Abstract
The subtype H9N2 avian influenza virus greatly threatens the Chinese poultry industry, even with annual vaccination. Waterfowl can be asymptomatically infected with the H9N2 virus. In this study, three H9N2 virus strains, designated A/Goose/Jiangsu/YZ527/2011 (H9N2, Gs/JS/YZ527/11), A/Goose/Jiangsu/SQ119/2012 (H9N2, Gs/JS/SQ119/12), and A/Goose/Jiangsu/JD564/2012 (H9N2, Gs/JS/JD564/12), were isolated from domestic geese. Molecular characterization of the three isolates showed that the Gs/JS/YZ527/11 virus is a double-reassortant virus, combining genes of A/Quail/Hong Kong/G1/97 (H9N2, G1/97)-like and A/Chicken/Shanghai/F/98 (H9N2, F/98)-like; the Gs/JS/SQ119/12 virus is a triple-reassortant virus combining genes of G1/97-like, F/98-like, and A/Duck/Shantou/163/2004 (H9N2, ST/163/04)-like. The sequences of Gs/JS/JD564/12 share high homology with those of the F/98 virus, except for the neuraminidase gene, whereas the internal genes of Gs/JS/YZ527/11 and Gs/JS/SQ119/12 are closely related to those of the H7N9 viruses. An infectivity analysis of the three isolates showed that Gs/JS/SQ119/12 and Gs/JS/YZ527/11 replicated well, with seroconversion, in geese and chickens, the Gs/JS/JD564/12 did not infect well in geese or chickens, and the F/98 virus only infected chickens, with seroconversion. Emergence of these new reassortant H9N2 avian influenza viruses indicates that these viruses can infect both chicken and goose and can produce different types of lesions in each species.
Keywords: H9N2, biological characteristics, chickens, geese, infection
Introduction
The subtype H9N2 low-pathogenic avian influenza viruses (AIVs) have been highly prevalent in the Chinese poultry industry since the first infection spread from waterfowl to chickens in 1994. Over the past 20 years, the H9N2 AIV has continuously evolved because the characteristics of its RNA genome make it susceptible to mutation and reassortment, as well as to selection pressure exerted by vaccine-induced antibodies. At present, H9N2 AIV has become common in chickens and has evolved into different antigenic groups. Importantly, aquatic birds, the natural reservoir of many influenza viruses [1,6,15,20,31], can carry the H9N2 AIV asymptomatically, increasing the risk of uncontrolled viral spread. However, H9N2 viruses isolated from wild geese have rarely been reported, until now.
Reassortment of the H9N2 virus with other influenza viral lineages has extended the range of host species infected by H9N2. The H9N2 virus can now infect land fowl, such as chickens, and waterfowl, such as ducks and geese, and is even transmitted to mammals, including swine and dogs [27,33,36]. As early as 1998, occasional human cases of H9N2 infection were reported in southern China [9,22,30], while in 2013, approximately 2.3% to 4.6% of asymptomatic poultry workers had antibodies against subtype H9 AIVs [14,37]. The H9N2 AIV is recognized as a donor of internal genes to other AIV subtypes, generating many AIV subtypes, including the H5N1, H7N9, and H10N8 viruses [2,16,25,35]. Therefore, investigation of the epidemiology and pathogenicity of the H9N2 AIVs in geese, an aquatic bird that is a natural reservoir of H9N2 AIVs [34], should have a high priority.
The H9N2 AIVs in Eurasia include three lineages: A/Chicken/Beijing/1/94 (H9N2, BJ/94)-like, A/Quail/Hong Kong/G1/97 (H9N2, G1/97)-like, and A/Duck/Hong Kong/Y439/97 (H9N2, Y439/97)-like [10]. The BJ/94-like and G1-like lineages are predominant in China. A/Chicken/Shanghai/F/1998 (H9N2, F/98)-like belongs to the BJ/94-like lineage and has become the common H9N2 virus in mainland China [38]. In this study, we isolated three F/98-like H9N2 AIVs from domestic geese at the Veterinary Hospital of Yangzhou University in 2011 and 2012 and analyzed the genomic sequences and infectivity of the three viruses in specific-pathogen-free (SPF) chickens and geese. This study extends the elucidation of the evolution of H9N2 subtype AIVs in landfowl and waterfowl.
Materials and Methods
Ethics statement
All animal experiments were approved by the Jiangsu Administrative Committee for Laboratory Animals (No. SYXK-SU-2007-0005) and complied with the Jiangsu Laboratory Animal Welfare and Ethics guidelines of the Jiangsu Administrative Committee for Laboratory Animals.
Isolation and identification of viruses
Tracheal or cloacal swabs, collected between 2011 and 2012 from sick geese with influenza-like symptoms of diarrhea, excess mucus in the sinuses, and nasal discharge in the Veterinary Hospital of Yangzhou University, were initially placed in phosphate buffered saline containing antibiotics. After freezing and subsequent centrifugation at 1,000 × g for 10 min at 4℃, the supernatant was collected. A 200 µL aliquot of the supernatant was inoculated into the allantoic cavities of 10-day-old SPF embryonated chicken eggs. After 72 h of incubation at 37℃, the viruses were tested for hemagglutinin (HA) activity. The three isolated viruses were identified as subtype H9e by performing hemagglutination inhibition (HI) assays using antisera against subtype H9 AIV, subtype H5 AIV, and Newcastle disease virus (NDV), respectively. After two passages in SPF embryonated eggs, the 50% egg infectious doses (EID50) of the isolates were determined by propagating serial dilutions of viruses in eggs (0.2 mL per embryo) and calculated as described by Reed and Muench [23]. The HI assays of the isolates were performed with an initial dilution of 1:2 as previously described [4]. The Ck/SH/F/98 virus, which was the generation obtained from the original virus after three passages in SPF embryonated eggs, was used in this study.
Genetic and phylogenetic analysis
Total viral RNA was extracted from allantoic fluids infected with purified isolates by using TRIzol Reagent (Invitrogen Life Technologies, USA). Viral RNA was reverse-transcribed into cDNA by using 12 base universal primers for influenza A viruses (U12 A/G:AGCG/AAAAGCAGG). The gene fragments were amplified by polymerase chain reaction (PCR) following reverse transcription using specific primers [3,13], and were sequenced and assembled by Takara Biotechnology (China). Sequences were edited by using the Lasergene software package (DNASTAR, USA). Phylogenetic trees were generated by using the MEGA6.0 software suite [29]. The neighbor-joining method with 1,000 bootstrap replicates was applied for phylogenetic and molecular evolutionary analyses; values lower than 70% were hidden. Dong et al. [3] proposed a precise nomenclature to define all lineages, and they performed a large-scale genomic sequence analysis of 571 H9N2 influenza viruses isolated from 1966 to 2009 in the world, and, as a result, the 571 H9N2 viral genomes were classified into 74 separate lineages including all eight genes of the BJ/1/94 and HK/G1/97 viruses, PB2, PB1, PA, and nucleoprotein (NP) genes of the SH/F/98 virus, and the PB2 gene of the ST/163/04 virus.
Cross-reactivity analysis
Antisera from SPF chickens vaccinated with the inactivated strain Ck/SH/F/98 were used to compare the antigenic characterizations of the three isolates with the vaccine strain Ck/SH/F/98 by performing the HI assay as previously described [4]. Briefly, 3-week-old SPF chickens were subcutaneously vaccinated twice by subcutaneous injection of 0.3 mL oil-emulsion of inactive whole virus vaccine of the Ck/SH/F/98 virus, which was inactived by adding 0.2% formalin (v/v) for 24 h at 37℃. The antisera were collected from vaccinated SPF chickens three weeks after the booster vaccination and used to characterize the antigenicity of Ck/SH/F/98, Gs/JS/YZ527/11, Gs/JS/SQ119/12, and Gs/JS/JD564/12 viruses. The HI assay result was expressed as the reciprocal of the highest serum dilution in which hemagglutination was inhibited.
Infectivity to geese or chickens
To identify the infectivity of the three isolates in geese or chickens, thirty healthy 3-week-old geese or chickens with negative antibody against H9 were assigned to 5 groups, with 6 geese or chickens per group. As a control, one group was uninfected. The other four groups were inoculated with 106 EID50/200 µL of Ck/SH/F/98 virus, Gs/JS/YZ527/11 virus, Gs/JS/SQ119/12 virus, or Gs/JS/JD564/12 virus. Three geese or chickens per group were euthanized with CO2 and necropsied at day four after infection, and the tracheas and lungs of the infected geese or chickens were collected, fixed in 10% formalin, embedded in paraffin, and then sectioned to prepare histological specimens. Antisera from another three infected geese or chickens per group were harvested on days 7, 14, and 21 after infection; subsequently, HI assays were performed to measure seroconversion against the corresponding viruses. This experiment was performed three times, with each group receiving approximately the same dose of virus; the results from the three experiments were similar and were included in the analyses.
Nucleotide sequence accession numbers
The nucleotide sequences for the three H9N2 isolates and the crucial reference viruses used in this study are available from GenBank under accession No. AY253750.1–AY253756.1, AY743216.1, KY608228–KY608235, KY608236–KY608243, KY608244–KY608251, AF156381.1, AF156399.1, AF156424.1, AF156410.1, AF156465.1, AF156479.1, KF188291.1–KF188298.1, AF156378.1, AF156396.1, AF156435.1, AF156421.1, AF156449.1, AF156407.1, AF156463.1, AF156477.2, KF188366.1, KF188366.1, KF188267.1, KF188264.1, KF188266.1, KF188269.1, CY024013.1, DQ997519.1, DQ997518.1, DQ997517.1, DQ997520.1, EF155293.1, CY191955.1–CY191962.1, KF609508.1–KF609515.1, KJ411975.1–KJ411982.1, KU143281.1, KU143332.1, KU143374.1, KU143419.1, KU143460.1, KU143505.1, KU143549.1, and KU143594.1.
Results
Isolation, identification, and antigenic analysis of viruses
Allantoic fluids infected by using tracheal or cloacal swabs taken from geese were harvested from embryonated SPF chicken eggs. The three viruses were identified as H9 with HA and a HI assay using chicken H9 antiserum and were identified as N2 with PCR and specific primers. The isolated viruses did not react with H5 or NDV antiserum. The three H9N2 isolates from geese were designated A/Goose/Jiangsu/YZ527/2011 (H9N2, Gs/JS/YZ527/11), A/Goose/Jiangsu/SQ119/2012 (H9N2, Gs/JS/SQ119/12), and A/Goose/Jiangsu/JD564/2012 (H9N2, Gs/JS/JD564/12). The isolates were purified and propagated in embryonated SPF chicken eggs by applying the infinite dilution method. The EID50/0.2 mL of Gs/JS/YZ527/11, Gs/JS/SQ119/12, and Gs/JS/JD564/12 were 10−6.83, 10−7.5, and 10−6.67, respectively. The EID50 of the Gs/JS/SQ119/12 virus was 21-fold higher than that of strain Ck/SH/F/98, which had an EID50 of 10−6.17. The HI titers of the isolated viruses Gs/JS/YZ527/11, Gs/JS/SQ119/12, and Gs/JS/JD564/12 against antiserum raised to Ck/SH/F/98 were 16, 16, and 64, respectively. The HI titer of virus Ck/SH/F/98 against antiserum raised against itself was 128.
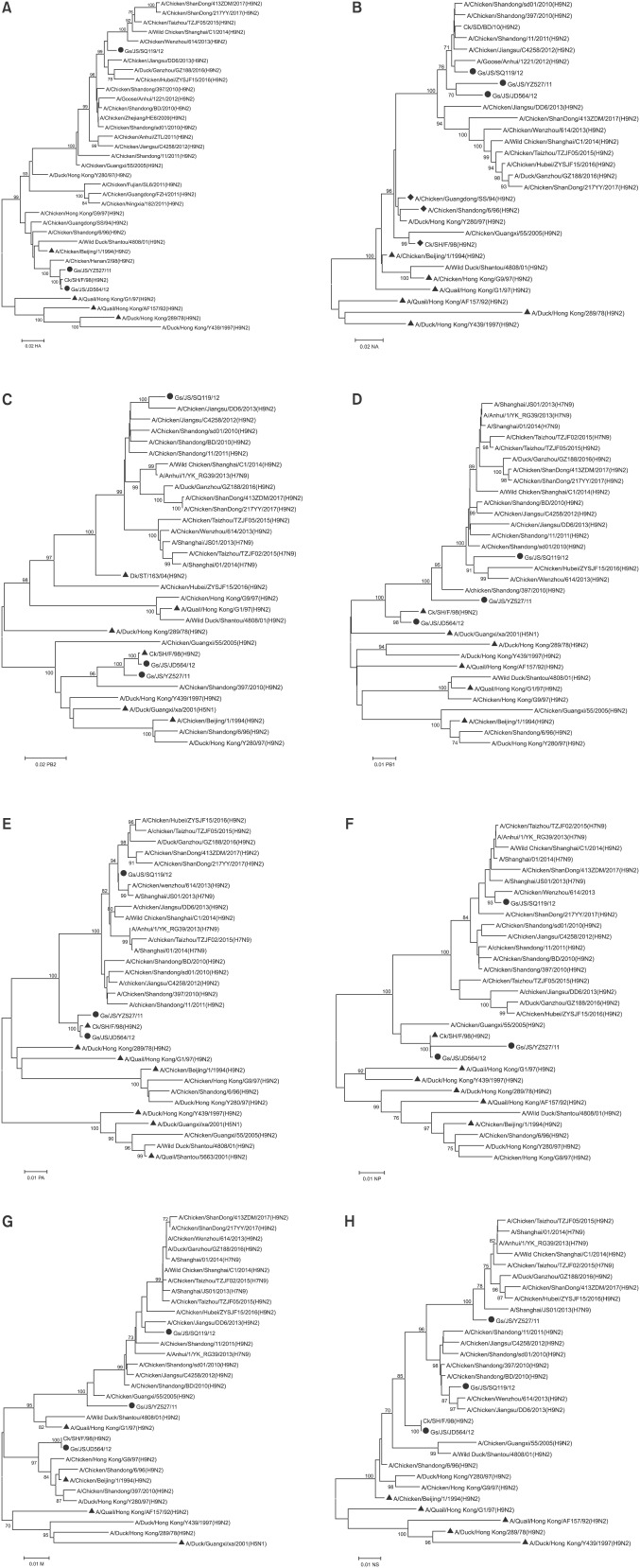
Phylogenetic analyses
Because of the complex evolutionary trajectories of the H9N2 AIVs, the viral genes encoding their surface and internal proteins have been divided into different lineages. Phylogenetic lineages of the eight assessed genes were defined by gene phylogeny as above described based on this system, a phylogenetic analysis of the surface genes showed that the HA genes of the three isolates all clustered in the BJ/94 lineage, represented by the SH/F/98 virus (panel A in Fig. 1). The HA genes of the Gs/JS/YZ527/11 and Gs/JS/JD564/12 viruses shared 99.2% and 99.7% nucleotide identities with that of Ck/SH/F/98, respectively. The HA gene of the Gs/JS/SQ119/12 virus clustered with representatives of the secondary lineage, A/Chicken/Shandong/BD/2010 (H9N2, Ck/SD/BD/10), with 97.7% nucleotide identity. The neuraminidase (NA) genes of the three isolates clustered on a previously unknown branch belonging to the Ck/BJ/1/94-like lineage, represented by the SH/F/98 virus (panel B in Fig. 1), and shared 95.3%, 98.2%, and 96.5% sequence identities with the NA gene of A/Chicken/Shandong/BD/2010 (H9N2, Ck/SD/BD/10), but only 92.9%, 93.4% and 92.7% nucleotide identities with the NA gene of the F/98 virus (panel B in Fig. 1). Therefore, the virus Ck/SD/BD/10 can be regarded as a novel representative on an undefined branch, based on the typical representative F/98-like branch in the Ck/BJ/1/94-like lineage. The phylogenetic tree of the NA genes of the H9N2 AIVs closely reflects the isolation timing of the isolates. The novel sequences of the NA genes indicate the emergence of a new previously undefined cluster since 2010.
Fig. 1. Phylogenetic trees for all genes of avian influenza viruses (AIVs) collected from Eastern China between 2011 and 2015. Representative strains are indicated by solid triangles; the solid circles indicate strains isolated in this study; the solid rhombus indicates three commercial vaccine strains. Unrooted phylogenetic trees were generated by the distance-based neighbor-joining method using the MEGA6.0 software suite [29]. Nucleotide positions: (A) HA, 128–1540; (B) NA, 87–1263; (C) PB2, 28–2262; (D) PB1, 31–2218; (E) PA, 25–2129; (F) NP, 35–1456; (G) M, 56–960; (H) NS, 75–800. The percentage of the replicate trees in which the associated taxa clustered together in a bootstrap test with 1,000 replications are shown next to the branches; values lower than 70% are hidden.
Phylogenetic analysis of the polymerase PB2 gene showed that the PB2 genes of Gs/JS/YZ527/11 and Gs/JS/JD564/12 clustered in the Ck/SH/F/98-like lineage, whereas the PB2 gene of Gs/JS/SQ119/12 clustered in the Dk/ST/163/04-like lineage (panel C in Fig. 1). The polymerase PB1, polymerase PA, and NP genes of isolates Gs/JS/YZ527/11 and Gs/JS/JD564/12 grouped with the Ck/SH/F/98 lineage, whereas all three genes of the Gs/JS/SQ119/12 virus were closely related to those of the Ck/SD/BD/10 virus, which occurred on the same small branches as those of isolate Gs/JS/SQ119/12 (panels D–F in Fig. 1). The M genes of isolates Gs/JS/YZ527/11 and Gs/JS/SQ119/12 clustered in the G1/97-like lineage, and the M gene of isolate Gs/JS/JD564/12 clustered in the Ck/BJ/1/94-like lineage, represented by the SH/F/98 virus (panel G in Fig. 1). The nonstructural (NS) genes of the three isolates all clustered within the Ck/BJ/1/94-like lineage, represented by the SH/F/98 virus, but on three different branches (panel H in Fig. 1).
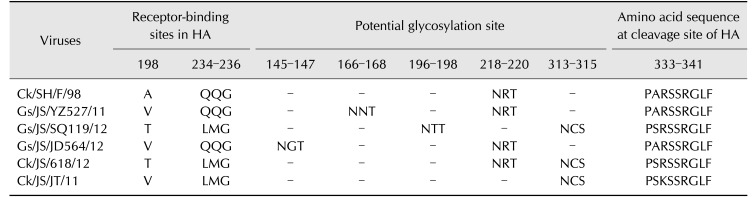
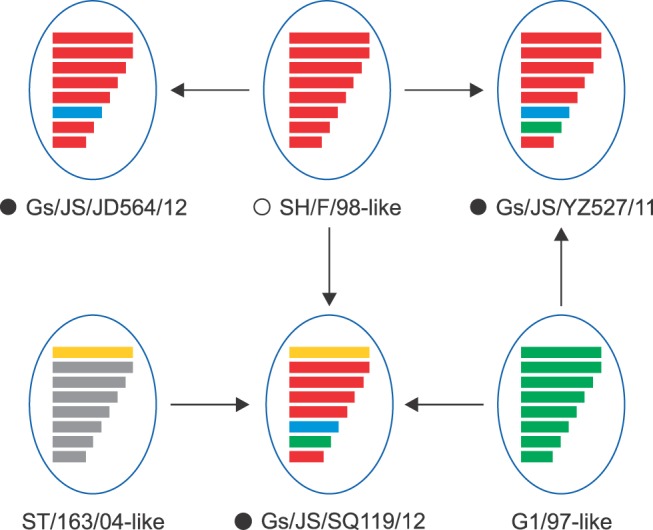
These results indicate that the Gs/JS/YZ527/11 virus is a double-reassortant virus, including G1/97-like and F/98-like sequences; the Gs/JS/JD564/12 virus is almost the prototype F/98-like virus, except for its NA gene; and the Gs/JS/SQ119/12 virus is a triple-reassortant virus, containing G1/97-like, F/98-like, and ST/163/04-like sequences (Fig. 2).
Fig. 2. Analysis of close viral relationships of each gene from the three isolates in this study and their nucleotide homology. The colors represent the different nucleotide homology with each gene of the closest virus. The nucleotide homology in the same lineage between the isolates in this study and chosen H7N9 viruses. Eight gene segments in each of the schematic virus particles are arranged from top to bottom to represent polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acidic (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), matrix (M), and nonstructural (NS) genes, and are indicated in same color with representative viruses for each lineage. The red indicates SH/F/98 lineage; the green indicates HK/G1/97 lineage; the yellow indicates ST/163/04 lineage; the blue indicates an undefined lineage, and the gray indicates another lineage.
Molecular analysis
The Ck/SH/F/98 virus has an alanine at position 198 of the HA protein, whereas Gs/JS/SQ119/12 has a threonine, and Gs/JS/YZ527/11 and Gs/JS/JD564/12 have valine (Table 1). Unexpectedly, at residues 234 to 236 in the antigenic site of HA, Gs/JS/YZ527/11 and Gs/JS/JD564/12 have a glutamine-glutamine- glycine (QQG) sequence, similar to that of the F/98 virus, whereas Gs/JS/SQ119/12 has a leucine-methionine-glycine (LMG) sequence (Table 1). Relative to the F/98 virus, one additional potential glycosylation site was observed at position 145–147 in Gs/JS/JD564/12 and at position 166–168 in Gs/JS/YZ527/11. Interestingly, two new potential glycosylation sites at positions 196–198 and 313–315 were detected in strain Gs/JS/SQ119/12, whereas the potential glycosylation site at position 218–220 in HA of Gs/JS/SQ119/12 was absent (Table 1). These changes in the HA receptor-binding site and its nearby potential glycosylation site also affect the binding of the virus to its host cell and, therefore, the pathogenicity of the influenza viruses. The HA cleavage-site sequence of all the isolates was arginine-serine-serine-arginine (RSSR)↓glycine-leucine-phenylalanine (GLF), which is characteristic of low-pathogenic AIVs (Table 1). Compared with the Ck/SH/F/98 and Gs/JS/JD564/12 viruses, the NS proteins of Gs/JS/YZ527/11 and Gs/JS/SQ119/12 contain a glutamic acid→lysine substitution at residue 227 (E227K), which is closely associated with increased pathogenicity of AIVs. Additionally, the Gs/JS/SQ119/12 virus possessed a leucine at position 226 in HA, as well as a PB2 gene from the ST/163/04-like virus, a gene that had been reported to be closely related with pathogenicity in mammals.
Table 1. Molecular characterization of hemagglutinin (HA) from virus isolates in this study (H9 numbering).
A, alanine; V, valine; T, threonine; QQG, glutamine-glutamine-glycine; LMG, leucine-methionine-glycine; NGT, asparagine-glycine-threonine; NNT, asparagine-asparagine-threonine; NRT, asparagine-arginine-threonine; NCS, asparagine-cysteine-serine; PARSSRGLF, proline-alanine-arginine-serine-serine-arginine-glycine-leucine-phenylalanine; PSRSSRGLF, proline-serine-arginine-serine-serine-arginine-glycine-leucine-phenylalanine; PSKSSRGLF, proline-serine-phenylalanine-serine-serine-arginine-glycine-leucine-phenylalanine.
Infectivity in geese and chickens
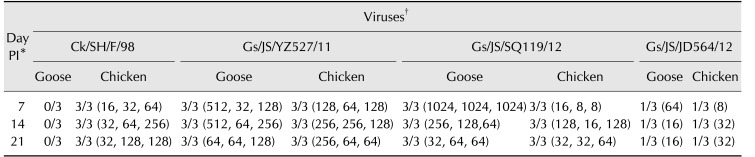
Asymptomatically infected aquatic birds are the natural repositories of potentially spreading viruses. In such cases, the influenza virus may or may not induce an immune response in the host. Seroconversion in waterfowl is considered to reflect the infectivity of influenza viruses. To investigate the infection characteristics of the three isolates in geese and chickens, SPF chickens and 3-week-old geese negative for antibodies against H9 were inoculated with the isolates; subsequently, goose and chicken sera were collected. The Gs/JS/YZ527/11 and Gs/JS/SQ119/12 viruses seriously infected the geese, with high HI titers against the corresponding virus, whereas the Gs/JS/JD564/12 virus caused seroconversion in only one of the three experimental geese tested. The Ck/SH/F/98 virus did not infect the geese (Table 2). Contrary to expectation, similar infection characteristics were observed in all three isolates in SPF chickens. The Gs/JS/YZ527/11 and Gs/JS/SQ119/12 viruses replicated well in chickens, but the Gs/JS/JD564/12 virus infected only one chicken; thus, it had not adapted well to chickens. By contrast, the Ck/SH/F/98 virus infected chickens (Table 2).
Table 2. Analysis of antisera responses in geese and chickens obtained via hemagglutination inhibition (HI) assay.
Data are presented as No. of positive HI antisera responses/No. of birds (HI titers in each bird). HI titers obtained with homologous virus. *The days post-infection (PI), when anti-H9N2 sera from birds infected with the homologous virus were collected (e.g., anti-SH/F/98 virus sera from birds infected with SH/F/98 virus). †Viruses used in animal experiments.
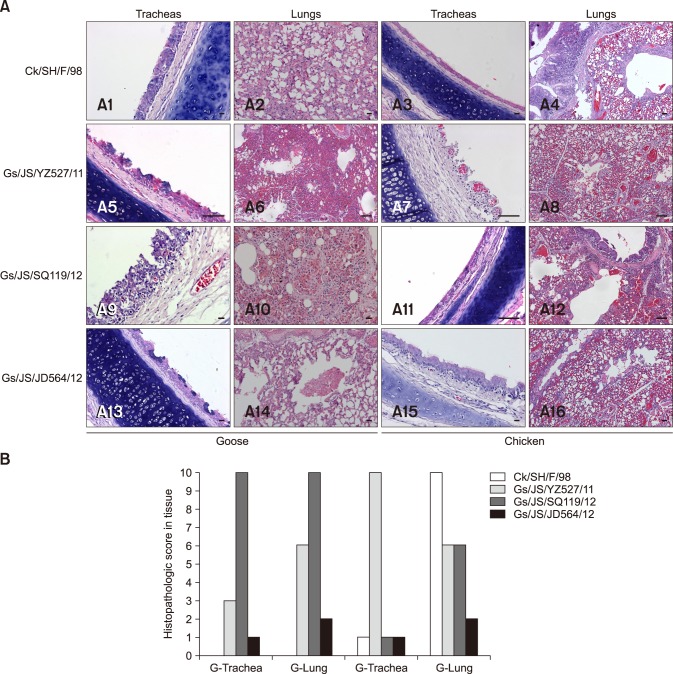
Only minor clinical signs with slight apathy and nasal discharge were observed in all infected geese or chickens except for geese infected with the Ck/SH/F/98 virus. However, the extent of the lesions in the geese and chickens confirmed the infectivity of the three isolates. On day four after inoculation with the isolated viruses, the tracheae and lungs of the geese and chickens showed significant lesions in the groups inoculated with Gs/JS/YZ527/11 or Gs/JS/SQ119/12, with congestion, loss of cilia, and inflammatory cell infiltration. Gs/JS/YZ527/11 induced more pathological changes in chickens than in geese, whereas Gs/JS/SQ119/12 caused more serious lesions in geese than in chickens (Table 2, Fig. 3). Gs/JS/JD564/12 induced only slight lesions, with congestion, in the lungs of geese and chickens (Table 2, Fig. 3), whereas no lesions were observed in tracheal or lung tissues of the geese infected with the Ck/SH/F/98 virus, although much inflammatory cell infiltration was detected in the lungs of the infected chickens (Table 2, Fig. 3). These results indicate that the goose-origin isolates Gs/JS/YZ527/11 and Gs/JS/SQ119/12 stably infected both geese and chickens, whereas strain Gs/JS/JD564/12 isolated from a goose failed to infect geese or chickens.
Fig. 3. (A) Histopathology of SPF chickens and Ck/SH/F/98, Gs/JS/YZ527/11, Gs/JS/SQ119/12, and Gs/JS/JD564/12 infected geese. Lungs and tracheas were collected at 4 days post inoculation fixed in 10% formalin, embedded in paraffin, and sectioned. (B) Pathological changes in trachea and lung in chickens at 4 days after inoculation with the isolated viruses. Tissue sections were observed, and histopathological changes were scored as follows. For trachea, 0, normal; 1, congestion; 2, cilia loss; 3, a few inflammatory cell infiltration; 7, a lot of inflammatory cell infiltration. For lung, 0, normal; 1, congestion; 2, hemorrhage; 3, inflammatory cell infiltration in bronchial submucosa; 7, a lot of inflammatory cell infiltration in bronchial submucosa and alveolus. Average values for three birds are shown. Data are representative of three independent experiments. G, goose; C, chicken. Scale bars = 5 µm (A9 and A10), 10 µm (A1, A3, and A15), 20 µm (A2, A14, and A16), 50 µm (A4–8, A11–13).
Discussion
Since the mid-1990s, the BJ/94-like and G1-like lineages of the subtype H9N2 AIVs have been the significant epidemic strains in China, and the F/98-like and G9-like sublineages derived from the BJ/94-like lineage have recently become new representatives [38]. Strain Ck/SH/F/98, which has been one of the most critical vaccine strains of H9N2 in the poultry industry in eastern China, was chosen, in this study, as the control virus and used in the analysis of the evolution and pathogenicity of the three H9N2 strains isolated from geese.
The amino acid sequence of the cleavage site in the HA protein is one of the most critical factors determining the pathogenicity and virulence of AIVs. However, it has been reported that the amino acid at position 627 in the PB2 protein also has a vital role in the virulence of the influenza viruses [11,12,18]. The three isolates examined in this study, as well as the CK/SH/F/98 virus, have characteristics of low-pathogenic AIVs and include the RSSR↓GLF sequence at the HA cleavage site [17] and glutamic acid at position 627 of the PB2 protein. Changes in the HA receptor-binding site and its nearby potential glycosylation site also affect the binding of the virus to its host cell and, therefore, the pathogenicity of the influenza viruses [8]. HA with the Q226L substitution (H3 numbering; residue 234 in H9 numbering) binds specifically to α-2, 6-linked sialic acid, which is mainly distributed in the human upper respiratory tract (Table 1) [19,26,32]. The receptor-binding sites in HA were quite conserved in our three isolates, except at positions 198 and 233–234. The three isolates from geese contain nine potential glycosylation sites in the HA protein, in contrast to the eight potential glycosylation sites in the control virus Ck/SH/F/98 (positions 29, 82, 141, 218, 298, 305, 492, and 551) (Table 1). Changes in potential glycosylation sites and receptor-binding sites confer virus-specific antigenic characteristics. This site diversity may also be related to differences in the binding characteristics of viruses to their host cells and the pathogenesis of the three isolates examined in this study. However, the molecular mechanism remains to be clarified. The three isolates and the vaccine strain Ck/SH/F/98 all contain drug-resistance mutations at position 13 (proline) in the PB1 protein, position 15 (isoleucine) in the M1 protein, and positions 31 (asparagine) and 55 (phenylalanine) in the M2 protein [21,24,28]. The NS proteins of Gs/JS/YZ527/11 and Gs/JS/SQ119/12 contain the E227K substitution associated with increased pathogenicity in AIVs, which is consistent with the observed EID50 titers of the isolates [21].
The NA genes of the three isolates in this study differed from that of Ck/SH/F/98, exhibiting only 92.9%, 93.4%, and 92.7% nucleotide identity (<95%), but they all shared high nucleotide identity at this gene with the Ck/SD/BD/10 (H9N2) virus. Our analysis also showed that the NA genes of the three isolates contain many mutations and can be considered a new lineage based on the NA gene, represented by the Ck/SD/BD/10 virus. All the genes of Gs/JS/SQ119/12 share high sequence identity with those of the Ck/SD/BD/10 virus. Five genes of Gs/JS/YZ527/11 share high sequence similarity with those of the Ck/SH/F/98 virus, whereas its other genes share high sequence identity with those of the Ck/SD/BD/10 virus, including the NA gene. We postulate that strain Ck/SD/BD/10 had a role in the transmission of the virus across the species barrier from chicken to goose. Our data suggest that different lineages of H9N2 viruses coexist among different species of domestic birds in live poultry markets, thereby accelerating the evolution of the AIVs [39] and providing opportunities for the viruses to spread across different bird species. In this context, the NA genes of the H9N2 viruses may have a key role in facilitating AIV spread across species barriers. In addition, we observed that three internal genes (PB1, M, and NS) of strain Gs/JS/YZ527/11 and all six internal genes of strain Gs/JS/SQ119/12 shared high identity with the corresponding genes of some H7N9 subtype AIVs; specifically, three genes of isolate Gs/JS/SQ119/12 share >98% nucleotide identity with some H7N9 reference viruses, indicating that these two isolates (Gs/JS/YZ527/11 and Gs/JS/SQ119/12) and the H7N9 viruses are closely related. In particular, the NP genes of strain Gs/JS/SQ119/12 showed high nucleotide identity with the NP genes of recently reported H7N9 viruses. These results are consistent with a previous report that H9N2 viruses can donate internal gene segments to the H7N9 subtype [5].
Our infectivity experiment showed that Gs/JS/JD564/12 could infect some geese and chickens, causing tissue lesions, whereas Ck/SH/F/98 only replicated in chickens. Gs/JS/JD564/12 and Ck/SH/F/98 have very different NA genes, suggesting that the NA protein may contribute to the adaptation of influenza viruses to new hosts. The Gs/JS/YZ527/11 and Gs/JS/SQ119/12 viruses also stably infected geese and chickens, implying that the H9N2 AIV can spread between terrestrial birds and waterfowl without hindrance. Through reassortment with other avian viruses, the Goose/Guangdong/1/96 (H5N1, Gs/Gd)-like viruses have generated multiple genotypes of H5N1 viruses, which have crossed from geese to chickens and other terrestrial poultry [7]. The three F/98 prototype isolates also overcame any obstacles to move from their reservoir in geese to chickens. The Gs/JS/SQ119/12 virus, which has a leucine at position 226 in HA and a PB2 gene from the ST/163/04-like virus, and which clustered close to the H7N7 human isolate A/Netherlands/219/2003 [3], may become potentially dangerous to humans.
The H9N2 influenza virus has recently drawn much attention in China because it is a donor of internal genes of the H7N9 virus. However, most studies have focused on the H9N2 AIVs from domestic poultry and mammals, such as chickens, pigs, and ducks, and there have been few reports of H9N2 AIV isolates from geese. In this study, we characterized the sequences and infectivity of three H9N2 AIVs, Gs/JS/YZ527/11, Gs/JS/SQ119/12, and Gs/JS/JD564/12, isolated from geese in Jiangsu Province, China. Our findings show that these H9N2 viruses present pathogenicity either in geese or chickens via the reassortment of segments.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31672516, 31172300, 30670079), supported by grant No. BE2016343 from Jiangsu Province, the Jiangsu University and College Natural Science Foundation (12KJA230002), the Doctoral Program of Higher Education of China (20133250110002) and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Causey D, Edwards SV. Ecology of avian influenza virus in birds. J Infect Dis. 2008;197(Suppl 1):S29–S33. doi: 10.1086/524991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun. 2014;5:3142. doi: 10.1038/ncomms4142. [DOI] [PubMed] [Google Scholar]
- 3.Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, Nolte DL, Ji G, He H. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS One. 2011;6:e17212. doi: 10.1371/journal.pone.0017212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards S. OIE laboratory standards for avian influenza. Dev Biol (Basel) 2006;124:159–162. [PubMed] [Google Scholar]
- 5.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs SE. Avian biology, the human influence on global avian influenza transmission, and performing surveillance in wild birds. Anim Health Res Rev. 2010;11:35–41. doi: 10.1017/S1466252310000058. [DOI] [PubMed] [Google Scholar]
- 7.Guan Y, Peiris JS, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci U S A. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Li J, Cheng X. [Discovery of men infected by avian influenza A (H9N2) virus] Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999;13:105–108. Chinese. [PubMed] [Google Scholar]
- 10.Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 11.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 12.Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 14.Huang R, Wang AR, Liu ZH, Liang W, Li XX, Tang YJ, Miao ZM, Chai TJ. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis. 2013;32:1347–1351. doi: 10.1007/s10096-013-1888-7. [DOI] [PubMed] [Google Scholar]
- 15.Krauss S, Webster RG. Avian influenza virus surveillance and wild birds: past and present. Avian Dis. 2010;54(1 Suppl):394–398. doi: 10.1637/8703-031609-Review.1. [DOI] [PubMed] [Google Scholar]
- 16.Liu D, Shi W, Gao GF. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet. 2014;383:869. doi: 10.1016/S0140-6736(14)60386-X. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Liu X, Cheng J, Peng D, Jia L, Huang Y. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996-2001. Avian Dis. 2003;47:116–127. doi: 10.1637/0005-2086(2003)047[0116:PAOTHG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 18.Mase M, Tanimura N, Imada T, Okamatsu M, Tsukamoto K, Yamaguchi S. Recent H5N1 avian influenza A virus increases rapidly in virulence to mice after a single passage in mice. J Gen Virol. 2006;87:3655–3659. doi: 10.1099/vir.0.81843-0. [DOI] [PubMed] [Google Scholar]
- 19.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munster VJ, Fouchier RA. Avian influenza virus: of virus and bird ecology. Vaccine. 2009;27:6340–6344. doi: 10.1016/j.vaccine.2009.02.082. [DOI] [PubMed] [Google Scholar]
- 21.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 22.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 23.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27:493–497. [Google Scholar]
- 24.Shanmuganatham KK, Jones JC, Marathe BM, Feeroz MM, Jones-Engel L, Walker D, Turner J, Rabiul Alam SM, Kamrul Hasan M, Akhtar S, Seiler P, McKenzie P, Krauss S, Webby RJ, Webster RG. The replication of Bangladeshi H9N2 avian influenza viruses carrying genes from H7N3 in mammals. Emerg Microbes Infect. 2016;5:e35. doi: 10.1038/emi.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi W, Li W, Li X, Haywood J, Ma J, Gao GF, Liu D. Phylogenetics of varied subtypes of avian influenza viruses in China: potential threat to humans. Protein Cell. 2014;5:253–257. doi: 10.1007/s13238-014-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Xu X, Liu Q, Liang D, Li C, He Q, Jiang J, Cui Y, Li J, Zheng L, Guo J, Xiong Y, Yan J. Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect Genet Evol. 2013;20:471–475. doi: 10.1016/j.meegid.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010;146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uyeki TM, Chong YH, Katz JM, Lim W, Ho YY, Wang SS, Tsang TH, Au WW, Chan SC, Rowe T, Hu-Primmer J, Bell JC, Thompson WW, Bridges CB, Cox NJ, Mak KH, Fukuda K. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg Infect Dis. 2002;8:154–159. doi: 10.3201/eid0802.010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandegrift KJ, Sokolow SH, Daszak P, Kilpatrick AM. Ecology of avian influenza viruses in a changing world. Ann N Y Acad Sci. 2010;1195:113–128. doi: 10.1111/j.1749-6632.2010.05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72:7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, Liu X, Shu Y, Peng D, Jiang T. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe. 2013;14:446–452. doi: 10.1016/j.chom.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003 (H9N2) virus. Microbes Infect. 2004;6:919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Liu L, Pu J, Zhao J, Sun Y, Shen G, Wei H, Zhu J, Zheng R, Xiong D, Liu X, Liu J. Risk perceptions for avian influenza virus infection among poultry workers, China. Emerg Infect Dis. 2013;19:313–316. doi: 10.3201/eid1902.120251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998-2002) J Gen Virol. 2008;89:3102–3112. doi: 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Wu J, Zeng X, Huang G, Zou L, Song Y, Gopinath D, Zhang X, Kang M, Lin J, Cowling BJ, Lindsley WG, Ke C, Peiris JS, Yen HL. Isolation of H5N6, H7N9 and H9N2 avian influenza A viruses from air sampled at live poultry markets in China, 2014 and 2015. Euro Surveill. 2016;21:30331. doi: 10.2807/1560-7917.ES.2016.21.35.30331. [DOI] [PMC free article] [PubMed] [Google Scholar]