ABSTRACT
Lymphocytic choriomeningitis mammarenavirus (LCMV) is an enveloped, negative-strand RNA virus that causes serious disease in humans but establishes an asymptomatic, lifelong infection in reservoir rodents. Different models have been proposed to describe how arenaviruses regulate the replication and transcription of their bisegmented, single-stranded RNA genomes, particularly during persistent infection. However, these models were based largely on viral RNA profiling data derived from entire populations of cells. To better understand LCMV replication and transcription at the single-cell level, we established a high-throughput, single-molecule fluorescence in situ hybridization (smFISH) image acquisition and analysis pipeline and examined viral RNA species at discrete time points from virus entry through the late stages of persistent infection in vitro. We observed the transcription of viral nucleoprotein and polymerase mRNAs from the incoming S and L segment genomic RNAs, respectively, within 1 h of infection, whereas the transcription of glycoprotein mRNA from the S segment antigenome required ∼4 to 6 h. This confirms the temporal separation of viral gene expression expected due to the ambisense coding strategy of arenaviruses and also suggests that antigenomic RNA contained in virions is not transcriptionally active upon entry. Viral replication and transcription peaked at 36 h postinfection, followed by a progressive loss of viral RNAs over the next several days. During persistence, the majority of cells showed repeating cyclical waves of viral transcription and replication followed by the clearance of viral RNA. Thus, our data support a model of LCMV persistence whereby infected cells can spontaneously clear infection and become reinfected by viral reservoir cells that remain in the population.
IMPORTANCE Arenaviruses are human pathogens that can establish asymptomatic, lifelong infections in their rodent reservoirs. Several models have been proposed to explain how arenavirus spread is restricted within host rodents, including the periodic accumulation and loss of replication-competent, but transcriptionally incompetent, viral genomes. A limitation of previous studies was the inability to enumerate viral RNA species at the single-cell level. We developed a high-throughput, smFISH assay and used it to quantitate lymphocytic choriomeningitis mammarenavirus (LCMV) replicative and transcriptional RNA species in individual cells at distinct time points following infection. Our findings support a model whereby productively infected cells can clear infection, including viral RNAs and antigen, and later be reinfected. This information improves our understanding of the timing and possible regulation of LCMV genome replication and transcription during infection. Importantly, the smFISH assay and data analysis pipeline developed here is easily adaptable to other RNA viruses.
KEYWORDS: LCMV, arenavirus, cyclical, gene probes, genome replication and transcription, kinetics, persistence, smFISH
INTRODUCTION
Several members of the arenavirus family are significant threats to human health. Lassa virus (LASV) and Junín virus cause hemorrhagic fever syndromes, while lymphocytic choriomeningitis virus (LCMV), the prototypic member of the family, is a well-known cause of severe birth defects and is highly lethal in immunocompromised individuals (1, 2). A critical imperative to better understand the key steps of the arenavirus life cycle is made evident by the fact that there are no FDA-approved vaccines to prevent arenavirus transmission and only a very limited repertoire of antivirals (3, 4). New strategies to prevent and treat arenavirus infections will likely hinge upon an improved understanding of key phases of the life cycle of these important human pathogens.
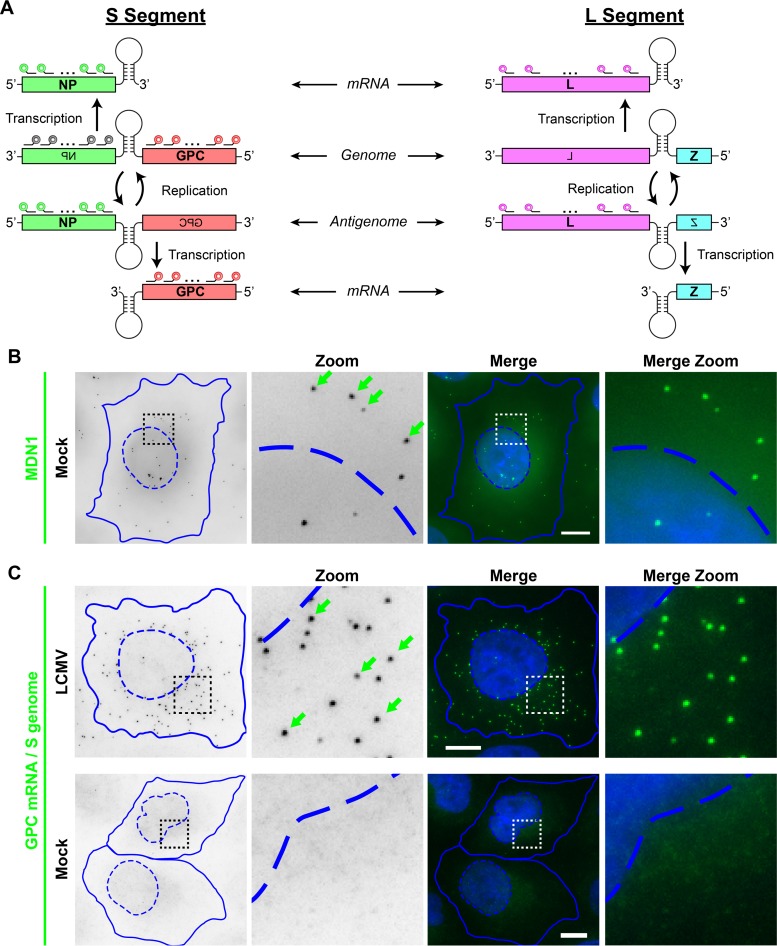
Arenaviruses are enveloped viruses that have a single-stranded, bisegmented, negative-sense RNA genome. The ∼3.5-kb small (S) and ∼7.2-kb large (L) genomic RNA segments each harbor two viral open reading frames in an ambisense orientation (Fig. 1A) (1). The nucleoprotein (NP) and polymerase (L) genes are carried in a typical negative-sense orientation on genomic RNA, while the glycoprotein (GPC) and matrix protein (Z) genes are carried in a pseudo-positive-sense orientation. The canonical sequence of genetic events following the release of arenavirus genomic RNA into the cytoplasm of a newly infected cell is (i) primary transcription of the NP and L mRNAs from the viral S and L genomic segments, respectively, followed by (ii) full-length replication of the S and L segment antigenomic RNAs and subsequent transcription of the GPC and Z mRNAs from the S and L antigenomic RNAs, respectively, and (iii) the replication of additional full-length genomic RNAs from the antigenomic RNA templates (Fig. 1A) (1, 5).
FIG 1.
LCMV RNA species can be specifically visualized by using multiple, singly labeled oligonucleotide smFISH probes. (A) Overview of the scheme used by arenaviruses to transcribe and replicate their single-stranded, ambisense, bisegmented genome. smFISH probes that recognize the S segment genomic RNA are shown in gray, probes that recognize the S segment genome and GPC mRNA are shown in red, probes that recognize the S segment antigenome and NP mRNA are shown in green, and probes that recognize the L segment antigenome and L mRNA are shown in pink. smFISH probe sets consist of pools of 48 individual 20-mer oligonucleotides, each labeled with a single fluorophore at their 3′ termini. (B) Uninfected cells were stained with a control smFISH probe set specific for MDN1 cellular mRNA labeled with Cy3. (C) Cells either were infected with LCMV at an MOI of 0.01 or, as a control, remained uninfected (mock). Cells were fixed at 24 hpi and stained with a Cy5-labeled smFISH probe set specific for S segment genomic RNA and GPC mRNA. Boxed regions of the cell are magnified and shown in columns labeled “Zoom.” Green arrows indicate example smFISH-stained spots most likely representing single labeled RNAs. Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries are shown in blue. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells to permit comparisons. Bars, 10 μm.
While rodent-borne arenaviruses cause severe diseases in humans, they are thought to be asymptomatic in their sylvatic hosts, where they can establish a persistent, lifelong infection (1). LCMV is carried by the common house mouse and can be transmitted vertically from mother to pup (6–8). The pups are born infected but never mount an effective immune response to clear the virus, as viral proteins are seen as self-antigens by the pups' developing immune system (6–8). Paradoxically, while LCMV can infect most cells in the host rodent, it tightly regulates its spread and therefore does not overrun its host. Several hypotheses have been proposed for how LCMV restricts its spread, including through (i) the production of defective interfering (DI) particles (9–11), which can enter susceptible host cells and make them refractory to productive infection (12, 13), and (ii) the accumulation of transcriptionally defective genomic and antigenomic RNAs, which limit viral protein expression and the production of infectious virus (5, 14, 15). It has also been proposed that LCMV can establish a cyclical, transient pattern of infection such that susceptible cells are productively infected for a short time before clearing the virus and once again becoming susceptible to reinfection by neighboring cells that remain productively infected (16–19).
A current gap in our knowledge of how arenaviruses restrict their dissemination is that we lack a detailed understanding of how the events of viral genome replication and transcription are regulated during the acute and persistent phases of infection. Previous studies examining the genetic events of arenavirus replication and transcription, including those described above regarding the accumulation of transcriptionally defective RNAs (5, 14, 15), relied on techniques such as Northern blotting or quantitative reverse transcription-PCR (RT-PCR). Both are powerful techniques used to examine RNA. Quantitative RT-PCR is exquisitely sensitive (20), and Northern blotting is able to specifically distinguish between each of the viral RNA species (15, 21–29). However, both techniques measure RNA at the population level and thus provide population-averaged data. Variability in RNA expression between individual cells in a heterogeneous population cannot be evaluated by using these approaches. Single-molecule RNA fluorescence in situ hybridization (smFISH) can bridge this technical gap to allow the detection of RNAs with single-copy sensitivity in individual cells by fluorescence microscopy (30). In the present study, we designed specific smFISH probe sets to fluorescently label different LCMV RNA species (Fig. 1A) and to quantitatively characterize their expression in single cells at discrete time points throughout the acute and persistent phases of arenavirus infection in an in vitro model. Our studies confirm the temporal separation of LCMV negative-sense and pseudo-positive-sense gene expression and show a pattern of cyclical loss and reappearance of viral RNA in most cells during persistence in a cell culture model of infection. Our studies provide insight into the functional genetic composition of infectious virions and the kinetics of transcription and replication in the hours immediately following initial infection and support a model of cyclical viral replication and transcription during persistence. Furthermore, the image acquisition and analysis pipeline developed here is easily adaptable to other viruses.
RESULTS
Visualization of LCMV RNA species in infected cells.
To visualize LCMV RNAs in cells by fluorescence microscopy, we designed smFISH probe sets complementary to different viral RNA species (see overview in Fig. 1A). An important feature of small-molecule RNA FISH is the ability to detect single RNA molecules using multiple, singly labeled oligonucleotide probes (30). The binding of the probe set to a specific target RNA causes single RNAs to appear as bright spots. To validate our ability to specifically label arenavirus RNAs, we used a cellular mRNA smFISH probe set specific for the housekeeping gene MDN1 as a control (Fig. 1B) for comparison with a smFISH probe set designed to target both viral S genome RNA and GPC mRNA (Fig. 1C). MDN1 probes detect cytoplasmic mRNAs as well as sites of active transcription in the nucleus (Fig. 1B). Next, we confirmed that the viral RNA smFISH probe set is highly specific, as a fluorescent signal was absent in uninfected cells, but bright spots were detected in LCMV-infected cells fixed at 24 h postinfection (hpi) (Fig. 1C). Moreover, similar to the smFISH staining obtained with our control, MDN1, individual smFISH spots were homogeneous in size, shape, and fluorescence intensity (Fig. 1B and C), consistent with the detection of single RNAs, as shown previously (30, 31). Furthermore, in contrast to the nucleus-transcribed MDN1 mRNAs, viral RNAs were largely excluded from the nucleus, consistent with the cytoplasmic viral life cycle (Fig. 1B and C).
smFISH probes complementary to viral mRNA species provide high signal-to-noise staining.
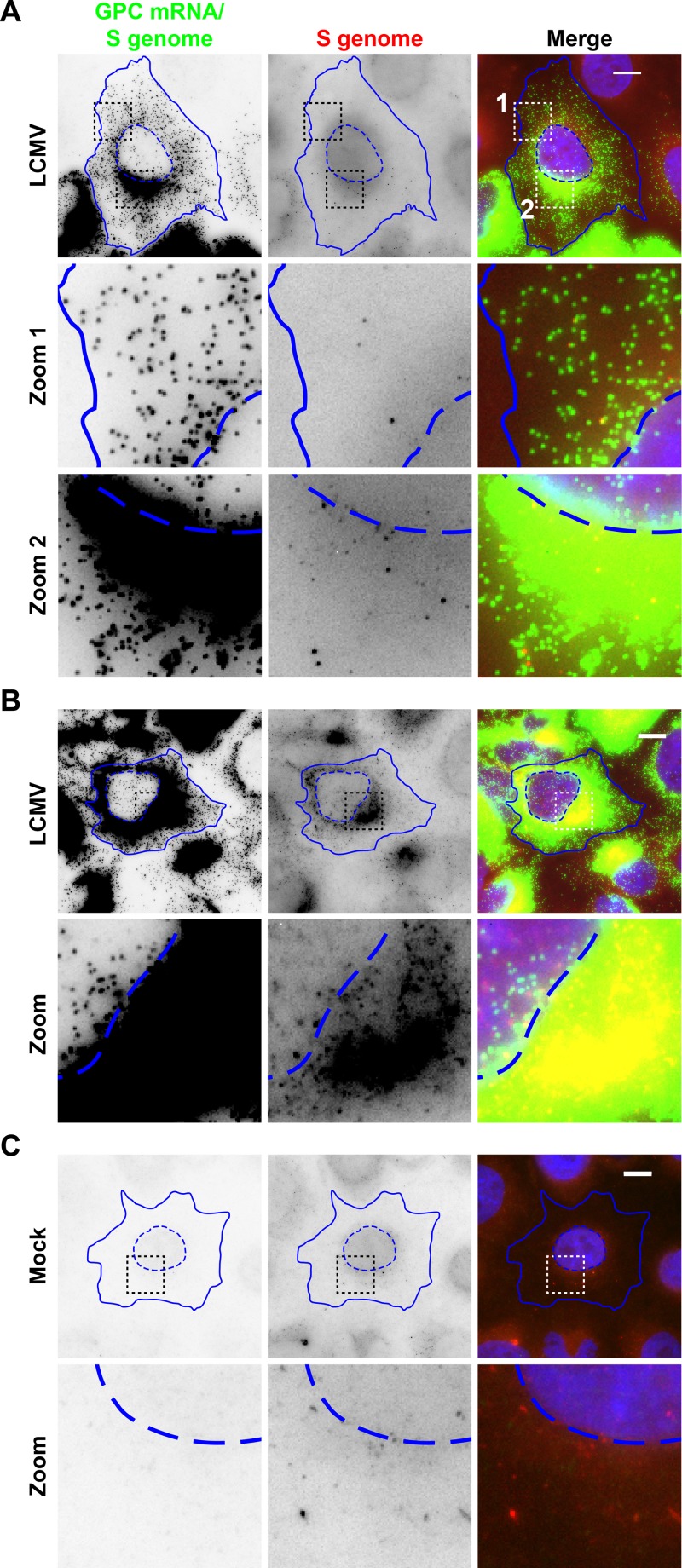
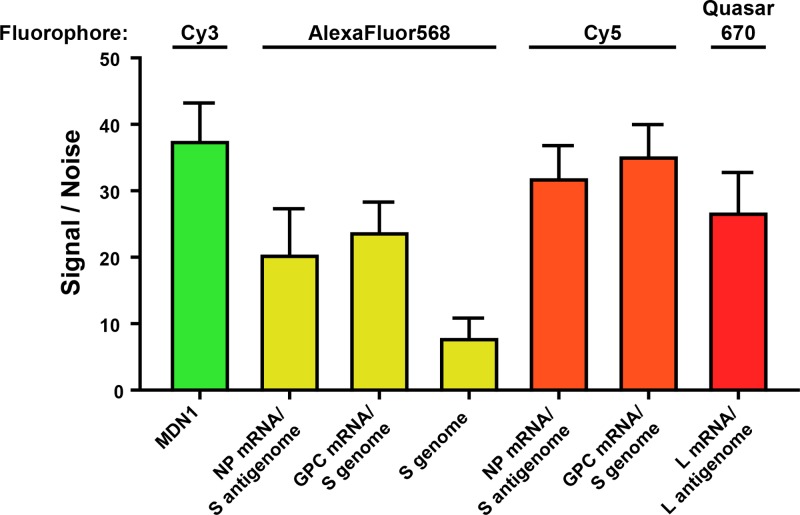
We designed multiple smFISH probe sets to have specificity for different RNA species produced during the course of the LCMV life cycle (Fig. 1A). Specifically, these probe sets target (i) the S genome only, (ii) GPC mRNA and the S genome, (iii) NP mRNA and the S antigenome, or (iv) L mRNA and the L antigenome. When infected cells were stained with probe sets complementary to the S genome and GPC mRNA (referred to as “GPC mRNA/S genome” here), we noted high-quality staining with the GPC mRNA/S genome probes, as evidenced by the homogeneity in spot size, shape, and intensity (Fig. 2A) and the high signal-to-noise ratio (Fig. 3). The NP mRNA/S antigenome and L mRNA/L antigenome probe sets yielded similar high-quality staining, as evidenced by the high signal-to-noise ratios (Fig. 3). However, we noted lower-quality staining with the “S-genome-only” probes, as evidenced by the dim staining (Fig. 2) and low signal-to-noise ratio (Fig. 3). Moreover, the S-genome-only probes yielded greater nonspecific staining in uninfected cells, potentially leading to the detection of false-positive spurious events (Fig. 2C), perhaps an artifact of the long exposure times and high light intensity needed to detect the binding of this less-sensitive probe set to its target. Similarly low signal-to-noise ratios were observed with probe sets specific for the S antigenome only or the L genome only (data not shown). It is possible that the encapsidation of the genome and antigenome by viral nucleoprotein partially occludes smFISH probe hybridization with these target RNA sequences and thus leads to the lower signal-to-noise ratios observed with these probe sets. Therefore, the use of these probe sets with cells containing small numbers of viral RNAs would be problematic due to the level of background staining observed (Fig. 2C). However, these probe sets are effective when paired with cells containing abundant copies of the viral genome or antigenome (Fig. 2B and data not shown) (32), which easily exceeds the quantity of background spots observed in mock-infected control cells (Fig. 2C). Because the probe sets that targeted an mRNA plus either the genome or antigenome provided the highest-quality staining and the highest sensitivity, we elected to use these probe sets to monitor the kinetics of viral transcription and replication events in infected cells.
FIG 2.
smFISH probe sets recognizing viral mRNA species exhibit high signal-to-noise staining. Mock- or LCMV-infected cells (24 hpi) were simultaneously stained with smFISH probe sets specific for either GPC mRNA and the S genome (Cy5) (green) or the S genome only (Alexa Fluor 568) (red). Representative LCMV-infected cells with moderate (A) or high (B) levels of viral RNA as well as a representative mock-infected cell (C) are displayed. Multiple z-stacks spanning the thickness of the cell were acquired, and maximum-intensity projections are displayed. Boxed regions of the cell are magnified and shown in rows labeled “Zoom.” Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries are shown in blue. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells to permit comparisons. Bars, 10 μm.
FIG 3.
smFISH probe sets recognizing viral mRNA species exhibit high signal-to-noise staining. Shown are signal-to-noise ratios of different smFISH probe sets labeled with the indicated fluorophores. Signal-to-noise ratios were calculated as the average amplitude of detected smFISH spots divided by the standard deviation of the signal in a region of the cell with no detected spots. The signal-to-noise ratio of 20 cells per smFISH probe set labeled with the indicated fluorophore was calculated, and the means and standard deviations are graphed.
smFISH spot detection and quantification in individual LCMV-infected cells.
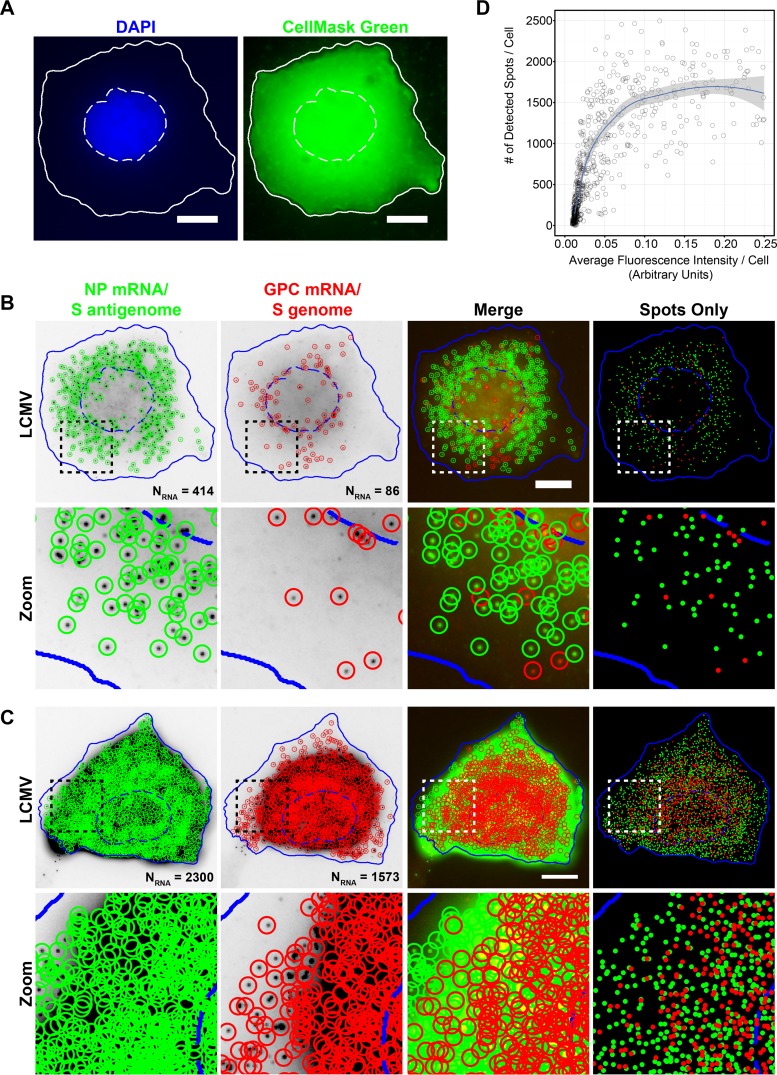
A primary goal of our study was to globally describe the kinetics of transcription and replication of the LCMV genome from the early hours following viral entry through the late stages of persistence. Ideally, we would be able to infect cells at a high multiplicity of infection (MOI) and take snapshots of a population of synchronously infected cells at time points throughout the course of arenavirus infection. However, we were obliged to infect cells at a low MOI due to the characteristic high prevalence of DI particles present in LCMV stocks (33). Because only a small proportion of cells would be productively infected upon virus inoculation, we needed to image a large population of cells at each time point tested to provide an accurate portrait of the heterogeneity present in a population of asynchronously infected cells. Thus, it was important for this study to both image and quantitatively characterize the smFISH staining of viral RNAs in a high-throughput fashion. To accomplish this goal, we automatically segmented the nuclei using 4′,6-diamidino-2-phenylindole (DAPI) and cell outlines using CellMask green fluorescent staining with CellProfiler software (34) (Fig. 4A). Next, smFISH-labeled viral RNAs were detected by using FISH-quant software (35) (Fig. 4B). We were able to image two distinct RNA smFISH probe sets labeled with spectrally distinct fluorophores in individual cells. This allowed us to characterize relative viral RNA expression levels and compare the localizations of different viral RNAs (Fig. 4B and C). We were able to robustly quantify viral RNAs across a range of expression levels using FISH-quant. We observed a direct relationship between the number of detected spots and the total fluorescence signal in the smFISH channel up to approximately 1,000 RNAs/cell, after which the number of detected viral RNAs reached a plateau (Fig. 4D). This represents the point at which smFISH spots were so dense that we were no longer able to accurately distinguish closely spaced RNAs. Examples of a cell displaying moderate levels of viral RNAs where the identification of diffraction limited spots was robust (Fig. 4B) and a cell with very high levels of expression of viral RNAs where overcrowded spots are unable to be effectively spatially resolved (Fig. 4C) are shown for reference. Thus, when viral RNA levels are relatively low (less than a thousand copies per cell), we have high confidence in the accuracy of the quantification provided by FISH-quant. However, when viral RNA levels are at their peak and RNAs are very dense, quantification leads to an underestimate of RNA expression levels and may complicate our ability to assess relative levels of different RNA species when both species are expressed at high levels.
FIG 4.
Automated detection and quantitation of LCMV RNAs labeled with spectrally distinct fluorophores. (A) Cell nuclei and cytoplasms were automatically segmented by using focus-based projections of DAPI (nuclei) or CellMask green (cytoplasm) z-stacks acquired through the thickness of the cell. Note that pixel intensities of the CellMask green projection displayed here have been log transformed to aid visualization. Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries are shown in white. Bar, 10 μm. (B and C) Maximum-intensity projections of LCMV-infected cells were fixed 24 hpi and stained with smFISH probe sets for the NP mRNA/S antigenome (Cy5) (green) and GPC mRNA/S genome (A568) (red). The boxed region of each cell is magnified and shown in the row labeled “Zoom.” Cells were segmented based on DAPI and CellMask green staining (see panel A), and spots were detected and localized in 3D by using FISH-quant. Individually detected RNAs are circled in green (NP mRNA/S antigenome) or red (GPC mRNA/S genome). The “Spots Only” column shows only the positions of the detected spots in relation to the cell boundaries defined by segmentation. Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries are shown in blue. The same intensity levels for a particular probe set were applied to both images of LCMV-infected cells to permit comparisons. Bar, 10 μm. (D) Scatter plot showing the relationship between the fluorescence intensity in the smFISH channel in the maximum-intensity projection of smFISH images and the number of smFISH spots detected by FISH-quant for LCMV-infected cells fixed at 24 hpi and stained with the Cy5-labeled smFISH probes specific for the NP mRNA/S antigenome.
Viral RNA transcription and replication following virus entry.
We next aimed to monitor the early events of viral genomic transcription and replication immediately following virus entry. Cells were infected with LCMV at an MOI of 0.1, fixed at multiple time points, and stained for the NP mRNA/S antigenome, GPC mRNA/S genome, or L mRNA/L antigenome, and several hundred cells were imaged and analyzed at each time point. As discussed above in relation to Fig. 2, our probe sets specific for only the genome or antigenome (but not an additional complementary viral mRNA) have low signal-to-noise ratios and sensitivity compared to those of probe sets that also target a viral mRNA. Importantly, FISH-quant was unable to detect viral genome or antigenome spots in cells that had been infected with LCMV for less than 8 h (Fig. 2 and data not shown). However, by 8 hpi and later, genome and antigenome spots become detectable with these probe sets (Fig. 2) (32). Therefore, in this set of experiments, smFISH spots detected with the NP mRNA/S antigenome, GPC mRNA/S genome, or L mRNA/L antigenome probe sets prior to 8 hpi are presumed to represent only the designated mRNA target in each case, whereas at 8 hpi and later, it is possible that a mixture of the targeted RNAs is detected.
Representative images of cells infected from 0 to 6 hpi are shown in Fig. 5A and B. Notably, transcription of the NP mRNA and L mRNA is detected as early as 1 h following infection (Fig. 5 and 6A and B), indicating that the primary transcription of the S and L genomic RNAs occurs soon after the entry and uncoating of arenavirus virions. The GPC mRNA, on the other hand, is first detected 6 h following infection (Fig. 5A and 6A and C). This delayed appearance of the GPC mRNA suggests that transcriptionally competent S antigenomic RNA is not delivered into cells by incoming virions. Furthermore, it suggests that a 4- to 6-h lag is required for the production of S antigenomic RNA, which serves as the template for the transcription of GPC mRNA (Fig. 1A). This result is in agreement with data from previous studies that examined arenavirus mRNA synthesis via Northern blotting (5, 26).
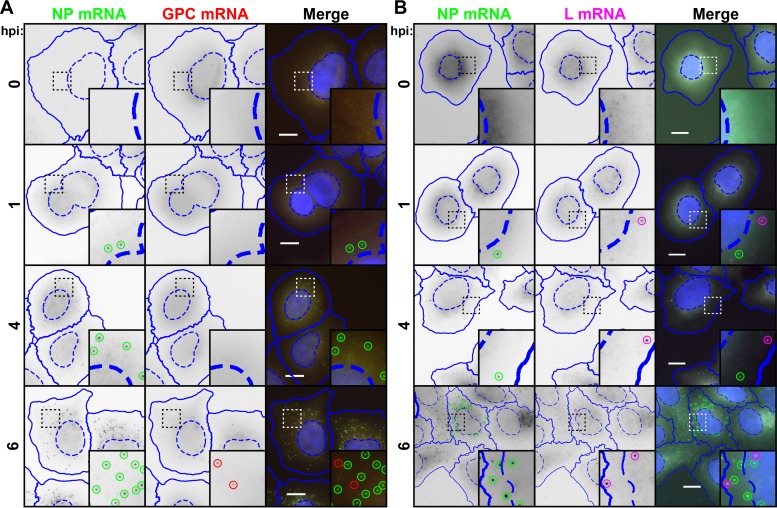
FIG 5.
Transcription of NP and L genes is detectable soon after infection, while GPC transcription occurs exclusively after a several-hour lag. Cells were infected with LCMV at an MOI of 0.1, fixed at various times following infection, and stained for NP mRNA (green) using a Cy5-labeled NP mRNA/S antigenome probe set and GPC mRNA (red) using an A568-labled GPC mRNA/S genome probe set (A) or for NP mRNA (green) using an A568-labeled NP mRNA/S antigenome probe set and L mRNA (magenta) using a Quasar 670-labeled L mRNA/L antigenome probe set (B). Note that for the time points shown (prior to 8 hpi), genomic and antigenomic RNAs are not detectable by smFISH probe sets with exclusive specificity for these RNAs (data not shown). Therefore, spots detected in this figure are presumed to represent only the mRNAs, but not the genome or antigenome, targeted by each respective probe set. Nuclear (hatched lines) and cytoplasmic (solid lines) boundaries, as determined by CellProfiler, are shown in blue. Identified spots are outlined by circles, which are green for NP mRNA, red for GPC mRNA, and magenta for L mRNA. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells across the time course to permit comparisons. Representative maximum-intensity projections from 1 of 2 independent experiments are shown. Bars, 10 μm.
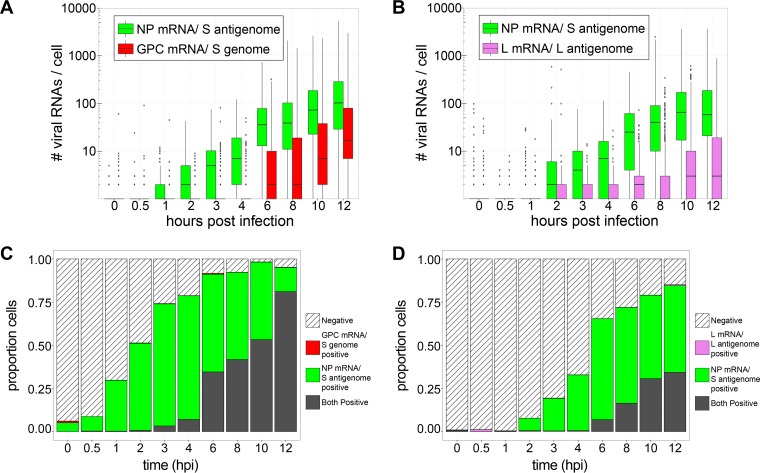
FIG 6.
Transcription of NP and L genes is detectable immediately upon infection, while GPC transcription occurs exclusively after a several-hour lag (Fig. 5). (A and B) Box plots representing the number of viral RNAs detected in cells at early time points following infection with LCMV (Fig. 5). (C and D) Stacked bar graphs showing the proportions of cells expressing RNAs detected by one, both, or neither viral RNA smFISH probe set. Between 620 and 1,316 cells were examined at each time point. In each case, RNAs identified by specific probe sets are designated by color. Note that for time points prior to 8 hpi, genomic and antigenomic RNAs are not detectable by smFISH probe sets with exclusive specificity for these RNAs (data not shown). Therefore, spots detected before 8 hpi are presumed to represent only the mRNAs, but not the genome or antigenome, recognized by each respective probe set. Spots detected at 8 hpi or later are presumed to be a mixture of all RNAs recognized by a particular probe set (e.g., mRNA and the genome or antigenome).
When the subcellular localization of pairs of NP mRNA and GPC mRNA or NP mRNA and L mRNA at 6 hpi or earlier was examined, no overt colocalization between viral mRNAs was noted (Fig. 5A and B).
Disproportionate transcription of S segment genes early after infection.
For each probe set used in the experiments shown in Fig. 5 and 6, the number of false-positive viral RNAs detected in mock-infected cells was used to establish a threshold to classify cells as either “positive” or “negative” for each of the tested viral RNA species. At 6 hpi (a time point before the virus in initially infected cells could have completed its life cycle and spread to adjacent, initially uninfected cells [36–39]), we observed that 65 to 90% of cells were positive for NP mRNA and that 40% were positive for GPC mRNA (Fig. 6C and D). This high frequency of cells containing S segment-derived transcripts was surprising given the fact that we initially infected cells at an MOI of 0.1 and thus would have expected only ∼10% of cells to have been expressing viral RNAs at this early time point. However, at this same early time point, only 8% of cells were positive for L segment-derived L mRNA (Fig. 6D), which is consistent with the expected frequency of viral RNA-positive cells based on the initial MOI. This result may suggest that a high proportion of viral particles either fails to package the L genome or, alternatively, delivers a transcriptionally defective L genome.
Viral RNA replication and transcription at the peak of acute infection.
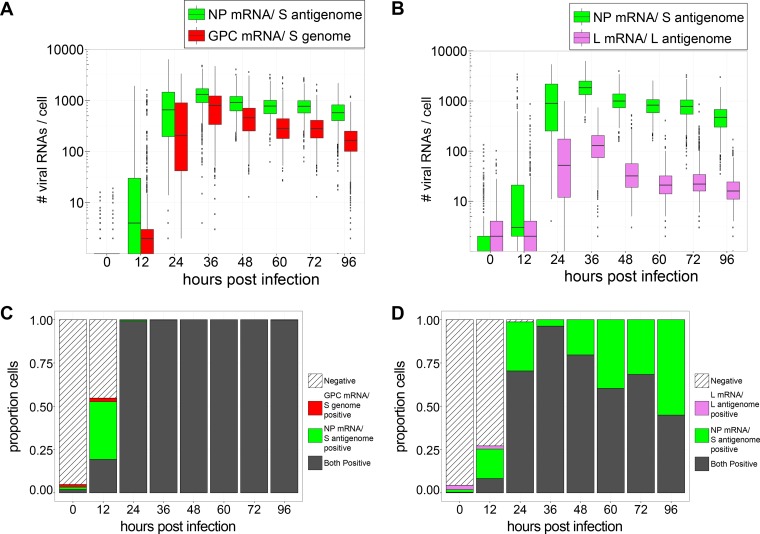
To profile LCMV RNAs at the peak of release of infectious virus during acute infection, cells were infected with LCMV at an MOI of 0.01, fixed at various time points between 12 and 96 hpi, and stained for the NP mRNA/S antigenome, GPC mRNA/S genome, or L mRNA/L antigenome, and several hundred cells were imaged and analyzed at each time point. Levels of viral RNAs detected by each probe set rapidly increased over the first 24 h of infection (Fig. 7A and B and 8A and B). The proportion of cells positive for these viral RNAs also rapidly increased over the first 24 h of infection such that almost all cells had substantial levels of all viral RNAs (Fig. 8C and D). Peak viral transcription and replication occurred at 36 h postinfection (Fig. 7A and B and 8A and B). At this time point, the viral smFISH signal was very dense, and true levels of viral NP mRNA and GPC mRNA were likely underestimated due to the inability of FISH-quant to accurately count tightly packed viral RNAs in the cytoplasm of infected cells (Fig. 4B and C, 7A and B, and 8A and B). Nevertheless, it is clear that the numbers of RNAs detected by the S segment-specific probe sets greatly exceeded those detected by the L segment-specific probe set (at least 10- to 35-fold higher) between 12 and 96 hpi (Fig. 8A and B and Table 1). Following peak viral transcription and replication at 36 h postinfection, levels of viral RNAs began to decrease (Fig. 7A and B and 8A and B). The proportion of cells positive for L mRNA/L antigenome expression decreased steadily beginning at 48 h postinfection (Fig. 8D). In contrast, all cells maintained NP mRNA/S antigenome and GPC mRNA/S expression over this entire time period (Fig. 8C and D).
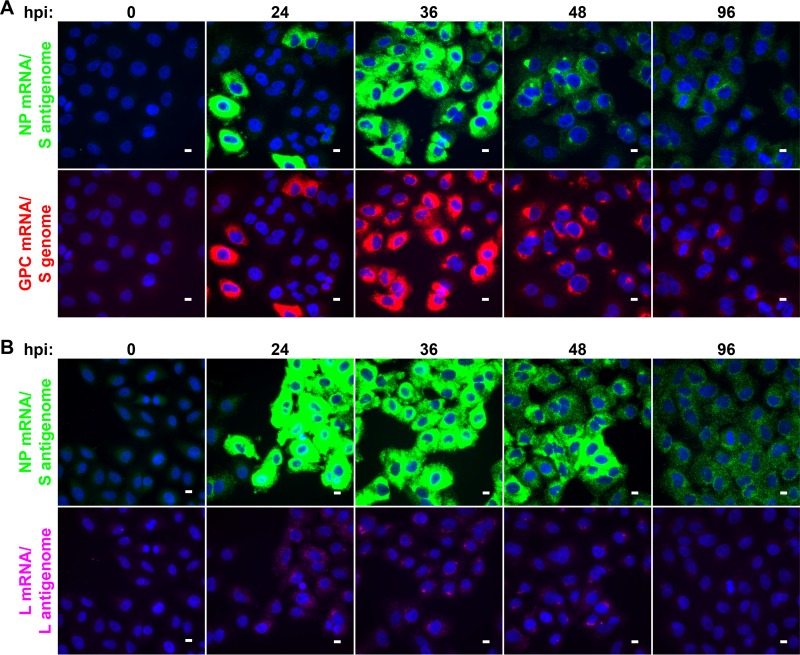
FIG 7.
Peak viral RNA replication and transcription occur at 36 hpi and are slowly lost from infected cells over the following days. Cells were infected with LCMV at an MOI of 0.01, fixed at various times following infection, and stained by using smFISH probe sets specific for the NP mRNA/S antigenome (Cy5) (green) and the GPC mRNA/S genome (A568) (red) (A) or for the NP mRNA/S antigenome (A568) (green) and the L mRNA/L antigenome (Quasar 670) (magenta) (B). Representative maximum-intensity projections of fields of infected cells at various time points from 1 of 2 independent experiments are shown. Each probe set is shown in its own row to highlight the difference in the levels to which these RNAs accumulate. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells across the time course to permit comparisons. Bars, 10 μm.
FIG 8.
Peak viral RNA replication and transcription occur at 36 hpi and are slowly lost from infected cells over the following days (Fig. 7). (A and B) Box plots representing the numbers of mRNAs detected in cells at time points during the peak period of LCMV infection. (C and D) Stacked bar graphs showing the proportions of cells expressing RNAs detected by one, both, or neither viral smFISH probe set. Between 480 and 1,659 cells were examined at each time point. RNAs identified by specific probe sets are designated by color.
TABLE 1.
Ratios of the expression levels of viral mRNAs in individual infected cellsb
| Time point | Mean ratio of NP mRNA and S antigenome/GPC mRNA and S genome ± SD | Mean ratio of NP mRNA and S antigenome/L mRNA and L antigenome ± SD |
|---|---|---|
| Early (hpi) (MOI = 0.1) | ||
| 0.5 | ND | ND |
| 1 | ND | ND |
| 2 | ND | ND |
| 3 | ND | ND |
| 4a | 4.3 ± 3.4 | ND |
| 6a | 6.0 ± 4.0 | 14.7 ± 9.5 |
| 8 | 4.9 ± 4.1 | 14.8 ± 12.1 |
| 10 | 5.6 ± 5.4 | 12.5 ± 9.7 |
| 12 | 5.0 ± 4.9 | 10.8 ± 9.5 |
| Peak (hpi) (MOI = 0.01) | ||
| 12 | 4.6 ± 4.1 | 8.8 ± 6.7 |
| 24 | 3.5 ± 3.4 | 14.8 ± 7.2 |
| 36 | 2.3 ± 1.9 | 18.0 ± 12.8 |
| 48 | 2.2 ± 1.2 | 28.7 ± 14.8 |
| 60 | 2.9 ± 1.2 | 34.1 ± 13.3 |
| 72 | 2.9 ± 1.1 | 30.1 ± 16.2 |
| 96 | 4.0 ± 1.9 | 24.5 ± 13.4 |
| Persistence (dpi) (MOI = 0.01) | ||
| 1.5 | 2.3 ± 1.6 | 10.2 ± 12.3 |
| 4 | 4.3 ± 2.0 | 10.5 ± 10.4 |
| 6 | 3.9 ± 2.4 | 4.9 ± 6.8 |
| 8 | 1.0 ± 0.7 | 2.0 ± 3.0 |
| 13 | 4.4 ± 2.8 | 24.1 ± 23.9 |
| 16 | 1.8 ± 1.7 | 2.4 ± 3.5 |
| 20 | 6.2 ± 3.7 | 28.1 ± 23.5 |
| 23 | 3.2 ± 2.0 | 5.8 ± 7.3 |
| 27 | 4.9 ± 4.2 | 2.7 ± 4.5 |
| 30 | 4.0 ± 2.3 | 9.1 ± 8.9 |
| 34 | 0.7 ± 1.2 | 2.2 ± 3.4 |
| 37 | 4.5 ± 3.8 | 6.1 ± 9.1 |
| 41 | 1.0 ± 1.3 | 7.3 ± 9.0 |
Note that for time points prior to 8 hpi, genomic and antigenomic RNAs are not detectable by smFISH probe sets with exclusive specificity for these RNAs (data not shown). Therefore, spots detected before 8 hpi are presumed to represent only the mRNAs, but not the genome or antigenome, recognized by each respective probe set. Spots detected at 8 hpi or later are presumed to be a mixture of all RNAs recognized by a particular probe set (e.g., mRNA and the genome or antigenome).
ND, not determined.
Cyclical patterns of infectious-virus production and antigen expression during persistent infection.
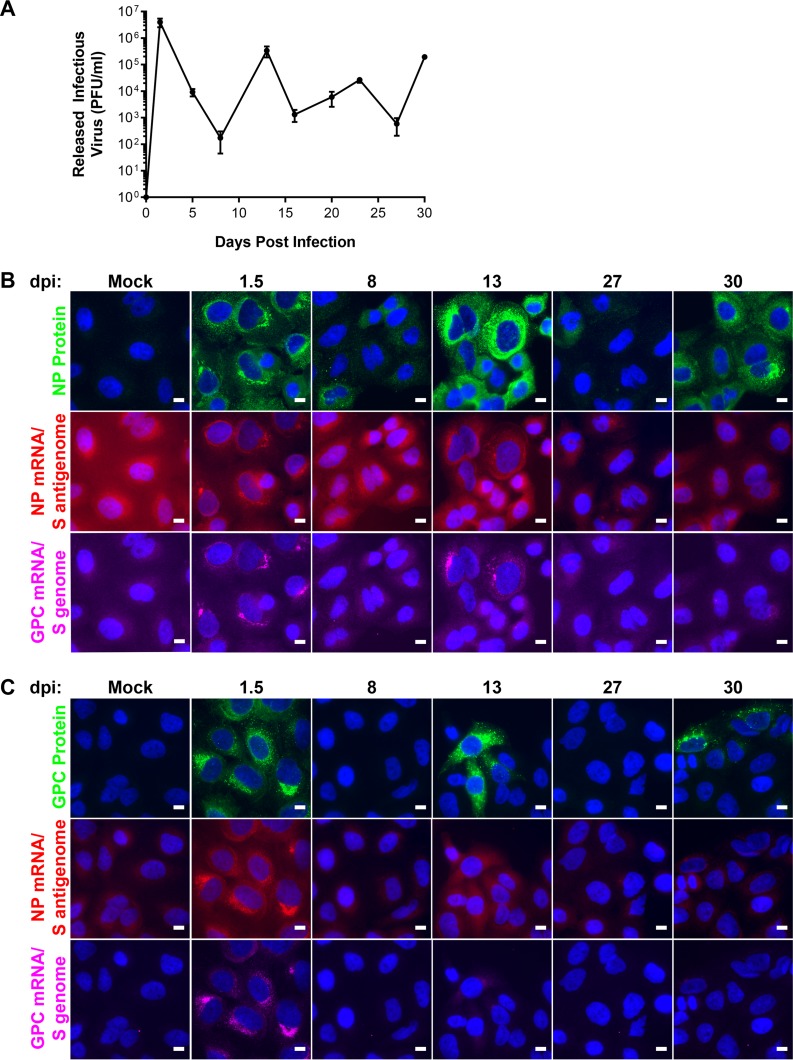
A key feature of arenavirus infection in cell culture is the cyclical pattern of release of infectious virus observed during the persistent phase of infection (16, 19, 40–42). We were particularly interested in using smFISH to assess how viral gene expression programs change during persistence and to examine how this correlated with the production of infectious virus and the translation of viral antigens. Toward this goal, cells were infected with LCMV at an MOI of 0.01, supernatants were collected, and cells were fixed at multiple time points between 1.5 and 30 days postinfection (dpi). As shown in Fig. 9A, A549 cells persistently infected with LCMV cyclically released waves of infectious virus over the first 30 days (Fig. 9A). We were curious how much viral antigen and viral RNA would be expressed in persistently infected cells at time points when cells were releasing high levels of infectious viral particles but also how much viral material was expressed at time points when cells released very little infectious virus. Therefore, we performed immunofluorescence analysis (IFA) to visualize NP or GPC and smFISH to visualize the NP mRNA/S antigenome and GPC mRNA/S in cells at multiple time points that corresponded to high and low points of infectious-virus release (Fig. 9B and C). At 1.5 dpi, all cells expressed NP and GPC proteins (Fig. 9B and C). Viral antigen expression mirrored the cycles of infectious-virus release, as shown in Fig. 9A. Viral antigen was expressed in fewer cells and/or at lower levels at 8 and 27 dpi (time points with low levels of infectious-virus release) than at 13 and 30 dpi (time points with higher levels of infectious-virus release) (Fig. 9B and C). While our goal was to visualize both viral RNA and viral antigen in infected cells by fluorescence microscopy, the sensitivity of smFISH was greatly reduced when combined with IFA, as evidenced by the higher levels of background and lower signal intensity (Fig. 9B and C). However, taking into account the lower sensitivity of smFISH in this particular experiment, we saw a correlation between the presence of viral RNA within cells and the expression of viral proteins (Fig. 9B and C). Thus, these results suggest that cells infected with LCMV can clear the infection, as evidenced by the majority of cells at specific time points (e.g., 8 and 27 dpi) that lack viral RNA or protein and produce little infectious virus.
FIG 9.
Cyclic periods of infectious virus particle release and antigen expression during persistence. Cells were infected with LCMV at an MOI of 0.01. (A) Supernatants from infected cells were collected at the indicated time points postinfection, the titers were determined, and the data are presented as mean PFU per milliliter ± standard deviations from 3 independent experiments. (B and C) Cells were fixed at the indicated time points following infection and stained by using smFISH probe sets specific for the NP mRNA/S antigenome (A568) (red), the GPC mRNA/S genome (Cy5) (magenta), and NP (Alexa 488) (green) (B) or GPC (Alexa 488) (green) (C). A single z-slice of a representative field of infected cells at various time points is shown. Each antibody or probe set is shown in its own row, and the same intensity levels were applied to all images of mock- and LCMV-infected cells across the time course to permit comparisons. Bars, 10 μm.
Cyclical patterns of genome transcription and replication during persistent infection.
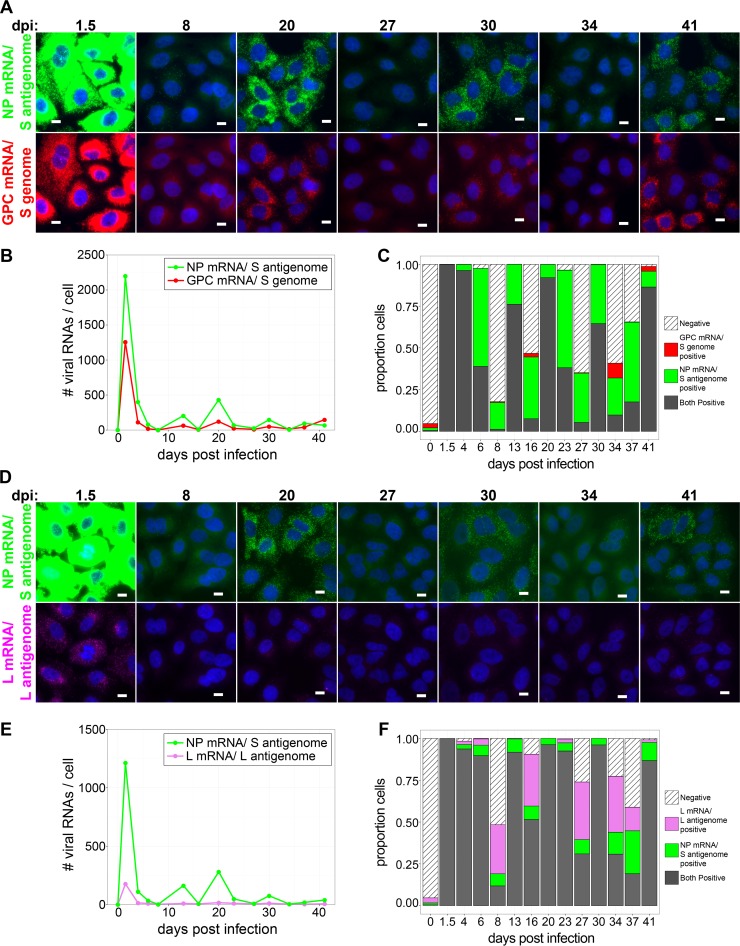
As the sensitivity of smFISH in our hands is diminished when it is performed in conjunction with immunofluorescent staining, we next used smFISH alone to examine the transcription and replication dynamics of arenavirus genomic RNA during the persistent phase of infection at the level of individual RNA molecules. Cells were infected with LCMV at an MOI of 0.01, fixed at multiple time points between 1.5 and 41 dpi, and stained for the NP mRNA/S antigenome, GPC mRNA/S genome, or L mRNA/L antigenome, and several hundred cells were imaged and analyzed at each time point. Note that our smFISH assay does not allow us to monitor particular cells over time but instead allows us to capture the viral RNA signature of individual cells at specific time points during infection. Following peak RNA transcription and replication at 36 hpi, we observed decreased levels of the NP mRNA/S antigenome, GPC mRNA/S genome, or L mRNA/L antigenome over the next several days such that at 8 dpi, the majority of cells examined were negative for all of these viral RNAs (Fig. 10A to F). However, by 13 dpi, the levels of viral RNAs detected by each probe set were increased, and the majority of cells were again positive for all viral RNAs (Fig. 10C and F). Viral RNA levels again dropped, and many cells were no longer positive for viral RNA by 16 dpi (Fig. 10C and F). This pattern of viral RNA clearance followed by increased levels of viral RNA expression and increased frequencies of viral RNA-expressing cells in the population was repeated in a cyclical fashion multiple times over the first 41 days following infection (Fig. 10). In summary, the sequential loss and reappearance of viral gene expression observed in these studies may represent a genetic signature of the previously recognized phenomenon of the cyclical production of infectious LCMV virions over time in cell culture models of persistent infection (16–18).
FIG 10.
Cyclic periods of viral RNA production and viral RNA loss occur during persistence. (A and D) Cells were infected with LCMV at an MOI of 0.01, fixed at the indicated time points following infection, and stained by using smFISH probe sets specific for the NP mRNA/S antigenome (Cy5) (green) and the GPC mRNA/S genome (A568; red) (A) or for the NP mRNA/S antigenome (A568; green) and the L mRNA/L antigenome (Quasar 670) (magenta) (D). Representative maximum-intensity projections of fields of infected cells at various time points from 1 of 2 independent experiments are shown. Each probe set is shown in its own row to highlight the difference in levels to which these RNAs accumulate. The same intensity levels for a particular probe set were applied to all images of mock- and LCMV-infected cells across the time course to permit comparisons. Bars, 10 μm. (B and E) Line graphs showing the average numbers of the indicated viral RNAs detected in cells at time points during the persistent phase of LCMV infection. (C and F) Stacked bar graphs showing the proportions of cells expressing RNAs detected by one, both, or neither viral smFISH probe set. Between 316 and 1,218 cells were examined at each time point.
Throughout the time course of persistence examined in this study, the NP mRNA/S antigenome was generally expressed at higher levels than the GPC mRNA/S genome (up to 5-fold-higher levels) (Fig. 10B and Table 1). The ratios between levels of the NP mRNA/S antigenome and L mRNA/L antigenome over this time period were more variable. At time points such as 13 and 20 dpi, when most cells in culture are positively expressing all viral RNA species, the NP mRNA/S antigenome greatly outnumbered the L mRNA/L antigenome (∼25-fold-higher levels) (Fig. 10E and Table 1). However, at other times, such as 8, 16, 27, and 34 dpi, when substantial proportions of cells had lost the expression of one or more viral RNAs, the ratio between NP mRNA/S antigenome and L mRNA/L antigenome levels in double-positive cells was greatly reduced (∼2-fold-higher levels of NP than of L mRNA) (Fig. 10E and Table 1). Notably, the magnitude of viral RNA expression during persistence never reached the high levels observed at the peak of acute infection (Fig. 10).
DISCUSSION
In the present study, we developed a high-throughput smFISH assay that allowed us to visualize single copies of LCMV RNAs in individual cells. Taking advantage of the sensitivity and quantitative aspect of this assay, we tracked the dynamics of viral replication and transcription spanning early time points following initial virus entry to late times during persistent infection. We observed that the transcription of the negative-sense NP and L mRNAs preceded that of the pseudo-positive-sense GPC mRNA, confirming the temporal separation of gene expression predicted by the ambisense coding strategy of the arenaviruses and suggesting that antigenomic RNA in virions is not transcriptionally active following release into a newly infected cell. Our studies demonstrated a hierarchal pattern of expression among viral RNAs and indicate that many infecting virus particles may lack L genomic RNA. Finally, over the course of persistent infection, we observed repeated cycles whereby cells appeared to transition from supporting active viral replication and transcription to a state where viral RNA is undetectable by smFISH. Collectively, these studies improve our understanding of the natural history of arenavirus replication and transcription during acute and persistent infection.
The smFISH assay developed here provided us with an opportunity to build upon previous studies and examine arenavirus genome replication and transcription with greater sensitivity and detail. Previous studies aimed at elucidating the early events of arenavirus transcription and genome replication used Northern blotting to visualize individual viral RNA species (23, 25, 26). Analyses of RNA from cells infected with LCMV or the New World arenavirus Pichinde virus failed to detect viral RNA from infected cells prior to 9 hpi (23) or 12 hpi (25), respectively. In the setting of infection with the New World arenavirus Tacaribe virus, Franze-Fernandez et al. detected S genomic RNA and NP mRNA at 2 hpi and S antigenomic RNA at 4 hpi, while GPC mRNA appeared several hours following the synthesis of S antigenomic RNA (26). The earliest time that the viral L segment has been observed was at 12 hpi (23). In the present study, we are able to detect viral NP and L mRNAs at 1 hpi. Our data support previous observations that viral NP mRNA expression occurs immediately following infection and that GPC mRNA expression occurs following a lag of several hours (5, 26). By probing single cells, we build upon that previous work by demonstrating that GPC mRNA expression is not detected, even at low levels, in the first hours following infection. In light of previous observations that antigenomic L and S segment RNAs are packaged into viral particles (20, 26), our inability to observe GPC mRNA in cells immediately following virus entry suggests that S antigenomic RNA packaged into virions is unable to be transcribed. Furthermore, it suggests that GPC mRNAs are not packaged into viral particles, as has been suggested for Z mRNA (43).
An interesting observation from our study was that, despite infection of cells at an MOI of 0.1, ∼65 to 90% of cells expressed one or more genes on the S genomic RNA segment at 6 hpi. Because it takes ∼8 h for an infected cell to make new infectious progeny (36–39), we were surprised to see such a high frequency of cells expressing these viral mRNAs at a time when the originally infected cells could not yet have spread the virus to additional uninfected cells in the monolayer. Notably, at this same 6-hpi time point, approximately 8% of cells expressed viral L mRNAs, which is consistent with the MOI utilized. One possible explanation for this observation could be that, within the viral stock, there may be a significant population of incomplete viral particles that possess the S segment but lack the L segment genomic RNA or a functional copy of this RNA. Indeed, this phenomenon has been observed for influenza A and Rift Valley fever viruses (44–46). Considering that the genetic basis for how arenavirus DI particles block the propagation of infectious virus particles is unknown, these results may provide clues for future studies to define the mechanism at work. Further examination will be necessary to explain the functional significance of this observation.
A hallmark characteristic of LCMV infection is the ability to establish an asymptomatic, persistent infection in reservoir rodents (47). Furthermore, it is possible to recapitulate key aspects of this persistent infection in cell culture models of infection (5, 40, 41, 48). One notable characteristic of cell culture models of LCMV infection is the cyclical rise and fall of the release of infectious virus seen during persistence (16, 19, 40–42). Several models have been proposed to explain how LCMV restricts its spread to establish and maintain a noncytopathic persistent infection, both in vitro and in vivo. The first model suggests that DI particles, which are produced in abundance by LCMV, can enter permissive host cells and interfere with the ability of standard infectious virus particles to successfully infect and complete the viral life cycle (9–13). Hotchin et al. proposed a second model that they termed cyclical, transient infection. In this model, cells infected with LCMV are initially productive in making infectious virus particles but later become refractory to superinfection and ultimately clear the virus (as evidenced by the loss of antigen expression and infectious-virus production), only to once again become susceptible to reinfection by the small number of cells that remain productively infected (16–19). Southern and colleagues proposed a third model that was based upon the dynamics and genetic identity of viral RNA species profiled during acute and persistent infection. In particular, they demonstrated by Northern blotting that LCMV RNAs (genome, antigenome, and mRNAs) accumulate to high levels during persistence, both in vitro and in vivo (5, 14, 15, 22). Furthermore, they showed that a proportion of these genomic and antigenomic RNAs, but not mRNAs, contained short deletions in the untranslated regions at their termini (5, 14, 15, 49). They proposed that these deleted RNAs were replication competent but transcriptionally incompetent. These data suggested a model where, during persistence, viral protein expression and infectious-virus production are inhibited due to the accumulation of high levels of transcriptionally defective genomic and antigenomic RNAs. Furthermore, because these deleted RNAs were found in virions, it was proposed that they serve as the molecular basis for DI particle interference. Finally, it was proposed that these deleted RNAs can be repaired by the viral polymerase to initiate bursts of productive replication/infectious-virus production during persistence. Each of these models, whether acting independently or in combination, would presumably restrict virus spread, allowing the virus to minimize its impact on host fitness while retaining its ability to propagate and ultimately maintain itself in nature. A potential caveat to the model proposed by Southern et al. comes from the finding that the 3′- and 5′-terminal sequences of the LCMV and LASV genomic RNA species were critical for both the transcription and replication of RNAs in a minigenome reporter assay (50, 51). Thus, the functional significance of these terminally truncated viral RNAs remains to be further elucidated.
The smFISH approach employed in this study can provide insights to build on this previous work. First, our study suggests that cells do not maintain high levels of genomic and antigenomic RNAs throughout persistence. The most likely explanation for the observation of high levels of the genome/antigenome during persistence by Southern and colleagues (5, 14, 15, 22) is that they measured viral RNAs from a fraction of the total cells within a population at a particular time point. Second, our findings of large numbers of cells lacking viral RNAs at several persistence time points (which correlate with the loss of infectious-virus release and antigen expression) suggest that the antigen-negative cells observed by Hotchin et al. (16–19) either had cleared infection or contained undetectable levels of RNA (by smFISH) as opposed to carrying replication-competent, but transcriptionally incompetent, terminally deleted genomic/antigenomic RNAs (as the model of Southern et al. would suggest [5, 14, 15, 49]). Our smFISH probe sets consisted of pools of 48 short oligonucleotide probes complementary to different regions spanning entire viral gene sequences. Therefore, even if genomic segments contained short terminal deletions, the majority of smFISH probes would bind their targets, and obvious smFISH signals would be present by fluorescence microscopy. Our findings also raise several new questions that require further study. For example, it will be important to define the molecular mechanism by which cells clear virus following infection and the specific host machinery responsible. Additionally, it would be particularly interesting to investigate the possibility that cycles of reinfection are jump-started from cells maintaining a reservoir of intracellular infectious virus, as has been documented in the literature (52, 53).
In summary, we have used fluorescence microscopy to visualize fluorescently labeled arenavirus RNA molecules in infected cells. Furthermore, we have described a flexible labeling, imaging, and image analysis pipeline that could be easily adapted to interrogate the events of transcription or genomic replication of any RNA virus, particularly where it is critical to image and quantify RNA levels in hundreds to thousands of cells under each experimental condition. We have taken advantage of this pipeline to examine the transcription and replication kinetics of LCMV RNAs over the course of infection. In particular, our data support the transient, cyclical infection model originally proposed by Hotchin et al. (16–19) and suggest that, following a period of productive infection, cells can clear infection, including viral genetic material and antigen, before becoming susceptible to reinfection. Furthermore, our data provide some support for the ideas that viral antigenomic RNA in virions may not be transcriptionally functional upon virus entry and that a significant fraction of virus particles may lack functional L genomic RNA, an observation that remains to be fully explored. Developing the ability to label genomic and antigenomic RNAs with greater sensitivity will be an important next step toward the construction of a quantitative model of the regulation of viral RNA replication and transcription over time with the goal of explaining the oscillatory behavior of viral RNA synthesis during persistence. While the cell culture model of persistent infection has provided interesting observations, it will be particularly important to examine how the genetic events of transcription and replication are regulated in an in vivo model of persistent infection in future studies.
MATERIALS AND METHODS
Cells and viruses.
A549 (ATCC CCL-185) cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium (catalog number 11320-033; Thermo Fisher) containing 10% fetal bovine serum and 1% penicillin-streptomycin (catalog number 15140-163; Thermo Fisher). LCMV Armstrong 53b (Arm53b) was provided by J. L. Whitton (The Scripps Research Institute, La Jolla, CA). A549 cells were infected with LCMV Arm53b at an MOI of 0.1 (Fig. 5 and 6) or an MOI of 0.01 (Fig. 1, 2, 4, and 7 to 10). For experiments examining late, persistent time points following infections, a T25 tissue culture flask of A549 cells was infected. The flask of infected cells was trypsinized, and cells were plated onto glass coverslips 24 h prior to the reported time points where coverslips were fixed, stained, and imaged, as described below (Fig. 10). The remaining cells were diluted and replated in a T25 flask until 24 h before the next examined time point, where this process was repeated. A standard plaque assay on Vero E6 cells was used to determine the titer of infectious LCMV in the collected supernatants. No cytopathic effect was observed in persistently infected cultures.
Single-molecule RNA FISH.
Cells were plated onto 14-mm round #1 glass coverslips. Following infection, cells were briefly washed in room-temperature Dulbecco's phosphate-buffered saline (DPBS) (with calcium and magnesium) (catalog number 14040133; Thermo Fisher) and fixed in 4% paraformaldehyde (PFA) in 1× phosphate-buffered saline (PBS) for 10 min at room temperature. Coverslips were washed twice in room-temperature PBS and fixed again at −20°C with 70% ethanol for at least 2 h. Coverslips were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (catalog number AM9770; Thermo Fisher) and washed once with 2× SSC and 10% formamide (catalog number BP227; Fisher Scientific). smFISH probes for different viral RNA species (Fig. 1A) were designed by using the Stellaris probe designer (Biosearch Technologies) (see Table S1 in the supplemental material). Unlabeled smFISH probes had a 3′ modified base with an amine functional group. Pools of 48 individual smFISH probes for a particular target RNA were combined at equimolar ratios and covalently labeled with Cy3 (catalog number PA23001; GE Healthcare), Alexa Fluor 568 (A568) (catalog number A20003; Thermo Fisher), or Cy5 (catalog number PA25001; GE Healthcare), as described previously (54). Coverslips were placed facedown on a 100-μl drop of hybridization mix containing 75 ng of the smFISH probe dissolved in hybridization buffer composed of 10% dextran sulfate (catalog number D8906; Sigma-Aldrich), 2× SSC, and 10% formamide. Hybridization occurred in a humidified chamber at 37°C overnight. Coverslips were washed twice in a solution containing 2× SSC and 10% formamide at 37°C for 30 min. Coverslips were then washed once in 1× PBS. For cellular segmentation, cells were stained with HCS CellMask green stain (catalog number H32714; Thermo Fisher) diluted at 50 ng/ml in PBS for 5 min at room temperature (note that this is significantly more dilute than recommended by the manufacturer, but we found it necessary in order to prevent overstaining of cells and thus to prevent spectral bleed-through into the Alexa Fluor 568 fluorescence channel). Nuclei were stained with DAPI (catalog number D9542; Sigma-Aldrich) at 1 μg/ml in PBS for 5 min at room temperature. Cells were washed a final time in PBS, briefly washed in water, dried, and mounted with ProLong Gold antifade reagent (catalog number P36934; Thermo Fisher).
Combined immunofluorescence and smFISH staining protocols were performed as described previously (55). LCMV NP was labeled with antibody 1-1.3, and GPC was labeled with antibody 33.6 (both from M. Buchmeier, University of California—Irvine). Primary antibodies were visualized with secondary Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) (catalog number A-11029; Thermo Fisher).
Image acquisition.
Wide-field fluorescent z-stacks were acquired by using a Nikon Ti Eclipse microscope with a 60× 1.4-numerical-aperture (NA) objective. Samples were illuminated with a light-emitting diode (LED) light source (Lumencor Spectra X light engine) with appropriate filter sets, and images were captured with a Hamamatsu Orca flash 4.0 LT scientific complementary metal oxide semiconductor (sCMOS) camera. z-stacks were captured at 300-nm increments, and the microscope was controlled by Nikon NIS Elements software. Captured ND2 images were converted to Tiff files by using the open-source Bio-formats toolkit (http://www.openmicroscopy.org/) (56).
Image segmentation and analysis.
DAPI and CellMask green z-stacks were projected by using a focus-based projection method, as described previously (57). Projected DAPI images were used for automatic nuclear segmentation in CellProfiler (Broad Institute) (34) and served as the seed for automatic secondary cellular segmentation using the projected CellMask green images (Fig. 4A). Statistics, including average pixel intensity, within the regions defined by primary and secondary segmentation were extracted from maximum-intensity projections of smFISH z-stacks by using CellProfiler (Fig. 4B to D).
Single smFISH-labeled RNAs were detected and localized in three dimensions (3D) by using FISH-quant (35). Briefly, smFISH z-stacks were filtered by using the “dual Gaussian filter,” and spots were detected by using the “local maximum” method. As a large number of acquired images required analysis, images were analyzed in “batch mode” with settings determined to give low rates of false-positive detections. The signal-to-noise ratios of different smFISH probe sets were determined as the average signal amplitude of the identified smFISH spots in an individual cell divided by the standard deviation of the fluorescent signal in a region of the same cell where smFISH spots were absent (Fig. 3).
Box-and-whisker plots were created by using the ggplot2 package in R. The box represents the interquartile range of the data, with the center line representing the median. Individual dots represent cells that are more than 1.5 times the interquartile range away from the median of the data.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge J. Lindsay Whitton for providing us with LCMV strain Arm53b and Samir Rahman, Philippe Clerc, Christian Weber, and Sophie Abélanet for technical assistance. We thank Pablo Navarro and Jason Stumpff for graciously offering the use of their microscopy equipment and for providing their expertise and Jean-Michel Arbona and Wei Ouyang for helpful discussions.
We also gratefully acknowledge funding support from NIH grants T32 HL076122-10 (B.R.K.), T32 AI055402 (C.M.Z.), R21 AI088059 (J.B.), and P20RR021905 and P30GM118228 (Immunobiology and Infectious Disease COBRE awards) (J.B.). D.Z. is supported by the Canadian Institute for Health Research (project grant-366682), the Fond de Recherche du Quebec (Chercheur-boursier Junior 2), and the Canadian Foundation for Innovation. C.Z., F.M., and A.S. were supported by the Institut Pasteur and the Fondation pour la Recherche Médicale (FRM). B.R.K. was supported by a Chateaubriand fellowship from the Office for Science and Technology at the Embassy of France in the United States. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02241-17.
REFERENCES
- 1.Buchmeier MJ, de la Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1827. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, LCMV in Transplant Recipients Investigation Team. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med 354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 3.Enria DA, Briggiler AM, Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res 78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Elliott LH, Belmont-Williams R. 1986. Lassa fever. Effective therapy with ribavirin. N Engl J Med 314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 5.Meyer BJ, de la Torre JC, Southern PJ. 2002. Arenaviruses: genomic RNAs, transcription, and replication. Curr Top Microbiol Immunol 262:139–157. [DOI] [PubMed] [Google Scholar]
- 6.Buchmeier MJ, Zajac AJ. 1999. Lymphocytic choriomeningitis virus, p 575–605. In Ahmed R, Chen I (ed), Persistent viral infections. John Wiley & Sons, New York, NY. [Google Scholar]
- 7.Buchmeier MJ, Welsh RM, Dutko FJ, Oldstone MB. 1980. The virology and immunobiology of lymphocytic choriomeningitis virus infection. Adv Immunol 30:275–331. doi: 10.1016/S0065-2776(08)60197-2. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann-Grube F, Martinez Peralta LM, Bruns M, Lohler J. 1983. Persistent infection of mice with the lymphocytic choriomeningitis virus, p 43–103. In Fraenkel-Conrat H, Wagner RR (ed), Virus-host interactions, receptors, persistence, and neurological diseases. Plenum Press, New York, NY. [Google Scholar]
- 9.Burns JW, Buchmeier MJ. 1993. Glycoproteins of the arenaviruses, p 17–35. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY. [Google Scholar]
- 10.Huang AS, Baltimore D. 1970. Defective viral particles and viral disease processes. Nature 226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 11.Oldstone MB. 1998. Viral persistence: mechanisms and consequences. Curr Opin Microbiol 1:436–441. doi: 10.1016/S1369-5274(98)80062-3. [DOI] [PubMed] [Google Scholar]
- 12.Huang AS. 1973. Defective interfering viruses. Annu Rev Microbiol 27:101–117. doi: 10.1146/annurev.mi.27.100173.000533. [DOI] [PubMed] [Google Scholar]
- 13.Welsh RM, O'Connell CM, Pfau CJ. 1972. Properties of defective lymphocytic choriomeningitis virus. J Gen Virol 17:355–359. doi: 10.1099/0022-1317-17-3-355. [DOI] [PubMed] [Google Scholar]
- 14.Meyer BJ, Southern PJ. 1994. Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J Virol 68:7659–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer BJ, Southern PJ. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J Virol 71:6757–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotchin J. 1974. Cyclical phenomena in persistent virus infection. J Reticuloendothel Soc 15:304–311. [PubMed] [Google Scholar]
- 17.Hotchin J. 1974. The role of transient infection in arenavirus persistence. Prog Med Virol 18:81–93. [PubMed] [Google Scholar]
- 18.Hotchin J. 1973. Transient virus infection: spontaneous recovery mechanism of lymphocytic choriomeningitis virus-infected cells. Nat New Biol 241:270–272. doi: 10.1038/newbio241270a0. [DOI] [PubMed] [Google Scholar]
- 19.Hotchin J, Kinch W, Benson L, Sikora E. 1975. Role of substrains in persistent lymphocytic choriomeningitis virus infection. Bull World Health Organ 52:457–463. [PMC free article] [PubMed] [Google Scholar]
- 20.Haist K, Ziegler C, Botten J. 2015. Strand-specific quantitative reverse transcription-polymerase chain reaction assay for measurement of arenavirus genomic and antigenomic RNAs. PLoS One 10:e0120043. doi: 10.1371/journal.pone.0120043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis SJ, Southern PJ. 1988. Deleted viral RNAs and lymphocytic choriomeningitis virus persistence in vitro. J Gen Virol 69(Part 8):1893–1902. doi: 10.1099/0022-1317-69-8-1893. [DOI] [PubMed] [Google Scholar]
- 22.Francis SJ, Southern PJ. 1988. Molecular analysis of viral RNAs in mice persistently infected with lymphocytic choriomeningitis virus. J Virol 62:1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller-Pace FV, Southern PJ. 1988. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology 162:260–263. doi: 10.1016/0042-6822(88)90419-9. [DOI] [PubMed] [Google Scholar]
- 24.Southern PJ, Singh MK, Riviere Y, Jacoby DR, Buchmeier MJ, Oldstone MB. 1987. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology 157:145–155. doi: 10.1016/0042-6822(87)90323-0. [DOI] [PubMed] [Google Scholar]
- 25.Shivaprakash M, Harnish D, Rawls W. 1988. Characterization of temperature-sensitive mutants of Pichinde virus. J Virol 62:4037–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franze-Fernandez MT, Zetina C, Iapalucci S, Lucero MA, Bouissou C, Lopez R, Rey O, Daheli M, Cohen GN, Zakin MM. 1987. Molecular structure and early events in the replication of Tacaribe arenavirus S RNA. Virus Res 7:309–324. doi: 10.1016/0168-1702(87)90045-1. [DOI] [PubMed] [Google Scholar]
- 27.Auperin DD, Romanowski V, Galinski M, Bishop DH. 1984. Sequencing studies of Pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J Virol 52:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polyak SJ, Zheng S, Harnish DG. 1995. Analysis of Pichinde arenavirus transcription and replication in human THP-1 monocytic cells. Virus Res 36:37–48. doi: 10.1016/0168-1702(94)00107-N. [DOI] [PubMed] [Google Scholar]
- 29.Fuller-Pace FV, Southern PJ. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J Virol 63:1938–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King BR, Kellner S, Eisenhauer PL, Bruce EA, Ziegler CM, Zenklusen D, Botten JW. 27 September 2017. Visualization of the lymphocytic choriomeningitis mammarenavirus (LCMV) genome reveals the early endosome as a possible site for genome replication and viral particle pre-assembly. J Gen Virol doi: 10.1099/jgv.0.000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziegler CM, Eisenhauer P, Bruce EA, Weir ME, King BR, Klaus JP, Krementsov DN, Shirley DJ, Ballif BA, Botten J. 2016. The lymphocytic choriomeningitis virus matrix protein PPXY late domain drives the production of defective interfering particles. PLoS Pathog 12:e1005501. doi: 10.1371/journal.ppat.1005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller F, Senecal A, Tantale K, Marie-Nelly H, Ly N, Collin O, Basyuk E, Bertrand E, Darzacq X, Zimmer C. 2013. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods 10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 36.Buchmeier MJ, Elder JH, Oldstone MB. 1978. Protein structure of lymphocytic choriomeningitis virus: identification of the virus structural and cell associated polypeptides. Virology 89:133–145. doi: 10.1016/0042-6822(78)90047-8. [DOI] [PubMed] [Google Scholar]
- 37.Dutko FJ, Pfau CJ. 1978. Arenavirus defective interfering particles mask the cell-killing potential of standard virus. J Gen Virol 38:195–208. doi: 10.1099/0022-1317-38-2-195. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann-Grube F, Popescu M, Schaefer H, Gschwender HH. 1975. LCM virus infection of cells in vitro. Bull World Health Organ 52:443–456. [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann-Grube F. 1971. Lymphocytic choriomeningitis virus, vol 10 Springer-Verlag, Vienna, Austria. [Google Scholar]
- 40.Lehmann-Grube F. 1967. A carrier state of lymphocytic choriomeningitis virus in L cell cultures. Nature 213:770–773. doi: 10.1038/213770a0. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann-Grube F, Slenczka W, Tees R. 1969. A persistent and inapparent infection of L cells with the virus of lymphocytic choriomeningitis. J Gen Virol 5:63–81. doi: 10.1099/0022-1317-5-1-63. [DOI] [PubMed] [Google Scholar]
- 42.Staneck LD, Trowbridge RS, Welsh RM, Wright EA, Pfau CJ. 1972. Arenaviruses: cellular response to long-term in vitro infection with Parana and lymphocytic choriomeningitis viruses. Infect Immun 6:444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salvato MS, Shimomaye EM. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1–10. doi: 10.1016/0042-6822(89)90216-X. [DOI] [PubMed] [Google Scholar]
- 44.Fonville JM, Marshall N, Tao H, Steel J, Lowen AC. 2015. Influenza virus reassortment is enhanced by semi-infectious particles but can be suppressed by defective interfering particles. PLoS Pathog 11:e1005204. doi: 10.1371/journal.ppat.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wichgers Schreur PJ, Kortekaas J. 2016. Single-molecule FISH reveals non-selective packaging of Rift Valley fever virus genome segments. PLoS Pathog 12:e1005800. doi: 10.1371/journal.ppat.1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. 2013. Most influenza a virions fail to express at least one essential viral protein. J Virol 87:3155–3162. doi: 10.1128/JVI.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis SJ, Southern PJ, Valsamakis A, Oldstone MB. 1987. State of viral genome and proteins during persistent lymphocytic choriomeningitis virus infection. Curr Top Microbiol Immunol 133:67–88. [DOI] [PubMed] [Google Scholar]
- 48.Oldstone MB, Buchmeier MJ. 1982. Restricted expression of viral glycoprotein in cells of persistently infected mice. Nature 300:360–362. doi: 10.1038/300360a0. [DOI] [PubMed] [Google Scholar]
- 49.Meyer BJ, Southern PJ. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J Virol 67:2621–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez M, de la Torre JC. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 77:1184–1194. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hass M, Westerkofsky M, Muller S, Becker-Ziaja B, Busch C, Gunther S. 2006. Mutational analysis of the Lassa virus promoter. J Virol 80:12414–12419. doi: 10.1128/JVI.01374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Zeijst BA, Noyes BE, Mirault ME, Parker B, Osterhaus AD, Swyryd EA, Bleumink N, Horzinek MC, Stark GR. 1983. Persistent infection of some standard cell lines by lymphocytic choriomeningitis virus: transmission of infection by an intracellular agent. J Virol 48:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanwick TL, Kirk BE. 1976. Analysis of baby hamster kidney cells persistently infected with lymphocytic choriomeningitis virus. J Gen Virol 32:361–367. doi: 10.1099/0022-1317-32-3-361. [DOI] [PubMed] [Google Scholar]
- 54.Zenklusen D, Singer RH. 2010. Analyzing mRNA expression using single mRNA resolution fluorescent in situ hybridization. Methods Enzymol 470:641–659. doi: 10.1016/S0076-6879(10)70026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song T, Zheng Y, Wang Y, Katz Z, Liu X, Chen S, Singer RH, Gu W. 2015. Specific interaction of KIF11 with ZBP1 regulates the transport of beta-actin mRNA and cell motility. J Cell Sci 128:1001–1010. doi: 10.1242/jcs.161679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldberg IG, Allan C, Burel JM, Creager D, Falconi A, Hochheiser H, Johnston J, Mellen J, Sorger PK, Swedlow JR. 2005. The Open Microscopy Environment (OME) data model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol 6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsanov N, Samacoits A, Chouaib R, Traboulsi AM, Gostan T, Weber C, Zimmer C, Zibara K, Walter T, Peter M, Bertrand E, Mueller F. 2016. smiFISH and FISH-quant—a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res 44:e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.