ABSTRACT
During entry, polyomavirus (PyV) is endocytosed and sorts to the endoplasmic reticulum (ER), where it penetrates the ER membrane to reach the cytosol. From the cytosol, the virus moves to the nucleus to cause infection. How PyV is transported from the cytosol into the nucleus, a crucial infection step, is unclear. We found that upon reaching the cytosol, the archetypal PyV simian virus 40 (SV40) recruits the cytoplasmic dynein motor, which disassembles the viral particle. This reaction enables the resulting disassembled virus to enter the nucleus to promote infection. Our findings reveal how a cytosolic motor can be hijacked to impart conformational changes to a viral particle, a process essential for successful infection.
IMPORTANCE How a nonenveloped virus successfully traffics from the cell surface to the nucleus to cause infection remains enigmatic in many instances. In the case of the nonenveloped PyV, the viral particle is sorted from the plasma membrane to the ER and then the cytosol, from which it enters the nucleus to promote infection. The molecular mechanism by which PyV reaches the nucleus from the cytosol is not entirely clear. Here we demonstrate that the prototype PyV SV40 recruits dynein upon reaching the cytosol. Importantly, this cellular motor disassembles the viral particle during cytosol-to-nucleus transport to cause infection.
KEYWORDS: disassembly, dynein, motor proteins, polyomavirus, virus-host interaction
INTRODUCTION
Polyomaviruses (PyVs) are the causative agents for a myriad of debilitating human diseases, especially in immunocompromised patients. Prominent human PyVs include BK virus, which causes hemorrhagic cystitis and nephropathy, JC virus, which induces progressive multifocal leukoencephalopathy (PML), and the Merkel cell PyV, which triggers Merkel cell carcinoma (1). Simian virus 40 (SV40) is the archetypal PyV, possessing both structural and genetic similarities to human PyVs, as well as sharing the same infection pathway with its human counterparts (1). Not surprisingly, studies on SV40 entry have provided significant insights into the cellular basis of human PyV infection.
Structurally, SV40 consists of 72 pentamers of the major structural protein VP1 that encases its DNA genome, with each pentamer harboring an internal hydrophobic minor protein, VP2 or VP3. When properly assembled, the viral particle has a diameter of approximately 45 nm (2, 3). To infect cells, SV40 binds to the ganglioside GM1 receptor on the plasma membrane, is endocytosed, and is targeted to the endolysosome (4, 5). The virus then sorts to the endoplasmic reticulum (ER), where it penetrates the ER membrane to access the cytosol (6–9). From the cytosol, SV40 mobilizes to the nucleus, where transcription and replication of the viral genome lead to lytic infection or cellular transformation (10). The molecular basis by which this virus reaches the nucleus from the cytosol, a critical infection step, is not entirely clear.
Using an unbiased protein-protein interaction approach, we identified cytoplasmic dynein (referred to below as dynein) as a binding partner of cytosol-localized SV40. Ensuing biochemical, cell-based, and microscopy studies in the context of loss-of-function conditions demonstrated that dynein promotes SV40 infection by disassembling the viral particle in the cytosol, an event that likely enables its subsequent entry into the nucleus. Our data reveal how a cellular motor can be exploited to induce structural changes to a viral particle that are critical for productive infection.
RESULTS
Dynein is important for SV40 infection.
To clarify how SV40 in the cytosol is transported into the nucleus, we used an unbiased biochemical approach to identify cellular factors that bind to cytosol-localized SV40. To this end, simian CV-1 fibroblasts were infected with SV40, and a cytosolic extract containing cytosol-localized SV40 was isolated using a detergent-based, semipermeabilized-membrane fractionation method described previously (11, 12). SV40 was precipitated from this extract by using an antibody directed against the viral VP1 protein, and the identities of SV40-interacting partners were revealed via “shotgun” mass spectrometry analysis of the precipitated material. As a negative control, a cytosolic extract derived from SV40-infected CV-1 cells pretreated with brefeldin A (BFA), a drug that blocks the trafficking of SV40 from the cell surface to the ER and hence the cytosol, was also prepared and was subjected to the same precipitation procedure. Because this extract should not contain any SV40, factors identified in this sample by mass spectrometry represent false-positive results.
Host proteins with at least three unique peptides that were at least 2-fold enriched over levels in the BFA-treated sample were considered potential SV40-interacting proteins (see the supplemental material). Included in this list are several cytosolic proteins, such as Hsc70, Hsp105, and kinesin-1, that have been shown previously to play important roles during SV40 infection (13–15). These observations demonstrated that this approach can be used successfully to identify host components involved in SV40 entry. Strikingly, the microtubule minus-end-directed cytosolic motor dynein and its associated components were also identified (Table 1). Because a role of dynein during SV40 entry has not been fully elucidated (16), we focused on this protein.
TABLE 1.
Cytosolic SV40-interacting dynein components
| Protein | Annotation | Role |
|---|---|---|
| DYNC1H1 | Cytoplasmic dynein 1 heavy chain 1 | Promotes minus-end trafficking of cargoes |
| DCTN1/p150 | Dynactin subunit 1 | Recruits dynein to microtubules and cargo; regulates dynein motility |
| NDE1/nudE | Nuclear distribution protein homolog 1 | A dynein activator that helps Lis1 interact with p150 |
| CCDC88A/GRDN | Girdin | A probable dynein activator |
| RANBP2/NUP358 | E3 SUMO-protein ligase | Interacts with the adaptor BICD2 and targets dynein/dynactin to the nuclear pore complex |
| DYNC1l2 | Cytoplasmic dynein 1 intermediate chain 2 | A noncatalytic dynein component that binds to cargoes and adaptors |
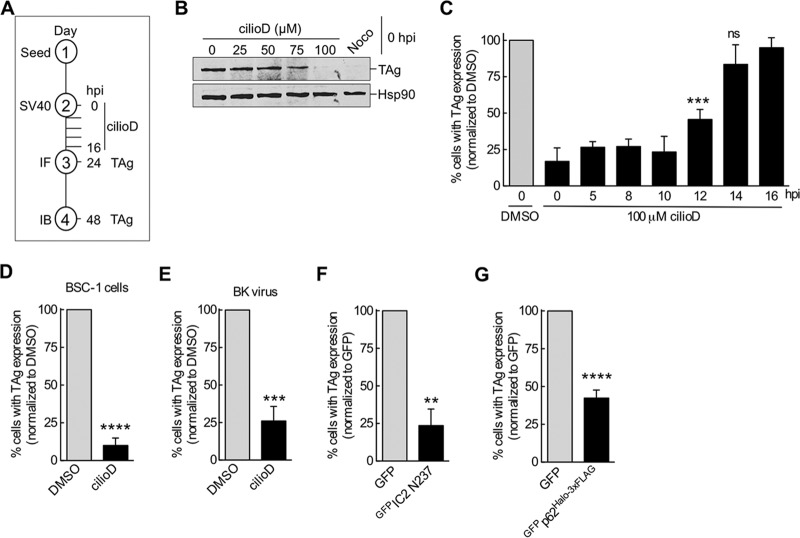
To evaluate the importance of dynein during SV40 infection, we asked if the dynein inhibitor ciliobrevin D (cilioD) blocks virus infection. We investigated this question by assessing the level of the virally encoded large T antigen (TAg), which is expressed only upon arrival of the virus in the host nucleus. CilioD is a cell-permeant, reversible, and specific blocker of the dynein AAA+ ATPase motor (17). By immunoblotting, we found that when different concentrations of cilioD were added at infection (0 h postinfection [hpi]) (the experimental setup is shown in Fig. 1A), 100 μM (and, to a lesser extent, 75 μM) cilioD blocked TAg expression, in contrast to expression in cells treated with the control, dimethyl sulfoxide (DMSO) (0 μM cilioD) (Fig. 1B). This is similar to the effect of the microtubule-disrupting agent nocodazole (Fig. 1B), as reported previously (18). When we performed a time course experiment using an immunofluorescence approach to score for TAg expression, we found that cilioD in fact maintained its ability to block SV40 infection even if the inhibitor was added 10 to 12 hpi (Fig. 1C). At these time points, a majority of SV40 has successfully trafficked from the plasma membrane to the ER and penetrated across the ER membrane to reach the cytosol (7, 11). However, at time points after 14 hpi, when the virus has entered the nucleus from the cytosol, cilioD no longer blocks infection (Fig. 1C). These results demonstrate that dynein executes an important function during SV40 infection, likely at a post–cytosol arrival step but prior to nuclear entry (see below). Dynein's role during SV40 entry is not confined to CV-1 cells; cilioD markedly impaired virus infection of simian epithelial BSC-1 cells (Fig. 1D). Additionally, this motor's role is not selective for SV40; cilioD also impaired infection of CV-1 cells by the related human BK virus (Fig. 1E).
FIG 1.
Dynein is important for SV40 infection. (A) Experimental setup used for panels B to E. IF, immunofluorescence; IB, immunoblotting. (B) CV-1 cells were infected with SV40 (MOI, ∼1) at 0 hpi and were treated either with different concentrations of cilioD or with 1 μM nocodazole (Noco). At 48 hpi, cells were lysed, and the resulting whole-cell extract was immunoblotted with the indicated antibodies. (C) CV-1 cells infected with SV40 were treated with 100 μM cilioD at different times postinfection. At 24 hpi, the extent of infection was analyzed by immunofluorescence microscopy by scoring 100 cells for each experiment. Data were normalized to those for the DMSO control (shaded bar). Values are averages of the means (n = 3) ± SD. ***, P < 0.001; ns, not significant. (D) BSC-1 cells were infected with SV40, and at 5 hpi, cells were treated with 100 μM cilioD. Values are averages of the means (n = 3) ± SD. ****, P < 0.0001. (E) CV-1 cells were infected with BK virus, and at 5 hpi, cells were treated with 100 μM cilioD. Values are averages of the means (n = 3) ± SD. ***, P < 0.001. (F) CV-1 cells expressing the indicated constructs were infected with SV40 (MOI, ∼1) for 24 h, fixed, and immunostained for TAg. Only GFP-expressing cells were scored for a TAg-positive signal by immunofluorescence microscopy. Data are normalized to those for the GFP control (shaded bar). Values are averages of the means (n = 3) ± SD. **, P < 0.01. (G) The experiment was similar to that for panel F, except that CV-1 cells were transfected with GFPp62Halo-3×FLAG. Values are averages of the means (n = 3) ± SD. ****, P < 0.0001.
To complement the chemical inhibitor approach, we used a dominant negative overexpression strategy. In this case, overexpression of the N-terminal 237 amino acids of dynein's intermediate chain 2 (IC2) tagged with GFP (GFPIC2 N237) has been shown previously to act in a dominant negative manner and disrupt the activity of dynein (19). Indeed, relative to the expression of control GFP, GFPIC2 N237 overexpression markedly blocked SV40 infection in CV-1 cells (Fig. 1F), further supporting the idea that dynein is essential for SV40 infection.
In mammalian cells, processive dynein activity requires the formation of a three-member protein complex comprising the dynein motor, the dynactin activator, and a cargo adaptor (20–22). Because our mass spectrometry analysis suggested that dynactin subunit 1 (p150) binds to cytosol-localized SV40, we asked if dynactin also played a role in SV40 infection. Since overexpression of dynactin subunit 4 (p62) has been shown previously to disrupt dynein activity (23, 24), we assessed if overexpression of a p62 construct (harboring N-terminally-tagged GFP and C-terminally-tagged Halo-3×FLAG [GFPp62Halo-3×FLAG]) affects SV40 infection. Compared to that with expression of the GFP control in CV-1 cells, GFPp62Halo-3×FLAG expression decreased the level of SV40 infection (Fig. 1G). This result indicates that in addition to the dynein motor, the action of dynactin supports SV40 entry into the nucleus.
Dynein is not required for SV40-induced focus formation.
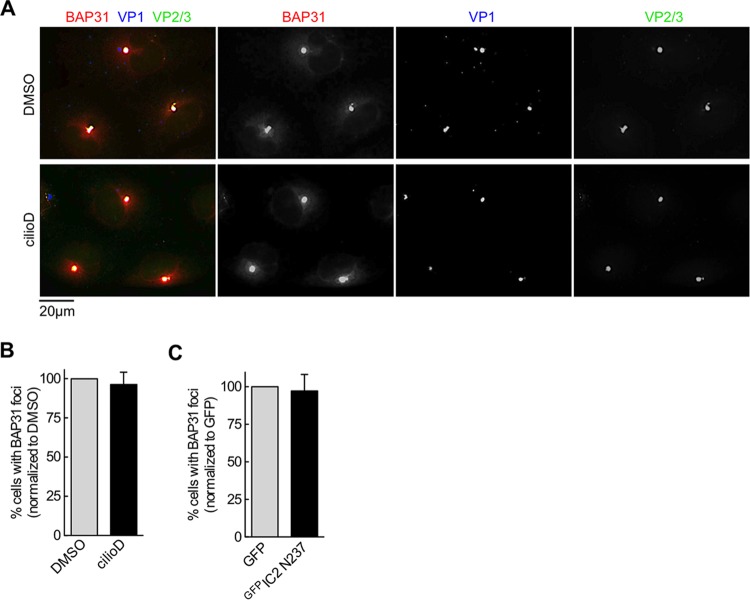
We next probed the requirement for dynein at specific SV40 entry steps. The time course analysis using cilioD (Fig. 1C) suggested that dynein likely operates at a step after the virus has reached the cytosol but before the entry of the viral particle into the nucleus. We therefore directly tested whether events that prime SV40 for cytosol entry from the ER, as well as arrival in the cytosol from the ER, are regulated by dynein. Upon arrival in the ER from the cell surface, SV40 reorganizes selective ER membrane proteins, such as BAP31, to specific sites, called foci, that act as cytosol entry sites for the virus (12, 25); the foci also harbor membrane penetration-competent SV40 (i.e., VP1 with exposed VP2 or VP3). The formation of SV40-induced foci thus reflects the ability of the virus to generate ER-to-cytosol membrane penetration sites that prepare the virus for cytosol entry. However, we found that cilioD treatment at 5 hpi did not affect SV40-induced focus formation (Fig. 2A; quantified in Fig. 2B), nor did expression of the dominant negative GFPIC2 N237 construct (quantified in Fig. 2C). These data suggest that dynein does not affect the formation of ER-to-cytosol membrane penetration sites that enable the successful arrival of the virus in the cytosol.
FIG 2.
Dynein is not required for SV40-induced focus formation. (A) Representative images of CV-1 cells infected with SV40 (MOI, ∼20) that were treated with either the control (DMSO) or 100 μm cilioD (at 5 hpi). After 16 h, cells were fixed and were immunostained with the indicated antibodies. (B) Quantification of the results shown in panel A. Cells harboring at least 1 BAP31-positive focus were scored as positive, and at least 100 cells were scored for each experiment. The data are normalized to those for the DMSO control (shaded bar). Values are averages of the means (n = 3) ± SD. (C) Quantification of focus formation in SV40-infected CV-1 cells expressing the indicated construct. The data are normalized to those for the GFP control (shaded bar). Values are averages of the means (n = 3) ± SD.
Dynein does not promote the arrival of SV40 from the ER in the cytosol.
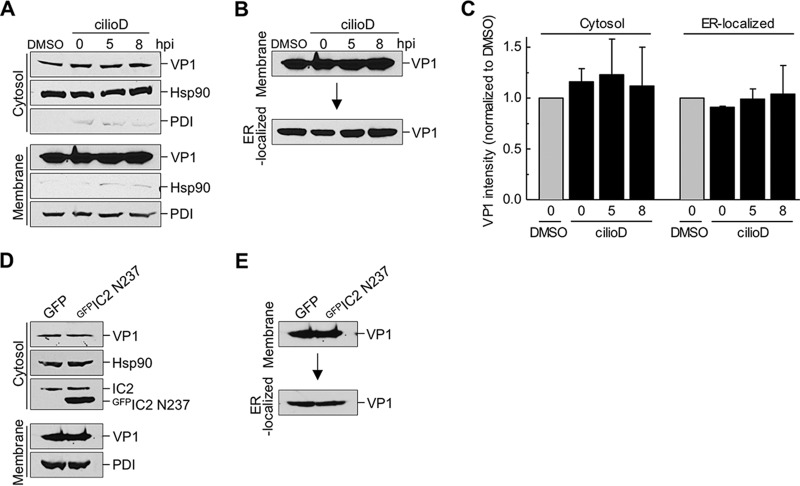
After focus formation, SV40 is physically delivered to the cytosol from the ER. To determine if the arrival of SV40 in the cytosol is controlled by dynein, we used a well-established cell-based, semipermeabilized-membrane assay of ER-to-cytosol transport (11, 12). In this assay, SV40-infected CV-1 cells that were either left untreated or treated with cilioD (at different postinfection time points) were first harvested and then incubated with a low concentration of digitonin to permeabilize the plasma membrane without damaging internal membranes. The cells were then centrifuged to generate two fractions: a supernatant fraction, which contains cytosolic proteins and virus that reaches the cytosol (“cytosol” fraction), and a pellet fraction, which harbors membranes including the ER, as well as virus associated with membranes (“membrane” fraction). The integrity of the fractionation procedure can be monitored by the release of the cytosolic marker Hsp90 to the cytosolic fraction and by the pelleting of the ER-resident protein disulfide isomerase (PDI) with the membrane fraction (Fig. 3A).
FIG 3.
Dynein does not promote the arrival of SV40 from the ER in the cytosol. (A) CV-1 cells were infected with SV40 (MOI, ∼5). Cells were either treated with the control (DMSO) or with 100 μM cilioD at the time of infection (0 hpi) or at 5 or 8 hpi. After 12 hpi, cells were harvested and were processed using the semipermeabilized-membrane cytosol arrival assay (see Materials and Methods). Hsp90 and PDI serve as markers for the cytosol and membrane fractions, respectively. (B) The membrane fraction (shown in panel A) was solubilized in a buffer containing 1% Triton X-100. After centrifugation, the extracted supernatant material containing ER-localized SV40 was analyzed by immunoblotting with VP1 antibodies. (C) Relative VP1 band intensities in the cytosol fraction (from panel A) and the ER-localized fraction (from panel B) were quantified. Data are normalized to those for DMSO (control). Values are averages of the means (n = 3) ± SD. (D) COS-7 cells expressing either GFPIC2 N237 or a GFP control were infected with SV40 (MOI, ∼5), and samples were processed as described for panel A. (E) The membrane fraction (from panel D) was processed as described for panel B.
Using this assay, we found that cilioD had no effect on the arrival of SV40 VP1 in the cytosol, regardless of when this inhibitor was added relative to the time of infection (Fig. 3A, top; VP1 band intensity is quantified in Fig. 3C). Thus, dynein does not regulate the arrival of SV40 from the ER in the cytosol. We also took advantage of this assay and used a biochemical protocol to extract ER-localized SV40 from the membrane fraction (see Materials and Methods). Using this method, we found that cilioD did not affect the level of ER-localized SV40 (Fig. 3B, bottom; quantified in Fig. 3C), confirming that dynein does not affect the arrival of SV40 from the cell surface in the ER (Fig. 1C).
Expression of the GFPIC2 N237 construct, a condition that impaired SV40 infection (Fig. 1F), did not affect the arrival of SV40 in the cytosol or the ER (Fig. 3D and E), in agreement with the effects of cilioD. These findings further support the premise that dynein does not regulate the transport of SV40 from the plasma membrane to the ER or the cytosol but instead acts at a post–cytosol arrival step leading to successful nuclear entry.
Dynein associates with cytosol-localized SV40.
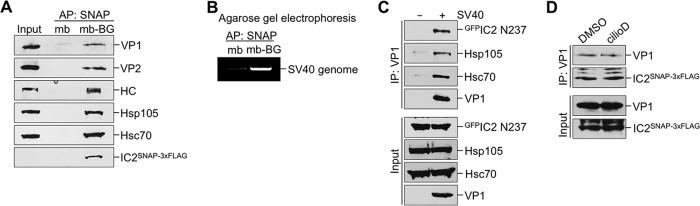
The observation that inhibition of dynein blocked SV40 infection without impacting the arrival of the virus from the cell surface in the ER or the cytosol suggests that dynein controls the transport of SV40 from the cytosol to the nucleus. In this scenario, we envision that dynein should bind to SV40, a possibility raised by the mass spectrometry data (Table 1). To confirm this interaction, a whole-cell extract derived from SV40-infected CV-1-derived COS-7 cells expressing a full-length IC2 construct that contains SNAP and 3×FLAG tags at its C terminus (IC2SNAP-3×FLAG) (26) was subjected to precipitation using either magnetic beads that capture the SNAP tag (mb-BG) (27) or, as a negative control, magnetic beads that do not (mb). COS-7 cells were used in this experiment because they support higher transfection efficiency than CV-1 cells. We found that precipitation of IC2SNAP-3×FLAG pulled down VP1 and VP2, as well as dynein's heavy chain (HC) and the cytosolic chaperones Hsc70 and Hsp105, which have been shown previously to promote SV40 infection (Fig. 4A) (13, 15). Importantly, the SV40 genome was also found in the precipitated material (Fig. 4B), indicating that the infectious form of the virus interacts with dynein. Precipitation of cytosol-localized SV40 coprecipitated the dominant negative construct GFPIC2 N237 expressed in COS-7 cells (Fig. 4C, top). Moreover, cilioD did not disrupt the SV40-dynein interaction (Fig. 4D), indicating that impairing the ATPase activity of the dynein motor did not affect cargo binding. Together, these findings demonstrate that upon reaching the cytosol from the ER, SV40 recruits the dynein motor.
FIG 4.
Dynein associates with cytosol-localized SV40. (A) A whole-cell extract derived from COS-7 cells expressing IC2SNAP-3×FLAG was subjected to a pulldown assay using either control magnetic beads (mb) or benzylguanine-conjugated magnetic beads (mb-BG). The precipitated material and the input were subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. AP, affinity purification. (B) The experimental setup was similar to that for panel A, except that DNA was extracted from the material pulled down using either mb or mb-BG. The extracted material was subjected to PCR amplification of the SV40 genome (corresponding to the TAg), and the product was analyzed by agarose gel electrophoresis. (C) CV-1 cells expressing GFPIC2 N237 were infected with SV40 (MOI, ∼25) (lane marked “+”). After 16 hpi, cells were treated with the DSP cross-linker, and the cytosol-localized SV40 fraction was isolated. From this fraction, SV40 was immunoprecipitated (IP) using an antibody against VP1. Bound proteins and the input were analyzed by immunoblotting using the indicated antibodies. (D) COS-7 cells expressing IC2SNAP-3×FLAG were treated with either DMSO or 100 μM cilioD. At 16 hpi, the cells were harvested, and IC2 was affinity purified and was analyzed as described for panel A.
Dynein disassembles cytosol-localized SV40.
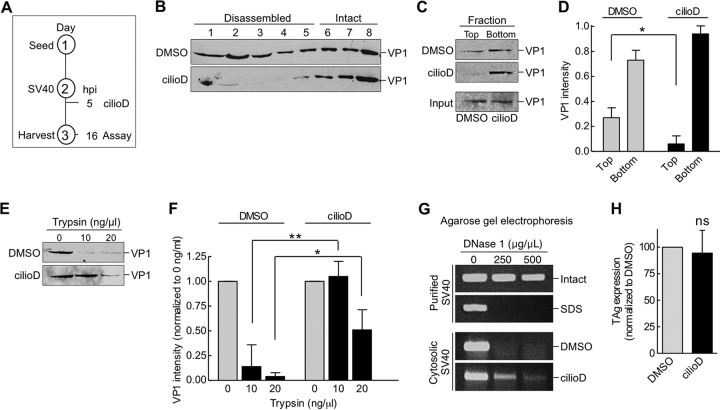
We next investigated the functional consequence of the SV40-dynein interaction. By discontinuous sucrose density sedimentation, we demonstrated previously that SV40 is disassembled in the cytosol (11, 13), a reaction that likely enables the resulting disassembled virus to gain entry into the nucleus (28–30). To test if dynein promotes the disassembly of SV40, we used three approaches. First, cytosol-localized SV40 was layered over a discontinuous sucrose gradient, and the sample was centrifuged. Large and intact viral particles sediment to the bottom fractions, whereas smaller viral particles, representing disassembled virus, remain on the top fractions. Using this method in the context of the experimental setup depicted in Fig. 5A, we found that cytosol-localized SV40 derived from control cells appeared throughout the gradient (Fig. 5B, top), with the virus in fractions 1 to 5 corresponding to disassembled virus and the virus in fractions 6 to 8 representing large and intact SV40, in agreement with our previous analysis (11, 13). However, when cytosol-localized SV40 derived from cilioD-treated cells was analyzed, the level of virus in fractions 1 to 5 decreased (Fig. 5B). These results indicate that disrupting dynein activity prevents SV40 disassembly.
FIG 5.
Dynein disassembles cytosol-localized SV40. (A) Depiction of the experimental setup used in panels B to F. (B) CV-1 cells were infected with SV40, and at 5 hpi, cells were treated with DMSO or 100 μM cilioD. At 16 hpi, cells were harvested, and SV40 in the cytosolic fraction was isolated. The cytosolic fraction was layered over a discontinuous 20%-to-40% sucrose gradient. After centrifugation, fractions were collected and were analyzed for the presence of VP1 by immunoblotting. (C) The cytosolic fractions analyzed in panel B were layered over a 20% sucrose cushion and were centrifuged, and the top and bottom fractions were collected. (D) The relative intensities of the VP1 bands shown in panel C were quantified. Values are averages of the means (n = 3) ± SD. *, P < 0.05. (E) The cytosolic fractions analyzed in panel B were treated with trypsin at the indicated concentrations. Samples were subjected to SDS-PAGE, followed by immunoblotting with an antibody against VP1. (F) The relative intensities of the VP1 bands shown in panel E were quantified. Data were normalized to those for DMSO (control). Values are averages of the means (n = 3) ± SD; *, P < 0.05; **, P < 0.01. (G) Intact and SDS-treated SV40, and the cytosol-localized SV40 shown in panel B, were incubated with increasing concentrations of DNase I. DNA was subjected to PCR amplification and was analyzed by agarose gel electrophoresis. (H) CV-1 cells transfected with the SV40 genome were treated with DMSO or 100 μM cilioD. At 24 hpi, cells were fixed and were immunostained for TAg. Values are averages of the means (n = 3) ± SD; ns, not significant.
As a second approach, cytosol-localized SV40 (from control DMSO-treated cells or cilioD-treated cells) were layered on a 20% sucrose cushion, and after centrifugation, the top and bottom fractions were collected and analyzed. In this assay, the virus in the top fraction corresponds to disassembled virus, whereas that in the bottom fraction represents large and intact virus. Again, we found that cilioD decreased the level of SV40 in the top fraction (Fig. 5C; quantified in Fig. 5D), further supporting the hypothesis that dynein disassembles SV40.
Using a third independent strategy, we assessed the structural state of the virus by limited proteolysis. In this assay, a conformationally altered viral particle displays more sensitivity to proteolytic digestion than an intact particle. When cytosol-localized SV40 was isolated from control DMSO-treated CV-1 cells according to the experimental protocol depicted in Fig. 5A, and the virus was treated with increasing concentrations of trypsin, VP1 was efficiently degraded (Fig. 5E; the VP1 intensity is quantified in Fig. 5F). In contrast, in cilioD-treated cells, the cytosol-localized virus displayed more resistance to trypsin digestion than the virus in the control sample (Fig. 5E; the VP1 intensity is quantified in Fig. 5F). Thus, impairing dynein activity renders the virus more structurally stable and therefore less accessible to the action of protease. These findings are consistent with those of the sucrose density sedimentation analyses, suggesting that dynein imparts conformational changes to SV40, leading to virus disassembly.
One possible consequence of dynein-mediated SV40 disassembly is exposure of the virus genome hidden in the native viral particle. To test this possibility, we asked if the viral genome is sensitive to DNase digestion, and we used PCR-based gene amplification to detect the presence or absence of the genome. As a control, we found that purified SV40 was resistant to increasing concentrations of DNase treatment, whereas SV40 treated with SDS (to artificially induce virus disassembly) was highly sensitive to digestion (Fig. 5G), as expected. When cytosol-localized SV40 was incubated with DNase, it was highly sensitive to digestion (Fig. 5G, DMSO), indicating that the virus genome is exposed in the cytosol. Importantly, when cytosol-localized SV40 was isolated from cilioD-treated cells, the virus genome became less sensitive to DNase digestion (Fig. 5G). These results suggest that dynein-mediated disassembly of SV40 plays a role in exposing the virus genome in the cytosol.
Our results suggest that using cilioD to inhibit the activity of dynein impairs SV40 disassembly. We posit that this step is essential in the delivery of a subviral particle harboring the genome into the nucleus to enable large T antigen expression. To ensure that cilioD is not affecting additional steps between SV40 disassembly in the cytosol and T antigen expression in the nucleus, we transfected the SV40 DNA genome and found that T antigen expression was unaffected by cilioD (Fig. 5H), indicating that dynein-induced disassembly of SV40 is likely the step targeted by cilioD that limits the delivery of the genome into the nucleus.
DISCUSSION
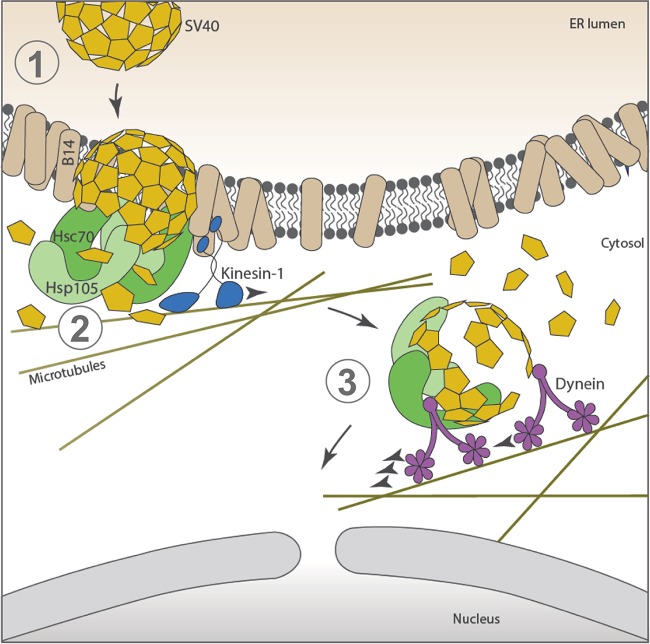
Like other nonenveloped viruses, PyVs undergo stepwise disassembly in order to enter host cells and cause infection. This study demonstrates how a PyV exploits the activity of a molecular motor to disassemble the viral particle, a step that is essential for productive infection. Placing our findings in context, the archetype SV40 infects cells by trafficking from the plasma membrane to the ER. In this compartment, protein disulfide isomerase (PDI) family members trigger the first step of structural changes to SV40, generating a hydrophobic viral particle that is integrated into the ER membrane (Fig. 6, step 1) (6–9). Selective ER membrane proteins, including transmembrane J-proteins, such as B14 (which recruits cytosolic chaperones, including Hsc70 and Hsp105), are reorganized within the ER lipid bilayer at distinct sites called foci in a process that depends on the molecular motor kinesin-1 (14, 25). At the foci, the cytosolic chaperones extract the membrane-embedded SV40 from the ER and move it into the cytosol, a step that is likely coupled to uncoating of the virus (Fig. 6, step 2) (13, 25). Importantly, our analyses here revealed that upon reaching the cytosol, SV40 recruits dynein, whose motor activity is hijacked to drive viral disassembly (Fig. 6, step 3). The final product of this series of events is a disassembled virus that is now poised and competent to cross the nuclear pore complex (NPC) and enter the nucleus.
FIG 6.
Dynein binds to and disassembles cytosol-localized SV40. To infect cells, SV40 traffics in a retrograde manner from the cell surface to reach the ER. Here, PDI family members induce conformational changes to SV40, creating a hydrophobic viral particle that inserts itself into the ER membrane (step 1). Specific ER membrane proteins, including transmembrane J-proteins such as B14 (which recruits cytosolic chaperones, including Hsc70 and Hsp105), are reorganized within the lipid bilayer of the ER at distinct sites called foci in a kinesin-1-dependent manner. At the foci, the cytosolic chaperones Hsc70 and Hsp105 extract the membrane-integrated SV40 from the ER and move it into the cytosol in a reaction that is likely linked to the uncoating of the virus (step 2). In this study, we found that when SV40 reaches the cytosol, it recruits the dynein motor to drive the disassembly of the virus (step 3). These concerted reactions generate a disassembled virus that can be transported across the NPC, entering the nucleus to cause infection.
How might the dynein-mediated viral disassembly reaction be important for nuclear entry? One possibility is that this reaction generates a disassembled viral particle containing a partially exposed genome that can more easily fit through the NPC (31). In fact, previous studies have suggested that SV40 gains nuclear entry via the NPC (28, 30). Alternatively, dynein-mediated disassembly may expose the nuclear localization signals (NLS) present in VP2 and VP3 (32, 33), which have been shown to target the virus to the NPC (34, 35). Obviously, these two possibilities do not have to be mutually exclusive.
After dynein-dependent disassembly, the resulting disassembled SV40 particle must be transported to the NPC in order to reach the nucleus. It is conceivable that dynein also carries out this transport function. Whether viral disassembly occurs first in the cytosol, followed by subsequent transport of the disassembled virus to the NPC, or whether SV40 is first targeted to the NPC, where viral disassembly ensues, remains unknown. In this regard, it is noteworthy that during the entry of nonenveloped adenovirus into the nucleus, the virus is transported by dynein to the NPC, where it is disassembled via the action of the kinesin-1 motor (36).
An important question raised by our study is how dynein disassembles SV40. We envision three scenarios. First, multiple dynein motors may simultaneously bind to one single viral particle, with some motors moving with a greater velocity than others (37). Hence, although different dynein motors move toward the same negative end of the microtubule, the difference in velocity between them generates a “drag and tug” on the viral particle, which causes it to disassemble. Second, a kinesin motor might interact with the same viral particle as the dynein motor. Because kinesin is a plus-end-directed microtubule motor that typically carries cargoes in the direction opposite that of dynein, the opposing force generated in this situation would cause viral disassembly. Third, it is possible that cytosol-localized SV40 is physically anchored to an unidentified cellular component, and the unidirectional pulling of the virus by dynein would create a mechanical stress that would disassemble the viral particle. Further experiments are clearly required to clarify these possibilities.
Our mass spectrometry data (Table 1) pinpointed subunits of the dynein motor (heavy and intermediate chains) and a dynactin subunit as SV40-interacting proteins. This list also identified two dynein activators (nudE and girdin) and a binding partner of the dynein adaptor BICD2, called RanBP2, as potential interacting partners of cytosol-localized SV40. Because these components regulate dynein's activity in cells (38), they might also mediate the interaction and disassembly of SV40 during entry. On the other hand, they may play a different role during SV40 entry, possibly in transporting the virus to the NPC. A systematic approach is required to delineate the precise role of these factors, if any, during SV40 infection.
Finally, our results demonstrated that the use of dynein during SV40 infection extends to other PyV family members, because impairing dynein function similarly disrupted human BK virus infection. This is in contrast to the suggestion in a previous report that dynein does not play a significant role in BK virus entry (39). The reason for this discrepancy is not clear, but it could be due to the different dynein inhibitor or cell type used. In agreement with our study, dynein was shown to be important during cell entry of the murine PyV (40), although it is not known whether this motor acts to disassemble the virus. Beyond the PyV family, other viruses, including human immunodeficiency virus type 1 (41, 42), influenza A virus (43), rhinoviruses (44), herpes simplex virus (45), and murine leukemia virus (46), exploit dynein's function to support some aspect of their entry processes. Thus, our study further underscores the concept that exploiting cellular motors is a common strategy used by viruses during infection.
MATERIALS AND METHODS
Cell lines and reagents.
CV-1, COS-7, and BSC-1 cells were purchased from the ATCC. Cells were grown in complete Dulbecco's modified Eagle medium (cDMEM, comprising 10% fetal bovine serum, 10 U/ml penicillin, and 10 μg/ml streptomycin [Gibco, Grand Island, NY]). Opti-MEM and 0.25% trypsin-EDTA were purchased from Gibco. The generation of the GFPIC2 N237 plasmid is described in reference 19, and the source of IC2SNAP-3×FLAG and GFPp62HALO-3×FLAG is Michael Cianfrocco (University of Michigan).
Chemicals and antibodies.
Nocodazole, Triton X-100, dithiothreitol (DTT), and phenylmethanesulfonyl fluoride (PMSF) were purchased from Sigma; SNAP capture magnetic beads, from New England BioLabs (Ipswich, MA); protein G-conjugated magnetic beads and dithiobis(succinimidyl propionate) (DSP), from Thermo Fisher (Rockford, IL); ciliobrevin D and digitonin, from EMD Millipore (San Diego, CA). The antibodies used in this study are listed in Table 2.
TABLE 2.
Antibodies used in this study
| Antibodya | Dilutionb | Source (product no.) |
|---|---|---|
| SV40 large TAg, mouse monoclonal | 1:500; 1:100 (IF) | Santa Cruz (sc147) |
| Hsp90, rabbit polyclonal | 1:3,000 | Santa Cruz (sc7947) |
| BAP31, rat monoclonal | 1:500 (IF) | Pierce (MA3-002) |
| VP1, mouse monoclonal | 1:2,000; 1:500 (IF) | Walter Scott, University of Miami |
| VP2/3, rabbit polyclonal | 1:2,000; 1:5,00 (IF) | Abcam (ab53983) |
| PDI, mouse monoclonal | 1:10,000 | Abcam (ab2792) |
| GFP, mouse monoclonal | 1:10,000 | Proteintech (66002-1-IG) |
| Hsp105, rabbit polyclonal | 1:2,500 | Santa Cruz (sc6241) |
| Hsc70, rabbit polyclonal | 1:2,500 | Pierce (PA5-27337) |
| BK virus large TAg, mouse monoclonal | 1:100 | Millipore (PAb416) |
| Dynein IC1/2, mouse monoclonal | 1:5,000 | Santa Cruz (sc13524) |
| Dynein HC, mouse monoclonal | 1:500 | Santa Cruz (sc514579) |
| Rabbit IgG, goat (HRP) | 1:3,000 | Sigma (A4914) |
| Mouse IgG, goat (HRP) | 1:3,000 | Sigma (A4416) |
| Rat IgG, goat (HRP) | 1:3,000 | Sigma (A5795) |
| Rat IgG, donkey polyclonal (Alexa Fluor 594) | 1:1,000 | Invitrogen (A21209) |
| Mouse IgG, goat polyclonal (Alexa Fluor 350) | 1:250 | Invitrogen (A11045) |
| Rabbit IgG, goat polyclonal (Alexa Fluor 488) | 1:1,000 | Invitrogen (A11008) |
Given as targeted protein, antibody type (label).
IF, immunofluorescence.
Preparation of SV40.
SV40 was purified as described previously (11). Briefly, CV-1 cells were transfected with pUCSV40 encoding the SV40 genome (GenBank accession no. J02400.1), a gift from Hiroshi Handa, Tokyo Medical University. Cells were harvested and were lysed in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5% Brij 58 for 30 min on ice, and the supernatant was collected after centrifugation at 20,000 × g for 10 min. A discontinuous 20% and 40% OptiPrep gradient (60% stock solution of iodixanol in water; Sigma) was prepared, and the supernatant was placed on top of the gradient. Tubes were centrifuged at 49,500 rpm for 2 h at 4°C in an SW55 Ti rotor (Beckman Coulter, Indianapolis, IN). A white interface that formed between 20% and 40% OptiPrep was collected, and aliquots were stored at −80°C for future use. Purified BK virus and an antibody against BK virus large TAg (pAB416) were generous gifts from Michael Imperiale (University of Michigan).
Immunopurification and identification of cytosolic SV40 binding partners.
Near-confluent CV-1 cells were infected with SV40 (multiplicity of infection [MOI], ∼25) for 16 h. Cells treated at 0 hpi with 5 μg/μl BFA were used as a negative control. Postinfection, cells were harvested with cold phosphate-buffered saline (PBS; Gibco) and were centrifuged at 500 × g for 5 min. Washed cells were incubated with freshly prepared DSP at 2 mM for 30 min at room temperature with intermittent shaking. This membrane-permeant, amine-reactive, and thiol-cleavable cross-linker was used to stabilize transient or weak protein-protein interactions. Excess cross-linker was quenched with 200 mM Tris (pH 7.5). Cells were then permeabilized with 0.05% digitonin in HNP buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, and 1 mM PMSF) at 4°C for 15 min. The cell lysate was clarified by centrifugation at 16,100 × g for 10 min at 4°C. The resulting supernatant was incubated with anti-VP1 for 3 h at 4°C in an end-to-end rotor. Samples were incubated with protein G-conjugated magnetic beads for 2 h at 4°C. Beads were washed three times with HNP buffer, and samples were boiled for 10 min at 95°C before being subjected to SDS-PAGE and silver staining (Invitrogen). For mass spectrometry analysis, the entire lane representing all proteins was excised from the silver-stained gel and was analyzed by “shotgun” mass spectrometry at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School). Host proteins with at least 3 unique peptides and 2-fold enriched compared to the BFA-treated sample were considered potential SV40-interacting proteins (see the supplemental material).
DNA transfection.
For transfection in CV-1 cells, 50% confluent cells in 6-well-plate dishes (diameter, 6 cm or 15 cm) were transfected with plasmid using the FuGENE HD (Promega, Madison, WI) transfection reagent at a ratio of 1:4, wt/vol (plasmid to transfection reagent). Cells were allowed to express the protein for at least 24 h before experimentation. For COS-7 cells, polyethylenimine (PEI; Polysciences, Warrington, PA) was used as the transfection reagent.
Immunofluorescence microscopy.
CV-1 cells grown on sterile coverslips were infected with SV40 at an MOI of ∼1 (for TAg expression studies) or ∼20 (for focus formation studies) for 24 h or 16 h, respectively. Infected cells were fixed with 1% formaldehyde for 15 min at room temperature, followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min. Cells were then blocked with blocking buffer containing 5% milk in TBST (Tris-buffered saline with 0.02% Tween 20) for 15 min. Immunostaining was performed with a primary antibody diluted in a blocking buffer for 1 h at room temperature, followed by five washes with the blocking buffer. Cells were incubated with a fluorescent-dye-conjugated secondary antibody for 30 min at room temperature. Cells were subsequently washed three times with blocking buffer, PBS, and water before air drying and mounting on glass slides (Fisher) using ProLong gold (Invitrogen) with or without 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, OR). Slides were allowed to dry in the dark at room temperature for at least 12 h before imaging. Images were taken using an inverted epifluorescence microscope (Nikon Eclipse TE2000-E; Nikon, Melville, NY) equipped with 10×, 40×, 60×, and 100× objectives (numerical aperture [NA], 1.40) and standard DAPI (blue), fluorescein isothiocyanate (FITC) (green), and tetramethyl rhodamine isocyanate (TRITC) (red) filter cubes. Images were processed using ImageJ software, version 1.48i (NIH). At least 100 cells were scored for each experiment.
Semipermeabilized-membrane cytosol arrival assay.
The semipermeabilized-membrane cytosol arrival assay was performed as described previously (11, 12) with minor modifications. Briefly, CV-1 cells were prechilled at 4°C for 20 min before infecting with SV40 (MOI, ∼5) for 2 h at 4°C. Cells were washed and incubated at 37°C. After 5 hpi, cells were treated with cilioD or the DMSO control. Postinfection, cells were permeabilized in HNP buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, and 1 mM PMSF) containing 0.05% digitonin at 4°C for 10 min. The sample was centrifuged at 16,100 × g for 10 min at 4°C, and the supernatant (cytosol) and pellet (membrane) fractions were collected. ER-localized SV40 was isolated by further treatment of the pellet fraction with HNP buffer containing 1% Triton X-100 for 10 min at 4°C, followed by centrifugation at 16,100 × g for 10 min at 4°C. The resulting supernatant fraction was analyzed by reducing SDS-PAGE.
Binding assays.
Cells expressing the transfected proteins were harvested, and the cell pellets were washed three times with cold PBS. Cells were then lysed with 0.05% digitonin in an HNP buffer at 4°C for 15 min. The cell lysate was clarified by centrifugation at 16,100 × g for 10 min at 4°C. The resulting supernatant was incubated with an antibody against SV40 VP1 overnight at 4°C in an end-to-end rotor. Samples were incubated with protein G-conjugated magnetic beads for 2 h at 4°C. Beads were washed three times with an HNP buffer, and samples were eluted with 1× SDS sample buffer containing 1.25% β-mercaptoethanol (Sigma) and were boiled for 10 min at 95°C before being subjected to SDS-PAGE and immunoblotting. For the purification of SNAP-tagged protein, washed cells were incubated with freshly prepared DSP at 2 mM for 30 min at room temperature with intermittent shaking. Excess cross-linker was quenched with 200 mM Tris (pH 7.5). Cells were lysed with 1% Triton X-100 in an HNP buffer containing 1 mM DTT. The lysis supernatant was incubated with prewashed benzylguanine (BG)-conjugated magnetic beads (mb) for an hour at 37°C. Beads were washed three times with 0.1% Triton X-100 in a buffer containing the reducing agent TSEP [Tris(2-carboxyethyl)phosphine]. Samples were eluted and processed as described above before being subjected to SDS-PAGE and immunoblotting.
Disassembly assay.
Cytosolic fractions were isolated as described above, layered on top of a discontinuous 40%, 30%, and 20% sucrose gradient, and subjected to centrifugation at 50,000 rpm for 30 min at 4°C. From the top, 25-μl fractions were sequentially collected and subjected to SDS-PAGE and immunoblotting. For the sucrose cushion assay, samples were layered over a 20% sucrose solution and were centrifuged, and two fractions corresponding to the top and bottom layers were subjected to SDS-PAGE and immunoblotting.
Protease and DNase sensitivity assays.
Protease sensitivity assays were performed by treating cytosolic fractions with different concentrations of trypsin (Sigma) at 4°C for 30 min before subjecting the fractions to SDS-PAGE and immunoblotting. For DNase sensitivity assays, the virus was incubated with various concentrations of DNase I (Roche, Switzerland) for 30 min at room temperature. DNA was then isolated and was amplified using PCR with primers to the SV40 genome (Fwd, 5′-GCAGTAGCAATCAACCCACA-3′; Rev, 5′-CTGACTTTGGAGGCTTCTGG-3′).
Statistical analysis.
Data presented in figures are averages of the mean values from at least three independent experiments (n ≥ 3), and error bars represent standard deviations (SD). Data were plotted using GraphPad Prism software, version 5.0b. A two-tailed Student t test was performed where indicated to compare experimental data sets to the control. Values marked with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) were considered to be significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Tsai and Verhey labs, especially Martin Engelke and Jeremy Welsch, for discussions and suggestions throughout this work.
This work was supported by NIH grants R01AI064296 and R01GM113722 to B.T. and R01GM070862 to K.J.V.
M.S.R., C.C.S., K.J.V., and B.T. conceived and designed the experiments. M.S.R. and C.C.S. performed the experiments. M.S.R., C.C.S., and B.T. analyzed the data. M.S.R., C.C.S., K.J.V., and B.T. contributed reagents/materials/analysis tools. M.S.R., C.C.S., K.J.V., and B.T. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00353-18.
REFERENCES
- 1.White MK, Gordon J, Khalili K. 2013. The rapidly expanding family of human polyomaviruses: recent developments in understanding their life cycle and role in human pathology. PLoS Pathog 9(3):e1003206. doi: 10.1371/journal.ppat.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stehle T, Gamblin SJ, Yan Y, Harrison SC. 1996. The structure of simian virus 40 refined at 3.1 Å resolution. Structure 4:165–182. doi: 10.1016/S0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 3.Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 4.Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, Helenius A. 2011. Role of endosomes in simian virus 40 entry and infection. J Virol 85:4198–4211. doi: 10.1128/JVI.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian M, Cai D, Verhey KJ, Tsai B. 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog 5(6):e1000465. doi: 10.1371/journal.ppat.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rainey-Barger EK, Magnuson B, Tsai B. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol 81:12996–13004. doi: 10.1128/JVI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Kartenbeck J, Stukenbrok H, Helenius A. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol 109(6 Part 1):2721–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, DiMaio D. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2(3):e00101-11. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanning E, Zhao K. 2009. SV40 DNA replication: from the A gene to a nanomachine. Virology 384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue T, Tsai B. 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog 7(5):e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- 13.Ravindran MS, Bagchi P, Inoue T, Tsai B. 2015. A non-enveloped virus hijacks host disaggregation machinery to translocate across the endoplasmic reticulum membrane. PLoS Pathog 11(8):e1005086. doi: 10.1371/journal.ppat.1005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravindran MS, Engelke MF, Verhey KJ, Tsai B. 2017. Exploiting the kinesin-1 molecular motor to generate a virus membrane penetration site. Nat Commun 8:15496. doi: 10.1038/ncomms15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupzyk A, Williams JM, Bagchi P, Inoue T, Tsai B. 2017. SGTA-dependent regulation of Hsc70 promotes cytosol entry of simian virus 40 from the endoplasmic reticulum. J Virol 91:e00232-17. doi: 10.1128/JVI.00232-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashok A, Atwood WJ. 2003. Contrasting roles of endosomal pH and the cytoskeleton in infection of human glial cells by JC virus and simian virus 40. J Virol 77:1347–1356. doi: 10.1128/JVI.77.2.1347-1356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O'Donnell M, Gelfand VI, Kapoor TM, Chen JK. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damm EM, Pelkmans L, Kartenbeck J, Mezzacasa A, Kurzchalia T, Helenius A. 2005. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J Cell Biol 168:477–488. doi: 10.1083/jcb.200407113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. 2003. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell 14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Foster HE, Rondelet A, Lacey SE, Bahi-Buisson N, Bird AW, Carter AP. 2017. Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell 169:1303–1314. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97–107. doi: 10.1016/S0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 24.Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. 2012. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell 23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walczak CP, Ravindran MS, Inoue T, Tsai B. 2014. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog 10(3):e1004007. doi: 10.1371/journal.ppat.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redwine WB, DeSantis ME, Hollyer I, Htet ZM, Tran PT, Swanson SK, Florens L, Washburn MP, Reck-Peterson SL. 2017. The human cytoplasmic dynein interactome reveals novel activators of motility. Elife 6:e28257. doi: 10.7554/eLife.28257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiger R, Luisoni S, Johnsson K, Greber UF, Helenius A. 2013. Investigating endocytic pathways to the endoplasmic reticulum and to the cytosol using SNAP-trap. Traffic 14:36–46. doi: 10.1111/tra.12018. [DOI] [PubMed] [Google Scholar]
- 28.Kuksin D, Norkin LC. 2012. Disassociation of the SV40 genome from capsid proteins prior to nuclear entry. Virol J 9:158. doi: 10.1186/1743-422X-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butin-Israeli V, Ben-nun-Shaul O, Kopatz I, Adam SA, Shimi T, Goldman RD, Oppenheim A. 2011. Simian virus 40 induces lamin A/C fluctuations and nuclear envelope deformation during cell entry. Nucleus 2:320–330. doi: 10.4161/nucl.2.4.16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi A, Li PP, Qu Q, Jafri QH, Kasamatsu H. 2007. Molecular dissection of nuclear entry-competent SV40 during infection. Virus Res 124:226–230. doi: 10.1016/j.virusres.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. 2015. Structure and gating of the nuclear pore complex. Nat Commun 6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gharakhanian E, Takahashi J, Kasamatsu H. 1987. The carboxyl 35 amino acids of SV40 Vp3 are essential for its nuclear accumulation. Virology 157:440–448. doi: 10.1016/0042-6822(87)90286-8. [DOI] [PubMed] [Google Scholar]
- 33.Nakanishi A, Shum D, Morioka H, Otsuka E, Kasamatsu H. 2002. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J Virol 76:9368–9377. doi: 10.1128/JVI.76.18.9368-9377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada M, Kasamatsu H. 1993. Role of nuclear pore complex in simian virus 40 nuclear targeting. J Virol 67:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clever J, Yamada M, Kasamatsu H. 1991. Import of simian virus 40 virions through nuclear pore complexes. Proc Natl Acad Sci U S A 88:7333–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. 2011. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 10:210–223. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Levi V, Serpinskaya AS, Gratton E, Gelfand V. 2006. Organelle transport along microtubules in Xenopus melanophores: evidence for cooperation between multiple motors. Biophys J 90:318–327. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriyama T, Sorokin A. 2008. Intracellular trafficking pathway of BK virus in human renal proximal tubular epithelial cells. Virology 371:336–349. doi: 10.1016/j.virol.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zila V, Difato F, Klimova L, Huerfano S, Forstova J. 2014. Involvement of microtubular network and its motors in productive endocytic trafficking of mouse polyomavirus. PLoS One 9(5):e96922. doi: 10.1371/journal.pone.0096922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawlica P, Berthoux L. 2014. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, Matthias P, Helenius A, Yamauchi Y. 2014. Influenza A virus uses the aggresome processing machinery for host cell entry. Science 346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 44.Ganjian H, Zietz C, Mechtcheriakova D, Blaas D, Fuchs R. 2017. ICAM-1 binding rhinoviruses enter HeLa cells via multiple pathways and travel to distinct intracellular compartments for uncoating. Viruses 9(4):E68. doi: 10.3390/v9040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye GJ, Vaughan KT, Vallee RB, Roizman B. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J Virol 74:1355–1363. doi: 10.1128/JVI.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opazo T, Garces A, Tapia D, Barraza F, Bravo A, Schwenke T, Cancino J, Arriagada G. 2017. Functional evidence of the involvement of the dynein light chain DYNLRB2 in murine leukemia virus infection. J Virol 91:e00129-17. doi: 10.1128/JVI.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.