Fig. 3).
Spatial pattern of EEEV evolution in Florida, New York, and Massachusetts.
The time-scaled maximum clade credibility tree of the complete genome data set from the BMCMC analysis (Fig. 3) showed a highly consistent topology to the nontemporal ML phylogeny (Fig. 4). Again, the extensive genetic diversity, the lack of large monophyletic groups, and long terminal branches of Florida sequences suggested that numerous viral populations circulated in the region for many years before being detected or sampled. Molecular clock dating of the seven Florida monophyletic groups provided evidence of multiple viral strains circulating in Florida since 1990s even though most viruses were sampled in the 2000s. For example, viruses in FL6 were collected during 2002 to 2003 and FL7 were collected during 2003 to 2010; however, the mean tMRCAs of FL6 and FL7 were dated to 1990 (95% HPD = 1987 to 1992) and 1991 (95% HPD = 1988 to 1993) individually, representing more than 10 years of unsampled circulation of FL6 and FL7 in the region (Fig. 3). In contrast, the mean tMRCAs of seven New York monophyletic groups and five Massachusetts monophyletic groups were dated to 2000s, indicative of recent viral introductions and shorter local persistence in the population. The mean periods on unsampled circulation for these monophyletic groups were estimated at <1 year to 2 years. Furthermore, there was evidence of dominant strain extinctions and replacements in New York and Massachusetts after a few years in circulation. In New York, the dominant strains introduced in 2002 were NY1 strains during 2003 to 2008, which were replaced by NY3 during 2009 to 2013. NY4 and NY5 were introduced in 2012 and cocirculated in 2013, while NY6 and NY7 were introduced in 2013 and cocirculated in 2014 (Fig. 3). In Massachusetts, MA1 strains, which were introduced in 2002, predominated during 2004 to 2007. There were multiple introductions during 2007 to 2009; MA2, MA3, and MA4 cocirculated during 2008 to 2013 and died out in 2013 (Fig. 3).
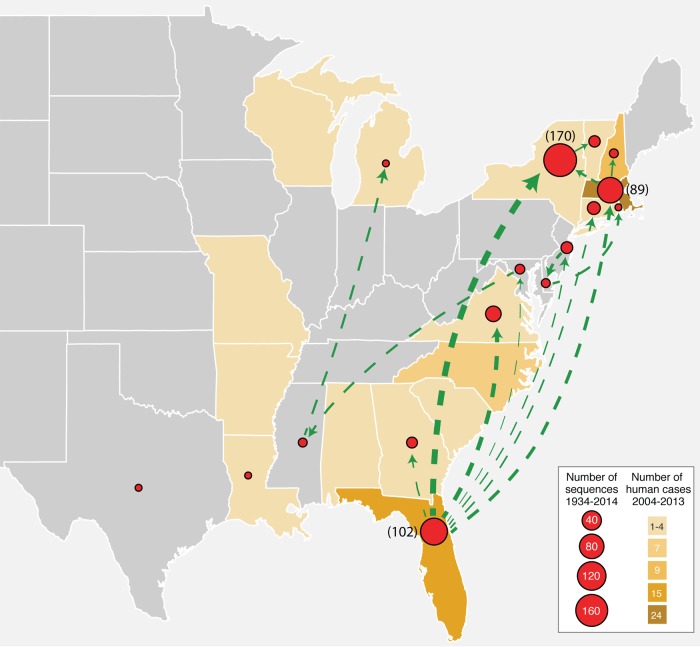
To better determine patterns of viral movement, we performed a Bayesian phylogeographic analysis. Accordingly, the ancestral geographic state at the backbone of the phylogeny of the EEEV sequences analyzed was estimated to be primarily Florida with state probabilities over 0.9 at most nodes (Fig. 6). Table 2 shows the route of virus movement with significant Bayes factors (BFs; cutoff = 3) and their transmission rates. This revealed that virus flow from Florida to the north (e.g., New York, Virginia, and Massachusetts) showed both the highest BFs and the highest rates compared to other states in the south. However, it is important to note that the highest rates are in part likely due to the bias of the large number of viral isolates from Florida, New York, and Massachusetts. Virus gene flow from Florida to Georgia and from New Jersey to Delaware also showed high BFs but relatively low rates, partly due to the limited number of sequences from Georgia, New Jersey, and Delaware. Strikingly, however, we did not find evidence of any significant gene flow from other states back to Florida in these data. These spatial diffusion patterns remain consistent among the repeated analyses with the data sets randomly subsampled for balanced sample sizes between New York, Massachusetts, and Florida. Overall, the data suggest that the spread of EEEV strains in the United States was characterized by strong outward movement from FL to other states that subsequently maintained occasional, local transmission of the viruses themselves (Fig. 7).
FIG 6.
Inference of the ancestral states in the MCC tree of EEEV. Sequences from different states are colored and described in the key. The ancestor geographic states with state probabilities over 0.9 at the backbone of the EEEV phylogeny are labeled in the tree.
TABLE 2.
Significant viral transmission between locations estimated by Bayesian analysisa
| Virus movement | Bayes factor (cutoff = 3) | Rate of state transition (mean) |
|---|---|---|
| Florida to New York | 1.16 × 106 | 3.29 |
| Florida to Virginia | 1.16 × 106 | 2.32 |
| Florida to Georgia | 38,577.14 | 0.85 |
| Florida to Massachusetts | 7,915.45 | 1.74 |
| New Jersey to Delaware | 1,487.32 | 1.59 |
| New York to Vermont | 661.17 | 0.86 |
| Florida to New Jersey | 210.08 | 0.76 |
| Florida to Connecticut | 204.93 | 0.91 |
| Florida to Maryland | 102.57 | 0.49 |
| Delaware to Rhode Island | 94.23 | 1.27 |
| Massachusetts to New York | 74.47 | 1.26 |
| Massachusetts to New Hampshire | 74.47 | 0.89 |
| Mississippi to Michigan | 66.91 | 1.21 |
| Maryland to Mississippi | 21.98 | 1.24 |
| Massachusetts to Vermont | 14.89 | 0.89 |
| New York to Virginia | 7.11 | 0.89 |
| Connecticut to New York | 6.82 | 1.18 |
| Florida to Texas | 5.57 | 0.29 |
| Virginia to Connecticut | 4.83 | 1.64 |
| Virginia to Texas | 4.83 | 0.97 |
| Virginia to New York | 3.63 | 1.24 |
| Connecticut to Virginia | 3.46 | 1.28 |
| New York to Massachusetts | 3.37 | 1.42 |
| Maryland to Michigan | 3.09 | 0.94 |
The Bayes factors and mean rates of the significant transitions between geographical states are shown.
FIG 7.
Map of the United States showing the EEEV cases and the virus movement inferred from the Bayesian phylogeographic analysis. Red circles in each state represent the EEEV isolates collected during 1931 to 2014 (circle size is proportional to the number of sequences analyzed). The numbers of human cases are indicated by the yellow shade in each state. Green arrow lines indicate the significant virus movement between states as reported by Bayes factor > 20 in the phylogeographic analysis using BEAST program. The line width is proportional to the transition rate between the states, as shown in Table 2.
DISCUSSION
This is the first large-scale genomic study of an important, neuroinvasive alphavirus pathogen in North America. Using high-throughput sequencing methods, we obtained a total of 433 complete genome sequences of EEEV strains collected from many states in the United States between 1934 and 2014, particularly from Florida, New York, and Massachusetts. These new EEEV sequence data significantly increased the number of publicly available genome sequences from 16 to more than 400 (a >20-fold increase) and, for the first time, allowed a comprehensive study of the genomic diversity and evolution of EEEV in North America. Our analyses show that the EEEV genome is highly conserved in general, and the evolution of EEEV is strongly clock-like. Notably, our phylogenetic analyses suggest different geographic regions in the United States are experiencing different epidemiological dynamics of EEEV. Most importantly, the phylogeography of EEEV in the United States appears to be compatible with a source-sink model in which the viruses generally move from Florida (source region, frequent persistence) to the North (e.g., New York, Massachusetts, and other regions as the sinks with less persistence).
The genetic analyses reveal that EEEV is a generally well-conserved virus with over 99% nucleotide/amino acid similarity across the genome and only two diverse amino acid positions detected by entropy analysis. The most variability is in the nsP3 gene, supporting previous studies based on partial genomes (6, 8, 28). In addition, EEEV exhibited strong, clock-like evolution with a mean evolutionary rate of 1.81 × 10−4 subs/site/year. Although such striking rate consistency over 8 decades is difficult to explain, it may in part reflect the closed mode of EEEV evolution in mosquitoes and birds (primary host) and the limited action of adaptive evolution, which was only identified at a single site (6K/TF) in our analysis (see below). In this context it is also notable that EEEV is seemingly evolving slower than most other arboviruses over the period that we studied, such as West Nile virus (mean of 5 × 10−4 subs/site/year) (29), dengue virus (mean of 1 × 10−3 subs/site/year) (30), western equine encephalitis virus (mean of 2.8 × 10−4 subs/site/year) (31), and Saint Louis encephalitis virus (mean of 5 × 10−4 subs/site/year) (32).
Although it is believed the 6K/TF protein is involved in envelope protein processing, membrane permeabilization, virion assembly and virus budding, its role is still relatively poorly understood (33), and the biological consequence of the 6K/TF mutation is uncertain. Notably, this 6K protein mutation—from alanine to valine—sporadically occurred multiple times (n = 10) at or near the terminal branches of the phylogenetic tree (Fig. 3, yellow circles), such that some are transiently deleterious mutations (34). This 6K mutation does not seem to associate with the sampling time (1990 to 2013) and animal sources of the virus isolates (eight from mosquitoes, three from humans, and one from birds of 13 isolates with 45Val on 6K protein). Furthermore, there is a well-documented ribosomal −1 frameshifting in 6K, resulting in the TF protein (likely a virulence factor involved in assembly), and this 6K mutation is located at the beginning of the conserved UUUUUUA motif (33). The last nucleotide coding alanine/valine is the first U in the motif.
A key result was that the genetic diversity of EEEV in Florida was much greater than in New York and Massachusetts. Together with the phylogenetic evidence of the multiple-year local persistence of the virus lineages, as well as the widespread EEEV activity in Florida found in previous epidemiologic studies, this strongly suggests that EEEV is enzootic in Florida and has evolved locally for many years. The observed level of genetic diversity and multiple-year local persistence could be due to the year-round activity of EEEV in Florida supported by a subtropical climate and bird migration from temperate northeastern regions of the United States to Florida every winter. Hence, EEEV transmission between birds and mosquitoes can occur at any time of the year. This year-round transmission pattern might help facilitate the generation and maintenance of extensive viral genetic diversity and, in turn, lead to cocirculation of multiple strains and the lack of predominant strains. However, it is clear that more sequences are needed to accurately determine the geographic patterns of EEEV movement.
Another notable finding of the phylogenetic analyses is the local EEEV persistence over some seasons in New York and Massachusetts (also previously suggested by epidemiological studies [7, 8, 11]), which is shorter than the persistence seen in Florida. EEEV prevalence in these two states is seasonal because of the temperate climate. Birds migrate to the southern regions, and mosquito activity ceases every winter. Thus, the persistence of EEEV in temperate regions could be explained in two ways: the reimportation of the same strain by infected migrating birds from the South or that EEEV-infected overwintering mosquitoes carry the virus through the winter. Adult females of some mosquito species overwinter by finding holes where they wait for warmer weather to emerge and lay eggs (35); however, EEEV has never been isolated from these insects. Recently, birds and reptiles (such as snakes) may represent overwintering hosts for EEEV (36–38). Although our analyses show evidence suggestive of EEEV overwintering in New York and Massachusetts, we did not detect local persistence of more than 10 years, as was observed in Florida. Indeed, New York and Massachusetts were characterized by a pattern in which the dominant strains in the region became extinct and were replaced by new strains after 1 to 5 years of circulation.
There has been a long-standing hypothesis that EEEV in Florida may be the source for epidemics in northeastern states that do not appear to maintain year-round transmission (6–8, 24, 39, 40). We tested this hypothesis using a phylogeographic approach, which provided evidence for a source-sink model of EEEV transmission in the United States, with Florida representing an important source location. Enzootic EEEV viruses in Florida appear to migrate and seed epidemics in northern states, such as New York and Massachusetts (sink regions). Such disseminations seem to be occasional, as in the phylogenetic tree the Northern viral strains were only originated from some lineages in Florida (Fig. 3), instead of overwhelmingly branching off from every different Florida lineage (also demonstrated by the significant phylogenetic structure in Table 1). This explains why sink regions have lower diversity instead of a representative sample of the genetic variation in the source region. Our analysis also suggests other states subsequently may act as occasional sources of viral spread, such as New Jersey to Delaware and New York to Vermont, although definitive results would require a more widespread sampling of EEEV. Indeed, since we have few samples from other states, such as Georgia, South Carolina, North Carolina, Alabama, and Louisiana, our phylogeographic analysis has little power to rule out the possibility that EEEV from these poorly or not-sampled locations could also seed transmission in the Northeast, which is a common pitfall for such a closed-system phylogeographic analysis (41).
Although there is an inherent sampling bias in our study, this work will establish a foundation for understanding the evolution and spread of EEEV in the eastern corridor of the United States. Clearly, a more complete understanding of the transmission and evolution of EEEV in the United States will require additional sampling, and such data may have important implications for the control and prevention of other mosquito-borne viruses in this geographic region.
MATERIALS AND METHODS
Virus isolation.
Our study is based on four collections of EEEV samples. Collection 1 consists of 76 EEEV strains collected from different states in the United States during 1934 to 2009, with the exception of three strains collected from outside the United States. Collection 2 consists of 88 isolates collected from Florida during 1986 to 2014. Collection 3 consists of 184 EEEV isolates collected from New York state during 1971 to 2014. Collection 4 consists of 85 EEEV isolates collected from Massachusetts during 2004 to 2014.
For collection 1, archived isolates were obtained from the World Reference Center for Emerging Viruses and Arboviruses, as well as other collections at the University of Texas Medical Branch, Galveston, TX. Viral isolates were selected to represent a broader temporal and geographic distribution and a spectrum of genetic diversity based on the state and year of collection, as well as the host/vectors known to contribute to EEEV's enzootic and epizootic transmission. Viruses were passaged once in African green monkey kidney (Vero) cells prior to cDNA preparation.
For collection 2, Florida isolates were selected from the archive of the Florida Department of Health, Bureau of Public Health Laboratories (BPHL) in Tampa. The BPHL has an extensive arbovirus surveillance program that includes testing for arboviral antibodies in sentinel chickens supplemented with isolation of arboviruses in nonchicken birds, mosquitoes, and other mammals. Submission of surveillance specimens for arbovirus detection and isolation is passive in that birds or mammals are submitted most often when exhibiting disease symptoms and mosquito pools are collected and submitted when increased viral activity is observed, such as seroconversions in the sentinel chickens or EEE cases identified in birds, animals, or humans. Viral isolates were previously cultured in Vero or Buffalo green monkey kidney (BGM) cells with one or two passages or by passage from suckling mouse brain to BGM and/or Vero cultures. The EEEV isolates in this study were selected to provide a broad range and diversity based on the county of detection, the year of collection, and the host source (avian, equine, mosquito, or other mammals).
For collection 3, New York isolates were selected from archived samples collected as part of the New York State Department of Health surveillance program, for which isolation and identification is performed at the Wadsworth Center Arbovirus Laboratory. Submission of surveillance specimens for arbovirus detection and isolation is passive in that mammals, predominantly horses and camelids, are submitted when exhibiting neurologic symptoms, and mosquitoes are collected and submitted as part of the surveillance program annually from May through October. Mosquitoes were pooled by sex, date, and location for testing. Original isolation was completed on Vero cells, and one additional amplification on Vero cells was completed prior to RNA extraction. Isolates were selected to reflect the temporal, geographic, and host diversity of EEEV in New York State.
For collection 4, samples from the Massachusetts Department of Public Health's Arbovirus Surveillance Program were selected from archived, frozen RNA isolated from original mosquito pools from 2004 to 2014 that were positive for two different EEEV targets by real-time detection PCR (42). The Arbovirus Surveillance Program is maintained as a passive surveillance system to monitor EEEV detection in mosquitoes from long-term, historic traps sites going back to 1957 and includes surveillance of new or expanding habitats where EEEV human, mammalian, and avian cases have been identified. Mosquito samples were selected to represent the temporal range of transmission each year (including early and late detections each season) from the same long-term trap sites, as well as the inclusion of other sites to ensure geographic diversity. Mosquito pools (≤50 mosquitoes per pool) collected between June and October and were sorted by species, date, and location. Although clinical, mammalian, and avian samples were not included in this study, positive mosquito pools from areas where EEEV transmission was likely to have occurred were included, if available.
RNA extraction and cDNA synthesis.
RNA was extracted from 250 μl of cell culture supernatant using TRIzol and chloroform, followed by ethanol precipitation, and rehydration with 32 μl of DNase and RNase-free water. cDNA was synthesized using a SuperScript III first-strand synthesis system for RT-PCR (Invitrogen/Life Technologies, Carlsbad, CA). The cDNA synthesis protocol was modified for a 25-μl reaction volume by using 50 μM oligo(dT), random hexamers (50 ng/μl), and 5 μl of RNA. To synthesize cDNA, samples were incubated for 10 min at 25°C, followed by 30 min at 43°C, 20 min at 48°C, and 30 min at 55°C. The concentration of EEEV-derived cDNA was estimated by EEEV-specific qPCR.
For the Massachusetts samples only, viral RNA was extracted from the mosquito pools by using a Qiagen QIAmp RNA minikit, and cDNA was prepared following a slight modification of the SuperScript III first-strand synthesis system for RT-PCR as described above, except that 10 μl of mosquito pool RNA was added, and the 25-μl reaction mixture was incubated for 80 min with a ramped temperature of 43 to 55°C in a 96-well plate. RNase H was added to each sample, followed by incubation for 20 min at 37°C. cDNA was then frozen prior to shipment for subsequent sequencing.
Library preparation and next-generation sequencing.
Sequencing libraries were prepared using two independent library preparation methods to get the best coverage for both coding regions and termini: (i) a Nextera DNA sample preparation kit (Illumina, Inc., San Diego, CA) with half reaction volumes as described previously (43, 44) and (ii) a modified RT-PCR sequence independent single-primer amplification (SISPA) procedure. SISPA employs a primer containing random hexamers and a 5′ tail that serves as a barcode (index) for the sample to simultaneously amplify nucleic acids and barcode the samples (45). Two independent SISPA reactions were performed on each sample using two different barcoded primers for better coverage. Subsequently, the SISPA amplicons were purified, pooled, and size selected (ca. 300 to 600 bp). In order to prepare SISPA pools for sequencing, a NEBNext Ultra II DNA Library Prep kit (Illumina) was used to construct the final sequencing libraries (New England Biolabs). Samples were sequenced on the Illumina HiSeq2000 platform. For samples requiring extra coverage at certain regions of the genome and for gap-filling, Ion Torrent PGM (Thermo Fisher Scientific) was used, in addition to Illumina sequencing. For Ion Torrent sequencing, for each sample, 100 ng of pooled DNA amplicons was sheared for 7 min, and Ion-Torrent-compatible barcoded adapters were ligated to the sheared DNA using the Ion Xpress Plus fragment library kit (Thermo Fisher Scientific) to create 400-bp libraries. Sequencing was performed on the Ion Torrent PGM using 316v2 or 318v2 chips (Thermo Fisher Scientific).
Genome assembly and annotation.
Sequence reads were sorted by barcode, trimmed, and de novo assembled using CLC Bio's De_novo_assembly program (http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/852/index.php?manual=De_novo_assembly.html). The resulting contigs were searched against custom full-length EEEV nucleotide databases to identify the closest reference sequence. All sequence reads were then mapped to the selected reference EEEV sequence using the CLC Bio clc_mapper_legacy program (http://resources.qiagenbioinformatics.com/manuals/clcassemblycell/current/index.php?manual=Options_clc_mapper_legacy.html). At loci where both Illumina and Ion Torrent sequence data agreed on a variation compared to the reference sequence, it was updated to reflect the difference. A final mapping of all sequence reads to the updated reference sequences was performed with CLC Bio's clc_mapper_egacy program. Curated assemblies were validated and annotated with the viral annotation software—Viral Genome ORF Reader, VIGOR 3.0 (46)—before submission to GenBank. VIGOR was used to predict genes, perform alignments, ensure the fidelity of open reading frames, associate nucleotide polymorphisms with amino acid changes, and detect any potential sequencing errors. The annotation was subjected to manual inspection and quality control before submission to GenBank (see below).
Data sets and genetic analyses.
We combined all 433 complete genomes of EEEV sequenced by us (76 from different states in the United States and outside the US, 88 from Florida, 184 from New York state, and 85 from Massachusetts) with 16 EEEV genome sequences generated by other research groups available in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) and generated a data set of 437 complete EEEV genome sequences (denoted as 437_genome), after excluding 12 duplicate sequences of the same viral isolates collected in Florida. We also generated 11 smaller data sets by polyprotein and individual genes in the genome, which are denoted as follows: 437_completeNSP, 437_completeSP, 437_NSP1, 437_NSP2, 437_NSP3, 437_NSP4, 437_Capsid, 437_E3, 437_E2, 437_6K, and 437_E1. Sequences were aligned separately using the MUSCLE program in MEGA 6.0 with manual adjustment (47).
Potential recombination within 437 EEEV complete genome sequences was screened using seven methods (RDP, GENECONV, Chimaera, MaxChi, SiScan, 3Seq, and BootScan) implemented in the Recombination Detection Program version 4.46 (RDP4) (48). Any phylogenetic incongruence between different regions and with P values less than 10−4 in at least RDP, GENECONV, and Bootscan is reported as evidence of recombination. Nucleotide and amino acid similarities across the genome and in each gene were also calculated. Nucleotide and amino acid entropy analyses of complete genome were performed using Shannon Entropy tool in Los Alamos HIV database (http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html). Threshold values for nucleotide and amino acid analyses were determined as previously described (26). A sliding window (window size, 30 codons; step size, 10 codons) pairwise analysis of the ratios of nonsynonymous versus synonymous substitutions per site (dN/dS) was performed using the method of Nei and Gojobori (49). Individual amino acid sites putatively under positive selection were identified by three different codon-based ML methods—SLAC, FEL, and IFEL—using the Datamonkey webserver (www.datamonkey.org) (50–52). A significance level of P < 0.05 was used in all methods.
Phylogenetic analyses.
We performed phylogenetic analyses on the 437_genome data set and 11 region/gene data sets. ML phylogenetic trees of all data sets were inferred using PhyML v3.0 (53). A general time reversal (GTR) nucleotide substitution model with a gamma distribution of among-site rate variation (GTR+Γ) was selected as the best-fit model by Modeltest in MEGA 6.0 and used in all tree inference methods. Phylogenetic trees were also inferred using the BMCMC method available in MrBayes version 3.2.5 (54) and were run for 1 × 108 steps. Trees were sampled every 1 × 104 steps, with the first 1,000 trees discarded as burn-in. The robustness of the ML tree was assessed by bootstrap analyses of at least 500 pseudoreplicates and by comparison with the topologies sampled in the Bayesian analysis. All phylogenies were rooted with the oldest EEEV strain (Ten Broeck, collected in 1933 in Virginia), which was the root suggested by BEAST analysis.
The posterior distribution of 437_genome trees generated by BMCMC was also used to assess the strength of geographic clustering in the data by using the phylogeny trait association test available in the Bayesian tip association significance testing (BaTS) program (55). Each sequence was therefore assigned to a character state by its sampling location, i.e., different states in the United States or others. The overall statistical significance of the geographic clustering of taxa in the EEEV phylogenies was determined using two phylogeny-trait association tests, the PS, and the AI tests, where the null hypothesis is that clustering by geographic information is not more than that expected by chance. In addition, the maximum clade statistic was used to compare the strength of clustering at each group by calculating the expected and observed mean clade size from each group. All three statistics were implemented in BaTS program and a significance level of P < 0.05 was used. A null distribution of these statistics was determined using the posterior distribution of BMCMC phylogenies.
Evolutionary dynamics and epidemic history.
To understand viral evolutionary dynamics and to infer the epidemiological history of EEEV in the United States, especially in Florida, New York, and Massachusetts, the evolutionary history of the EEEV complete genome data sets were also inferred using the BMCMC method available in the BEAST V1.8.2 package (http://tree.bio.ed.ac.uk/software/BEAST) (56). In the 437_genome data set, the sequences without sampling time or location information were excluded, as were a small number of sequences from the isolates outside the United States. This resulted in a data set of 422 EEEV complete genome sequences (422_genome). Nine individual gene data sets were also generated. ML phylogenies for these data sets were inferred using the methods described above.
First, to determine the extent of temporal structure of the sequences and consequently the reliability of the estimates of substitution rates and times since most recent common ancestors (tMRCAs), we performed a regression of root-to-tip genetic distances against sampling dates on the ML trees using TempEst (http://tree.bio.ed.ac.uk/software/tempest/) (57). As this revealed significant temporal structure (see Results), we next performed temporal phylogenetic analyses of the 422 coding sequence data set partitioned by nine genes in BEAST. This analysis utilized the GTR+Γ substitution model and the Gaussian Markov random fields (GMRF) Bayesian skyride coalescent tree prior. Both a strict molecular clock and a relaxed molecular clock (uncorrelated lognormal [UCLN]) were attempted in separate runs. Despite the clear root-to-tip regression slope (see Results), UCLN presented a better fit than the strict clock model based on Bayes factor (log Bayes factor = 39.57). Finally, we performed a discrete phylogeographic BEAST analysis (58) to help reveal the spatial diffusion of EEEV among the localities sampled. This utilized the 422_genome data set in which each sequence was assigned character state based on its sampling location at the state level (Fig. 7).
To ensure sufficient mixing and convergence in parameter samples in each BEAST run (i.e., effective sampling size parameters [ESS] > 200), a Markov chain Monte Carlo (MCMC) was run for 500 million generations, and a 10% burn-in was removed. The results were accessed using the Tracer program v1.6.0 (http://tree.bio.ed.ac.uk/software/tracer) to ensure that stationarity was achieved. The posterior distribution of BMCMC trees was summarized as the maximum clade credibility (MCC) tree and generated by TreeAnnotator v1.8.2 (available in the BEAST v1.8.2 package) with the first 10% of trees removed as burn-in. MCC trees were visualized by using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/FigTree). Bayes factors for gene flows between the sampled locations in phylogeographic analysis were estimated using the SPREAD program (http://tree.bio.ed.ac.uk/software/SPREAD).
Addressing sampling bias in sequence data.
Phylogeographic analyses were repeated 10 times on randomly subsampled data sets controlled for balanced sample sizes between Florida, New York, and Massachusetts (although this cannot account for locations that have not been sampled or only poorly sampled). However, this randomly equal-size subsampling would allow more robust comparison of the relative levels and directionalities of EEEV transmissions between Florida, New York, and Massachusetts. These subsampled BEAST runs used the GTR+Γ substitution model, the GMRF Bayesian skyride coalescent tree prior, and the UCLN relaxed molecular clock.
Accession number(s).
All sequences were submitted to GenBank as part of the BioProject IDs PRJNA183000 and PRJNA263186. The GenBank accession numbers of 76 historical EEEV sequences collected from different states in the United States and outside the United States are given in Table S1 in the supplemental material. The sequences of Florida samples have accession numbers KU840291 to KU840311 and KU840313 to KU840379. The sequences from Massachusetts have accession numbers KX029230 to KX029319, while the sequences from New York have accession numbers KX000047 to KX000231.
Supplementary Material
ACKNOWLEDGMENTS
This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under award U19AI110819. This study was also supported by a grant from the National Institutes of Health to T.R.U. (grant R56AI01372). This publication was supported by Cooperative Agreement no. NU50CK000423, funded by the Centers for Disease Control and Prevention. S.R.D. is also supported by the NIH-funded Tennessee Center for AIDS Research (P30 AI110527). The content is solely the responsibility of the authors and does not represent official views of the Centers for Disease Control and Prevention, the National Institutes of Health, the Florida State Department of Health, the Massachusetts Department of Public Health, or the New York State Department of Health, USA.
We thank the New York State Bureau of Communicable Diseases, especially Bryon Backenson, and local mosquito units for mosquito collections in New York State. We acknowledge the contributions of Joseph Maffei and Susan Jones for technical assistance, as well as the medium and tissue culture facility of the Wadsworth Center of the Department of Health, State of New York. We thank the entire array of local and state stakeholders within Massachusetts who contribute to its long-standing mosquito surveillance efforts. We thank Susmita Srivastava for sequence submission to GenBank. We thank Edward C. Holmes for critical reading and constructive input.
Y.T., A.J.A., S.C.W., T.R.U., S.C.S., A.T.C., L.D.K., and S.R.D. conceived the study. L.A.H.-L., A.J.A., S.C.W., S.H., P.M.A., R.B.T., T.A., T.R.U., S.C.S., A.T.C., and L.D.K. identified samples from historical collections, propagated virus in cell lines, extracted viral RNA, and prepared cDNA. R.A.H., V.P., and M.H.S. performed viral sequence-independent amplification, library preparation, and viral sequencing. N.F. and T.B.S. assembled and analyzed the genomes and finished genome sequences as needed. Y.T. and T.T.-Y.L. analyzed the data. Y.T., T.T.-Y.L., L.A.H.-L., and S.R.D. wrote the manuscript, and all authors reviewed and edited the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00074-18.
REFERENCES
- 1.Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. 2001. Evolutionary relationships and systematics of the alphaviruses. J Virol 75:10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver SC, Winegar R, Manger ID, Forrester NL. 2012. Alphaviruses: population genetics and determinants of emergence. Antiviral Res 94:242–257. doi: 10.1016/j.antiviral.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feemster RF. 1938. Outbreak of encephalitis in man due to the eastern virus of equine encephalomyelitis. Am J Public Health Nations Health 28:1403–1410. doi: 10.2105/AJPH.28.12.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fothergill LD, Dingle JH, Fellow JJ. 1938. A fatal disease of pigeons caused by the virus of the eastern variety of equine encephalomyelitis. Science 88:549–550. doi: 10.1126/science.88.2293.549-a. [DOI] [PubMed] [Google Scholar]
- 5.Webster LT, Wright FH. 1938. Recovery of eastern equine encephalomyelitis virus from brain tissue of human cases of encephalitis in Massachusetts. Science 88:305–306. doi: 10.1126/science.88.2283.305. [DOI] [PubMed] [Google Scholar]
- 6.White GS, Pickett BE, Lefkowitz EJ, Johnson AG, Ottendorfer C, Stark LM, Unnasch TR. 2011. Phylogenetic analysis of eastern equine encephalitis virus isolates from Florida. Am J Trop Med Hyg 84:709–717. doi: 10.4269/ajtmh.2011.10-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young DS, Kramer LD, Maffei JG, Dusek RJ, Backenson PB, Mores CN, Bernard KA, Ebel GD. 2008. Molecular epidemiology of eastern equine encephalitis virus, New York. Emerg Infect Dis 14:454–460. doi: 10.3201/eid1403.070816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong PM, Andreadis TG, Anderson JF, Stull JW, Mores CN. 2008. Tracking eastern equine encephalitis virus perpetuation in the northeastern United States by phylogenetic analysis. Am J Trop Med Hyg 79:291–296. [PubMed] [Google Scholar]
- 9.Arrigo NC, Adams AP, Weaver SC. 2010. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol 84:1014–1025. doi: 10.1128/JVI.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brault AC, Powers AM, Chavez CL, Lopez RN, Cachon MF, Gutierrez LF, Kang W, Tesh RB, Shope RE, Weaver SC. 1999. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg 61:579–586. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- 11.Weaver SC, Hagenbaugh A, Bellew LA, Gousset L, Mallampalli V, Holland JJ, Scott TW. 1994. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol 68:158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilar PV, Robich RM, Turell MJ, O'Guinn ML, Klein TA, Huaman A, Guevara C, Rios Z, Tesh RB, Watts DM, Olson J, Weaver SC. 2007. Endemic eastern equine encephalitis in the Amazon region of Peru. Am J Trop Med Hyg 76:293–298. [PubMed] [Google Scholar]
- 13.Kondig JP, Turell MJ, Lee JS, O'Guinn ML, Wasieloski LP Jr. 2007. Genetic analysis of South American eastern equine encephalomyelitis viruses isolated from mosquitoes collected in the Amazon Basin region of Peru. Am J Trop Med Hyg 76:408–416. [PubMed] [Google Scholar]
- 14.Carrera JP, Forrester N, Wang E, Vittor AY, Haddow AD, Lopez-Verges S, Abadia I, Castano E, Sosa N, Baez C, Estripeaut D, Diaz Y, Beltran D, Cisneros J, Cedeno HG, Travassos da Rosa AP, Hernandez H, Martinez-Torres AO, Tesh RB, Weaver SC. 2013. Eastern equine encephalitis in Latin America. N Engl J Med 369:732–744. doi: 10.1056/NEJMoa1212628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Novaes Oliveira R, Iamamoto K, Silva ML, Achkar SM, Castilho JG, Ono ED, Lobo RS, Brandao PE, Carnieli P Jr, Carrieri ML, Kotait I, Macedo CI. 2014. Eastern equine encephalitis cases among horses in Brazil between 2005 and 2009. Arch Virol 159:2615–2620. doi: 10.1007/s00705-014-2121-4. [DOI] [PubMed] [Google Scholar]
- 16.Luciani K, Abadia I, Martinez-Torres AO, Cisneros J, Guerra I, Garcia M, Estripeaut D, Carrera JP. 2015. Madariaga virus infection associated with a case of acute disseminated encephalomyelitis. Am J Trop Med Hyg 92:1130–1132. doi: 10.4269/ajtmh.14-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva ML, Auguste AJ, Terzian AC, Vedovello D, Riet-Correa F, Macario VM, Mourao MP, Ullmann LS, Araujo JP Jr, Weaver SC, Nogueira ML. 2015. Isolation and characterization of Madariaga virus from a horse in Paraiba State, Brazil. Transbound Emerg Dis 64:990–993. doi: 10.1111/tbed.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CDC. 2006. Eastern equine encephalitis–New Hampshire and Massachusetts, August-September 2005. MMWR Morb Mortal Wkly Rep 55:697–700. [PubMed] [Google Scholar]
- 19.Massachusetts Department of Public Health. 2012. Arbovirus surveillance summary, 2012. Massachusetts Department of Public Health, Boston, MA: http://www.mass.gov/eohhs/docs/dph/cdc/arbovirus/2012-summary.pdf. [Google Scholar]
- 20.Centers for Disease Control and Prevention. 2015. Eastern equine encephalitis epidemiology and geographic distribution. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/EasternEquineEncephalitis/tech/epi.html. [Google Scholar]
- 21.Bigler WJ, Lassing EB, Buff EE, Prather EC, Beck EC, Hoff GL. 1976. Endemic eastern equine encephalomyelitis in Florida: a twenty-year analysis, 1955-1974. Am J Trop Med Hyg 25:884–890. doi: 10.4269/ajtmh.1976.25.884. [DOI] [PubMed] [Google Scholar]
- 22.Howard JJ, Morris CD, Emord DE, Grayson MA. 1988. Epizootiology of eastern equine encephalitis virus in upstate New York, USA. VII. Virus surveillance 1978-85, description of 1983 outbreak, and series conclusions. J Med Entomol 25:501–514. [DOI] [PubMed] [Google Scholar]
- 23.Morris CD. 1988. Eastern equine encephalomyelitis, p 1–31. In Monath TP. (ed), The arboviruses: epidemiology and ecology, vol III. CRC Press, Boca Raton, FL. [Google Scholar]
- 24.Molaei G, Armstrong PM, Graham AC, Kramer LD, Andreadis TG. 2015. Insights into the recent emergence and expansion of eastern equine encephalitis virus in a new focus in the Northern New England, USA. Parasit Vectors 8:516. doi: 10.1186/s13071-015-1145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen JC, Moore FR, Williams AJ, Stark L, Miller EA, Morley VJ, Krohn AR, Garvin MC. 2011. Test of recrudescence hypothesis for overwintering of eastern equine encephalomyelitis virus in gray catbirds. J Med Entomol 48:896–903. doi: 10.1603/ME10274. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis MC, Lam HC, Zhang Y, Wang L, Hesse RA, Hause BM, Vlasova A, Wang Q, Zhang J, Nelson MI, Murtaugh MP, Marthaler D. 2016. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev Vet Med 123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo TC, Johansson DX, Haugland O, Liljestrom P, Evensen O. 2014. A 6K-deletion variant of salmonid alphavirus is nonviable but can be rescued through RNA recombination. PLoS One 9:e100184. doi: 10.1371/journal.pone.0100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allison AB, Stallknecht DE, Holmes EC. 2015. Evolutionary genetics and vector adaptation of recombinant viruses of the western equine encephalitis antigenic complex provides new insights into alphavirus diversity and host switching. Virology 474:154–162. doi: 10.1016/j.virol.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anez G, Grinev A, Chancey C, Ball C, Akolkar N, Land KJ, Winkelman V, Stramer SL, Kramer LD, Rios M. 2013. Evolutionary dynamics of West Nile virus in the United States, 1999-2011: phylogeny, selection pressure, and evolutionary time-scale analysis. PLoS Negl Trop Dis 7:e2245. doi: 10.1371/journal.pntd.0002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu PY, Ke GM, Chen PC, Liu LT, Tsai YC, Tsai JJ. 2013. Spatiotemporal dynamics and epistatic interaction sites in dengue virus type I: a comprehensive sequence-based analysis. PLoS One 8:e74165. doi: 10.1371/journal.pone.0074165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergren NA, Auguste AJ, Forrester NL, Negi SS, Braun WA, Weaver SC. 2014. Western equine encephalitis virus: evolutionary analysis of a declining alphavirus based on complete genome sequences. J Virol 88:9260–9267. doi: 10.1128/JVI.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues SG, Nunes MR, Casseb SM, Prazeres AS, Rodrigues DS, Silva MO, Cruz AC, Tavares-Neto JC, Vasconcelos PF. 2010. Molecular epidemiology of Saint Louis encephalitis virus in the Brazilian Amazon: genetic divergence and dispersal. J Gen Virol 91:2420–2427. doi: 10.1099/vir.0.019117-0. [DOI] [PubMed] [Google Scholar]
- 33.Firth AE, Chung BY, Fleeton MN, Atkins JF. 2008. Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol J 5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pybus OG, Rambaut A, Belshaw R, Freckleton RP, Drummond AJ, Holmes EC. 2007. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol 24:845–852. doi: 10.1093/molbev/msm001. [DOI] [PubMed] [Google Scholar]
- 35.Burkett-Cadena ND, White GS, Eubanks MD, Unnasch TR. 2011. Winter biology of wetland mosquitoes at a focus of eastern equine encephalomyelitis virus transmission in Alabama, USA. J Med Entomol 48:967–973. doi: 10.1603/ME10265. [DOI] [PubMed] [Google Scholar]
- 36.Bingham AM, Graham SP, Burkett-Cadena ND, White GS, Hassan HK, Unnasch TR. 2012. Detection of eastern equine encephalomyelitis virus RNA in North American snakes. Am J Trop Med Hyg 87:1140–1144. doi: 10.4269/ajtmh.2012.12-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham SP, Hassan HK, Chapman T, White G, Guyer C, Unnasch TR. 2012. Serosurveillance of eastern equine encephalitis virus in amphibians and reptiles from Alabama, USA. Am J Trop Med Hyg 86:540–544. doi: 10.4269/ajtmh.2012.11-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White G, Ottendorfer C, Graham S, Unnasch TR. 2011. Competency of reptiles and amphibians for eastern equine encephalitis virus. Am J Trop Med Hyg 85:421–425. doi: 10.4269/ajtmh.2011.11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver SC, Scott TW, Rico-Hesse R. 1991. Molecular evolution of eastern equine encephalomyelitis virus in North America. Virology 182:774–784. doi: 10.1016/0042-6822(91)90618-L. [DOI] [PubMed] [Google Scholar]
- 40.Weaver SC, Hagenbaugh A, Bellew LA, Netesov SV, Volchkov VE, Chang GJ, Clarke DK, Gousset L, Scott TW, Trent DW, et al. . 1994. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 202:1083. doi: 10.1006/viro.1994.1445. [DOI] [PubMed] [Google Scholar]
- 41.Lam TT, Zhu H, Guan Y, Holmes EC. 2016. Genomic analysis of the emergence, evolution, and spread of human respiratory RNA viruses. Annu Rev Genomics Hum Genet 17:193–218. doi: 10.1146/annurev-genom-083115-022628. [DOI] [PubMed] [Google Scholar]
- 42.Lambert AJ, Martin DA, Lanciotti RS. 2003. Detection of North American eastern and western equine encephalitis viruses by Nucleic acid amplification assays. J Clin Microbiol 41:379–385. doi: 10.1128/JCM.41.1.379-385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stucker KM, Schobel SA, Olsen RJ, Hodges HL, Lin X, Halpin RA, Fedorova N, Stockwell TB, Tovchigrechko A, Das SR, Wentworth DE, Musser JM. 2015. Haemagglutinin mutations and glycosylation changes shaped the 2012/13 influenza A(H3N2) epidemic, Houston, Texas. Euro Surveill 20:21122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geoghegan JL, Tan le V, Kuhnert D, Halpin RA, Lin X, Simenauer A, Akopov A, Das SR, Stockwell TB, Shrivastava S, Ngoc NM, Uyen le TT, Tuyen NT, Thanh TT, Hang VT, Qui PT, Hung NT, Khanh TH, Thinh le Q, Nhan le NT, Van HM, Viet do C, Tuan HM, Viet HL, Hien TT, Chau NV, Thwaites G, Grenfell BT, Stadler T, Wentworth DE, Holmes EC, Van Doorn HR. 2015. Phylodynamics of enterovirus A71-associated hand, foot, and mouth disease in Viet Nam. J Virol 89:8871–8879. doi: 10.1128/JVI.00706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djikeng A, Halpin R, Kuzmickas R, DePasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. 2008. Viral genome sequencing by random priming methods. BMC Genomics 9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Sundaram JP, Stockwell TB. 2012. VIGOR extended to annotate genomes for additional 12 different viruses. Nucleic Acids Res 40:W186–W192. doi: 10.1093/nar/gks528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 49.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426. [DOI] [PubMed] [Google Scholar]
- 50.Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 51.Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol 2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 54.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.