Abstract
Clostridium perfringens causes diarrhea and other diseases in animals and humans. We investigated the prevalence, toxin gene profiles, and antibiotic resistance of C. perfringens isolated from diarrheic dogs (DD) and non-diarrheic dogs (ND) in two animal hospitals in Seoul, Korea. Fecal samples were collected from clinically DD (n = 49) and ND (n = 34). C. perfringens was isolated from 31 of 49 DD (63.3%) and 21 of 34 ND dogs (61.8%). All C. perfringens strains were positive for the α toxin gene, but not for the β, ε, or ι toxin genes; therefore, all strains were identified as type A C. perfringens. All isolates were cpe-negative, whereas the β2 toxin gene was identified in 83.9% and 61.9% of isolates from DD and ND, respectively. Most isolates were susceptible to ampicillin (94%), chloramphenicol (92%), metronidazole (100%), moxifloxacin (96%), and imipenem (100%). However, 25.0% and 21.2% of isolates were resistant to tetracycline and clindamycin, respectively. Molecular subtyping of the isolated strains was performed by using pulsed-field gel electrophoresis. Fifty-two isolates were classified into 48 pulsotypes based on more than 90% similarity of banding patterns. No notable differences were observed among the isolates from DD and ND.
Keywords: Clostridium perfringens, bacterial toxins, dogs, drug resistance, pulsed-field gel electrophoresis
Introduction
Clostridium perfringens causes numerous gastrointestinal infections, such as necrotic enteritis, enterotoxemia, and hemorrhagic gastroenteritis, in many mammalian species [26,28,33]. Toxins of C. perfringens have received considerable attention as critical virulence factors for C. perfringens-associated diseases in animals and humans [28]. Most diseases caused by C. perfringens are mediated by one or more toxins [26]. C. perfringens is classified into five types (A–E) based on its production of four major toxins: α (CPA), β (CPB), ε (ETX), and ι (IAP) [26,29]. In addition to these major toxins, C. perfringens strains can express other toxins, such as perfringolysin O (PFO), enterotoxin (CPE), and CPB2 (β2) toxin [33]. CPE is suspected to be associated with diarrhea in dogs, humans, and other animals [5,29,33].
In companion animals, such as dogs and cats, C. perfringens can cause conditions ranging from mild diarrhea to fatal necrohemorrhagic enteritis [28]. Evaluation of the distribution of C. perfringens strains and assessment of their traits, such as toxin profiles, are important to elucidating the role of C. perfringens in canine diarrhea. Although there have been numerous reports on the prevalence of C. perfringens strains and their toxin profiles in fecal samples from diarrheic dogs (DD) and non-diarrheic dogs (ND) [2,9,10,15,20,22,29,32,34], studies detailing the prevalence of toxin genes, relatedness between DD and ND strains, and their antimicrobial susceptibility profiles have not been conducted in Korea.
As limited information about the in vitro or in vivo susceptibilities of C. perfringens strains from canine feces is available, the drug of choice for treating canine diseases associated with this bacterium has been determined based on results from human cases [12,14]. Antimicrobials recommended for the treatment of C. perfringens-associated diarrhea in canines include beta-lactams, macrolides, metronidazole, and tetracycline [14]. Administration of antibiotics below their minimum inhibitory concentration (MIC) may increase the resistance of C. perfringens toward them, making treatment difficult [14,27].
The objective of this study was to investigate the prevalence of toxin genes and antibiotic resistance of C. perfringens isolates obtained from DD and ND in animal hospitals in Korea. In addition, pulsed-field gel electrophoresis (PFGE) was performed in order to compare subtype patterns between the C. perfringens isolates from dogs.
Materials and Methods
Collecting samples
Stool samples were collected from 83 dogs, 49 DD and 34 ND, between 2011 and 2013. All samples were obtained from hospitalized dogs at two animal hospitals: A (university animal hospital) and B (local animal hospital). Fecal samples were collected directly from the rectum by using a swab method. A sterilized cotton swab was inserted into the rectum and rotated gently to collect a sample of fecal matter. All feces were stored at 4℃ and were used for the isolation of C. perfringens within 24 h after collection.
Isolation of C. perfringens
A small portion of feces on a cotton swab was directly inoculated onto tryptose sulfite cycloserine (TSC; Oxoid, UK) agar with 5% egg yolk (Oxoid). The remaining sample was enriched in 10 mL of cooked meat media (Oxoid) followed by incubation at 37℃ for 24 h. After enrichment, a loopful of culture was inoculated onto TSC agar. All plates were incubated under anaerobic conditions at 37℃ for 24 to 48 h. Suspicious black colonies with lecithinase production from each plate were subcultured under aerobic and anaerobic conditions on blood agar. Colonies showing double hemolysis and growing only under anaerobic conditions were regarded as presumptive C. perfringens strains, and were selected and stored at −70℃.
Polymerase chain reaction (PCR) amplification
A colony subcultured on blood agar was removed and genomic DNA was extracted by applying the boiling method [4]. Table 1 shows primer sequences and cycling conditions for PCR amplification of toxin genes cpa, cpb, cpe, cpb2, etx, and iap, encoding α, β, entero, β2, ε, and ι toxin, respectively [1,3,21]. The amplification was conducted by using a PCR reaction mixture (20 µL) consisting of Frenche PCR PreMix (iNtRON Biotechnology, Korea), 50 ng of template DNA, and 500 nM of each primer. Each PCR product was analyzed on 1.5% agarose gel containing ethidium bromide (EtBr; Promega, USA). Gel images were visualized by using a GelDoc XR gel analyzer (Bio-Rad, USA) under ultraviolet light. Strains ATCC 3624, NCTR 3626, KNIH_CL_D, KNIH_CL_E, KNIH_Kim_1, and PM_S44 were used as the control strains for cpa, cpb, ext, iap, cpe, and cpb2 genes, respectively.
Table 1. Polymerase chain reaction primers and reaction conditions used in this study.
For, forward; Rev, reverse.
Antibiotic susceptibility test
MIC values of antibiotics against C. perfringens were determined by using E-test kits (bioMérieux, France). Each isolate was suspended in Mueller-Hinton broth (Sigma-Aldrich, USA) to achieve a McFarland turbidity of 0.5 and then inoculated onto Brucella agar with 5% sheep blood, hemin, and vitamin K1 (BD Difco, USA). An E-test strip of each antibiotic (ampicillin, chloramphenicol, metronidazole, moxifloxacin, tetracycline, clindamycin, and imipenem) was placed on the plate and followed by incubation at 37℃ for 48 h under anaerobic conditions. The MIC value was noted according to the manufacturer's instructions, and susceptibility and resistance were determined according to the Interpretive Standards for Anaerobes in Clinical and Laboratory Standards Institute [6]. C. perfringens ATCC 13124 was used as the quality control strain.
PFGE subtyping
PFGE was performed for subtyping of the 52 C. perfringens isolates from 49 DD and 34 ND according to the procedure described by Matushek et al. [18] with a few modifications. A single colony was grown in 10 mL of brain heart infusion broth (Sigma-Aldrich) at 37℃ for 48 h under anaerobic conditions and centrifuged. Cultures were suspended in 1 mL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to achieve an absorbance level of 1.5 to 2.0 at 600 nm. The cells (100 µL) were mixed with 80 µL of 2× lysis buffer (10 mM Tris [pH 8.0], 0.2 M EDTA [pH 8.0], 1 M NaCl, 0.5% sodium lauryl sarcosine, 0.2% sodium deoxycholate, and 0.5% polyethylene glycol hexadecyl ether) and 200 µL of 1.5% low melting point agarose (Life Technologies, USA), and the mixture was then placed in a rectangular mold and solidified. The solid plugs were incubated overnight in 5 mL of lysis buffer with lysozyme (1 mg/mL; Sigma-Aldrich) and mutanolysin (20 U/mL; Sigma-Aldrich) at 50℃ with agitation at 95 rpm. The lysis buffer was removed and each plug was incubated at 37℃ for 4 h in 5 mL of buffer (0.5 M EDTA, pH 8.0, 1% sodium lauryl sarcosine) with proteinase K (1 mg/mL; Research Products International, USA). Plugs were rinsed twice with TE buffer and then incubated at 50℃ for 30 min in TE buffer containing 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Each plug was rinsed twice in TE buffer and sliced into small pieces. The plugs were digested overnight in restriction buffer with 40 units of SmaI (Fermentas, USA). Electrophoresis was carried out in 1% of SeaKem Gold agarose gels (Lonza, Switzerland) prepared in electrophoresis buffer (0.5% Tris-borate-EDTA buffer; Sigma-Aldrich). The digested DNA was separated by using a CHEF Mapper (Bio-Rad) with the following conditions: low molecular weight (MW) = 30 kb, high MW = 600 kb, initial switch time = 0.5 sec, final switch time = 40 sec, included angle = 120°, and running time = 21 h. The gels were stained with EtBr for 30 min and then washed in distilled water for 20 min. The gels were visualized on the Gel Doc (Bio-Rad) and the images were saved as TIFF files. Banding patterns and dendrograms were created by determining the distance matrices and using an unweighted pair group method with arithmetic mean clustering method. Subtyping patterns were analyzed by using BioNumerics software (ver. 7; Applied Maths, USA). For the construction of dendrograms, a 1% position tolerance shift between similar bands was used.
Results
Prevalence of C. perfringens in DD and ND
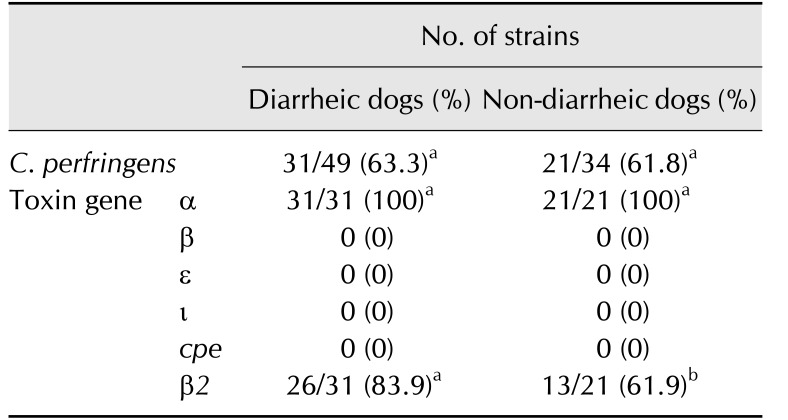
The prevalences of C. perfringens in samples from 49 DD and 34 ND and their toxin gene profiles are presented in Table 2. The prevalence of C. perfringens was not statistically different (p > 0.05) between the DD (31 of 49, 63.3%) and ND (21 of 34, 61.8%) groups. All presumptive C. perfringens strains were positive for cpa, and none was positive for cpb, iap, or etx. Thus, all isolated strains were identified as type A C. perfringens. All isolates screened by PCR were cpe-negative. However, cpb2 was identified in 83.9% (26 of 31) and 61.9% (13 of 21) of isolates from DD and ND, respectively, and the difference between the groups was statistically significant (p < 0.05).
Table 2. The prevalence of Clostridium perfringens and toxin gene profiles.
Different superscript letters (a, b) within a row indicate a significant difference (p < 0.05) in the number of positive samples. The number of positives in diarrheic and non-diarrheic dogs was compared by using Fisher's exact test in GraphPad InStat software (ver. 3.05; GraphPad Software).
Antibiotic susceptibility
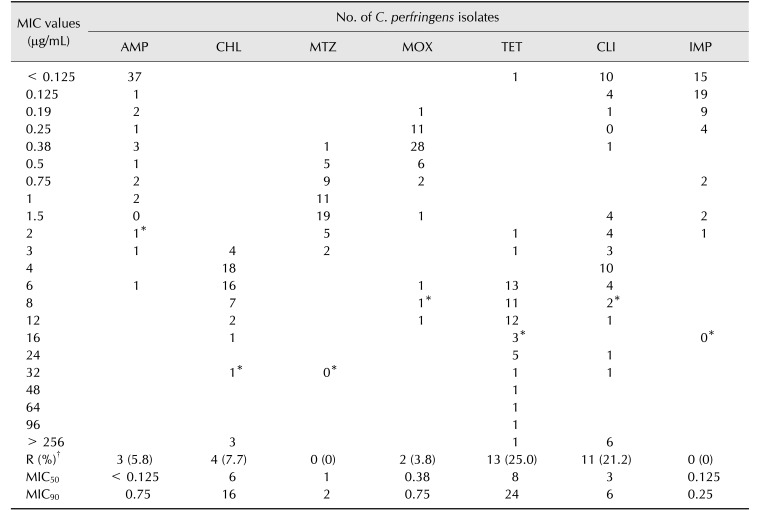
The MIC values of antibiotics against C. perfringens are summarized in Table 3. The antibiotic susceptibility profile of each strain is presented in Fig. 1. Most isolates were susceptible to the tested antibiotics, but 25.0% and 21.2% of isolates were resistant to tetracycline and clindamycin, respectively (Table 3). Among the 52 isolates, four strains from hospital A (KDD_3, 12, 19, and KND_19; Fig. 1) were resistant to three different classes of antibiotics, indicating multidrug resistance. No notable differences were observed in terms of antibiotic resistance between strains from DD and ND.
Table 3. Comparison of antimicrobial susceptibility profiles and minimum inhibitory concentrations (MIC) for 52 Clostridium perfringens isolates from 83 dogs.
AMP, ampicillin; CHL, chloramphenicol; MTZ, metronidazole; MOX, moxifloxacin; TET, tetracycline; CLI, clindamycin; IMP, imipenem; MIC50, 50% MIC; MIC90, 90% MIC. *Breakpoint for each antibiotic. †No. of resistant strains (%).
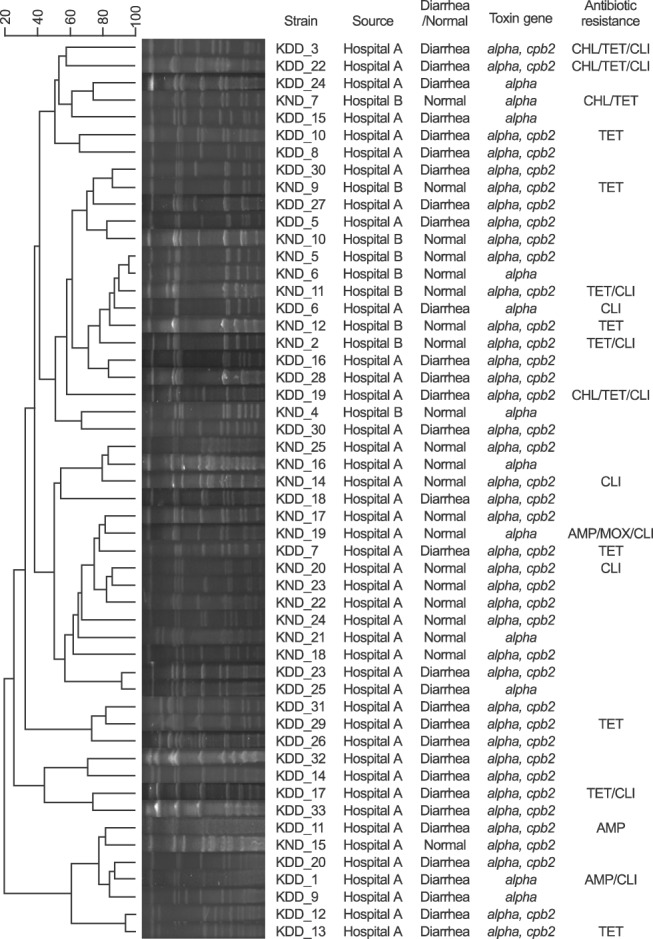
Fig. 1. Pulsed-field gel electrophoresis profiles of Clostridium perfringens from diarrheic and non-diarrheic dogs. Cut-off value was 90% similarity.
Molecular subtyping
The dendrogram generated from the obtained PFGE patterns, virtual gel images, and profiles of each strain is summarized in Fig. 1. It has been reported that PFGE analysis can strongly discriminate phenotypically similar C. perfringens isolated from human clinical samples, food products, and veterinary samples [23,24]. Based on the PFGE banding patterns with ≥ 90% similarity, the strains were classified into 48 pulsotypes (Fig. 1). The PFGE banding patterns of the isolates were not associated with the source hospital, distribution of toxin genes, or antibiotic resistance patterns (Fig. 1). Moreover, there was no remarkable difference in PFGE banding patterns between the isolates from DD and ND. Although some strain pairs, such as KND_5 and 6 and KDD_12 and 13, showed high similarity (> 95%) in banding patterns, most strains were genetically unrelated, indicating genetic diversity among the C. perfringens isolates from fecal samples of the DD and ND groups (Fig. 1). Each strain pair showing high similarity, KND_5 and 6 (96.3%) and KDD_12 and 13 (96.5%), could be distinguished because they had different antibiotic resistance and toxin profiles.
Discussion
The prevalence of C. perfringens found in this study is similar to those reported in other studies, which demonstrated that 61% to 94% DD subjects and 56% to 88% ND subjects had C. perfringens in their feces [2,9,10,15,20,22,29,32,34]. Although some researchers have found a significant difference between C. perfringens prevalence in DD and ND [15,34], it has been reported that a high occurrence frequency of the bacterium is not an indicative or diagnostic sign of C. perfringens-associated disease [9].
In this study, no isolate had the cpe gene. In previous studies, the percentage of C. perfringens strains harboring cpe or related genes was 0% to 34% in DD and 0% to 14% in ND [15,16,34,35]. Although some studies reported non-significant associations between the detection of cpe and the presence of diarrhea [9,10,15,35], the CPE toxin has been strongly implicated as a cause of canine diarrhea [15,32,34]. However, the pathogenesis of C. perfringens-associated diarrhea in dogs has not been fully described, and the diagnostic criteria for establishing CPE-mediated animal disease are lacking, because CPE is also detected in ND [16,30,33]. In this study, the results revealed that diarrhea in dogs is not CPE-mediated.
However, the information generated in this study is limited because the quantity of cells in each fecal sample was not considered. Although most previous studies have focused on finding a correlation between presence of bacteria (or toxin) and diarrhea [9,15,16,20,22,32], the pathogenesis of C. perfringens-associated diarrhea in dogs has remained unclear. In future research, the correlation between quantity of bacteria in feces and diarrhea should be considered, since the number of bacteria could affect elucidation of the mechanism involved in C. perfringens-associated diarrhea.
It has been reported that C. perfringens-associated diarrhea in dogs may not be mediated by CPE alone and other toxin genes might help in propagation of the disease [31]. Interestingly, the prevalence of cpb2 reported in this study was considerably higher than previously reported rates of 1% to 22.7% in ND and 5% to 32% in DD [9,29,31]. Moreover, the number of cpb2-positive isolates from DD (83.9%) was significantly higher than that of cpb2-positive isolates from ND (61.9%). Some studies have found an association between type A C. perfringens positive for cpb2 and the occurrence of diarrhea in pigs and horses [11,13]. Although the role of β2-toxigenic C. perfringens in dogs is not well described [29], it has been suggested that β2 toxin alone, or in combination with enterotoxin, probably contributes to diarrhea in dogs [31]. The high rate of occurrence of cpb2 among isolates from DD indicates that the toxin may play a role in pathogenesis of diarrheal disease in dogs. However, the observation that its presence in ND was also very high raises some concerns about whether cpb2 is responsible for diarrheal disease in dogs. More studies are necessary to elucidate the role and importance of β2 toxin in C. perfringens-associated diarrhea in dogs.
Although C. perfringens is one of the most common canine pathogens, few studies have evaluated the susceptibility profile of this bacterium to antibiotics used to treat diarrhea related to C. perfringens. Marks and Kather [14] tested antibiotic resistance of 131 C. perfringens strains from feces of DD and ND. They reported that most strains were highly susceptible to ampicillin and metronidazole, and that 21% of the strains had a MIC of over 16 mg/L for tetracycline. These results were consistent with the results of our antimicrobial susceptibility tests. Kather et al. [12] reported in their follow-up study that 41% of tested canine isolates were positive for both tetracycline genes, tetA and tetB, although they found only 21% of strains were phenotypically resistant to tetracycline. Based on our results, the use of tetracycline and clindamycin, especially at low doses, should be avoided because the exposure of microorganisms to antibiotics below the MIC can result in an increase in resistance, making treatment difficult [14]. Multidrug resistant (MDR) C. perfringens strains were resistant to three different classes of antibiotics used in this study. Several studies have documented MDR strains of C. perfringens in different animals, but few reports are available on canine isolates [7,8,14,25]. The in vitro antimicrobial susceptibility of the C. perfringens isolates described in this study could help optimize antibiotic treatment for C. perfringens-associated diarrhea in dogs, particularly in cases of increased resistance of anaerobic bacteria isolated from both humans and animals [12,19]. Moreover, the discovery of MDR C. perfringens and its high rate of resistance toward tetracycline and clindamycin indicates the importance of continuous surveillance of antibiotic resistance of C. perfringens isolated from dogs [14].
There were no identical strains detected in this study, but cross-contamination of pathogenic C. perfringens between hospitalized dogs may occur through hospital environments and direct contact because C. perfringens persists as a spore for long periods even under unfavorable conditions [17]. Although C. perfringens can cause enteritis in humans and other animals, there are few studies on the epidemiology of C. perfringens in canines. Isolation of strains followed by subtyping can elucidate transmission patterns and risk factors. Thus, our results could serve as a reference for further epidemiological research to trace the source of C. perfringens in outbreaks caused by transmission between companion animals and humans.
In the present study, we genotypically and phenotypically characterized C. perfringens in fecal samples from DD and ND by investigating the isolates toxin profiles, antibiotic resistance, and molecular subtyping patterns. Our study has some limitations; for example, we could obtain limited information about the clinical history of the dogs in the study, such as hospitalization, onset of diarrhea, and antibiotic exposure within 6 months of fecal analysis, information that may influence the presence of C. perfringens and its toxicity and antibiotic susceptibility. However, as this is the first such study in Korea, the data in this study could provide important information about the occurrence and characteristics of C. perfringens isolated from dogs.
Acknowledgments
We thank Drs. Carl Cerniglia, John Sutherland, and Steven Foley for their critical reviews of the manuscript. The views expressed herein do not necessarily reflect those of the U.S. Food and Drug Administration or the U.S. Department of Health and Human Services. This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Agriculture, Food and Rural Affairs Research Center Support Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (716002-7).
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Baums CG, Schotte U, Amtsberg G, Goethe R. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet Microbiol. 2004;100:11–16. doi: 10.1016/S0378-1135(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 2.Berset-Istratescu CM, Glardon OJ, Magouras I, Frey CF, Gobeli S, Burgener IA. Follow-up of 100 dogs with acute diarrhoea in a primary care practice. Vet J. 2014;199:188–190. doi: 10.1016/j.tvjl.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Bueschel DM, Jost BH, Billington SJ, Trinh HT, Songer JG. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet Microbiol. 2003;94:121–129. doi: 10.1016/s0378-1135(03)00081-6. [DOI] [PubMed] [Google Scholar]
- 4.Chon JW, Hyeon JY, Park JH, Song KY, Kim JH, Seo KH. Improvement of mannitol-yolk-polymyxin B agar by supplementing with trimethoprim for quantitative detection of Bacillus cereus in foods. J Food Prot. 2012;75:1342–1345. doi: 10.4315/0362-028X.JFP-11-519. [DOI] [PubMed] [Google Scholar]
- 5.Chon JW, Park JS, Hyeon JY, Park C, Song KY, Hong KW, Hwang IG, Kwak HS, Seo KH. Development of real-time PCR for the detection of Clostridium perfringens in meats and vegetables. J Microbiol Biotechnol. 2012;22:530–534. doi: 10.4014/jmb.1107.07064. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Wayne: CLSI; 2014. [Google Scholar]
- 7.Dornbusch K, Nord CE, Dahlbäck A. Antibiotic susceptibility of Clostridium species isolated from human infections. Scand J Infect Dis. 1975;7:127–134. doi: 10.3109/inf.1975.7.issue-2.09. [DOI] [PubMed] [Google Scholar]
- 8.Dutta GN, Devriese LA. Macrolide-lincosamide-streptogramin resistance patterns in Clostridium perfringens from animals. Antimicrob Agents Chemother. 1981;19:274–278. doi: 10.1128/aac.19.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein MR, Kruth SA, Bersenas AM, Holowaychuk MK, Weese JS. Detection and characterization of Clostridium perfringens in the feces of healthy and diarrheic dogs. Can J Vet Res. 2012;76:161–165. [PMC free article] [PubMed] [Google Scholar]
- 10.Hackett T, Lappin MR. Prevalence of enteric pathogens in dogs of north-central Colorado. J Am Anim Hosp Assoc. 2003;39:52–56. doi: 10.5326/0390052. [DOI] [PubMed] [Google Scholar]
- 11.Herholz C, Miserez R, Nicolet J, Frey J, Popoff M, Gibert M, Gerber H, Straub R. Prevalence of β2-toxigenic Clostridium perfringens in horses with intestinal disorders. J Clin Microbiol. 1999;37:358–361. doi: 10.1128/jcm.37.2.358-361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kather EJ, Marks SL, Foley JE. Determination of the prevalence of antimicrobial resistance genes in canine Clostridium perfringens isolates. Vet Microbiol. 2006;113:97–101. doi: 10.1016/j.vetmic.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 13.Klaasen HL, Molkenboer MJ, Bakker J, Miserez R, Häni H, Frey J, Popoff MR, van den Bosch JF. Detection of the β2 toxin gene of Clostridium perfringens in diarrhoeic piglets in the Netherlands and Switzerland. FEMS Immunol Med Microbiol. 1999;24:325–332. doi: 10.1111/j.1574-695X.1999.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 14.Marks SL, Kather EJ. Antimicrobial susceptibilities of canine Clostridium difficile and Clostridium perfringens isolates to commonly utilized antimicrobial drugs. Vet Microbiol. 2003;94:39–45. doi: 10.1016/s0378-1135(03)00061-0. [DOI] [PubMed] [Google Scholar]
- 15.Marks SL, Kather EJ, Kass PH, Melli AC. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med. 2002;16:533–540. doi: 10.1892/0891-6640(2002)016<0533:gapcop>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Marks SL, Rankin SC, Byrne BA, Weese JS. Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med. 2011;25:1195–1208. doi: 10.1111/j.1939-1676.2011.00821.x. [DOI] [PubMed] [Google Scholar]
- 17.Márquez-González M, Cabrera-Díaz E, Hardin MD, Harris KB, Lucia LM, Castillo A. Survival and germination of Clostridium perfringens spores during heating and cooling of ground pork. J Food Prot. 2012;75:682–689. doi: 10.4315/0362-028X.JFP-11-409. [DOI] [PubMed] [Google Scholar]
- 18.Matushek MG, Bonten MJ, Hayden MK. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott PF, Walker RD, White DG. Antimicrobials: modes of action and mechanisms of resistance. Int J Toxicol. 2003;22:135–143. doi: 10.1080/10915810305089. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie E, Riehl J, Banse H, Kass PH, Nelson S, Jr, Marks SL. Prevalence of diarrhea and enteropathogens in racing sled dogs. J Vet Intern Med. 2010;24:97–103. doi: 10.1111/j.1939-1676.2009.0418.x. [DOI] [PubMed] [Google Scholar]
- 21.Meer RR, Songer JG. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res. 1997;58:702–705. [PubMed] [Google Scholar]
- 22.Minamoto Y, Dhanani N, Markel ME, Steiner JM, Suchodolski JS. Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Vet Microbiol. 2014;174:463–473. doi: 10.1016/j.vetmic.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Nauerby B, Pedersen K, Madsen M. Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet Microbiol. 2003;94:257–266. doi: 10.1016/s0378-1135(03)00118-4. [DOI] [PubMed] [Google Scholar]
- 24.Park M, Deck J, Foley SL, Nayak R, Songer JG, Seibel JR, Khan SA, Rooney AP, Hecht DW, Rafii F. Diversity of Clostridium perfringens isolates from various sources and prevalence of conjugative plasmids. Anaerobe. 2016;38:25–35. doi: 10.1016/j.anaerobe.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Rood JI, Maher EA, Somers EB, Campos E, Duncan CL. Isolation and characterization of multiply antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob Agents Chemother. 1978;13:871–880. doi: 10.1128/aac.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salari Sedigh H, Rajabioun M, Razmyar J, Kazemi Mehrjerdi H. An unusual necrotic myositis by Clostridium perfringens in a German Shepherd dog: a clinical report, bacteriological and molecular identification. Vet Res Forum. 2015;6:349–353. [PMC free article] [PubMed] [Google Scholar]
- 27.Schentag JJ, Gilliland KK, Paladino JA. What have we learned from pharmacokinetic and pharmacodynamic theories? Clin Infect Dis. 2001;32(Suppl 1):S39–S46. doi: 10.1086/319375. [DOI] [PubMed] [Google Scholar]
- 28.Silva RO, Lobato FC. Clostridium perfringens: a review of enteric diseases in dogs, cats and wild animals. Anaerobe. 2015;33:14–17. doi: 10.1016/j.anaerobe.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Silva RO, Santos RL, Pires PS, Pereira LC, Pereira ST, Duarte MC, de Assis RA, Lobato FC. Detection of toxins A/B and isolation of Clostridium difficile and Clostridium perfringens from dogs in Minas Gerais, Brazil. Braz J Microbiol. 2013;44:133–137. doi: 10.1590/S1517-83822013005000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Songer JG, Uzal FA. Clostridial enteric infections in pigs. J Vet Diagn Invest. 2005;17:528–536. doi: 10.1177/104063870501700602. [DOI] [PubMed] [Google Scholar]
- 31.Thiede S, Goethe R, Amtsberg G. Prevalence of β2 toxin gene of Clostridium perfringens type A from diarrhoeic dogs. Vet Rec. 2001;149:273–274. doi: 10.1136/vr.149.9.273. [DOI] [PubMed] [Google Scholar]
- 32.Tupler T, Levy JK, Sabshin SJ, Tucker SJ, Greiner EC, Leutenegger CM. Enteropathogens identified in dogs entering a Florida animal shelter with normal feces or diarrhea. J Am Vet Med Assoc. 2012;241:338–343. doi: 10.2460/javma.241.3.338. [DOI] [PubMed] [Google Scholar]
- 33.Uzal FA, Vidal JE, McClane BA, Gurjar AA. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinology J. 2010;2:24–42. [PMC free article] [PubMed] [Google Scholar]
- 34.Weese JS, Staempfli HR, Prescott JF, Kruth SA, Greenwood SJ, Weese HE. The roles of Clostridium difficile and enterotoxigenic Clostridium perfringens in diarrhea in dogs. J Vet Intern Med. 2001;15:374–378. [PubMed] [Google Scholar]
- 35.Zerbini L, Ossiprandi MC. Molecular typing of Clostridium perfringens strains isolated from dogs by toxin gene amplification. Ann Fac Med Vet Di Parma. 2009;29:115–128. [Google Scholar]