ABSTRACT
Analytic treatment interruption (ATI) studies are required to evaluate strategies aimed at achieving ART-free HIV remission, but the impact of ATI on the viral reservoir remains unclear. We validated a DNA size selection-based assay for measuring levels of integrated HIV DNA and applied it to assess the effects of short-term ATI on the HIV reservoir. Samples from participants from four AIDS Clinical Trials Group ATI studies were assayed for integrated HIV DNA levels. Cryopreserved peripheral blood mononuclear cells (PBMCs) were obtained for 12 participants with available samples pre-ATI and approximately 6 months after ART resumption. Four participants also had samples available during the ATI. The median duration of ATI was 12 weeks. Validation of the HIV integrated DNA size-exclusion (HIDE) assay was performed using samples spiked with unintegrated HIV DNA, HIV-infected cell lines, and participant PBMCs. The HIDE assay eliminated 99% of unintegrated HIV DNA species and strongly correlated with the established Alu-gag assay. For the majority of individuals, integrated DNA levels increased during ATI and subsequently declined upon ART resumption. There was no significant difference in the levels of integrated HIV DNA between the pre- and post-ATI time points, with a median ratio of post- to pre-ATI HIV DNA levels of 0.95. Using a new integrated HIV DNA assay, we found minimal change in the levels of integrated HIV DNA in participants who underwent an ATI, followed by 6 months of ART. This suggests that short-term ATI can be conducted without a significant impact on the levels of integrated proviral DNA in the peripheral blood.
IMPORTANCE Interventions aimed at achieving sustained antiretroviral therapy (ART)-free HIV remission require treatment interruption trials to assess their efficacy. However, these trials are accompanied by safety concerns related to the expansion of the viral reservoir. We validated an assay that uses an automated DNA size-selection platform for quantifying levels of integrated HIV DNA and is less sample- and labor-intensive than current assays. Using stored samples from AIDS Clinical Trials Group studies, we found that short-term ART discontinuation had minimal impact on integrated HIV DNA levels after ART resumption, providing reassurance about the reservoir effects of short-term treatment interruption trials.
KEYWORDS: assay, HIV, integrated DNA, reservoir, treatment interruption
INTRODUCTION
Antiretroviral therapy (ART) is effective in maintaining viral suppression but cannot eradicate the viral reservoir (1, 2). After ART initiation, the HIV reservoir size dramatically declines during the first year of treatment, but the decay rate subsequently slows with largely stable HIV DNA levels despite more than a decade of ART (3, 4). Consequently, the cessation of ART generally results in rapid viral rebound (5).
Strategies aimed at achieving sustained ART-free remission will ultimately require efficacy testing using analytic treatment interruption (ATI) studies. However, there are a number of safety concerns associated with ATI trials, including concern over whether short-term treatment interruption leads to irreversible reseeding and expansion of the HIV reservoir (6, 7). Given the growing interest in treatment interruption studies, addressing these concerns has implications on the safety and design of future ATI trials.
The assessment of the HIV reservoir changes in treatment interruption trials would benefit from a scalable assay that can quantify levels of integrated HIV DNA, especially as unintegrated forms of HIV DNA comprise a major proportion of total HIV DNA during ATI and for a period after ART reinitiation (8). However, the traditional Alu-gag integrated HIV DNA assay is challenging to perform and dependent on the presence of Alu elements in close proximity to the HIV integration sites (9–11). For these reasons, we validated a novel assay for measuring HIV integrated DNA levels that avoids these drawbacks. This assay uses a fully automated platform for the size-selection of genomic DNA to eliminate unintegrated HIV DNA, followed by quantitative PCR (qPCR) measurements of integrated DNA levels based on a previously proposed method (12). We termed this the HIV integrated DNA size-exclusion (HIDE) assay and used it to assess changes in the levels of integrated HIV DNA in participants of AIDS Clinical Trials Group (ACTG) ATI trials.
RESULTS
Participant characteristics.
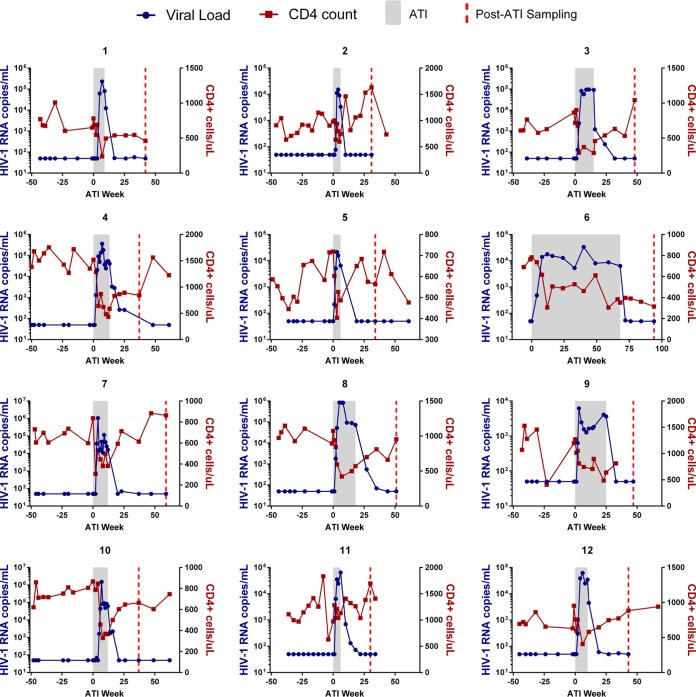
The 12 ACTG ATI participants had been on ART for a median of 3.9 years with a median CD4 count of 852 cells/mm3 (Table 1). All participants were on an ART regimen for at least 1 year prior to ATI, experienced viral rebound within 5 weeks of treatment interruption, and subsequently achieved viral suppression upon ART resumption (Fig. 1). Four participants had available samples during ATI at a median of 12 weeks after ART discontinuation and with a median plasma viral load of 51,664 copies/ml.
TABLE 1.
Study participant information
| Participant | ART duration prior to ATI (yrs) | ATI duration (wks) | ART duration post-ATI (wks) | Infection phase upon ART start | Plasma viral load prior to ATI (copies/ml) | CD4+ count prior to ATI (per mm3) |
|---|---|---|---|---|---|---|
| 1 | 8.7 | 9 | 33 | Chronic-treated | <50 | 780 |
| 2 | 1.1 | 6 | 25 | Early-treated | <50 | 979 |
| 3 | 3.7 | 16 | 33 | Chronic-treated | <50 | 718 |
| 4 | 7.2 | 13 | 24 | Chronic-treated | <50 | 1,520 |
| 5 | 1.1 | 6 | 28 | Early-treated | <50 | 719 |
| 6 | 3.2 | 68 | 26 | Chronic-treated | <50 | 783 |
| 7 | 12.1 | 12 | 47 | Chronic-treated | <50 | 836 |
| 8 | 6.4 | 18 | 33 | Chronic-treated | <50 | 1,073 |
| 9 | 6.2 | 25 | 22 | Chronic-treated | <50 | 1,270 |
| 10 | 3.3 | 12 | 25 | Chronic-treated | <50 | 867 |
| 11 | 1.0 | 6 | 24 | Early-treated | <50 | 972 |
| 12 | 4.1 | 10 | 33 | Chronic-treated | <50 | 758 |
| Median | 3.9 | 12.0 | 27.0 | 851.5 |
FIG 1.
Viral loads and CD4+ counts. Blue lines depict viral loads, and red lines depict CD4+ cell counts. The ATI period is shaded gray, and on-ART data are shown as white.
HIDE assay validation.
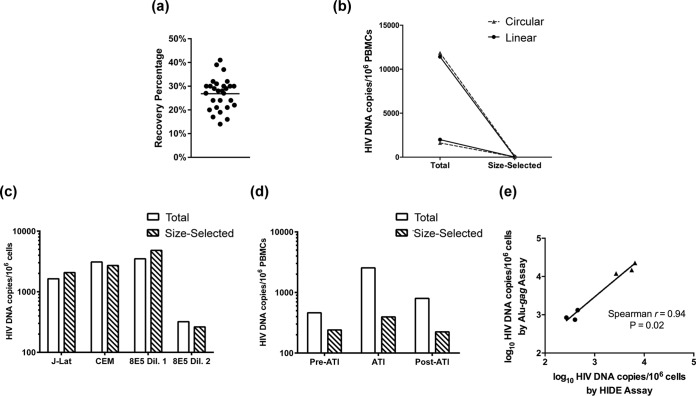
A median 28% of input DNA was recovered after automated size selection for DNA fragments larger than 20 kb (Fig. 2a). The HIDE assay eliminated approximately 99% of linear and circular unintegrated HIV DNA species at either high or low levels of spiked HIV DNA (Fig. 2b). We performed the HIDE assay on three cell lines containing integrated, replication-incompetent HIV DNA (Fig. 2c). The median ratio of HIV DNA levels before and after size selection in these cell lines was 1.07, demonstrating that the assay did not selectively eliminate HIV DNA. We performed concurrent measurements of total and size-selected HIV DNA in three participants. The levels of HIV DNA were higher in total versus size-selected DNA across all time points, but the difference was especially evident during the treatment interruption (Fig. 2d). In addition, in both cell lines and participant samples, the integrated HIV DNA levels measured by the HIDE assay correlated strongly with integrated DNA measurements by the Alu-gag method (Spearman r = 0.94, P = 0.02, Fig. 2e).
FIG 2.
HIDE assay validation. (a) DNA recovery percentage for each sample after 20-kb size selection on the BluePippin platform. (b) HIV DNA levels of uninfected DNA spiked with linear or circular HIV DNA before and after genomic DNA size selection. (c) HIV DNA levels in three cell lines (J-Lat, HIV-infected CEM, and two dilutions of 8E5) containing integrated, replication-deficient HIV provirus before and after size selection by HIDE assay. (d) Levels of total and size-selected HIV DNA in a representative participant (PID 4) at pre-ATI, during ATI, and post-ATI time points. (e) Correlation between integrated HIV DNA levels measured by the HIDE assay and by the Alu-gag assay for cell lines (triangles) and participant samples (circles).
Effect of short-term ATI on integrated HIV DNA levels.
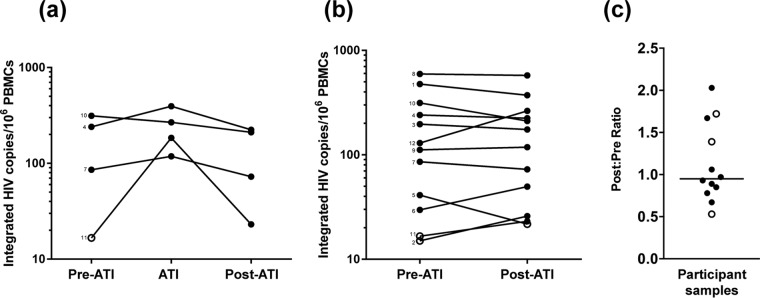
Participants with available samples were found to have a median increase in integrated HIV DNA levels during ATI of +94 HIV DNA copies/106 peripheral blood mononuclear cells (PBMCs), and the levels subsequently declined after ≥24 weeks of ART at a median of −109 HIV DNA copies/106 PBMCs (Fig. 3a). Among all 12 participants, there was no significant difference between the pre- and post-ATI time points in levels of HIV integrated DNA (median difference, −15 copies/106 PBMCs; Wilcoxon signed-rank P = 0.34; Fig. 3b). The median ratio of post- to pre-ATI HIV DNA levels was 0.95 (Q1, Q3 = 0.8, 1.6; Fig. 3c), which was not significantly different than a ratio of 1.0 (P = 0.79). Within the 42% of individuals that had higher HIV DNA levels post-ATI, the increase in integrated DNA levels was generally small, with a median change of 11 HIV DNA copies/106 PBMCs (Q1, Q3 = 7, 20 HIV DNA copies/106 PBMCs). There was no significant correlation between the duration of ATI and the ratio of post- to pre-ATI reservoir size.
FIG 3.
Treatment interruption had minimal effect on levels of integrated HIV DNA in PBMCs. (a) Levels of integrated HIV DNA pre-ATI, during treatment interruption, and post-ATI in four participants. Participant numbers are written next to the corresponding pre-ATI data point. (b) Pre- and post-ATI levels of integrated HIV DNA for 12 participants. For samples that are below the LOD, the LOD for that sample is used (open circle). (c) Ratio of post-ATI to pre-ATI levels of integrated HIV DNA. An open circle represents whether either the pre- or post-ATI time point was below the LOD. The line represents the median value.
DISCUSSION
In this study, we validated a new technique for quantifying levels of integrated HIV DNA, termed the HIDE assay. This technique uses a fully automated size selection of genomic DNA that is less sample- and labor-intensive than either the Alu-gag assay or manual gel-based selection of genomic DNA (8, 10, 13). We used this assay to assess the impact of short-term ATI on changes in integrated HIV DNA levels. The HIDE assay eliminated approximately 99% of unintegrated HIV DNA species, did not affect levels of integrated HIV DNA, and showed high correlation with the standard Alu-gag assay. We detected minimal change in the levels of integrated HIV DNA after short-term ATI that is followed by approximately 6 months of ART.
While quantifying intact full-length proviral numbers and the viral outgrowth assay are the gold standards for measuring the HIV reservoir, these assays are costly, labor- and time-intensive, dependent on large sample volumes, and challenging to implement for large-scale clinical studies (14–16). In HIV nonsuppressed individuals, it is difficult to interpret total HIV DNA levels given the presence of unintegrated forms of HIV DNA (8). In addition, unintegrated HIV DNA species can persist despite long-term ART, suggesting that measuring total HIV DNA, even in patients on long-term ART, may overestimate integrated HIV DNA levels (17, 18). Although assays for integrated HIV DNA do not differentiate between replication-competent and -incompetent proviruses, levels of integrated HIV DNA have been shown to strongly correlate with quantitative viral outgrowth assay (QVOA), suggesting that they can provide a valuable surrogate for the true reservoir size (14, 19). The HIDE assay overcomes two key hurdles intrinsic to the conventional Alu-gag assay: (i) the need for integrated DNA standards that are challenging to create and (ii) the reliance on the proximity of random Alu elements to the HIV provirus, requiring high numbers of replicates to account for the varying amplification efficiency (9, 20). By eliminating unintegrated DNA species and using only conserved HIV-specific primers, this assay, while using less replicates, can effectively measure both widely diverse and predominately clonal proviral populations (21). While our assay showed strong correlation with the Alu-gag assay, this analysis was performed on a limited set of participant samples given the amount of DNA needed to perform the Alu-gag assay. For participant samples, the conventional Alu-gag approach required 4- to 10-fold more DNA than the HIDE assay. The correlation may be driven by the difference in the participant samples and the cell lines used, but the trend remains if each subset is analyzed individually. This limitation highlights one of the barriers of using the Alu-gag assay in sample-limited settings or large clinical trials.
ATI trials remain an indispensable tool in the evaluation of strategies for HIV remission. However, there remains a concern among both potential participants and physicians about the possible reseeding or expansion of the HIV reservoir due to the ATI. A study by Montserrat et al. reported an increase in integrated HIV DNA levels after ATI that was not reversible with ART reinitiation (6). However, all participants in this study received repeated immunologic interventions, with the majority receiving infusions of HIV-pulsed dendritic cells. On the other hand, individuals included in our study were all selected from the nonintervention arm of their respective studies to avoid the potential confounding effects of any immunologic therapies. Using the HIDE assay, we found that in general, short-term ATI did not have a significant impact on the levels of integrated HIV DNA after ART resumption. Despite an increase in integrated DNA levels during the ATI, the viral reservoir subsequently was reduced to pre-ATI levels after approximately 6 months of ART, possibly suggesting that most newly infected cells during the ATI have relatively limited life spans. These findings suggest that short-term ATI studies can be conducted without an irreversible increase in integrated HIV DNA levels. Furthermore, several recent studies have reported results supporting our findings that the reservoir size does not significantly change after ART reinitiation as assessed by total HIV DNA, QVOA, and near-full-length sequencing (22–24). However, additional studies will be needed to confirm these findings among the tissue reservoirs of HIV.
In summary, this study validated the HIDE assay, an automated size-selection-based method to quantify levels of integrated HIV DNA. The HIDE assay showed that short-term ATI had minimal impact on levels of integrated HIV DNA after ART resumption. The HIDE assay could be a useful technique for evaluating the impact of strategies aimed at reducing the HIV reservoir, and our results provide a measure of reassurance on the possible reseeding of the HIV reservoir after short-term ATI.
MATERIALS AND METHODS
Study participants.
Cryopreserved PBMCs were obtained for 12 participants with available samples from previously completed ACTG ATI trials in the placebo arms (A5197 [25], A5068 [26], A371 [27], and A5170 [28]). The median duration of ATI was 12 weeks (Q1, Q3 = 7, 17). Samples were obtained at a time point immediately prior to ATI and a median of 27 weeks (Q1, Q3 = 24, 33) after ART reinitiation. Four participants had samples available during the ATI.
HIDE assay.
DNA was extracted from cryopreserved PBMCs using a QIAmp DNA minikit (Qiagen). For each sample, 10 μg of DNA was loaded into a 0.75% agarose gel cassette with an external S1 marker and size selected using a 20-kb high-pass protocol on a BluePippin pulsed-field gel electrophoresis system (Sage Science) to remove unintegrated viral DNA species. HIV DNA levels were quantified by qPCR with primer and probe targeting HIV LTR/gag and normalized to cellular input by qPCR targeting the CCR5 gene (21). The limit of detection (LOD) was calculated based on a 2 HIV DNA copies/well value adjusted to the cellular input per well.
HIDE assay validation.
To confirm the removal of unintegrated HIV DNA by the HIDE assay, we spiked HIV-negative DNA with either linear near-full-length HIV amplicons or a 12-kb HIV-encoding plasmid at various quantities. To ensure that measurements of integrated DNA did not change due to the size selection process, we also measured HIV DNA levels before and after size selection in three HIV-infected cell lines with integrated replication-defective HIV provirus. These included J-Lat cells (clone 9.2) containing an env-defective proviral copy, 8E5 cells containing an RT-defective proviral copy, and CEM cells infected with pseudotyped Δenv HIV virions allowing only one round of replication and propagated in vitro for 4 weeks (13). Prior to size selection and quantification, DNA extracted from the HIV-infected cell lines was diluted in HIV-negative DNA to approximate the HIV DNA levels in infected individuals. Integrated DNA levels in these cell lines and in cells from one participant were also assayed by the Alu-gag method (9, 11).
Statistical analysis.
Data analysis was performed using Prism 6 (GraphPad, La Jolla, CA). A Wilcoxon matched-pair signed-rank test was used in analysis of pre- and post-size selection values, as well as in analysis of pre- and post-ATI integrated DNA levels. The ratio of post- to pre-ATI integrated DNA levels was compared to the ratio of 1.0 by the Wilcoxon signed-rank test. The LOD value was used in the analysis for samples quantified below the LOD.
Ethics statement.
Written informed consent was provided by all study participants for use of stored samples in HIV-related research. This study was approved by the Partners Institutional Review Board.
ACKNOWLEDGMENTS
We are grateful for the contributions of the participants who made this study possible. We thank the staff and principal investigators of ACTG studies A371, A5068, A5170, and A5197. We thank Ronald Bosch and Evgenia Aga at the Harvard School of Public Health for help and advice. We appreciate the support of the MIT BioMicro Center and thank the Tsibris and Kuritzkes labs for their valuable feedback.
This study was supported in part by National Institute of Allergy and Infectious Diseases of the National Institutes of Health grants AI125109 (to J.Z.L.), AI068634 (Statistical and Data Management Center of the AIDS Clinical Trials Group), AI068636 (AIDS Clinical Trials Group), UM1 AI106701, UM1 AI126617 (to U.O.), AI120011 (to U.O.), and a subcontract from AI068636 to the Harvard Virology Support Laboratory (to D.R.K.). J.Z.L. has received research support and served as a consultant for Gilead Sciences and Merck.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Chun TW, Fauci AS. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci U S A 96:10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 3.Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. 2014. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, Rinaldo CR, Riddler SA, Hogg E, Godfrey C, Collier AC, Eron JJ, Mellors JW. 2017. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 13:1–21. doi: 10.1371/journal.ppat.1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, Kuritzkes DR, Lederman MM, Para M, Gandhi RT. 2016. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 30:343–353. doi: 10.1097/QAD.0000000000000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montserrat M, Plana M, Guardo AC, Andrés C, Climent N, Gallart T, Leal L, Gatell JM, Sánchez-Palomino S, García F. 2017. Impact of long-term antiretroviral therapy interruption and resumption on viral reservoir in HIV-1 infected patients. AIDS 31:1895–1897. doi: 10.1097/QAD.0000000000001560. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Bonet M, Puertas MC, Fortuny C, Ouchi D, Mellado MJ, Rojo P, Noguera-Julian A, Muñoz-Fernández MA, Martinez-Picado J. 2015. Establishment and replenishment of the viral reservoir in perinatally HIV-1-infected children initiating very early antiretroviral therapy. Clin Infect Dis 61:1169–1178. doi: 10.1093/cid/civ456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Günthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. 2008. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis 197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 9.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol 76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Spiegelaere W, Malatinkova E, Lynch L, Van Nieuwerburgh F, Messiaen P, O'Doherty U, Vandekerckhove L. 2014. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of Poisson statistics. Clin Chem 60:886–895. doi: 10.1373/clinchem.2013.219378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. 2007. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology 368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lada S, VanBelzen J, Ignacio C, Strain M, O'Doherty U, Richman DD. 2017. Novel assay to measure integrated HIV DNA in PBMC from ART-suppressed persons, abstr 300. Conference on Retroviruses and Opportunistic Infections (CROI), Seattle, WA. [Google Scholar]
- 13.Liszewski MK, Yu JJ, O'Doherty U, O'Doherty U. 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 16.Sharaf RR, Li JZ. 2017. The alphabet soup of HIV reservoir markers. Curr HIV/AIDS Rep 14:72–81. doi: 10.1007/s11904-017-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agosto LM, Liszewski MK, Mexas A, Graf E, Pace M, Yu JJ, Bhandoola A, O'Doherty U. 2011. Patients on HAART often have an excess of unintegrated HIV DNA: Implications for monitoring reservoirs. Virology 409:46–53. doi: 10.1016/j.virol.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray JM, Zaunders JJ, McBride KL, Xu Y, Bailey M, Suzuki K, Cooper DA, Emery S, Kelleher AD, Koelsch KK. 2014. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J Virol 88:3516–3526. doi: 10.1128/JVI.03331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. 2016. Integrated and total HIV-1 DNA predict ex vivo viral outgrowth. PLoS Pathog 12:e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O'Doherty U. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART-treated patients. Virology 379:78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. 2008. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc 3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 22.Williams JP, Hurst J, Stöhr W, Robinson N, Brown H, Fisher M, Kinloch S, Cooper D, Schechter M, Tambussi G, Fidler S, Carrington M, Babiker A, Weber J, Koelsch KK, Kelleher AD, Phillips RE, Frater J. 2014. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin R, Hamimi C, Lambert-Niclot S, Carcelain G, Bellet J, Assoumou L, Tubiana R, Calvez V, Dudoit Y, Costagliola D, Autran B, Katlama C, ULTRASTOP Study Group. 2016. Treatment interruption in chronically HIV-infected patients with an ultralow HIV reservoir. AIDS 30:761–769. doi: 10.1097/QAD.0000000000000987. [DOI] [PubMed] [Google Scholar]
- 24.Clarridge KE, Blazkova J, Einkauf K, Petrone M, Refsland EW, Justement JS, Shi V, Huiting ED, Seamon CA, Lee GQ, Yu XG, Moir S, Sneller MC, Lichterfeld M, Chun T-W. 2018. Effect of analytical treatment interruption and reinitiation of antiretroviral therapy on HIV reservoirs and immunologic parameters in infected individuals. PLoS Pathog 14:e1006792. doi: 10.1371/journal.ppat.1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schooley RT, Spritzler J, Wang H, Lederman MM, Havlir D, Kuritzkes DR, Pollard R, Battaglia C, Robertson M, Mehrotra D, Casimiro D, Cox K, Schock B. 2010. AIDS Clinical Trials Group 5197: a placebo-controlled trial of immunization of HIV-1-infected persons with a replication-deficient adenovirus type 5 vaccine expressing the HIV-1 core protein. J Infect Dis 202:705–716. doi: 10.1086/655468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson JM, Pat Bucy R, Spritzler J, Saag MS, Eron JJ, Coombs RW, Wang R, Fox L, Johnson VA, Cu-Uvin S, Cohn SE, Mildvan D, O'Neill D, Janik J, Purdue L, O'Connor DK, Vita CDi, Frank I. 2006. Evidence that intermittent structured treatment interruption, but not immunization with ALVAC-HIV vCP1452, promotes host control of HIV replication: the results of AIDS Clinical Trials Group 5068. J Infect Dis 194:623–632. doi: 10.1086/506364. [DOI] [PubMed] [Google Scholar]
- 27.Volberding P, Demeter L, Bosch RJ, Aga E, Pettinelli C, Hirsch M, Vogler M, Martinez A, Little S, Connick E. 2009. Antiretroviral therapy in acute and recent HIV infection: a prospective multicenter stratified trial of intentionally interrupted treatment. AIDS 23:1987–1995. doi: 10.1097/QAD.0b013e32832eb285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skiest DJ, Su Z, Havlir DV, Robertson KR, Coombs RW, Cain P, Peterson T, Krambrink A, Jahed N, McMahon D, Margolis DM. 2007. Interruption of antiretroviral treatment in HIV-infected patients with preserved immune function is associated with a low rate of clinical progression: a prospective study by AIDS Clinical Trials Group 5170. J Infect Dis 195:1426–1436. doi: 10.1086/512681. [DOI] [PubMed] [Google Scholar]