ABSTRACT
gp41, one of the baculovirus core genes, encodes the only recognized tegument (O-glycosylated) protein of the occlusion-derived virion (ODV) phenotype so far. A previous study using a temperature-sensitive Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) mutant showed that GP41 plays a crucial role in budded virion (BV) formation. However, the precise function of GP41 in the baculovirus replication cycle remains unclear. In this study, AcMNPV GP41 was found to accumulate around the ring zone (RZ) region within the infected nucleus and finally assembled into both BVs and ODVs. Deletion of gp41 from the AcMNPV genome showed that BVs were no longer formed and ODVs were no longer assembled, suggesting the essential role of this gene in baculovirus virion morphogenesis. In infected cells, besides the 42-kDa monomers, dimers and trimers were detected under nonreducing conditions, whereas only trimeric GP41 forms were selectively incorporated into BVs or ODVs. Mutations of all five cysteines in GP41 individually had minor effects on GP41 oligomer formation, albeit certain mutations impaired infectious BV production, suggesting flexibility in the intermolecular disulfide bonding. Single mutations of key leucines within two predicted leucine zipper-like motifs did not interfere with GP41 oligomerization or BV and ODV formation, but double leucine mutations completely blocked oligomerization of GP41 and progeny BV production. In the latter case, the usual subcellular localization, especially RZ accumulation, of GP41 was abolished. The above findings clearly point out a close correlation between GP41 oligomerization and function and therefore highlight the oligomeric state as the functional form of GP41 in the baculovirus replication cycle.
IMPORTANCE The tegument, which is sandwiched between the nucleocapsid and the virion envelope, is an important substructure of many enveloped viruses. It is composed of one or more proteins that have important functions during virus entry, replication, assembly, and egress. Unlike another large DNA virus (herpesvirus) that encodes an extensive set of tegument components, baculoviruses very likely exploit the major tegument protein, GP41, to execute functions in baculovirus virion morphogenesis and assembly. However, the function of this O-glycosylated baculovirus tegument protein remains largely unknown. In this study, we identified trimers as the functional structure of GP41 in baculovirus virion morphogenesis and showed that both disulfide bridging and protein-protein interactions via the two leucine zipper-like domains are involved in the formation of different oligomeric states. This study advances our understanding of the unique viral tegument protein GP41 participating in the life cycle of baculoviruses.
KEYWORDS: baculovirus, tegument protein, GP41, function, virion assembly, oligomerization, leucine zipper
INTRODUCTION
The viral tegument is a proteinaceous layer sandwiched between the nucleocapsid and envelope of virus particles. Tegument proteins (also called matrix proteins for some viruses) have been demonstrated to play important roles in the replication cycles of a wide variety of enveloped viruses, including herpesviruses (1–4), retroviruses (5, 6), orthomyxoviruses (7, 8), paramyxoviruses (9–11), filoviruses (12), and pneumoviruses (13). One of the best-studied examples in relation to tegument proteins is herpesvirus, a large double-stranded DNA virus in which more than 20 tegument proteins have been identified by mass spectrometry (MS). Many of these have also been characterized functionally as playing distinct roles in the virus replication cycle, such as promoting virus entry, activating gene expression, facilitating immune evasion, and regulating virion assembly and egress (14).
In baculovirus, a large double-stranded enveloped DNA virus, only a single protein, GP41, has been identified as an O-glycosylated tegument protein so far (15). Baculoviruses are specific for insects and have been used widely as biocontrol agents against insect pests, as well as being used as efficient gene expression and gene delivery vectors for human therapy (16–18). During the millions of years of evolution, baculoviruses developed a unique “life” cycle by producing two distinct virion phenotypes: budded virions (BVs) and occlusion-derived virions (ODVs) (19). ODVs infect midgut epithelial cells to initiate primary infection in insect larvae, while BVs are responsible for cell-to-cell spread of infection within the larval body, which is referred to as systemic infection (20, 21). Quantitative proteomic analysis has indicated that BVs and ODVs share similar nucleocapsid proteins; however, they are quite distinct in the composition of the viral envelope, which may result in the different tissue tropisms of these two types of virions for infection (22–24).
GP41 was originally identified as an O-linked glycoprotein most likely to occupy the space between the nucleocapsid and the envelope of the ODV, i.e., within the tegument (15). Whether GP41 is associated with BVs, however, remains controversial. GP41 was found to be associated exclusively with the ODVs, but not BVs, of Spodoptera litura multicapsid nucleopolyhedrovirus (SpltNPV) and Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) (15, 25, 26). However, a comparative proteomic study of Helicoverpa armigera nucleopolyhedrovirus (HearNPV) showed that GP41 was detected on both BVs and ODVs by mass spectrometry, although it was not detected on BVs by Western blotting (24).
There are several reports on GP41, but its function remains largely unclear. gp41 is one of the 38 baculovirus core genes. In AcMNPV and Spodoptera frugiperda multicapsid nucleopolyhedrovirus (SfMNPV), gp41 is expressed as a late viral gene and is transcribed from 12 h postinfection (p.i.) onwards (27, 28). The transcript of the gp41 gene does not seem to be spliced (29). In the case of SpltNPV, transcripts of gp41 were detected from 12 to 96 h p.i., and the protein was found to be distributed in both the cytoplasm and the nucleus (25). An AcMNPV temperature-sensitive (ts) mutant (tsB1074) with a single nucleotide mutation in the gp41 gene resulted in a single-cell infection phenotype, with which the infection does not spread and is restricted to the original transfected cells. Further electron microscopy (EM) observations showed that nucleocapsids failed to egress from the nuclear membrane to produce BVs at the nonpermissive temperature, suggesting an important role of GP41 in BV production (30). However, a gp41 knockout Bombyx mori NPV (BmNPV) was found to produce small amounts of progeny BVs (31). Since GP41 was detected exclusively on the ODV tegument of AcMNPV by liquid chromatography-tandem mass spectrometry (LC-MS/MS), we speculate that GP41 may also play an important role in ODV formation. However, the detailed function of GP41 during the baculovirus replication cycle remains to be investigated.
In this study, the gp41 gene of AcMNPV was knocked out by use of the λ Red homologous recombination method, and its function during the virus replication cycle was comprehensively investigated. GP41 was found to be not only essential for nucleocapsid egress from the nucleus to produce BVs but also indispensable for ODV morphogenesis, and therefore the nature of GP41 was further investigated. When GP41 was found as dimers and trimers in infected cells and as trimers in both BVs and ODVs, we set out to investigate whether disulfide bridging and protein-protein interactions were involved in the oligomerization of GP41 and what consequences specific mutations in GP41 to prevent oligomerization would have on the morphogenesis and assembly of BVs and ODVs.
RESULTS
Transcription, expression, and localization of AcMNPV GP41 in infected cells.
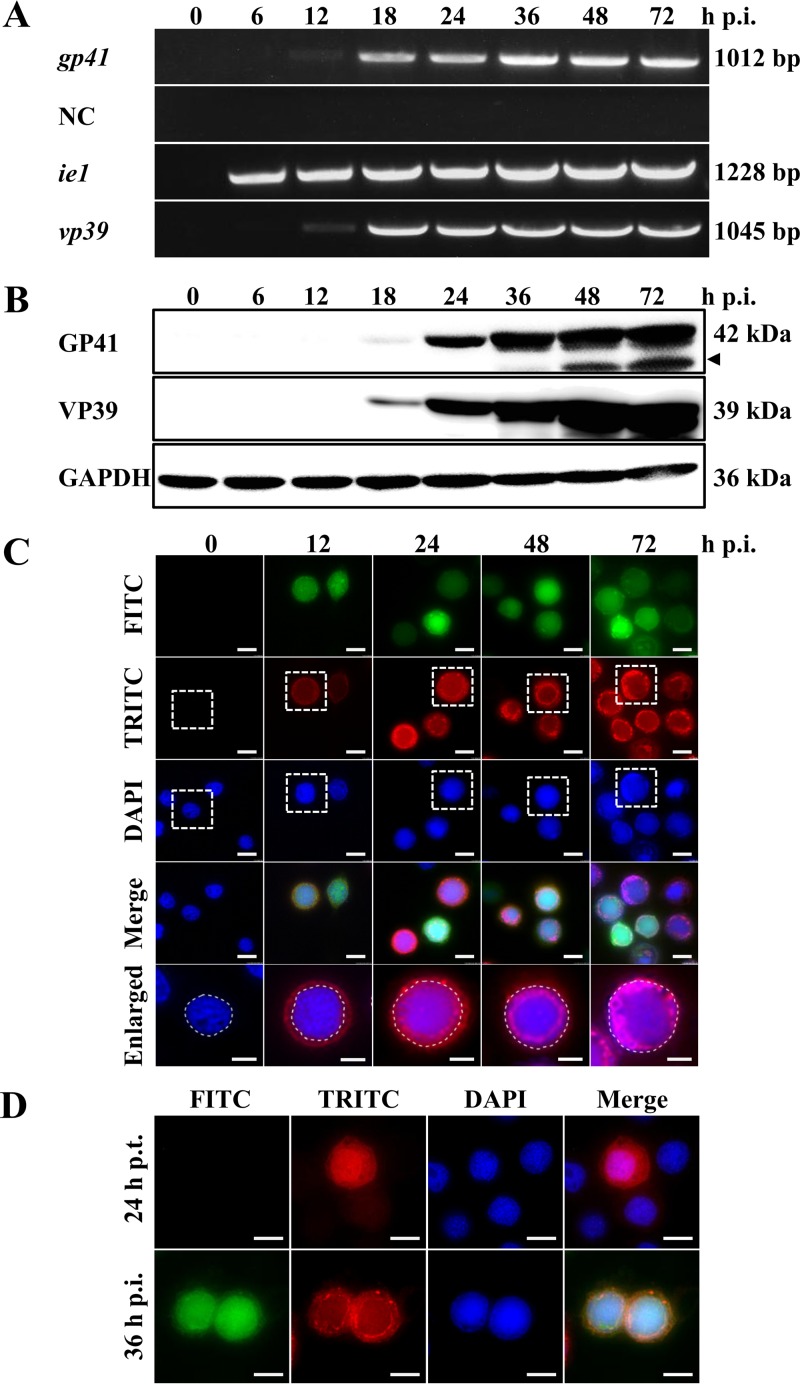
gp41 was reported to be a baculovirus late gene (28). In the present study, we determined the transcription and expression profile of gp41 in Sf9 cells (Fig. 1). Cells were infected with the control virus AcBac-egfp-ph at a multiplicity of infection (MOI) of 5 50% tissue culture infective dose (TCID50) units/cell, and transcription was detected by reverse transcription-PCR (RT-PCR) at the indicated time points. As shown in Fig. 1A, the earliest time that a gp41-specific transcript (1,012 bp) was detected was at 12 h p.i., and the detection lasted through 72 h p.i. No PCR product was detected in the negative-control group (NC), for which reverse transcriptase was omitted, excluding the possibility of contamination of genomic DNA. A viral immediate early gene and a late gene (ie1 and vp39, respectively) were also included as controls. The result was consistent with previous reports for gp41 transcripts in AcMNPV-infected Tnms42 and Sf21 cells (27, 28). An AcMNPV GP41-specific polyclonal antibody (pAb) was then produced as described in Materials and Methods, and the time course of protein expression in AcBac-egfp-ph-infected Sf9 cells was analyzed by Western blotting. As shown in Fig. 1B, a protein band migrating at 42 kDa, the expected size of GP41 (27), was first detected at 18 h p.i. and then increased gradually from 24 to 72 h p.i. An ∼35-kDa GP41-specific band was detected at later stages of infection, from 36 h p.i. onwards (indicated by a black triangle). This may be a specific breakdown product but may also be a functional protein domain of GP41 at late stages of infection. The viral capsid protein VP39 (32) and the host protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal controls. Taken together, the results of transcription and expression analyses confirmed that gp41 is a late gene of AcMNPV.
FIG 1.
Transcription, expression, and localization of AcMNPV GP41 in infected cells. (A) Time course analysis of AcMNPV gp41 transcription. Cells were infected with the control virus AcBac-egfp-ph and analyzed at the indicated time points. Total RNA was extracted, and gp41, ie1, and vp39 were detected by RT-PCR. The negative control (NC) was an RT-PCR mixture without reverse transcriptase. (B) Western blot analysis of the GP41 expression profile. Cells were infected with control virus and analyzed at the indicated time points. The expression profile of GP41 was detected by use of a specific anti-GP41 pAb. The major viral capsid protein VP39 and the host protein GAPDH were used as controls. (C) Subcellular localization of GP41 in infected cells by immunofluorescence microscopy. Sf9 cells were infected with control virus and fixed at the indicated time points. The boxed views in the tetramethyl rhodamine isocyanate (TRITC) and 4′,6-diamidino-2-phenylindole (DAPI) images were merged and enlarged and are shown in the last row. FITC, fluorescein isothiocyanate. (D) Subcellular localization of GP41 in transfected or infected cells. Sf9 cells were transfected with plasmid pIZ/V5-gp41. At 24 h p.t., cells were infected with the control virus AcBac-egfp-ph or mock infected. The localization of GP41 was detected by immunofluorescence microscopy at 24 h p.t. or 36 h p.i. Enhanced green fluorescent protein (EGFP) (green) was used as an indicator of successful virus infection, and an anti-GP41 pAb was used to detect the localization of GP41 in the infected cells (red). The nuclei were stained with Hoechst 33258 (blue). Bars, 5 μm for the enlarged images and 10 μm for the others.
To study the subcellular localization of GP41 during infection, Sf9 cells were infected with AcBac-egfp-ph at an MOI of 5 TCID50 units/cell, and the localization of GP41 was detected by immunofluorescence microscopy at the indicated time points (Fig. 1C). Upon virus infection, fluorescence signaling of GP41 was first detected at 12 h p.i. and appeared to be distributed mainly in the cytoplasm but was also evenly distributed in the nucleus. At 24 h p.i., the GP41 protein was detected around the ring zone (RZ) region of the nucleus, although fluorescence was also found spread throughout the cytoplasm and the nucleus. The RZ localization pattern lasted until the very late phase of infection, suggesting its importance for GP41 function. To observe the localization of GP41 in the absence of virus infection, a plasmid expressing gp41 alone under the control of the OpIE2 promoter was used to transfect Sf9 cells. As shown in Fig. 1D, GP41 alone was found spreading all over the cells in transfected cells (upper panels) but accumulated in the RZ upon virus infection (lower panels), suggesting that the RZ accumulation of GP41 was the result of chaperoning with a certain unknown viral or host protein. This observation could also be interpreted to mean that GP41 is incorporated into virions, which causes its concentration in the RZ.
gp41 knockout completely abolishes infectious BV production.
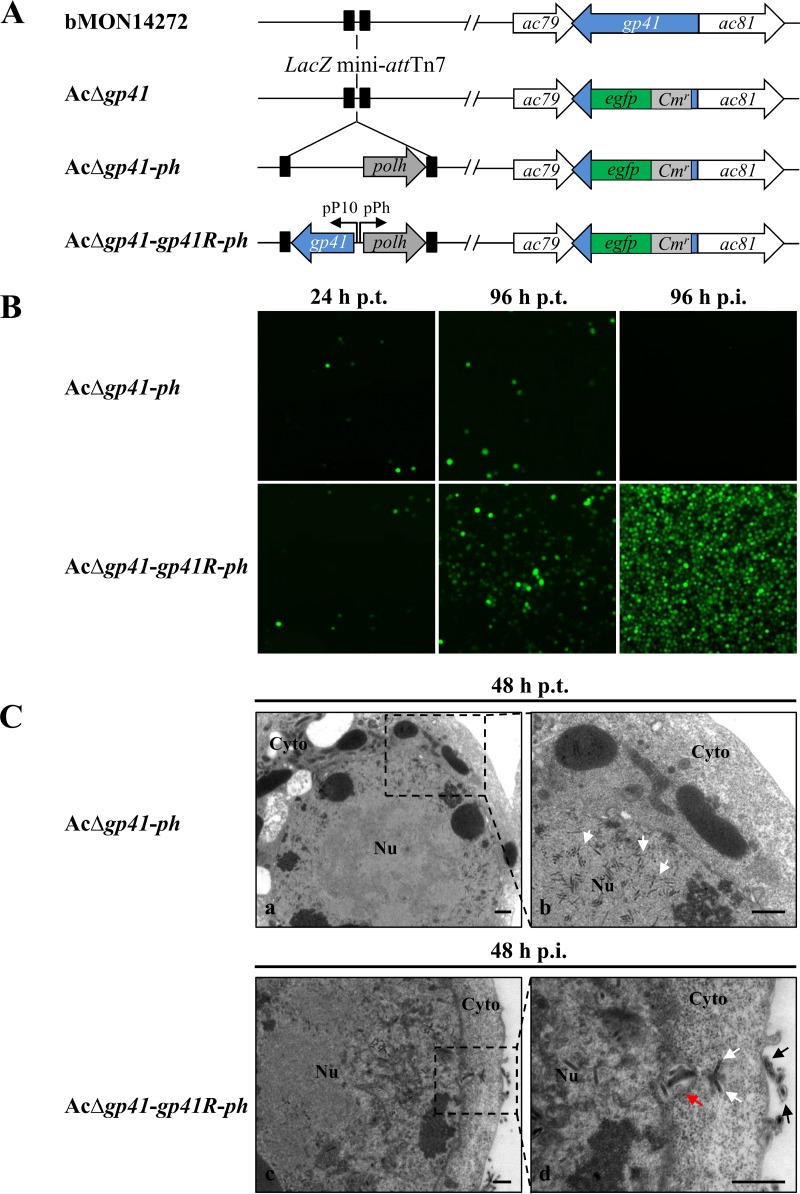
Although an important role for GP41 in BV formation has been claimed, the evidence was based on a ts mutant with a single nucleotide substitution in the AcMNPV gp41 gene (30). In addition, a more recent report showed that a gp41 knockout BmNPV could spread slowly in tissue culture (31). To better investigate the function of the gp41 gene (ac80) in the AcMNPV replication cycle, a gp41 knockout bacmid, AcΔgp41-ph, and a gp41 repaired bacmid (AcΔgp41-gp41R-ph) (Fig. 2A) were constructed as described in Materials and Methods.
FIG 2.
Construction and characterization of gp41 knockout and gp41 repaired AcMNPV bacmids. (A) Schematic representations of the recombinant bacmids AcΔgp41, AcΔgp41-ph, and AcΔgp41-gp41R-ph. (B) Transfection-infection assay. Sf9 cells were transfected with bacmid DNA of AcΔgp41-ph or AcΔgp41-gp41R-ph, and at 96 h p.t., the supernatants were used to infect uninfected Sf9 cells. Fluorescence was observed at the indicated time points. (C) EM analysis of nucleocapsid assembly and BV formation. Sf9 cells were transfected or infected with recombinant bacmid AcΔgp41-ph (a and b) or AcΔgp41-gp41R-ph (c and d). At 48 h p.t. or 48 h p.i., cells were fixed and observed by TEM. Panels b and d are enlarged pictures of the boxed regions in panels a and c. White arrows, nucleocapsids; red arrow, nucleocapsids enclosed within a cytoplasmic vesicle; black arrows, mature BVs. Nu, nucleus; Cyto, cytoplasm. Bars, 500 nm.
To determine the effect of the gp41 deletion on AcMNPV replication and propagation, Sf9 cells were transfected with the gp41 knockout bacmid (AcΔgp41-ph) and the gp41 repaired bacmid (AcΔgp41-gp41R-ph) separately. As shown in Fig. 2B, fluorescence was observed at 24 h posttransfection (p.t.) with both the gp41 deleted and repaired bacmids, which indicated that the respective transfections were successful. At 96 h p.t., fluorescence was found to be restricted to single cells within AcΔgp41-ph-transfected cells; in contrast, cell-to-cell spread of fluorescence was observed in AcΔgp41-gp41R-ph-transfected cells at 96 h p.t., signaling secondary infection. When the transfection supernatants were collected to infect another batch of healthy Sf9 cells, no infection was observed in cells infected with AcΔgp41-ph from the harvested supernatant, while obvious infection was detected in AcΔgp41-gp41R-ph-infected cells at 96 h p.i. The transfection-infection assay showed that the production of infectious BV was completely blocked as a result of the deletion of gp41. As such, this is consistent with the previous report using a ts mutant (30).
EM analysis further confirmed that in gp41 knockout bacmid-transfected cells, nucleocapsids appeared to be properly formed and gathered around the nuclear periphery at 48 h p.t. (Fig. 2C, panels a and b, white arrows); however, the egress of nucleocapsids from the nucleus to the cytoplasm for further BV budding was not observed (Fig. 2C, panels a and b). In contrast, in the repaired virus AcΔgp41-gp41R-ph-infected cells, nucleocapsids entrapped within a cytoplasmic vesicle (red arrow), trafficking in the cytoplasm (white arrows), and budding at the plasma membrane to form mature BVs (black arrows) were detected at 48 h p.i. (Fig. 2C, panels c and d). Collectively, these results confirmed an essential role of GP41 in nuclear egress of nucleocapsids and BV formation.
GP41 is required for ODV morphogenesis and is associated with nucleocapsids and the ODV envelope.
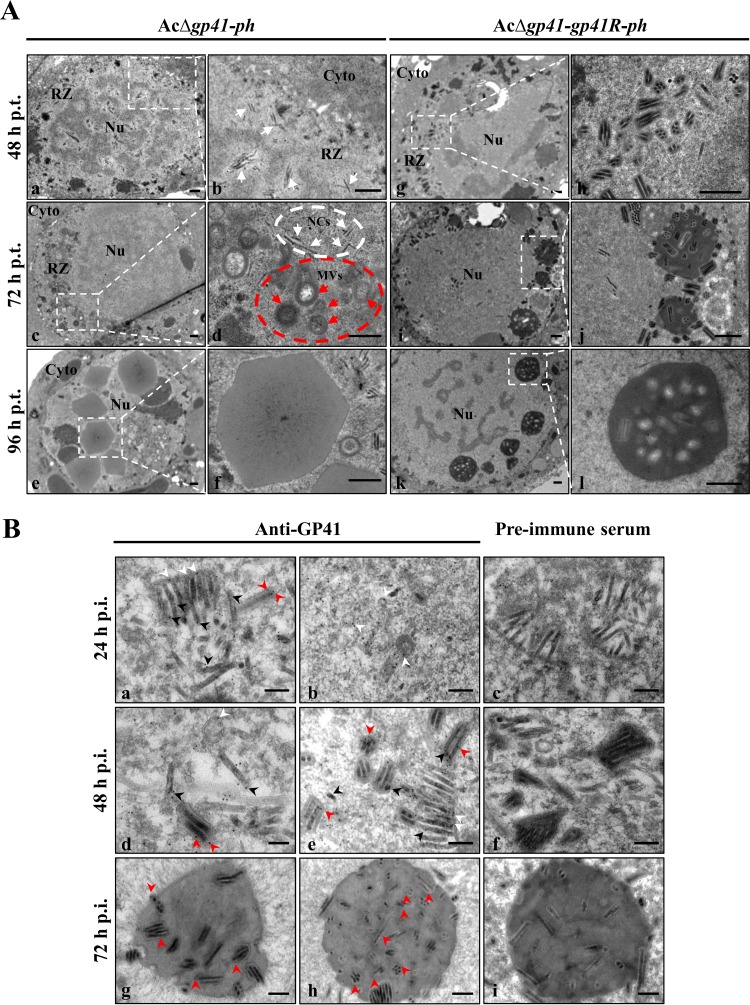
GP41 is the major tegument protein of baculovirus ODVs; however, its impact on ODV formation has not been reported before. Therefore, EM was used to monitor the effect of the gp41 deletion on virus infection, particularly on ODV morphogenesis. The RZ is a region near the margins of nuclei and surrounding the virogenic stroma where nucleocapsids are enveloped to form ODVs (33). The results showed that the transport of assembled nucleocapsids to the RZ within the nucleus was not obviously affected by the deletion of gp41 at 48 h p.t. (Fig. 3A, panels a and b, white arrows). However, the morphogenesis of ODVs in AcΔgp41-ph-transfected cells was also substantially affected (Fig. 3A, panels a and b). At 72 h p.t., nucleocapsids (Fig. 3A, panel d, white arrows in the white dashed circle) and microvesicles (MVs) (Fig. 3A, panel d, red arrows in the red dashed circle) were found aggregated in the RZ (Fig. 3A, panel c); however, the association between nucleocapsids and MVs was inhibited (Fig. 3A, panel d), and therefore ODVs could not form properly. Furthermore, at 96 h p.t., empty occlusion bodies (OBs) lacking ODVs were seen (Fig. 3A, panels e and f). In contrast, in the gp41 repaired bacmid-transfected cells, a large number of ODVs properly formed and aggregated around the RZ at 48 h p.t. (Fig. 3A, panels g and h). Along with infection progress, most ODVs were incorporated into OBs at 72 h p.t. (Fig. 3A, panels i and j). At 96 h p.t., mature OBs were observed in the nucleus (Fig. 3A, panels k and l). These results strongly indicate that GP41 is essential not only for BV formation but also for proper ODV morphogenesis and embedding.
FIG 3.
EM analysis of ODV morphogenesis and GP41 localization. (A) EM analysis of virion morphogenesis in recombinant virus-transfected cells. Sf9 cells were transfected with AcΔgp41-ph (a to f) or AcΔgp41-gp41R-ph (g to l) and fixed at 48, 72, and 96 h p.t. Ultrathin sections of the cells were observed by TEM. For each panel on the left, the right picture show an enlarged view of the boxed region. White arrows and red arrows indicate nucleocapsids and MVs, respectively. Nu, nucleus; Cyto, cytoplasm; NCs, nucleocapsids. Bars, 500 nm. (B) IEM analysis of GP41 localization in infected cells and virions. Sf9 cells were infected with control virus at an MOI of 5 TCID50 units/cell and harvested at 24, 48, and 72 h p.i. The cells were probed with anti-GP41 pAb as the primary antibody and goat anti-rabbit IgG coated with gold particles (10 nm) as the secondary antibody. Cell sections were also detected with preimmune rabbit serum as a control group. The samples were observed by TEM. Arrowheads indicate the location of GP41. Bars, 200 nm.
To further investigate the role of GP41 in ODV morphogenesis, immunoelectron microscopy (IEM) was used to detect the location of GP41 in the control virus AcBac-egfp-ph-infected cells. At 24 h p.i., nucleocapsids, MVs, and immature ODVs were observed in the nucleus. As shown in Fig. 3B, panels a and b, GP41 was localized on the nucleocapsids (black arrowheads), MVs, and ODV envelopes (white arrowheads) as well as in the tegument region of the ODVs (red arrowheads). At 48 h p.i., the localization of GP41 in the tegument of ODVs (red arrowheads) was more obvious, since mature ODVs were observed (Fig. 3B, panels d and e). GP41 was also found to be associated with MVs/ODV envelopes (white arrowheads) and nucleocapsids (black arrowheads) (Fig. 3B, panels d and e). At 72 h p.i., GP41 was also observed in the ODVs (red arrowheads) within OBs, and some GP41 was found in the matrix of OBs (Fig. 3B, panels g and h). In the control group, for which rabbit preimmune serum was used, few signals were detected on the nucleocapsids, MVs, ODVs, or OBs (Fig. 3B, panels c, f, and i). The IEM results confirmed that GP41 is a tegument protein and the association of GP41 with nucleocapsids and the ODV envelope fraction (or MVs). The results are compatible with a role for GP41 in linking ODV envelopes/MVs and nucleocapsids to mediate ODV formation.
GP41 proteins are selectively assembled as trimers into BVs and ODVs.
To further study the structure and localization of GP41 on AcMNPV virions, BVs and ODVs (from OBs) were harvested from infected cells and insect larvae, respectively, and subjected to Western blot analyses. To compare the relative amounts of GP41 on BVs and ODVs, the genomic DNA copy numbers of BVs and ODVs were determined by quantitative PCR (qPCR). Due to the presence of much smaller amounts of GP41 in BVs than in ODVs, Western blot analyses were performed by loading of 7.5 × 1012 and 3.75 × 1011 copies per lane for BVs and ODVs, respectively, to obtain approximately equivalent amounts of GP41 in the gel. The major viral capsid protein VP39 was used as a loading control, and the BV-specific envelope protein GP64 and the ODV-specific envelope protein per os infectivity factor 5 (PIF5) were used as controls to detect the purity of BVs and ODVs (Fig. 4). The results showed that GP41 could be detected in both BVs and ODVs of AcMNPV. However, the amount of GP41 present in ODVs far exceeded that in BVs (Fig. 4A). Actually, in many Western blot experiments, only with a huge amount of BV sample could a weak signal of GP41 be detected, which may explain why GP41 was not previously found in BVs of AcMNPV (26).
FIG 4.
Oligomerization of GP41 in infected cells and virions. (A) Detection of GP41 on BVs and ODVs. BVs harvested from control virus AcBac-egfp-ph-infected cells and ODVs from AcBac-egfp-ph-infected larvae were purified and analyzed by Western blotting using anti-GP41 pAb. The major viral capsid protein VP39 and the BV- and ODV-specific envelope proteins GP64 and PIF5, respectively, were used as controls. (B) Analysis of GP41 oligomerization in infected cells. Sf9 cells were infected with control virus (WT) or the gp41 repaired virus (Rep) at an MOI of 5 TCID50 units/cell. Cells were collected at 36 h p.i., treated with nonreducing buffer or reducing buffer, and subjected to Western blot analysis using anti-GP41 pAb. (C) Subcellular fractionation of virus-infected cells. Sf9 cells were infected with AcBac-egfp-ph at an MOI of 5 TCID50 units/cell. Cytoplasmic (Cy) and nuclear (Nu) components were separated at 36 h p.i., and GP41 was detected under nonreducing conditions. Lamin B and GAPDH were detected under reducing conditions to indicate the fractionation efficiency. Wc, whole-cell lysates. (D) Analysis of GP41 oligomerization in virions. BVs and ODVs were purified and subjected to Western blotting under nonreducing conditions, using a method similar to that described above.
To learn more about the function of GP41, we investigated whether or not AcMNPV GP41 could form oligomers in infected cells and/or BVs/ODVs. AcBac-egfp-ph (wild type [WT]) and the gp41 repaired virus (Rep) were used to infect Sf9 cells, and cells were harvested at 36 h p.i. and subjected to Western blot analyses under either nonreducing or reducing conditions (Fig. 4B). Under reducing conditions, only the monomeric form of GP41 (∼42 kDa) was detected for both viruses, which confirms the result shown in Fig. 1B. Under nonreducing conditions, besides the monomer band, two additional specific bands were detected: an ∼84-kDa band, which is indicative of a dimeric form of GP41, and an ∼130-kDa band, which may represent a trimeric form of GP41. Further fractionation of the infected cells into cytoplasmic and nuclear components showed that oligomers had already formed in the cytoplasm (Fig. 4C).
The BVs and ODVs were then subjected to Western blot analysis under nonreducing conditions. Note that since the amount of GP41 in BVs is relatively low compared to that in ODVs, we adjusted the loading amounts of BVs versus ODVs to obtain a representative image. Interestingly, only trimeric bands (∼130 kDa) of GP41 were detected on both BVs and ODVs (Fig. 4D). The results thus showed that GP41 formed oligomers in infected cells, while GP41 was selectively assembled as trimers into progeny virions. This suggests that a trimer is likely to be the functional form of GP41 in BVs and ODVs.
Impacts of single-point mutations of cysteines on GP41 oligomerization and function.
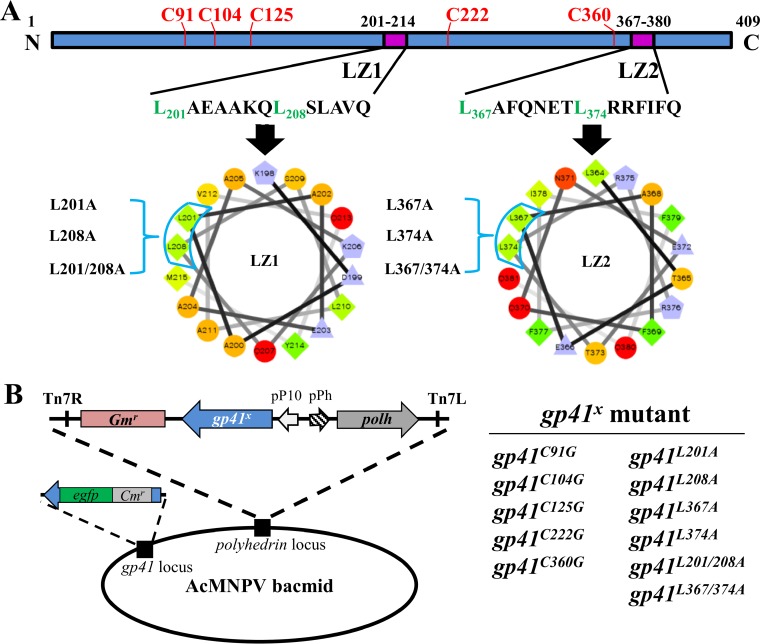
AcMNPV GP41 contains five cysteines that may be involved in forming disulfide bonds (Fig. 5A). The five cysteines of AcMNPV GP41 are found to be conserved only in group I alphabaculoviruses and are not completely conserved in the Baculoviridae family. To determine the possible roles of the cysteine residues in GP41, five single-point mutants, GP41C91G, GP41C104G, GP41C125G, GP41C222G, and GP41C360G, were generated in donor plasmids, and then a series of mutated bacmids, i.e., AcΔgp41-gp41C91G-ph, AcΔgp41-gp41C104G-ph, AcΔgp41-gp41C125G-ph, AcΔgp41-gp41C222G-ph, and AcΔgp41-gp41C360G-ph, were constructed (Fig. 5B).
FIG 5.
Construction of cysteine and leucine zipper mutant recombinant viruses. (A) Prediction of GP41 oligomerization motifs. The leucine zipper-like coiled-coil domain was predicted by PCOILS and is presented by use of helical wheel projection software. The key leucine residues are presented as green diamonds. (B) Construction of cysteine and leucine zipper-like motif mutant recombinant viruses. The cysteines and key leucines were mutated individually or in combination by site-directed mutagenesis. The ph gene and each of the mutant gp41 genes were coinserted into the ph locus of the AcΔgp41 bacmid to construct AcΔgp41-gp41x-ph.
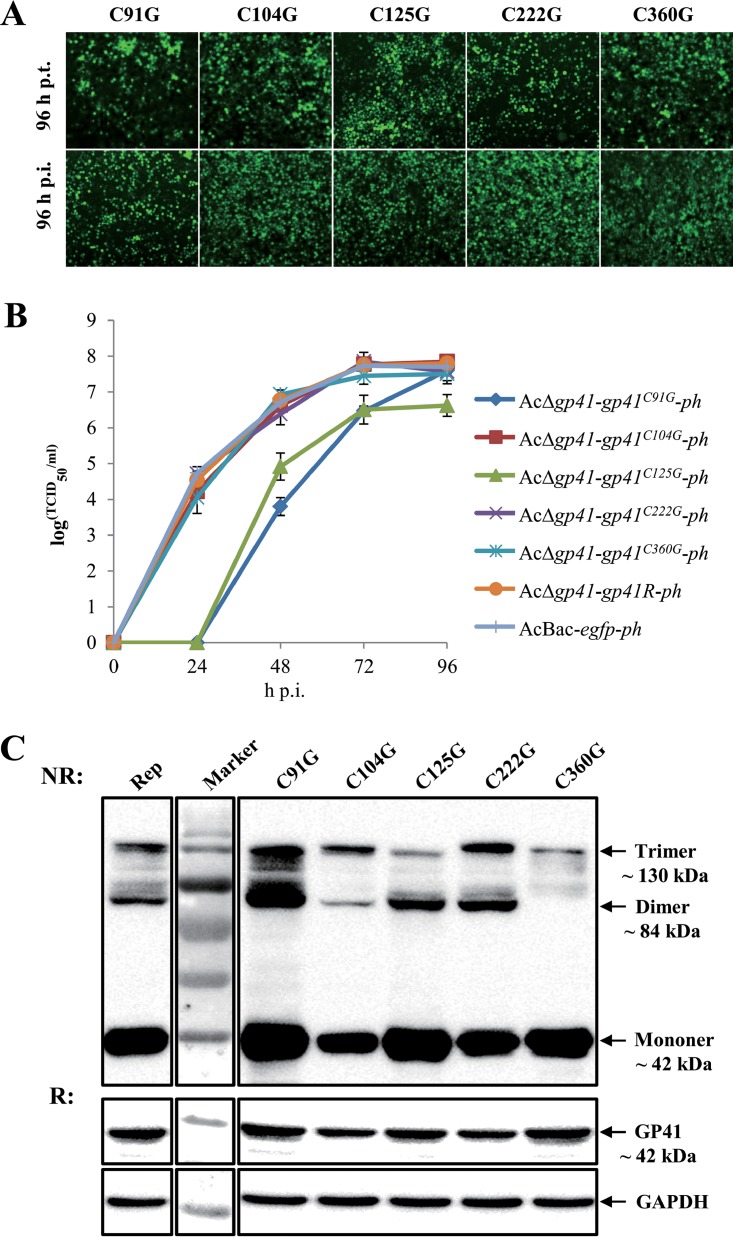
The above-described bacmids were used to transfect or infect Sf9 cells. As shown in Fig. 6A, fluorescence was observed at 96 h p.t., indicating successful transfection. All five single-point mutants were able to produce infectious BVs, as indicated by infection assay at 96 h p.i. Further one-step growth curve assays showed that the C91G and C125G mutants produced significantly smaller amounts of infectious BVs than those obtained with the gp41 repaired virus and the WT control virus (P < 0.001), although the BV titer of the C91G mutant was similar to that of the repaired virus at the final stage of infection (96 h p.i.). The other three mutants (C104G, C222G, and C360G) had a BV production capability comparable to that of the WT virus (P > 0.05) (Fig. 6B).
FIG 6.
Characterization of cysteine mutant recombinant viruses. (A) Transfection-infection assay. Sf9 cells were transfected with the five cysteine mutants, and fluorescence was observed at 96 h p.t. and 96 h p.i. (B) One-step growth curve analysis of cysteine mutants. Sf9 cells were infected separately with the five single cysteine mutants, WT virus, or repaired virus at an MOI of 5 TCID50 units/cell. Virus titers of supernatants collected at different time points were determined by endpoint dilution assay. The experiments were replicated twice. Error bars show standard deviations. (C) Sf9 cells were infected with the five cysteine mutants and the gp41 repaired recombinant virus and subjected to Western blot analysis under nonreducing (NR) or reducing (R) conditions.
To identify the role of cysteines in GP41 oligomerization, cysteine mutant-infected cells were subjected to Western blot analysis at 36 h p.i. under either nonreducing or reducing conditions. All five mutants showed oligomers, although some were found in smaller amounts than those for the gp41 repaired virus (Rep), in the infected cells under nonreducing conditions (Fig. 6C). GAPDH was detected as a cell sample control. The results suggest that no individual cysteine is essential for disulfide bridging of GP41; however, the C91G and C125G mutations may have affected the structure or functionality of GP41 and impaired BV production.
Two leucine zipper-like motifs are required for BV infectivity and ODV assembly.
To determine possible motifs responsible for GP41 oligomerization, bioinformatic analyses were performed. Two leucine zipper-like coiled-coil motifs were found in GP41 by use of PCOILS software, and they were designated LZ1 (amino acids [aa] 201 to 214) and LZ2 (aa 367 to 380) (Fig. 5A, purple boxes). Of the two, LZ1 is conserved among all baculoviruses except the deltabaculoviruses, while LZ2 is conserved only in alpha- and betabaculoviruses (data not shown). The leucine zipper motif is a common oligomerization domain that facilitates protein-DNA interactions or protein-protein interactions through the hydrophobic surface of the parallel/antiparallel coiled-coil motifs (34). The key leucines (or isoleucine/valine in some cases) are crucial for stabilizing the oligomers. Helical wheel representation of the two putative leucine zipper-like motifs showed that L201 and L208 in LZ1 as well as L367 and L374 in LZ2 may be crucial for formation of a hydrophobic surface in the coiled-coil structure (Fig. 5A).
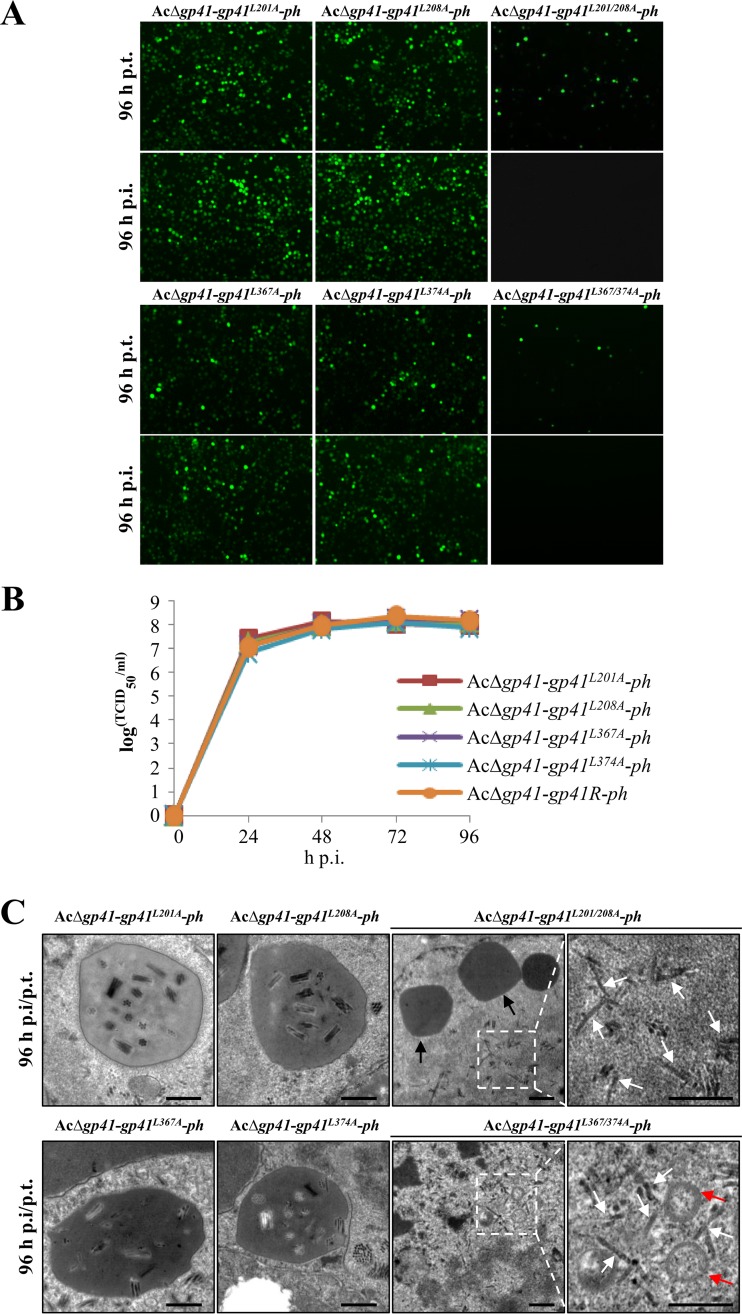
To investigate whether the leucine zipper-like motifs and the pertinent leucine residues are important for GP41 oligomerization and function, four single-point mutants, GP41L201A, GP41L208A, GP41L367A, and GP41L374A, and two double-site mutants, GP41L201/208A and GP41L367/374A, were generated in donor plasmids, and then a series of mutated bacmids, i.e., AcΔgp41-gp41L201A-ph, AcΔgp41-gp41L208A-ph, AcΔgp41-gp41L367A-ph, AcΔgp41-gp41L374A-ph, AcΔgp41-gp41L201/208A-ph, and AcΔgp41-gp41L367/374A-ph, was constructed (Fig. 5B). The above recombinant bacmids were used to transfect or infect Sf9 cells. As shown in Fig. 7A, all four single-point mutants were able to produce infectious BVs. In contrast, for the double-site mutants, AcΔgp41-gp41L201/208A-ph and AcΔgp41-gp41L367/374A-ph, although transfection was successful, no fluorescence was detected in the infection assay. This indicated that double mutation of leucine residues in either LZ1 or LZ2 is lethal for infectious BV production. To further quantify the effects of single-point mutations on BV production, one-step growth curve analysis was performed. The results showed that the infectious BV production levels of the four single-point mutant recombinant viruses were not significantly different from that of the gp41 repaired virus (AcΔgp41-gp41R-ph) (P > 0.05) (Fig. 7B).
FIG 7.
Characterization of leucine zipper-like motif mutant recombinant viruses. (A) Transfection-infection assay. Sf9 cells were transfected with leucine zipper-like motif mutant recombinant bacmids, and at 96 h p.t. the transfection supernatants were used to infect healthy Sf9 cells. Fluorescence was observed at 96 h p.t. and 96 h p.i. (B) One-step growth curve assay of mutated viruses. Sf9 cells were infected with single leucine mutant viruses or the gp41 repaired virus (AcΔgp41-gp41R-ph) at an MOI of 5 TCID50 units/cell. The supernatants of infected cells were harvested at the indicated time points, and virus titers were determined by endpoint dilution assay. The experiments were replicated twice. Error bars show standard deviations. (C) EM analysis of GP41 leucine zipper-like motif mutants. Cells were infected with single leucine mutant viruses or transfected with double leucine mutant bacmid DNA. At 96 h p.t. and 96 h p.i., cells were harvested for EM analysis. Black arrows indicate OBs, red arrows indicate MVs, and white arrows indicate nucleocapsids. Bars, 500 nm.
To further analyze ODV morphogenesis of the leucine zipper mutants, EM analyses were performed. At 96 h p.i., cells infected with single-point mutants showed the expected ODV formation and incorporation of ODVs into OBs, similar to the situation with repaired virus-infected cells (Fig. 7C). For the double-site mutants, AcΔgp41-gp41L201/208A-ph and AcΔgp41-gp41L367/374A-ph, since no infectious BVs were produced, transfected cells (96 h p.t.) were used for EM analyses. The results showed that nucleocapsids (white arrows) formed as in the WT. It seemed that these nucleocapsids were adjacent to MV-like structures (red arrows), but they may not be linked to form ODVs properly (Fig. 7C). In addition, empty OBs produced by these double mutants were found (black arrows), as was the case with gp41 knockout virus. Taken together, these results suggest that the double leucine mutations in GP41 severely affected BV formation, ODV envelopment, and embedding in OBs.
Leucine zipper-like motifs contribute to GP41 oligomerization and RZ localization.
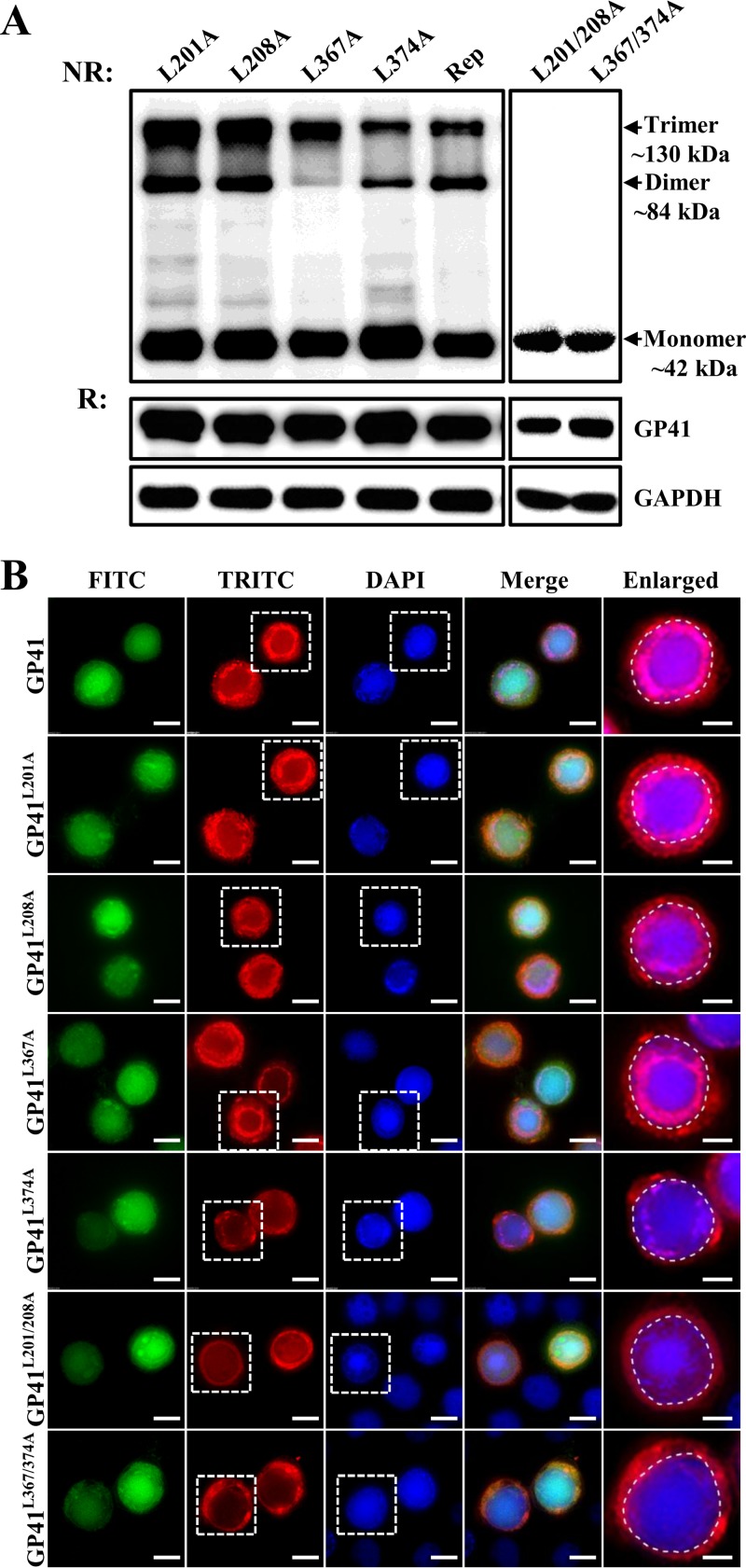
To see whether leucine zipper-like motifs contribute to the oligomerization of GP41, leucine zipper mutant-infected or -transfected cells were subjected to Western blotting to analyze oligomers under nonreducing or reducing conditions. As shown in Fig. 8A, when cell samples of the four single-point mutants were analyzed under nonreducing conditions, besides the monomeric form of ∼42 kDa, a dimeric form (∼84 kDa) and a trimeric form (∼130 kDa) could also be detected. This is similar to the results for the control virus and the gp41 repaired virus (Rep), although AcΔgp41-gp41L374A-ph seemed to have fewer oligomers than those of the other three single-point mutants in the infected cells (Fig. 8A, upper left panel). For cells transfected with the double leucine mutants, AcΔgp41-gp41L201/208A-ph and AcΔgp41-gp41L367/374A-ph, only an ∼42-kDa monomeric protein band was detected (Fig. 8A, upper right panel). When cell protein samples were treated under reducing conditions and separated by PAGE, GP41 appeared as a monomer in all cases, as expected (Fig. 8A, middle panels). GAPDH was detected as a sample loading control (Fig. 8A, lower panels). These experiments showed that single mutations of the key leucines in LZ1 and LZ2 did not have an obvious impact on the oligomerization of GP41 but that double mutations completely aborted the formation of oligomers.
FIG 8.
Oligomerization and subcellular localization of leucine zipper-like motif mutants. (A) Oligomerization of leucine zipper-like motif mutant GP41. Sf9 cells were infected with single-site mutants or transfected with double-site mutants and collected at 36 h p.i. or 48 h p.t. After being treated under nonreducing (NR) or reducing (R) conditions, cell samples were analyzed by Western blotting using anti-GP41 pAb. GAPDH was detected as a control. (B) Subcellular localization of leucine zipper-like motif mutants. Cells were infected or transfected with leucine zipper-like motif mutants and fixed at 36 h p.i. or 48 h p.t. The locations of GP41 mutants were detected by immunofluorescence microscopy with anti-GP41 pAb. The enlarged fields show the boxed regions of the TRITC and DAPI channels. Bars, 5 μm for the enlarged images and 10 μm for the others.
Immunofluorescence microscopy was used to detect localization of mutated GP41 proteins in the infected or transfected Sf9 cells. In single-point mutant-infected cells, most of the GP41 was transported to the RZ, although some GP41 was found to be diffused in the cytoplasm and the nucleus, which is similar to the localization of GP41 in the control virus-infected cells. The L374A mutant also showed a lower level of RZ localization (Fig. 8B, enlarged field). However, in double-site mutant-transfected cells, the RZ accumulation of GP41 disappeared, and the protein was found to be distributed evenly in the cytoplasm as well as in the nucleus (Fig. 8B, enlarged fields). These results are indicative that leucine zipper-like motifs are critical for GP41 oligomerization and accumulation at the RZ.
DISCUSSION
gp41 encodes the only known tegument protein of baculoviruses and is expected to play a crucial role during the virus replication cycle. However, so far, knowledge on the function and structure of GP41 remains largely unclear. In the present study, by construction of a gp41-deleted AcMNPV strain, we confirmed that GP41 is essential for infectious BV production (Fig. 2B). This is consistent with a previous report for AcMNPV (30). For BmNPV, gp41 was found to also play an important role in infectious BV production, although it is not essential (31). This indicates that the functions of GP41 may not be identical in different baculoviruses.
EM pictures showed that the nucleocapsids appeared to be properly formed and assembled but were retained at the nuclear periphery (Fig. 2C). This may indicate that GP41 is involved in nucleus-cytoplasm transportation of nucleocapsids to form BVs. This is similar to the role of the tegument protein M1 of influenza virus, which mediates nuclear export of the viral RNPs (vRNPs) (35) and, further, provides the driving force for virus budding (36, 37). We have shown here that GP41 is associated with AcMNPV BVs (Fig. 4), further underscoring the role of GP41 in BV formation. GP41 may be involved not only in nucleocytoplasmic transport, as shown here, but also (later) in cytoplasmic transport of nucleocapsids and BV egress, as for the influenza virus tegument protein M1 (35). Interestingly, two recent studies identified the interaction of GP41 with host endosomal sorting complex required for transport III (ESCRT-III) components or N-ethylmaleimide-sensitive factor (NSF), implying roles for GP41 in nucleocapsid egress through the nuclear envelope and in cytoplasmic transportation during BV budding (38, 39). We also noticed that the amount of GP41 in BVs was much lower than that in ODVs (Fig. 4). Therefore, the previous inability to detect GP41 in BVs may well have been due to the insufficient amounts of sample being tested or to low detection sensitivity by the metabolic labeling method for labeling of glycans in GP41 (15).
The abundant amount of GP41 in ODVs is indicative of an important role of GP41 in ODV formation. In our study, the RZ accumulation of GP41 within the infected nucleus at the late stage of infection supported a role for GP41 in ODV formation (Fig. 1). Deletion of GP41 and subsequent EM analyses showed that GP41 is indispensable for ODV envelopment and subsequent embedding into OBs (Fig. 3A). Further IEM analysis showed that GP41 is located not only in the tegument of ODV but also on both ODV envelopes/MVs and nucleocapsids (Fig. 3B), suggesting that GP41 may act as a matchmaker to bring the two together. Among the tegument proteins of herpesviruses, many have been verified as important for virus primary or secondary envelopment. For example, the pUL11-pUL16 complex of herpes simplex virus 1 (HSV-1) possibly bridges the nucleocapsid and envelope (40), and the pUL36-pUL37 complex of HSV-1 also acts as another bridge during virus assembly. In addition, the deletion of either complex results in a defect in secondary envelopment and in accumulation of nonenveloped capsids in the cytoplasm (3).
Tegument proteins of many viruses self-assemble into oligomers to exert their specific roles in virus assembly and release. For example, human cytomegalovirus (HCMV) pUL71 was found to form oligomers, and its oligomerization seems to be essential for virus maturation (41). For RNA viruses, it is also quite common to find their matrix proteins forming oligomers. Dimerization of the canine distemper virus (CDV) matrix (M) protein governs its cell membrane periphery localization and membrane budding activity (11). Studies have shown that Ebola virus VP40 (42, 43), respiratory syncytial virus M (44), vesicular stomatitis virus M (45), and influenza virus M1 (8) form oligomers or multimers in vivo or in vitro. The oligomerization of these matrix proteins is considered to be important for RNA virus assembly and may provide the driving force for virus budding (8, 44, 45). In our study, three kinds of molecular mass structures, representing monomers, dimers, and trimers of GP41, were detected in infected cells under nonreducing conditions (Fig. 4B), suggesting that GP41 is oligomerized under native conditions. Interestingly, only the trimeric structure was selectively assembled into both BVs and ODVs, implying that trimers are likely to be the major functional form of GP41. Coincidently, the filovirus matrix protein VP40 can form two different oligomeric structures that may function at distinct steps of virus assembly and budding (46). In the case of HIV-1, the dimers of the Gag protein are precursors for efficient oligomerization to form higher-order structures (47). Whether the dimeric structure of GP41 represents another functional form or just serves as an intermediate in trimer assembly remains to be investigated.
To determine the mechanism of GP41 oligomerization, we focused on a common and well-characterized protein interaction interface, the coiled-coil structure (48). Two leucine zipper-like coiled-coil motifs are located in the central and C-terminal regions of GP41 (Fig. 5A). Leucine zipper motifs are also very crucial for the oligomerization of other viral tegument proteins. For example, mutation of the putative leucine zipper motif of HCMV pUL71 abrogated its oligomerization and impaired virus growth (41). Deletion of the region containing the leucine zipper motifs of the pseudorabies virus tegument protein pUL36 impaired the replication of the virus, suggesting a role for the leucine zipper motif in the homo-oligomerization needed for interaction with other proteins (49). Our results showed that double mutations of the key leucines of either leucine zipper-like motif completely abolished oligomerization and RZ localization of GP41 (Fig. 8), ODV morphogenesis, and production of BV progeny (Fig. 7), providing a functional connection between the trimer structure of GP41 and ODV assembly/BV production. However, there is still a possibility that certain mutations may result in additional structural changes required for GP41 functionality.
Apart from the coiled-coil structure, disulfide bonds also play an important role in protein-protein interaction, and therefore the role of disulfide bonds in GP41 oligomerization was investigated. Under reducing conditions, GP41 ended up as a monomer, yet single-point mutations of all five cysteines individually had only minor influences on oligomer formation, although certain mutants had impaired BV formation (Fig. 6). It is possible that alternative disulfide bonds are formed upon single mutation or that there is more than 1 disulfide bond in GP41 and that blocking one does not affect oligomerization. It may be that intramolecular leucine zipper and disulfide bond interactions provide both compensatory stability to the GP41 trimer complex and flexibility in its multiple interactions with various proteins leading to diverse processes, such as BV and ODV envelopment. Vertebrate viruses have evolved multiple tegument proteins for virion assembly, and it is possible that GP41, the major tegument protein, is a highly multifunctional protein which is exposed on the outside of nucleocapsids and executes a very different envelopment process resulting in BVs and ODVs.
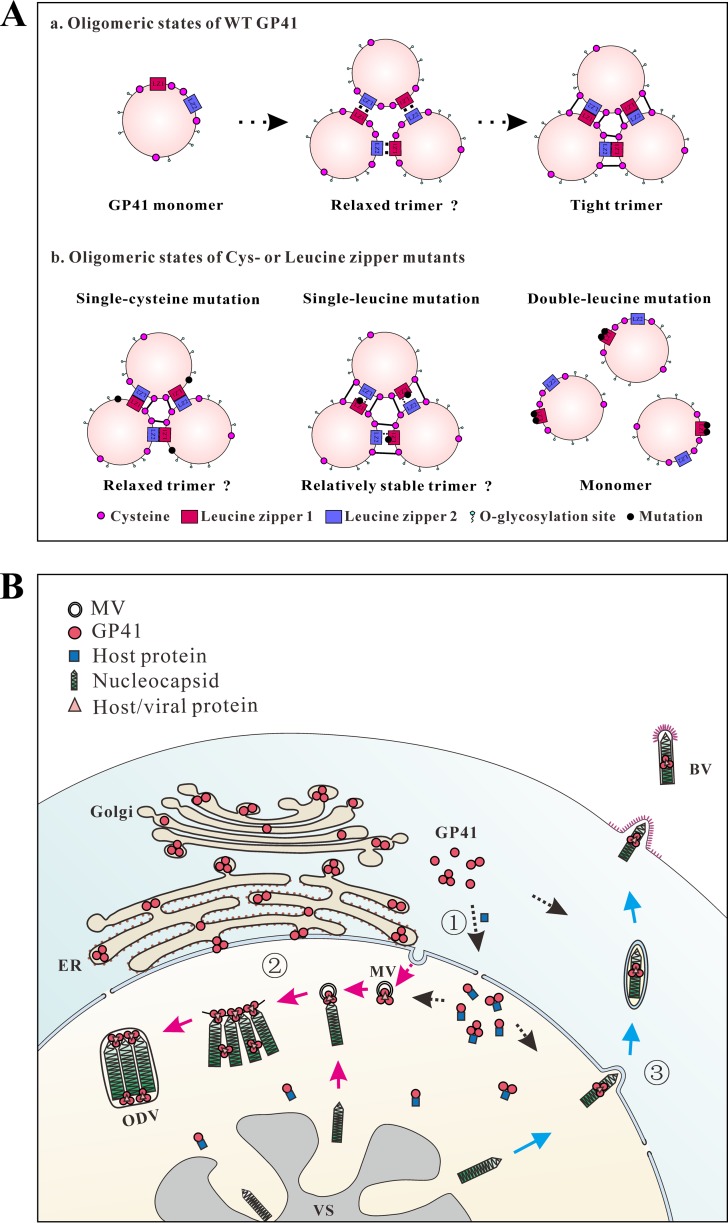
For the structural assembly of the trimers of WT GP41, we propose that the leucine zipper-like domains of GP41 are involved in the initial recruitment of monomers to form relaxed trimers, upon which disulfide bridging occurs to consolidate the trimer (Fig. 9A, panel a). This model is compatible with the observation that double leucine mutants abort trimer formation as well as BV and ODV production (Fig. 7 and 8). This suggests that intermolecular disulfide bonds may not form properly in the absence of leucine zippers. Since fewer trimers and decreased BV infectivity were observed with certain single cysteine mutations, we propose that relaxed GP41 trimers were formed (Fig. 9A, panel b). Single leucine mutations did not affect GP41 trimer formation and BV and ODV production, suggesting that relatively stable trimers were formed (Fig. 9A, panel b). This model predicts that double or more cysteine mutants or single cysteine and leucine mutants combined will result in less-stable trimers, possibly resulting in low or no BV or ODV formation.
FIG 9.
Proposed model for GP41 transportation, oligomerization, and function in the AcMNPV life cycle. (A) Working model for oligomerization of WT GP41 and different mutants. (a) Oligomeric states of WT GP41. GP41 recruits other GP41 monomers to form relaxed trimers via leucine zipper-like motifs. Afterwards, disulfide bonds form and bind the trimers tightly and stably. (b) Oligomeric states of cysteine or leucine zipper mutants. In single cysteine mutants, one disulfide bond is broken, and other disulfide bonds together with leucine zipper-like motifs loosely maintain the trimeric structure. In single leucine mutants, although the hydrophobic interaction is partially interrupted, disulfide bonds still maintain a relatively stable trimeric structure. However, a double leucine mutation totally interrupts the interactions of GP41 monomers. (B) Transportation and function of GP41 in infected cells. Three forms (monomers, dimers, and trimers) of GP41 are synthesized in the cytoplasm and transported into the nucleus (step 1; black arrows). The nuclear import and RZ accumulation of GP41 likely happen with the help of certain still-unknown viral/host proteins. Within the infected nucleus, GP41 trimers may interact with MVs and nucleocapsids to accomplish ODV envelopment and embedding (step 2; pink arrows), promote efficient nuclear egress of nucleocapsids, or facilitate cytoplasmic transport of nucleocapsids to form BVs (step 3; blue arrows). VS, virogenic stroma; ER, endoplasmic reticulum; Golgi, Golgi apparatus.
Based on the results so far, we propose a model for GP41 transportation, oligomerization, and function in the baculovirus life cycle. As shown in Fig. 9B (part 1), after being synthesized in the cytoplasm, GP41 may undergo O-glycosylation (15) in the endoplasmic reticulum (ER) or Golgi apparatus of the cytoplasm. By nuclear-cytoplasmic fractionation and Western blot analysis, oligomers were found to already be formed in the cytoplasm (Fig. 4C). Since GP41 itself lacks a recognizable nuclear localization signal, it might enter the nucleus (Fig. 1D, upper panels) chaperoned by certain host proteins or via an O-GlcNAc-dependent mechanism (50, 51). The RZ accumulation within the infected nucleus may be facilitated by certain, still-unknown viral/host proteins (Fig. 1D, lower panels, and 9B, black arrows). During ODV morphogenesis within the infected nucleus, trimeric GP41 may interact with MVs and nucleocapsids to mediate ODV envelopment and embedding (Fig. 9B, part 2, pink arrows). During BV assembly and formation, trimeric GP41 may promote efficient egress of nucleocapsids from the nuclear envelope to the cytoplasm. Alternatively, trimeric GP41 in the cytoplasm may facilitate cytoplasmic transport of nucleocapsids to finally form BV (Fig. 9B, part 3, blue arrows).
In summary, our data demonstrate that (i) GP41 is not only required for nucleocapsid egress to form BVs but also essential for proper ODV morphogenesis; (ii) GP41 is much more abundantly assembled into ODVs than into BVs; (iii) GP41 forms oligomers in infected cells, but only trimers are assembled into virions; and (iv) leucine zipper-like motifs, with the help of disulfide bonding, play a crucial role in facilitating GP41 oligomerization and localization and thus are essential for GP41 function. These findings enrich our understanding of the function of baculovirus tegument protein and virus assembly.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda 9 (Sf9) cells were cultured in Grace's insect medium (pH 6.0; Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL) at 27°C. The control virus AcBac-egfp-ph, a recombinant AcMNPV with an enhanced green fluorescent protein (egfp) reporter gene and the AcMNPV polyhedrin (ph) gene, was constructed previously (52).
Construction of the gp41 knockout bacmid.
The AcMNPV gp41 gene was replaced by a chloramphenicol resistance (Cmr) gene and an egfp gene through homologous recombination in Escherichia coli, as described previously (53). To avoid any influence on the transcription of the flanking genes, the 72-nucleotide (nt) 5′-terminal and 240-nt 3′-terminal regions of the gp41 open reading frame (ORF) were preserved. Briefly, a 618-bp sequence upstream of the gp41 ORF and a 686-bp sequence downstream of the gp41 gene were amplified with primers gp41UP-F/R and gp41DN-F/R (Table 1), using AcMNPV bacmid DNA as the template. The upstream and downstream homologous arms were inserted separately into plasmid pKS-egfp-Cmr (54), flanking an egfp-Cmr cassette. The resulting plasmid was designated pKS-gp41up-egfp-Cmr-gp41dn. Linear gp41up-egfp-Cmr-gp41dn fragments for homologous recombination were generated by KpnI and EcoRV digestion and then used to transform E. coli BW25113 competent cells containing the AcMNPV bacmid (bMON14272) and the λ Red recombinase-encoding plasmid pKD46 (53). Positive clones were selected with chloramphenicol/kanamycin and further verified by PCR. The resulting gp41 knockout bacmid was named AcΔgp41.
TABLE 1.
Primers used in this paper
| Application and primer name (restriction enzyme) | Sequence (5′-3′)a |
|---|---|
| Deletion of gp41 | |
| gp41UP-F (KpnI) | GCGGGTACCTTTTGTGAACGGGCTCGCTAA |
| gp41UP-R (XhoI) | GCGCTCGAGCAGCCCAGCATGTTCCAGAA |
| gp41DN-F (EcoRV) | GCGGATATCCGTTGCCGGATTAGAGGGAT |
| gp41DN-R (XbaI) | GCGTCTAGAAGATTAAATACGATAGTGAACTGT |
| Repair of gp41 and ph | |
| ph-F (EcoRI) | GGTGAATTCATGCCGGATTATTCATACCGTCCCA |
| ph-R (SalI) | ACTTGTCGACTTAATACGCCGGACCAGTGAAC |
| gp41-F (XhoI) | GCGCTCGAGATGACAGATGAACGTGGCAAT |
| gp41-R (NheI) | GCGGCTAGCTTATGCAGTGCGCCCTTTCGT |
| Mutagenesis of gp41 | |
| gp41C91G-F | GAATCTATTTGGTACAACAAAGGTGTGGATTTTGTTCAAAAGATTATTAG |
| gp41C91G-R | ATAATCTTTTGAACAAAATCCACACCTTTGTTGTACCAAATAGATTCACC |
| gp41C104G-F | CAAAAGATTATTAGATATTACAGGGGCAATGACATGTCAGAACTTAGTCC |
| gp41C104G-R | ACTAAGTTCTGACATGTCATTGCCCCTGTAATATCTAATAATCTTTTGAA |
| gp41C125G-F | TTTTTATAAACACTATTCGTGACATGGGCATCGACACGAACCCAATCAGC |
| gp41C125G-R | CGCTGATTGGGTTCGTGTCGATGCCCATGTCACGAATAGTGTTTATAAAA |
| gp41C222G-F | TACATGGTGGCGGAAGCGGTGACGGGCAACATTCCCATTCCGTTACCTTT |
| gp41C222G-R | GATTGAAAGGTAACGGAATGGGAATGTTGCCCGTCACCGCTTCCGCCACC |
| gp41C360G-F | CGGCCGCTCATTTGACCAAAAGCGCTGGCCAGGAGAGCTTGACCGAATTG |
| gp41C360G-R | ACGCCAATTCGGTCAAGCTCTCCTGGCCAGCGCTTTTGGTCAAATGAGCG |
| gp41L201A-F | CTGATATGTTGGGCAAAGACGCTGCCGCCGAGGCCGCCAAACAACTC |
| gp41L201A-R | GAGTTGTTTGGCGGCCTCGGCGGCAGCGTCTTTGCCCAACATATCAG |
| gp41L208A-F | TTGGCCGAGGCCGCCAAACAAGCCAGTCTGGCCGTCCAGTACATG |
| gp41L208A-R | CATGTACTGGACGGCCAGACTGGCTTGTTTGGCGGCCTCGGCCAA |
| gp41L367A-F | GCCAGGAGAGCTTGACCGAAGCGGCGTTCCAAAACGAAACTCTAAG |
| gp41L367A-R | CTTAGAGTTTCGTTTTGGAACGCCGCTTCGGTCAAGCTCTCCTGGC |
| gp41L374A-F | TTGGCGTTCCAAAACGAAACTGCCAGACGTTTTATTTTTCAACAAATAAAT |
| gp41L374A-R | ATTTATTTGTTGAAAAATAAAACGTCTGGCAGTTTCGTTTTGGAACGCCAA |
| gp41L201/208A-F | GACGCTGCCGCCGAGGCCGCCAAACAAGCCAGTCTGGCCGTCCAGTACATG |
| gp41L201/208A-R | GACTGGCTTGTTTGGCGGCCTCGGCGGCAGCGTCTTTGCCCAACATATCA |
| gp41L367/374A-F | CCGAAGCCGCGTTCCAAAACGAAACTGCCAGACGTTTTATTTTTCAACAAATAAAT |
| gp41L367/374A-R | CGTCTGGCAGTTTCGTTTTGGAACGCGGCTTCGGTCAAGCTCTCCTGGC |
| Prokaryotic expression | |
| gp41exp-F (BamHI) | GCGGGATCCATGACAGATGAACGTGGCAAT |
| gp41exp-R (XhoI) | GCGCTCGAGTTATGCAGTGCGCCCTTTCG |
| gapdhexp-F (EcoRI) | CCGGAATTCATGTCCAAAATCGGTATCAAC |
| gapdhexp-R (XhoI) | CCGCTCGAGTTAATCCTTGGTCTGGATGTACTTG |
| pif5exp-F (BamHI) | CGCGGATCCATGAGTTTTTTTTCAAATCTTCGCGCAGTC |
| pif5exp-R (XhoI) | CCGCTCGAGAGGCATTAACTTGCCGCTGACGCTGTC |
| laminBexp-F (BamHI) | GCGGGATCCATGTCGTCAAAAACGAAAAGGACT |
| laminBexp-R (NotI) | GCGGCGGCCGCTTACATGATACGACAGTTCTCTTC |
| Time course analysis of transcription | |
| Oligo(dT) | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT |
| gp41in-F | CAAGAGCAAAGAACCGACG |
| gp41in-R | TTATGCAGTGCGCCCTTTCGT |
| ie1in-F | ATGACGCAAATTAATTTTAACGCGTC |
| ie1in-R | CATATTTGTTTGGGGGATTGTCGG |
| vp39in-F | ATGGCGCTAGTGCCCGTGGGTATGG |
| vp39in-R | TTAGACGGCTATTCCTCCACCTGCTTC |
| Subcellular localization analysis | |
| gp41-F2 (EcoRI) | GCGGAATTCATGACAGATGAACGTGGCAA |
| gp41-R2 (XbaI) | GCGTCTAGATTATGCAGTGCGCCCTTTC |
| qPCR analysis | |
| vp80-F | GACGATGTCGTTAATCGTGC |
| vp80-R | ATCAGCATCGCTATTCAGATAA |
Underlining indicates restriction sites.
Construction of gp41 repaired and site-directed mutant recombinant bacmids.
To construct a gp41 repaired recombinant bacmid, the Bac-to-Bac baculovirus expression system was used. First, the AcMNPV ph gene was amplified by use of ph-F/R primers (Table 1) and cloned into the pFastBacDual transfer vector under the control of the ph promoter to produce pFBD-ph. The gp41 gene was then PCR amplified by using AcMNPV bacmid DNA as the template and further inserted into the NheI and XhoI sites of pFBD-ph and driven by the AcMNPV p10 promoter. Positive clones were verified by PCR and DNA sequencing. The resulting donor plasmid was named pFBD-gp41-ph. Donor plasmids pFBD-ph and pFBD-gp41-ph were used individually to transform DH10β competent cells containing the AcΔgp41 bacmid and a helper plasmid expressing transposase. Recombinant bacmids AcΔgp41-ph and AcΔgp41-gp41R-ph were selected by gentamicin and kanamycin resistance and blue-white screening and further identified by PCR with M13 primers, as previously described (55).
Site-directed mutagenesis of the cysteine or leucine residues of GP41 was performed by using overlap extension PCR (56). For example, to generate the GP41C91G mutant, two overlapped PCR fragments were first amplified by use of primers gp41-F/gp41C91G-R and gp41C91G-F/gp41-R. The two overlapped PCR fragments were then annealed, and the secondary round of PCR was carried out with the gp41-F/gp41-R primer pair. Primers for site-directed mutagenesis are listed in Table 1. The individually mutated genes were inserted into pFBD-ph to generate donor plasmids (pFBD-gp41x-ph) and further transposed into the AcΔgp41 bacmid to generate mutant recombinant viruses. The five recombinant bacmids containing cysteine mutations were named AcΔgp41-gp41C91G-ph, AcΔgp41-gp41C104G-ph, AcΔgp41-gp41C125G-ph, AcΔgp41-gp41C222G-ph, and AcΔgp41-gp41C360G-ph. The six recombinant bacmids containing mutations in the leucine zipper-like motifs were named AcΔgp41-gp41L201A-ph, AcΔgp41-gp41L208A-ph, AcΔgp41-gp41L367A-ph, AcΔgp41-gp41L374A-ph, AcΔgp41-gp41L201/208A-ph, and AcΔgp41-gp41L367/374A-ph. The PCR products for all site-directed mutations were sequenced to check for unintended mutations.
Transfection and infection assay.
To generate recombinant viruses, the DNAs of the gp41 knockout bacmid, the gp41 repaired bacmid, and a series of gp41 mutated recombinant bacmids were extracted and used to transfect Sf9 cells. Five micrograms of each bacmid DNA was used for transfection experiments using 10 μl of Cellfectin reagent, performed according to the instructions of the Cellfectin II manual (Invitrogen). Supernatants were harvested at 4 days p.t. The clarified transfection supernatant was used to infect a fresh batch of healthy Sf9 cells. The transfection or infection was monitored by fluorescence microscopy.
Time course analyses of transcription.
Sf9 cells were infected with the control virus AcBac-egfp-ph at an MOI of 5 TCID50 units/cell. Cells were harvested at 0, 6, 12, 18, 24, 36, 48, and 72 h p.i., and total RNA was extracted by use of TRIzol reagent (Invitrogen) according to the manufacturer's instructions. After treatment with RNase-free DNase I (TaKaRa) to eliminate potential contamination with genomic DNA, an oligo(dT) primer (Table 1) was used for reverse transcription to yield cDNA by use of a PrimeScript RT reagent kit according to the kit instructions (TaKaRa). The target fragments of gp41, ie1, and vp39 were amplified by PCR with the specific primer pairs gp41in-F/gp41in-R, ie1in-F/ie1in-R, and vp39in-F/vp39in-R (Table 1). For the negative-control (NC) group, RT-PCR was performed without reverse transcriptase in the reaction mixture.
Preparation of pAbs.
The whole ORF of gp41 was amplified by PCR with the gp41exp-F and gp41exp-R primers. AcMNPV bacmid DNA was used as the template. PCR products were inserted into the BamHI and XhoI sites of the pET32a+ vector for protein expression. The recombinant plasmid pET32a-gp41 was then used to electrotransform E. coli strain BL21. The expressed GP41 proteins were purified as described previously for other proteins (57) and then used for rabbit immunization to elicit pAbs against GP41 (anti-GP41). The entire ORF of the Helicoverpa armigera gapdh gene (GenBank accession number JF417983.1), which shares ∼97% aa identity with the Sf9 gapdh gene product (GenBank accession number KT218670.1), was PCR amplified from HzAM1 cells by use of primers gapdhexp-F and gapdhexp-R and inserted into the pET28a+ vector. The purified GAPDH proteins were used to produce anti-GAPDH pAb. The AcMNPV PIF5 pAb was produced similarly. The fragment encoding aa 1 to 324 of pif5 was amplified by use of the pif5exp-F and pif5exp-R primers and inserted into the pET28a+ vector. The purified PIF5 proteins were used for rabbit immunization. To generate anti-lamin B pAb, the ORF of the Sf9 lamin B gene (GenBank accession number KT318393) was PCR amplified by use of primers laminBexp-F/R (Table 1) and cloned into the pET28a+ vector. The anti-lamin B pAb was harvested from an immunized rabbit. All primers are listed in Table 1.
Cytoplasmic and nuclear fractionation assay.
Sf9 cells (2 × 107) were infected with the control virus AcBac-egfp-ph at an MOI of 5 TCID50 units/cell. At 36 h p.i., cells were harvested and separated into cytoplasmic or nuclear fractions according to the method of a previous report (58), with slight adjustments. Briefly, cells were lysed with 1% NP-40 for 30 min on ice and spun at 1,000 × g for 5 min to separate the cytoplasmic and nuclear fractions. The supernatant was collected as the cytoplasmic fraction, and the pellet was resuspended with NP-40 lysis buffer as the nuclear fraction. Western blot analyses were performed with anti-GP41 (1:5,000), anti-GAPDH (1:4,000), and anti-lamin B (1:4,000) pAbs.
Western blot analysis.
For the time course analysis of protein expression, Sf9 cells were seeded into 35-mm dishes, infected with the control virus AcBac-egfp-ph at an MOI of 5 TCID50 units/cell, and collected at 0, 6, 12, 18, 24, 36, 48, and 72 h p.i. To analyze the localization of GP41 in virions, BVs collected from AcBac-egfp-ph-infected cells were purified as previously described (26). OBs derived from AcBac-egfp-ph-infected Spodoptera exigua larvae were purified as previously described (59). ODVs were released from OBs by alkaline treatment and further purified by centrifugation (22). To analyze the oligomerization of gp41 mutant recombinant viruses, Sf9 cells were infected with the repaired virus or the mutated recombinant viruses at an MOI of 5 TCID50 units/cell (for double leucine mutants, transfected cells were used), and cells were collected at 36 h p.i. or 48 h p.t. for further analyses.
Transfected/infected cells or purified BVs or ODVs were suspended with phosphate-buffered saline. Before separation by 12% SDS-PAGE, samples were treated with SDS-PAGE loading buffer (50 mM Tris-HCl, 2% SDS, 0.1% bromophenol blue, 10% glycerol) under either reducing conditions (with 5% 2-mercaptoethanol) or nonreducing conditions (without 2-mercaptoethanol) and heated for 10 min at 95°C. The proteins were then transferred to a 0.22-μm nitrocellulose membrane. Western blot analyses were carried out with anti-GP41 (1:5,000 dilution), anti-GAPDH (1:4,000), anti-PIF5 (1:4,000), anti-VP39 (1:5,000) (26), or anti-GP64 (1:5,000) (32) pAb as the primary antibody and with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (1:5,000 dilution; Pierce) as the secondary antibody. A PageRuler prestained protein ladder (Thermo Scientific) was used to indicate protein sizes.
Quantification of BVs and ODVs by qPCR.
The total genomic DNAs of BVs and ODVs were extracted as described previously (32). After extraction, 1 μl BV/ODV genomic DNA was quantified by use of qPCR. qPCR was performed with SYBR premix Ex Taq II (TaKaRa) and the vp80-F/R primers (Table 1). AcBac-egfp-ph bacmid DNA was serially diluted for creation of a standard curve. qPCRs were carried out for 40 cycles of 95°C for denaturation and 59°C and 72°C for annealing/extension.
Electron microscopy.
For transmission EM (TEM) analysis, cells (2 × 106) were seeded into 35-mm dishes and infected with the repaired virus or mutated recombinant viruses. At different time points, cells were washed twice with phosphate-buffered saline, fixed with 2.5% (wt/vol) glutaraldehyde, and processed for TEM as described previously (60). For IEM, Sf9 cells were infected with the control virus AcBac-egfp-ph at an MOI of 5 TCID50 units/cell and harvested at 24, 48, and 72 h p.i. Cells were fixed with 1% paraformaldehyde-0.5% glutaraldehyde for 10 min at 4°C and refixed with 2% paraformaldehyde-2.5% glutaraldehyde for 1 h at 4°C. The cell samples were subsequently dehydrated and embedded according to previously published methods (61). Ultrathin sections were immunostained with anti-GP41 pAb (1:50) or preimmune rabbit serum (1:50) as the primary antibody. Goat anti-rabbit IgG coated with gold particles (10 nm) was used as the secondary antibody (1:50; Sigma). The samples were observed by use of a TEM (FEI Tecnai G2) operating at a 75-kV acceleration voltage.
Immunofluorescence microscopy.
Sf9 cells (1 × 106) were seeded into 35-mm glass-bottomed culture dishes and allowed to adhere for 2 h. Cells were infected with the control virus AcBac-egfp-ph or a recombinant virus at an MOI of 5 TCID50 units/cell or transfected with 5 μg of recombinant bacmid DNA. At the indicated time points, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 5% bovine serum albumin. After that, cells were treated with anti-GP41 pAb (1:500 dilution) in 1% bovine serum albumin for 2 h and then incubated with Alexa 555-labeled goat anti-rabbit (1:500; Abcam) for 1 h at room temperature. Nuclei were stained with Hoechst 33258 (Beyotime). The subcellular localization of proteins was detected by fluorescence microscopy.
To visualize the subcellular localization of GP41 in the absence of virus infection, the gp41 gene was amplified from the AcMNPV genome by use of the gp41-F2/R2 primers (Table 1) and inserted into the EcoRI and XbaI sites of the pIZ/V5-His vector (Invitrogen) to generate a transient-expression vector, pIZ/V5-gp41. One microgram of plasmid was transfected into Sf9 cells. At 24 h p.t., cells were infected with AcBac-egfp-ph at an MOI of 5 TCID50 units/cell or mock infected. Cells harvested at 24 h p.t. or 36 h p.i. were subjected to immunofluorescence microscopy as described above.
One-step growth curve analysis.
To perform one-step growth curve analysis, Sf9 cells (1 × 106) were infected with the control virus, the repaired virus, and the mutated recombinant viruses at an MOI of 5 TCID50 units/cell. After incubation for 1 h at 27°C, the viruses were removed and cells were washed three times with 1 ml of Grace's medium before addition of 2 ml of fresh Grace's medium (supplemented with 10% fetal bovine serum). Supernatants were collected at 0, 24, 48, 72, and 96 h p.i. and clarified by centrifugation. Infectious BV yields were determined by endpoint dilution assays. Statistical analysis was performed with SPSS software. Student's t test was used to analyze the significance of differences between gp41 mutant and repaired recombinant viruses.
ACKNOWLEDGMENTS
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB11030400), the National Natural Science Foundation of China (grants 31621061, 31400140 and 31370191), the Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (grant QYZDJ-SSW-SMC021), the Virology Key Frontier Science Program of the State Key Laboratory of Virology (grant klv-2016-03), and a grant to J.M.V. from the State Key Laboratory of Virology and the Wuhan Institute of Virology.
We acknowledge Ding Gao, Anna Du, Pei Zhang, Bichao Xu, and Juan Min, from the core facility and technical support facility of the Wuhan Institute of Virology, for technical assistance.
REFERENCES
- 1.Kalejta RF. 2008. Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72:249–265. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998. doi: 10.1007/s13238-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diefenbach RJ. 2015. Conserved tegument protein complexes: essential components in the assembly of herpesviruses. Virus Res 210:308–317. doi: 10.1016/j.virusres.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Owen DJ, Crump CM, Graham SC. 2015. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiorentini S, Marini E, Caracciolo S, Caruso A. 2006. Functions of the HIV-1 matrix protein p17. New Microbiol 29:1–10. [PubMed] [Google Scholar]
- 6.Bukrinskaya A. 2007. HIV-1 matrix protein: a mysterious regulator of the viral life cycle. Virus Res 124:1–11. doi: 10.1016/j.virusres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Nayak DP, Hui EK, Barman S. 2004. Assembly and budding of influenza virus. Virus Res 106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez-Puertas P, Albo C, Perez-Pastrana E, Vivo A, Portela A. 2000. Influenza virus matrix protein is the major driving force in virus budding. J Virol 74:11538–11547. doi: 10.1128/JVI.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watkinson RE, Lee B. 2016. Nipah virus matrix protein: expert hacker of cellular machines. FEBS Lett 590:2494–2511. doi: 10.1002/1873-3468.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pentecost M, Vashisht AA, Lester T, Voros T, Beaty SM, Park A, Wang YE, Yun TE, Freiberg AN, Wohlschlegel JA, Lee B. 2015. Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins. PLoS Pathog 11:e1004739. doi: 10.1371/journal.ppat.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bringolf F, Herren M, Wyss M, Vidondo B, Langedijk JP, Zurbriggen A, Plattet P. 2017. Dimerization efficiency of canine distemper virus matrix protein regulates membrane-budding activity. J Virol 91:e00521-17. doi: 10.1128/JVI.00521-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmins J, Ruigrok RW, Weissenhorn W. 2004. Structural studies on the Ebola virus matrix protein VP40 indicate that matrix proteins of enveloped RNA viruses are analogues but not homologues. FEMS Microbiol Lett 233:179–186. doi: 10.1111/j.1574-6968.2004.tb09480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghildyal R, Ho A, Jans DA. 2006. Central role of the respiratory syncytial virus matrix protein in infection. FEMS Microbiol Rev 30:692–705. doi: 10.1111/j.1574-6976.2006.00025.x. [DOI] [PubMed] [Google Scholar]
- 14.Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitford M, Faulkner P. 1992. A structural polypeptide of the baculovirus Autographa californica nuclear polyhedrosis virus contains O-linked N-acetylglucosamine. J Virol 66:3324–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makkonen KE, Airenne K, Yla-Herttulala S. 2015. Baculovirus-mediated gene delivery and RNAi applications. Viruses 7:2099–2125. doi: 10.3390/v7042099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kost TA, Condreay JP, Jarvis DL. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol 23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kost TA, Kemp CW. 2016. Fundamentals of baculovirus expression and applications. Adv Exp Med Biol 896:187–197. doi: 10.1007/978-3-319-27216-0_12. [DOI] [PubMed] [Google Scholar]
- 19.Rohrmann GF. 1992. Baculovirus structural proteins. J Gen Virol 73:749–761. doi: 10.1099/0022-1317-73-4-749. [DOI] [PubMed] [Google Scholar]
- 20.Volkman LE, Summers MD. 1977. Autographa californica nuclear polyhedrosis virus: comparative infectivity of the occluded, alkali-liberated, and nonoccluded forms. J Invertebr Pathol 30:102–103. doi: 10.1016/0022-2011(77)90045-3. [DOI] [PubMed] [Google Scholar]
- 21.Summers MD. 1971. Electron microscopic observations on granulosis virus entry, uncoating and replication processes during infection of the midgut cells of Trichoplusia ni. J Ultrastruct Res 35:606–625. doi: 10.1016/S0022-5320(71)80014-X. [DOI] [PubMed] [Google Scholar]
- 22.Braunagel SC, Summers MD. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 23.Slack J, Arif BM. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv Virus Res 69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou D, Zhang L, Deng F, Fang W, Wang R, Liu X, Guo L, Rayner S, Chen X, Wang H, Hu Z. 2013. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J Virol 87:829–839. doi: 10.1128/JVI.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan L, Li Z, Gong Y, Yu M, Yang K, Pang Y. 2005. Characterization of gp41 gene of Spodoptera litura multicapsid nucleopolyhedrovirus. Virus Res 110:73–79. doi: 10.1016/j.virusres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Wang R, Deng F, Hou D, Zhao Y, Guo L, Wang H, Hu Z. 2010. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J Virol 84:7233–7242. doi: 10.1128/JVI.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitford M, Faulkner P. 1992. Nucleotide sequence and transcriptional analysis of a gene encoding gp41, a structural glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol 66:4763–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu JC, Maruniak JE. 1995. Nucleotide sequence and transcriptional analysis of the gp41 gene of Spodoptera frugiperda nuclear polyhedrosis virus. J Gen Virol 76:1443–1450. doi: 10.1099/0022-1317-76-6-1443. [DOI] [PubMed] [Google Scholar]
- 29.Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J Virol 87:6391–6405. doi: 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olszewski J, Miller LK. 1997. A role for baculovirus GP41 in budded virus production. Virology 233:292–301. doi: 10.1006/viro.1997.8612. [DOI] [PubMed] [Google Scholar]
- 31.Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. 2012. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res 165:197–206. doi: 10.1016/j.virusres.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Wang M, Tan Y, Yin F, Deng F, Vlak JM, Hu Z, Wang H. 2008. The F-like protein Ac23 enhances the infectivity of the budded virus of gp64-null Autographa californica multinucleocapsid nucleopolyhedrovirus pseudotyped with baculovirus envelope fusion protein F. J Virol 82:9800–9804. doi: 10.1128/JVI.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohrmann GF. 2013. Baculovirus molecular biology, 3rd ed National Center for Biotechnology Information, Bethesda, MD. [PubMed] [Google Scholar]
- 34.Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88. doi: 10.1016/S0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 35.Akarsu H, Burmeister WP, Petosa C, Petit I, Muller CW, Ruigrok RW, Baudin F. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J 22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welsch S, Muller B, Krausslich HG. 2007. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett 581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Yue Q, Gao J, Wang Z, Chen YR, Blissard GW, Liu TX, Li Z. 2017. Roles of cellular NSF protein in entry and nuclear egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J Virol 91:e01111-17. doi: 10.1128/JVI.01111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue Q, Yu Q, Yang Q, Xu Y, Guo Y, Blissard GW, Li Z. 2018. Distinct roles of cellular ESCRT-I and ESCRT-III proteins in efficient entry and egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J Virol 92:e01636-17. doi: 10.1128/JVI.01636-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 41.Meissner CS, Suffner S, Schauflinger M, von Einem J, Bogner E. 2012. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J Virol 86:3370–3382. doi: 10.1128/JVI.06556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, Aman MJ. 2003. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc Natl Acad Sci U S A 100:15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, Weissenhorn W. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J 19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forster A, Maertens GN, Farrell PJ, Bajorek M. 2015. Dimerization of matrix protein is required for budding of respiratory syncytial virus. J Virol 89:4624–4635. doi: 10.1128/JVI.03500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaudier M, Gaudin Y, Knossow M. 2002. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J 21:2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. 2003. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312:359–368. doi: 10.1016/S0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- 47.Alfadhli A, Dhenub TC, Still A, Barklis E. 2005. Analysis of human immunodeficiency virus type 1 Gag dimerization-induced assembly. J Virol 79:14498–14506. doi: 10.1128/JVI.79.23.14498-14506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Busch SJ, Sassone-Corsi P. 1990. Dimers, leucine zippers and DNA-binding domains. Trends Genet 6:36–40. doi: 10.1016/0168-9525(90)90071-D. [DOI] [PubMed] [Google Scholar]
- 49.Bottcher S, Granzow H, Maresch C, Mohl B, Klupp BG, Mettenleiter TC. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J Virol 81:13403–13411. doi: 10.1128/JVI.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duverger E, Pellerin-Mendes C, Mayer R, Roche AC, Monsigny M. 1995. Nuclear import of glycoconjugates is distinct from the classical NLS pathway. J Cell Sci 108:1325–1332. [DOI] [PubMed] [Google Scholar]
- 51.Duverger E, Roche AC, Monsigny M. 1996. N-acetylglucosamine-dependent nuclear import of neoglycoproteins. Glycobiology 6:381–386. doi: 10.1093/glycob/6.4.381. [DOI] [PubMed] [Google Scholar]
- 52.Shang Y, Wang M, Xiao G, Wang X, Hou D, Pan K, Liu S, Li J, Wang J, Arif BM, Vlak JM, Chen X, Wang H, Deng F, Hu Z. 2017. Construction and rescue of a functional synthetic baculovirus. ACS Synth Biol 6:1393–1402. doi: 10.1021/acssynbio.7b00028. [DOI] [PubMed] [Google Scholar]
- 53.Hou S, Chen X, Wang H, Tao M, Hu Z. 2002. Efficient method to generate homologous recombinant baculovirus genomes in E. coli. Biotechniques 32:783, 784, 786, 788. [DOI] [PubMed] [Google Scholar]
- 54.Huang H, Wang M, Deng F, Hou D, Arif BM, Wang H, Hu Z. 2014. The ha72 core gene of baculovirus is essential for budded virus production and occlusion-derived virus embedding, and amino acid K22 plays an important role in its function. J Virol 88:705–709. doi: 10.1128/JVI.02281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin F, Du R, Kuang W, Yang G, Wang H, Deng F, Hu Z, Wang M. 2016. Characterization of the viral fibroblast growth factor homolog of Helicoverpa armigera single nucleopolyhedrovirus. Virol Sin 31:240–248. doi: 10.1007/s12250-016-3710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 57.Zou Z, Liu J, Wang Z, Deng F, Wang H, Hu Z, Wang M, Zhang T. 2016. Characterization of two monoclonal antibodies, 38F10 and 44D11, against the major envelope fusion protein of Helicoverpa armigera nucleopolyhedrovirus. Virol Sin 31:490–499. doi: 10.1007/s12250-016-3831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nie Y, Fang M, Erlandson MA, Theilmann DA. 2012. Analysis of the Autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication. J Virol 86:3985–3994. doi: 10.1128/JVI.06849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng F, Wang R, Fang M, Jiang Y, Xu X, Wang H, Chen X, Arif BM, Guo L, Wang H, Hu Z. 2007. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. J Virol 81:9377–9385. doi: 10.1128/JVI.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Lent JW, Groenen JT, Klinge-Roode EC, Rohrmann GF, Zuidema D, Vlak JM. 1990. Localization of the 34 kDa polyhedron envelope protein in Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. Arch Virol 111:103–114. doi: 10.1007/BF01310508. [DOI] [PubMed] [Google Scholar]
- 61.Guan Z, Zhong L, Li C, Wu W, Yuan M, Yang K. 2016. The Autographa californica multiple nucleopolyhedrovirus ac54 gene is crucial for localization of the major capsid protein VP39 at the site of nucleocapsid assembly. J Virol 90:4115–4126. doi: 10.1128/JVI.02885-15. [DOI] [PMC free article] [PubMed] [Google Scholar]