Abstract
Esophagitis, whether caused by acid reflux, allergic responses, graft-versus-host disease, drugs, or infections, is a common condition of the gastrointestinal tract affecting nearly 20% of the US population. The instigating agent typically triggers an inflammatory response. The resulting inflammation is a risk factor for the development of esophageal strictures, Barrett esophagus, and esophageal adenocarcinoma. Research into the pathophysiology of these conditions has been limited by the availability of animal and human model systems. Three-dimensional organotypic tissue culture (OTC) is an innovative three-dimensional multicellular in vitro platform that recapitulates normal esophageal epithelial stratification and differentiation. We hypothesized that this platform can be used to model esophagitis to better understand the interactions between immune cells and the esophageal epithelium. We found that human immune cells remain viable and respond to cytokines when cultured under OTC conditions. The acute inflammatory environment induced in the OTC significantly affected the overlying epithelium, inducing a regenerative response marked by increased cell proliferation and epithelial hyperplasia. Moreover, oxidative stress from the acute inflammation induced DNA damage and strand breaks in epithelial cells, which could be reversed by antioxidant treatment. These findings support the importance of immune cell–mediated esophageal injury in esophagitis and confirms the utility of the OTC platform to characterize the underlying molecular events in esophagitis.
Inflammatory conditions of the gastrointestinal tract, such as reflux esophagitis, Helicobacter pylori–induced gastritis, graft-versus-host disease, or inflammatory bowel disease, are common conditions. These inflammatory disorders cause pain and distress acutely, which adversely affects the quality of life,1, 2 but they can also lead to the induction of lumen-obstructing strictures or dysplasia and cancer.3, 4
Research into inflammatory conditions of the esophagus, including reflux esophagitis, eosinophilic esophagitis, and graft-versus-host disease, have been hampered by the limited availability of suitable approaches to model these conditions. Much of the past work has relied on the availability of human patient biopsies, which are difficult to obtain and primarily suitable for descriptive studies. Immortalized cell lines representing normal squamous, Barrett, and esophageal adenocarcinoma, are available5; however, two-dimensional cultured–immortalized cells cannot effectively model complex interactions between epithelial cells and their microenvironment. Animal models are another important tool of biomedical research.6, 7, 8 Although animal models have yielded insights,6 they are not ideal because of anatomic differences between mice and humans at the squamocolumnar junction.
Organotypic culture (OTC) systems allow for the co-culture of immortalized human epithelial cell lines together with primary fibroblasts in three-dimensional (3D) tissue reconstructions.9 Under OTC conditions, embryonic human esophageal fibroblasts are embedded in a collagen matrix, and epithelial cells are seeded on top. Exposure of the epithelial cells to the air–liquid interface triggers differentiation, producing a mature epithelium.9 A strength of the OTC approach is the ability to perform physiologically relevant in vitro studies under controlled conditions. In past studies, we have used OTC to model early molecular events that promote the development of Barrett esophagus (BE) and esophageal adenocarcinoma. We demonstrated that expression of the proinflammatory enzyme cyclooxygenase 2 in human telomerase reverse transcriptase–immortalized human esophageal epithelial (STR) cells led to prostaglandin E2 production and the induction of a mucin-secreting metaplasia with features of BE but only under OTC conditions.10 In addition, with the use of OTCs we determined that the onset of BE metaplasia is a multistep process that requires increased proliferation, senescence inhibition, and epigenetic alterations.11 Moreover, when evaluated under OTC conditions, the Barrett cell lines CP-A, CP-B, CP-C, and CP-D demonstrate distinct phenotypes.12 These new phenotypes emerged only as a result of the 3D OTC system.
Here, we modify an established 3D organotypic culture system to model esophageal inflammation.13 Under 3D OTC conditions, human esophageal keratinocytes undergo a complete differentiation and stratification to produce a fully mature epithelium.9 To this culture we added human peripheral blood mononuclear cells (PBMCs) and stimulated them to induce an acute inflammatory response. After the addition and appropriate stimulation of immune cells in OTCs, we studied the associated morphologic changes and the effect of the inflammatory environment on epithelial differentiation, proliferation, and apoptosis. Finally, we sought to identify the contribution and potential mechanism of immune response to DNA damage. This physiologically relevant human cell culture system thus provides a novel platform for the study of human esophagitis and the effects of inflammation on esophageal epithelial responses.
Materials and Methods
Cell Lines
Immortalized human primary normal esophageal epithelial/human telomerase reverse transcriptase9 cells designated as STR herein were maintained in keratinocyte serum-free medium (Invitrogen, Carlsbad, CA) supplemented with epidermal growth factor, pituitary bovine extract, and 1% penicillin/streptomycin. FEF3 human esophageal fibroblasts9 were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Immune Cells and Cytokine Treatment
PBMCs were collected from de-identified healthy volunteers and isolated freshly by the Human Immunology Core at the University of Pennsylvania under an Institutional Review Board–approved protocol. Proinflammatory cytokines IL-7 (10 ng/mL; Cell Signaling Technologies, Danvers, MA) and IL-15 (20 ng/mL; ProSpec, Rehovot, Israel) were added to the cell culture media; IL-2 (10 U/mL; BD Biosciences, San Jose, CA) was also added to support PBMC viability. The cytokines were replenished during each media exchange.
3D Organotypic Culture
Organotypic culture was performed as described previously.9 The fibroblast feeder layer and 6.75 × 105 PBMCs were embedded within a collagen/Matrigel matrix and was allowed to mature for 7 days, after which time 5 × 105 epithelial cells were seeded on top and allowed to grow to confluence for an additional 4 days. Then the culture media level was reduced, exposing the keratinocytes to the air–liquid interface, which stimulates epithelial differentiation into a multilayer epithelium typical for the esophagus. On day 15 OTCs were harvested for histologic examination, RNA, and/or protein isolation. N-acetyl cysteine (5 mmol/L; Sigma-Aldrich, St. Louis, MO) was added to the culture media as an antioxidant in some experiments as indicated, based on a prior published study.14
RNA Isolation, Reverse Transcription, and Real-Time Quantitative PCR
The epithelial layer was peeled off the collagen base, and total RNA was isolated (Nucleospin RNA II kit; Macherey-Nagel, Düren, Germany), according to the manufacturer's instructions. The quantity of isolated RNA samples was checked by spectrophotometry (NanoDrop 3.1.0; NanoDrop Technologies, Rockland, DE). Total RNA (1 μg) was used for cDNA synthesis using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). All real-time PCR reactions were performed using the StepOne Plus instrument, and the amplifications were performed using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase was used as the normalization control. The relative changes in gene expression were determined using the ΔΔCT method as described in Applied Biosystems User Bulletin No. 2 (P/N 4303859).
Western Blot Analysis
Denatured protein (20 μg) was fractionated on a NuPAGE Bis-Tris 4% to 12% gel (Life Technologies, Carlsbad, CA). After electrotransfer, Immobilon-P membranes (Millipore, Billerica, MA) were blocked with phosphate-buffered saline with Tween 20 (PBST) containing 5% milk, followed by overnight incubation with rabbit anti-interferon (IFN) regulatory factor 1 (IRF-1; dilution 1:1000; Abcam, Cambridge, MA) at 4°C. Mouse anti–α-tubulin (dilution 1:10,000; Sigma-Aldrich) was used as a loading control. Rabbit secondary antibody (Sigma-Aldrich) was used at a dilution of 1:10,000. Targeted proteins were visualized using a chemiluminescence detection system (Amersham ECL or ECL Prime; GE Healthcare Life Sciences, Pittsburgh, PA).
Immunohistochemistry
Five-micromolar paraffin-embedded sections were deparaffinized, and heat-mediated antigen retrieval was performed with 10 mmol/L citric acid buffer (pH 6.0). Endogenous peroxidases were quenched using hydrogen peroxide before sections were incubated in avidin D and biotin blocking reagents. Sections were incubated with primary rabbit CD45 (dilution 1:50; Abcam), rabbit Ki-67 (dilution 1:200; Abcam), mouse filaggrin (dilution 1:50; Abcam), rabbit loricrin (dilution 1:2000; Abcam), or anti-IRF (Cell Signaling Technologies; catalog no. 8478; dilution 1:150) antibodies and biotinylated secondary antibodies and an avidin-horseradish peroxidase conjugate (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) according to the manufacturer's protocol. The signal was developed using the 3, 39-diaminobenzidine substrate kit (Vector Laboratories). Sections were counterstained with hematoxylin. Slides were visualized under Nikon E600 brightfield microscope (Melville, NY), and digital images were taken with iVision software version 4.0.16 (Atlanta, GA). For Ki-67+ cell quantitation, images were taken to cover the entire OTC section. We analyzed three different tissue sections per OTC, and three separate OTCs for each treatment. All images were analyzed with ImageJ software version 1.47 (NIH, Bethesda, MD; http://imagej.nih.gov/ij), counting all of the Ki-67+ nuclei and positive cells.
Immunofluorescence
Frozen sections were fixed in 10% Neutral Buffered Formalin and blocked in Blocking Buffer. Sections were incubated with primary γH2AX (dilution 1:250; Cell Signaling Technologies), primary anti-CD45 (dilution 1:100; catalog no. 136-485; Cell Signaling Technologies), and fluorochrome-conjugated secondary antibodies. Nuclei were counterstained with DAPI. Slides were visualized under Nikon E600 fluorescence and Eclipse Ti-U confocal microscopes (Nikon). Digital images were taken with iVision and Metamorph software version 64 bit 7.7.5.0 (Sunnyvale, CA). All images were analyzed with ImageJ software (NIH).
TUNEL Assay
For the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay, apoptosis in paraffin-embedded sections was assessed using an In Situ Cell Death Detection Kit In (TMRM Red; Roche, Branchburg, NJ) according to the manufacturer's instructions. Slides were visualized under Nikon E600 brightfield microscope, and digital images were taken with iVision software. Images were taken to cover the entire OTC section. We analyzed three different tissue sections per OTC, and three separate OTCs for each treatment. All images were analyzed with ImageJ software (NIH), counting all of the nuclei and positive cells.
Comet Assay
After day 15, epithelial layers of OTCs were peeled off the collagen base and then gently trypsinized, resulting in a suspension of single epithelial cells. These cells were added to 1% low-melting agarose at 37°C at a cell density of 1 × 105/mL. The Low gelling 2-Hydroxyethylagarose (Sigma-Aldrich) and cell mixture was applied to Comet slides (Trevigen, Gaithersburg, MD) and then incubated in prechilled alkaline lysis buffer overnight at 4°C as described.15 After lysis, horizontal electrophoresis was performed for 25 minutes at 10 V. DNA content was stained with 10 μg/mL propidium iodide for 20 minutes in the dark at room temperature. Comet slides were visualized under a Nikon E600 fluorescence microscope. Digital images were taken with iVision software. Comets were analyzed with OpenComet software version 1.3.0.16
Flow Cytometry
Intracellular levels of reactive oxygen species (ROS) in epithelial cells were determined by flow cytometry with 2′,7′-dichlorodihydrofluorescein diacetate (DCF) dye (Life Technologies). In brief, DCF (final concentration 10 μmol/L) was added to the cell culture media for 60 minutes, then the media was changed and OTCs were further cultured for up to 3 hours before peeling the epithelial layer and trypsinizing into single cells. After staining, cells were washed and then analyzed in Dulbecco phosphate-buffered saline containing 1% bovine serum albumin using a FACS Calibur (BD Biosciences). FlowJo software version X 10.0.7r2 (TreeStar, Ashland, OR) was used for data analysis.
Statistical Analysis
GraphPad Prism software version 7.0 (GraphPad Software, Inc., San Diego, CA) software was used for all statistical analyses. Unpaired t-test was used for comparisons between two groups. Data from multiple groups were analyzed using one-way analysis of variance with Tukey's post hoc test. All data were expressed as means ± SEM. P values ≤ 0.05 were designated as significant.
Results
Organotypic Culture Environment Sustains Immune Cells and Permits Their Normal Activation When Stimulated by Cytokines
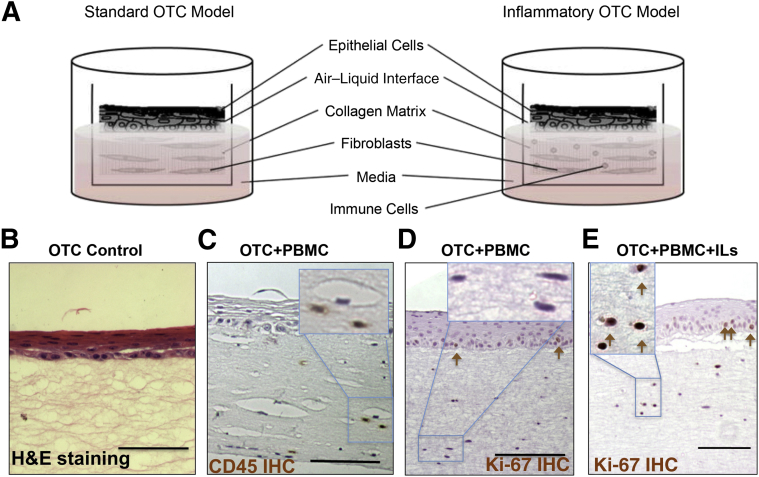
Organotypic culture systems allow for the co-culture of immortalized normal human esophageal epithelial cells together with primary human fibroblasts in a 3D tissue reconstruction that recapitulates normal esophageal epithelial stratification and differentiation (Figure 1, A and B).9 OTC is a novel means by which to perform physiologically relevant in vitro studies under controlled conditions.10, 11 To develop a human in vitro model of esophageal inflammatory conditions such as esophagitis, we explored adding human immune cells to the 3D OTC system to evaluate their behavior and function. Human PBMCs were included in the collagen/Matrigel extracellular matrix at the initiation of the culture, before the establishment of the epithelial cell layer (Figure 1, A and C). To ensure immune cell viability during the 12-to 14-day culture period, IL-2 was added to the culture media. PBMCs remained viable throughout the culture period, as indicated by CD45+-stained cells in the collagen matrix/stromal compartment of the OTCs (Figure 1C). Interestingly, no spontaneous activation of immune cells was observed, demonstrated by the absence of lymphocyte proliferation (Figure 1D), despite that the PBMCs, fibroblasts, and epithelial cells were derived from different donor sources and likely would have had a mismatch in their major histocompatibility complex antigens.
Figure 1.
Modeling esophageal inflammation. A: Illustration of the standard OTC method and the modification introduced by incorporating PBMCs into the collagen matrix to model inflammation. B: Representative images show H&E staining of normal human esophageal keratinocytes grown under standard OTC conditions. C: CD45 IHC staining of an OTC with human PBMCs and IL-2 added to maintain PBMC viability. CD45+ staining indicates the presence of immune cells in the collagen matrix. Inset: Higher power magnification of CD45+ cells. D: IHC staining for the cell proliferation marker Ki-67 in an OTC+PBMC culture with only IL-2 to maintain PBMC viability. Ki-67+ cells noted in the epithelium (brown arrows) but not in stromal cells. Inset: Higher power magnification of Ki-67− stromal cells. E: IHC staining for the cell proliferation marker Ki-67 in an OTC+PBMC+IL culture (IL-2, -7, and -15 included) to induce an acute inflammatory response. Ki-67+ cells noted in both the epithelium and stromal cells (brown arrows). Inset: Higher power magnification of Ki-67+ stromal cells. Scale bars: 75 μm (B–D). Original magnification: ×400 (B–D, insets). H&E, hematoxylin and eosin; IHC, immunohistochemical; OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
To elicit a robust T helper cell type 1 (Th1) acute inflammatory response, we elected to include the potent proinflammatory cytokines IL-7 and -15 in the culture media, in addition to IL-2. IL-7 and -15 are both tissue-derived cytokines most abundantly expressed by stromal and epithelial cells, including keratinocytes.17, 18, 19, 20 Both cytokines were potent inducers of Th1 acute inflammatory cytokines, including IFN-γ in T cells and monocytes, and both have profound effects on T-cell survival and proliferation, including helper (CD4+) and effector (CD8+) T cells and natural killer cells, during all phases of T-cell development.20, 21
Unlike IL-2 alone, these three cytokines together supported the induction of an acute inflammatory response in the OTC-PBMC co-culture environment. The addition of IL-7 and -15 induced proliferation in the immune cells, as indicated by the significant increase in the number of Ki-67+ cells in the stroma (Figure 1, D and E) and the increased number of CD45+ immune cells in the OTC-PBMC cultures (Figure 2A). This effect was not observed when PBMCs were co-cultured with only IL-2 (Figure 2A). In addition to the increased number of PBMCs in the subepithelial matrix, there was some infiltration of lymphocytes into the OTC epithelium, which is characteristic of esophagitis (Figure 2B). However, even under these favorable conditions, the CD45+ cells constituted a small fraction of the epithelial compartment in the OTC-PBMC cultures, about 0.6% based on counts of >6000 total cells.
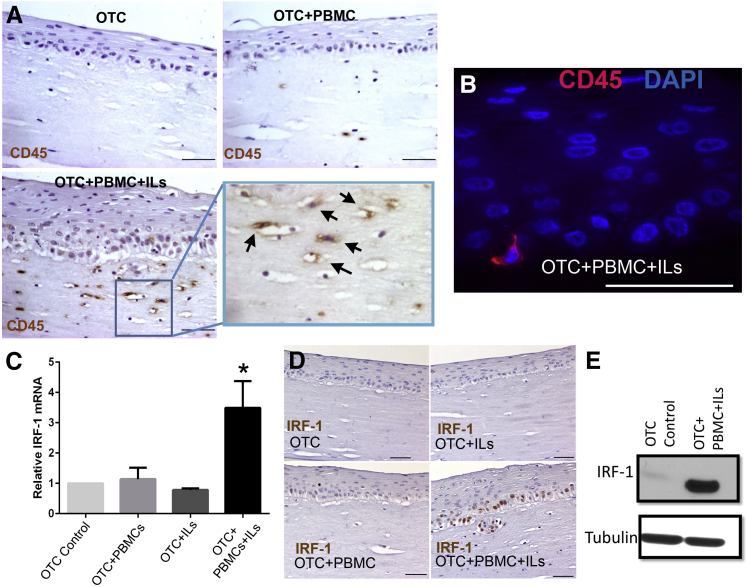
Figure 2.
Organotypic culture system is permissive for the proliferation and activation of immune cells. A: Representative images show immunohistochemical staining for CD45+ cells, a marker of leukocytes, in OTC control, OTC+PBMC (with IL-2 only for PBMC viability), and OTC+PBMC+IL (IL-2, -7, and -15) cultures. Boxed area is shown at higher magnification to the right. Arrows indicate positive CD45+ cells in the collagen matrix of the OTC+PBMC+IL culture. B: Representative immunofluorescence staining shows the presence of infiltrating CD45+ cells (red CD45+ cell) in the OTC+PBMC+IL epithelium (blue indicates DAPI-stained nuclei). C: Real-time quantitative PCR analysis of relative mRNA expression of IRF-1 in the epithelium of the OTCs. D: Pattern of immunohistochemical staining for IRF-1+ cells in OTC control, OTC+IL, OTC+PBMC (with IL-2 only), and OTC+PBMC+IL (IL-2, -7, and -15) cultures. E: Representative Western blot analysis for IRF-1 protein levels in the epithelium from the OTC control and OTC+PBMC+IL (IL-2, -7, and -15)-treated cultures. Blots were stripped and reprobed for the loading control tubulin. Data are expressed as means ± SEM (C). n = 3 for OTC+PBMC (+IL-2 only) and OTC+ILs (IL-2, -7, and -15); n = 7 for OTC control and OTC+PBMC+ILs (IL-2, -7, and -15). ∗P < 0.05, analysis of variance and Tukey rank mean testing. Scale bars: 50 μm (A, B, and D). Original magnification, ×400 (A, boxed area). IRF-1, interferon regulatory factor-1; OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
A general characteristic of immune cell activation is the increased production of cytokines. We were unable to directly detect the canonical Th1 cytokine IFN-γ in cell culture media from the OTC+PBMC+IL cultures despite multiple attempts (data not shown). We suspected this is because of the dense OTC collagen matrix in which the PBMCs are maintained in, because we readily detected increased IFN-γ levels in the cell culture media when PBMCs were cultured in a soft Matrigel matrix and exposed to the same Th1 proinflammatory cocktail IL-2, -7, and -15 (data not shown). As a surrogate marker, we examined the overlying epithelium in the OTCs for expression of IRF-1, a member of the IFN regulatory transcription factor family and a well-known target gene of IFN-γ signaling.22 We observed a highly significant increase in IRF-1 mRNA and protein levels detected in the epithelium from the OTC+PBMC+IL cultures (Figure 2, C–E). However, IRF-1 mRNA and protein were not increased in the epithelium from control cultures treated either with the Th1-promoting ILs or with PBMCs alone (Figure 2, C–E), indicating this response was specific to the OTC+PBMC+IL cultures. Moreover, IRF-1 was identified in the nucleus of the overlying epithelial cells (Figure 2D), confirming epithelial cells as the source of the increased IRF-1 mRNA and protein. Together these findings established that human PBMCs incorporated into the OTC environment remained viable and could proliferate and produce IFN-γ when provoked by proinflammatory cytokines.
Acute Inflammatory OTC Environment Induces a Hyperplasia of the Epithelium That Is Associated with Increased Expression of Markers for Mature Squamous Cells
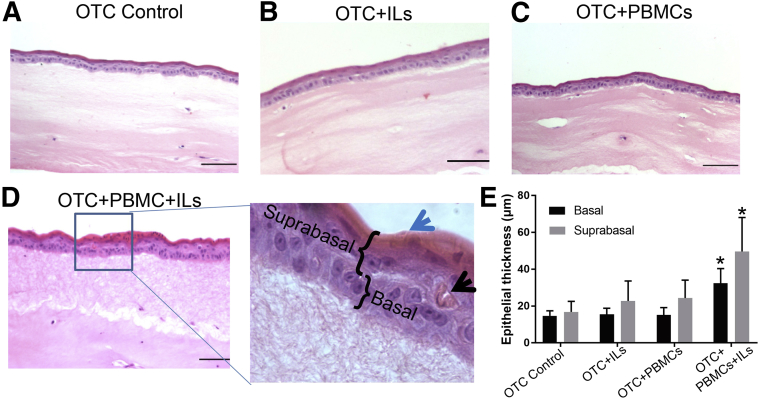
We next sought to identify the effects of activated immune cells on the overlying esophageal epithelial cells. To control for the effects of IL-2, -7, and -15, or the effects of PBMCs+IL-2 separately, OTCs with all of the ILs or immune cells with only IL-2 were established, in addition to the combined OTC+PBMCs+ILs. These OTCs were fixed and embedded and then stained with hematoxylin and eosin to examine for morphologic changes in the overlying epithelium. Cultures with cytokine cocktail–treated PBMCs displayed a highly significant increase in epithelial thickening, both in the basal (15 ± 2.8 to 32 ± 6 μm) and suprabasal (17 ± 5.8 to 50 ± 18 μm) compartments (Figure 2A, Figure 3, A–D). Some of the increase in the suprabasal compartment was from the noticeable increase in the uppermost layer. This doubling of epithelial thickness in the OTCs was significant and was confirmed by measurements across multiple cultures (Figure 3E).
Figure 3.
Morphologic effects of activated immune cells on esophageal epithelium. A–D: Representative images of hematoxylin and eosin staining show the morphologic differences of OTCs under different culture conditions, including Control OTC (A); OTC with only IL-2, -7, and -15 added (OTC+ILs) (B); OTC with only PBMCs and IL-2 added to maintain PBMC viability (OTC+PBMC) (C); and OTC with both PBMCs and the IL-2, -7, and -15 cocktail (OTC+PMBC+ILs) (D). Inset: Significant epithelial thickening was observed together with expansion of mature cell layers (blue arrow) and regenerative keratin pearls (black arrow) only in the OTC+PBMC+IL culture. These changes were not present in control cultures. Representative basal and suprabasal regions indicated. E: Quantification of epithelial thickness in microns, measured in four to five regions on each section and from three different tissue sections per culture, three separate cultures in total for each treatment. Data are expressed as means ± SD (E). n = 3 for all cultures. ∗P < 0.05, analysis of variance and Tukey rank mean testing. Scale bars: 50 μm (A–D). Original magnification, ×400 (D, insets). OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
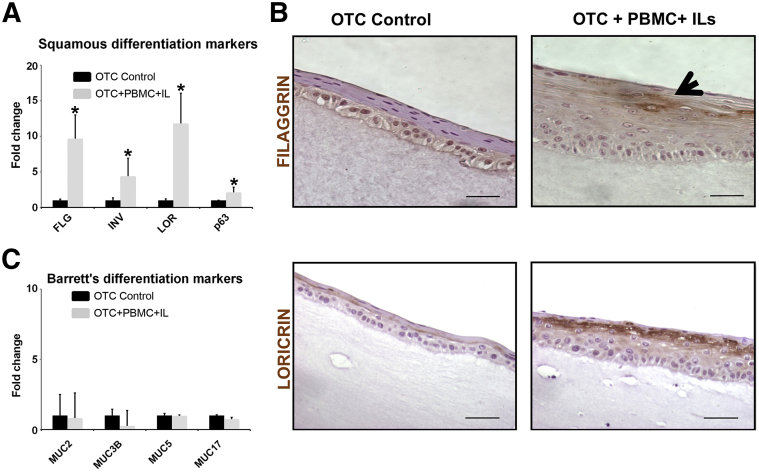
Morphologically, there was a noticeable expansion of the differentiated suprabasal epithelial compartment marked by eosinophilic keratin pearl formation (Figure 3D). Cultures were therefore examined for alterations in epithelial differentiation markers. mRNA from the epithelial layer was isolated after harvesting the OTCs and analyzed by real-time quantitative PCR for changes of squamous differentiation markers filaggrin, loricrin, and involucrin,23 and also for the esophageal epithelium basal cell marker p63. In the presence of the acute inflammatory environment (ILs+PBMCs), filaggrin, involucrin, and loricrin mRNA levels in the epithelium were increased 5- to 12-fold compared with control cultures, whereas p63 was increased only twofold (Figure 4A). The increase in the expression of the squamous differentiation markers was confirmed by immunohistochemical staining of the OTC+PBMC+IL culture compared with the control OTCs (Figure 4B). Because chronic reflux esophagitis in vivo is associated with induction of an intestinalized metaplasia known as BE,24 we also screened our cultures for changes in markers associated with BE, including mucin (MUC)2, MUC3B, MUC5, and MUC17.5 However, no significant changes were found in the expression of these markers in any of the OTCs (Figure 4C). Together these findings were consistent with the induction of epithelial hyperplasia in the OTC epithelium in response to an acute inflammatory environment.
Figure 4.
Acute inflammatory conditions promote expression of squamous cell but not Barrett esophagus gene markers. A: Real-time quantitative PCR analysis of mRNA levels for markers of squamous differentiation, FLG, INV, LOR, and basal cell marker p63. B: Representative immunohistochemical staining for squamous differentiation markers FLG (arrow) and LOR in OTC control and OTC+PBMC+IL (IL-2, -7, and -15) cultures. C: Real-time quantitative PCR analysis of mRNA levels for Barrett esophagus–associated markers. Data are expressed as means ± SD (A and C). n = 4. ∗P < 0.05, unpaired t-test. Scale bars: 50 μm (B). FLG, filaggrin; INV, involucrin; LOR, loricrin; MUC2, mucin 2; MUC3B, mucin 3B; MUC5, mucin 5, and MUC17, mucin 17; OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
Proinflammatory Environment Modeled with the OTC System Alters Cell Proliferation and Cell Death in the Epithelium
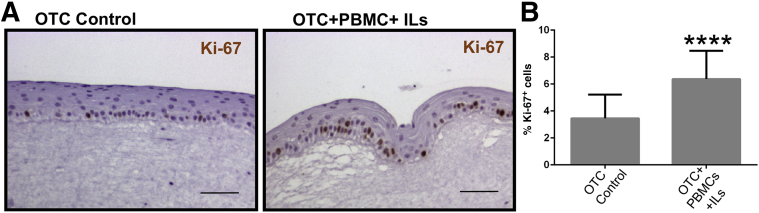
The thickening of the OTC epithelium in the acute inflammatory environment suggested there might be changes in epithelial cell proliferation. We examined for changes in proliferative rates by staining for the proliferation marker Ki-67 in the epithelial cell layer. There were significantly more Ki-67+ basal epithelial cells in the OTCs with PBMCs and the IL cocktail than control cultures (Figure 5A). When quantified across multiple cultures, cell proliferation was increased twofold (3.4 ± 1.8 Ki-67+ cells compared with 6.4 ± 2.1% Ki-67+ cells with PBMCs+ILs) (Figure 5B).
Figure 5.
The acute inflammatory environment induces OTC epithelial cell proliferation. A: Representative Ki-67 immunohistochemical staining in control OTC and the OTC treated with both PBMCs and the proinflammatory IL cocktail (IL-2, -7, and -15). B: Quantitation of Ki-67+ staining. The number of Ki-67+ cells was expressed as a percentage of the total number of keratinocytes in a counted field. Data are expressed as means ± SD (B). n = 3 cultures per condition, with 500 cells counted per culture. ∗∗∗∗P ≤ 0.0001, unpaired t test. Scale bars: 50 μm (A and B). OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
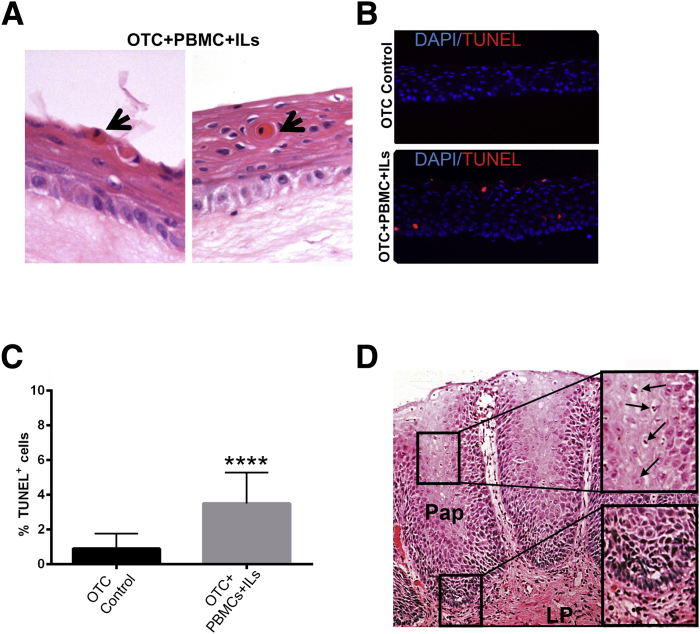
This elevated proliferative rate was offset in part by increased epithelial cell apoptosis. Dyskeratotic cells with hypereosinophilic cytoplasm and shrunken hyperchromatic nuclei were readily identified in the suprabasal and superficial epithelial compartments of the PBMC-IL–treated OTCs (Figure 6A) but not control cells. Because these cells might be undergoing apoptosis, we confirmed and quantified the onset of programed cell death in the OTCs by TUNEL staining. Although there were rare TUNEL+ cells in the control OTCs (Figure 6B), there was significantly increased numbers of TUNEL+ epithelial cells in the epithelium of the OTCs with the acute inflammatory environment (Figure 6B). Counts of apoptotic cells over multiple OTC+PBMC+IL cultures revealed a fourfold increase in TUNEL+ cells (3.5% ± 1.8% with PBMCs+ILs) compared with control OTCs (0.9% ± 0.8% control cultures) (Figure 6C). Together, the epithelial responses observed in the OTCs with PBMCs+ILs recapitulate several features associated with acid reflux esophagitis in humans (Figure 6D). Although we did not observe dilated intercellular spaces, elongated papillae, or ballooning degeneration of epithelial cells25 in the OTCs, we found increased inflammatory cell infiltration of subepithelial space, enhanced basal cell proliferation, general epithelial hyperplasia with expansion of the suprabasal compartment, and increased numbers of apoptotic cells in our engineered model system (Figure 6D).
Figure 6.
Epithelial cell apoptosis is significantly increased in OTCs treated with PBMCs and IL-2, -7, and -15. A: Representative H&E staining shows increased level of dyskeratotic cells in the suprabasal compartment of the OTC+PBMC+IL (IL-2, -7, and -15) cultures (black arrows). B: Representative epifluorescence images of combined nuclear DAPI (blue) and the apoptosis marker TUNEL (red) staining in OTC control and OTC+PBMC+IL cultures. C: Quantitative analysis of TUNEL+ cells. TUNEL+ cells were expressed as a percentage of the total number of DAPI-stained cells in a field. D: H&E staining of representative biopsy of human gastroesophageal reflux disease–induced esophagitis. Pathologic features include elongated papillae, intrapapillary vessel dilation, enhanced proliferating of basal zone cells, increased cell apoptosis, and dilation of intracellular spaces. Upper inset: Enlargement of suprabasal zone with apoptotic cells (arrows). Apoptotic cells are marked by nuclear and cytoplasmic condensation and fragmentation. Lower inset: Enlargement of basal zone with hyperplasia and increased subepithelial lymphoid infiltration. Data are expressed as means ± SD (C). n = 3 cultures per condition, with 500 cells counted per culture. ∗∗∗∗P ≤ 0.0001, unpaired t-test. Original magnification: ×400 (A and B); ×200 (D, main image); ×400 (D, insets). H&E, hematoxylin and eosin; LP, lamina propria; OTC, organotypic tissue culture; Pap, papilla; PBMC, peripheral blood mononuclear cell; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling.
Acute Inflammatory Conditions in the OTC Induces Epithelial DNA Damage
Esophagitis, and the elevated ROS associated with it, can damage the epithelial cells, including the induction of DNA damage and DNA strand breaks (DSBs).26, 27, 28 However, the primary source for the elevated ROS in the epithelial cells, and resulting DNA damage, is not fully understood. Gastric acid and bile have been directly implicated in ROS induction and DNA damage in epithelial cells by in vitro studies, in the absence of immune cells. However, in a recent study of 12 patients who had their antireflux medication withdrawn, endoscopic biopsies of the resulting esophagitis revealed that the acute epithelial response included basal cell hyperplasia and an influx of T cells but not loss of surface epithelial cells from the caustic injury.29 This work suggested that much of the epithelial injury in response to the reflux was caused by cytokine production and the resultant immune response and not the reluxate. Given this association of epithelial cell DNA damage and DSBs with the caustic acid and bile injury,14, 30, 31, 32 we determined whether in our model system we could similarly demonstrate that an acute inflammatory environment could similarly raise epithelial cell ROS and damage DNA.
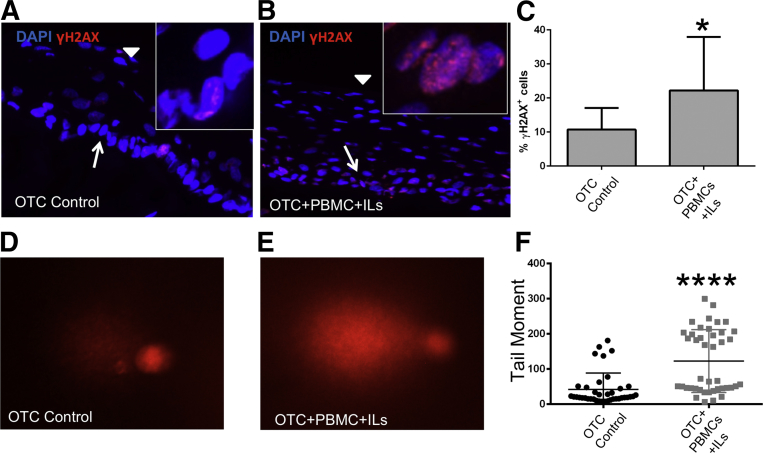
The onset of DNA damage, especially in the setting of DSBs, induced a complex cellular response. A key component of the response was the phosphorylation of histone 2A, yielding γH2AX. γH2AX is a key factor in the DNA-damage repair machinery.33 Detection of phosphorylated γH2AX, visualized as punctate staining in the nucleus, is a sensitive marker to identify DSBs. We explored for DNA damage and DSBs in the epithelial cells of the OTC epithelium by immunofluorescence staining for γH2AX. In the OTC controls, there were rare epithelial cells with weak nuclear γH2AX staining (Figure 7A). In the OTC+PBMC+IL cultures, however, γH2AX staining of the nuclei was more intense (Figure 7B), and the frequency of stained nuclei was increased twofold to 22% ± 15% compared with OTC controls (10% ± 6%) (Figure 7C). Together, this suggested a greater level of DNA damage in the OTC+PBMC+IL cultures compared with the OTC controls.
Figure 7.
The acute inflammatory environment in OTC+PBMC+IL cultures induces DNA damage and DNA strand breaks in the overlying epithelial cells. A: Representative low-power confocal microscopy images of nuclear DAPI (blue) and the DNA damage/DNA strands break marker γH2AX staining (red) in the squamous epithelium of OTC controls. Arrow indicates basal cell layer; arrowhead, superficial cell layers. Inset: High-power confocal image showing granular pattern of γH2AX nuclear staining in a rare positive cell. B: Same as in A except images of nuclear DAPI (blue) and γH2AX staining (red) in squamous epithelium from OTC+PBMC+IL (IL-2, -7, and -15) cultures. Arrow indicates basal cell layer; arrowhead, superficial cell layers. Inset: High-power confocal image showing granular pattern of γH2AX nuclear staining in multiple positive cells. C: Quantitative analysis of γH2AX+ cells. γH2AX+ cells were expressed as a percentage of the total number of DAPI-stained cells. D and E: Representative image from one of the experiments of stained nuclei from Comet assay for DNA strand breaks in overlying squamous epithelial cells from OTC control (D) or OTC+PBMC+IL (E) cultures. Nuclei with longer, brighter tails are an indicator of DNA strand breaks in Comet assays. F: Quantitation of Comet assay tail moment across the experiments. Data are expressed as means ± SD (C) and means ± SD of the mean (F). n = 3 cultures per condition, with at least 500 cells counted per culture (C); n = 3 cultures per condition, with 57 control and 50 PBMC+ILs Comet tails analyzed in total across the three cultures using OpenComet image analysis software version 1.3.016 in total from the three experiments (F). ∗P ≤ 0.05, U-test; ∗∗∗∗P < 0.0001, unpaired t-test. Original magnification: ×200 (A and B, main images); ×900 (A and B, insets); ×100 (D and E). OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell.
To further validate the DNA-damaging effects of the acute inflammatory environment on the epithelial cells, we subjected them to a Comet assay to detect DSBs at the single-cell level. After harvesting the OTCs, the squamous epithelial layers were trypsinized into single cells and resuspended in an agarose gel. The cells were lyzed under alkaline condition overnight and then subjected to electrophoresis. The level of DSBs was reflected in the length and intensity (moment) of DNA migration from the nucleus. This was quantified using image analysis software as described.16 In the OTC+PBMC+IL cultures, there were a greater number of cells with longer and brighter tails than the control OTC cells (Figure 7, D and E). The analysis of several dozen comets from multiple experiments revealed a significant increase in the number of cells with extended comet tail moments in the OTC+PBMC+IL cells compared with control cells (Figure 7F). When calculated, the average comet tail moment of the OTC+PBMC+IL cells was double that of control cells (Figure 7F). In summary, the acute inflammatory environment engineered into the OTCs resulted in significant DNA damage and DSBs in the overlying epithelial cells.
Acute Inflammatory Environment Increases Oxidative Stress in the Epithelium and Contributes to DNA Damage
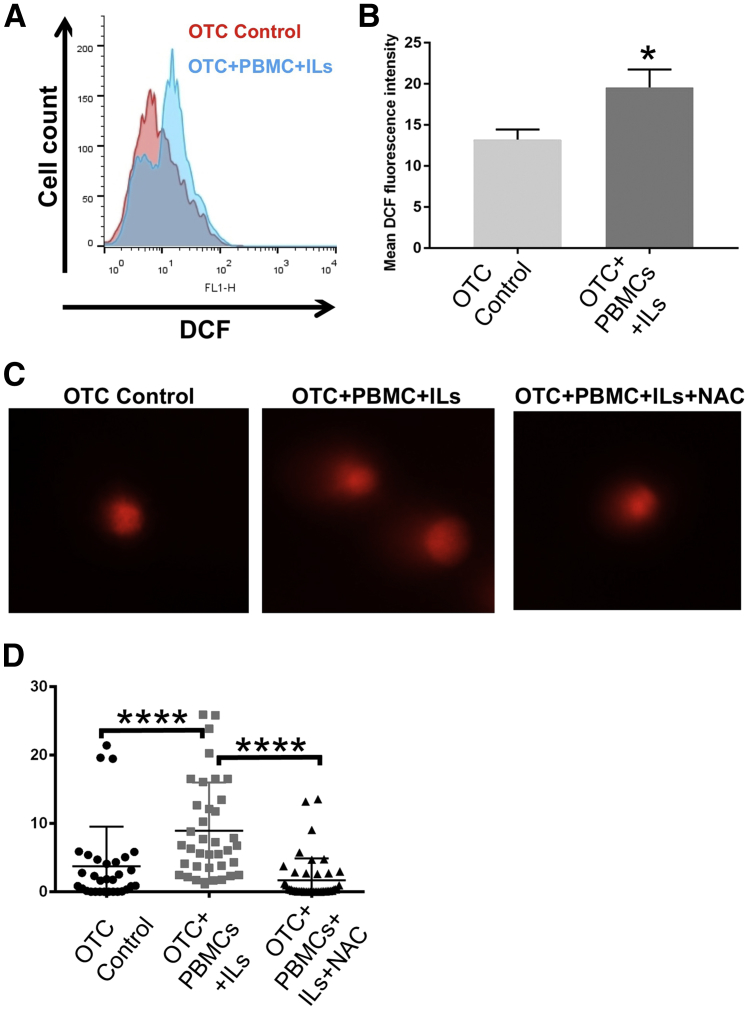
Inflammatory responses are known to contribute to tissue oxidative stress by the production of ROS. ROSs are produced in abundance by infiltrating immune cells and can damage proteins, lipids, and DNA and lead to DSNs,31 which is thought to be a key driver of carcinogenesis.30, 31 To directly quantify the level of oxidative stress in the OTC control and OTC+PBMC+IL cultures' epithelial cell compartment, we stained the cells with DCF fluorescence dye before termination of the cultures. DCF fluoresced on reaction with reactive oxygen compounds and this fluorescence was readily measured by flow cytometry. At the completion of the culture, the squamous epithelial layers were again trypsinized into single cells, and the DCF fluorescence intensity was measured by flow cytometry. We observed that the DCF fluorescence intensity was significantly increased in the OTC-PBMC-IL cells from a mean intensity of 13.2 ± 1.2 in control OTCs to a mean intensity of 19.6 ± 2.2 in the OTC+PBMCs+ILs (Figure 8, A–C). Finally, to determine whether the oxidative stress was contributing to the increased DNA damage and DSBs, we treated the OTC-PBMC-IL cells with the potent antioxidant N-acetyl cysteine throughout the culturing period, and then subjected the epithelium to Comet assays to quantify the DSBs. Treatment of the OTC-PBMC-IL cultures with N-acetyl cysteine significantly decreased DSBs in the esophageal epithelial cells, as evidenced by the reduction in Comet tail moments to control levels (Figure 8D). Taken together these findings suggest that the acute inflammatory environment in OTCs was associated with increased oxidative stress in the epithelial cells, resulting in DNA damage, and that treatment with a potent antioxidant has a protective effect, diminishing the severity of DNA-damage and DSBs.
Figure 8.
Oxidative stress is increased in squamous epithelial cells from OTCs with an acute inflammatory environment and it is responsible for the observed increase in DNA strand breaks. A: Intracellular ROS levels in epithelial cells from OTC control (red graph) and OTC+PBMC+IL (blue graph) cultures, as quantified by flow cytometry using the intracellular fluorescence dye DCF. One of the experiments is shown. B: Quantification of mean DCF fluorescence intensity measured by flow cytometry in epithelial cells from OTC control and OTC+PBMC+IL cultures. C: Representative image from one of the experiments of stained nuclei from Comet assay for DNA strand breaks in overlying squamous epithelial cells from OTC control, OTC+PBMC+IL, and antioxidant-treated OTC+PBMC+IL+NAC cultures. D: Quantitation of Comet assay tail moment across the experiments. Data are expressed as means ± SD (B) and means ± SD of the mean (D). n = 3 experiments (A and C); n = 3 (B); n = 3 cultures per condition, with 41 control, 37 PBMC+IL, and 44 PBMC+IL+NAC Comet tails analyzed across the three cultures using OpenComet image analysis software version 1.3.016 in total from the three experiments (D). ∗P ≤ 0.05, t-test; ∗∗∗∗P < 0.0005; analysis of variance and Tukey rank means testing. DCF, 2′,7′-dichlorodihydrofluorescein diacetate; NAC, N-acetyl cysteine; OTC, organotypic tissue culture; PBMC, peripheral blood mononuclear cell; ROS, reactive oxygen species.
Discussion
Research models are essential tools by which physicians and scientists explore disease mechanisms and develop novel targeted therapies. Although entirely necessary for research to progress, these same models also can constrain that effort. If no models exist, it is difficult for physicians and scientists to make significant progress in discerning mechanisms responsible for disease pathogenesis or to identify novel therapies to treat that condition. Equally important, are the concessions and assumptions inherent within any model, some of which are unrecognized, that can have significant implications for the conclusions drawn from studies using those models.
Cell culture methods have been established for more than half a century, and, although they have been highly successful and contributed to many significant advances, it has become increasingly clear that classic cell culture methods are artificial.2, 3 The research community has begun to appreciate a number of important limitations to these methods, including an overreliance on transformed and neoplastic cell lines, use of stiff plastic and glass culture plates, and a dependence on undefined serums to support and sustain cell growth. To address these recognized concerns, investigators have been adopting newer primary organoid34, 35 and organotypic9 culture systems for research applications. A strength of these culture systems is that they better replicate in vivo normal human tissue microenvironments in an in vitro setting.13, 36 Moreover, unexpected phenotypes have emerged from cells cultured under these more physiologically faithful culture systems10, 23, 37, 38
In the present study we expanded the original protocol of organotypic culture to model esophageal inflammation, a key feature of esophagitis, and to obtain a greater insight into interactions between the esophageal epithelium and acutely activated immune cells. We found that OTC environment can sustain immune cell viability and cytokine responsiveness, and we demonstrated that human PBMCs incorporated into OTCs respond appropriately to proinflammatory cytokines IL-7 and -15. This includes proliferation by the PBMCs in response to the proinflammatory ILs, increased infiltration of the epithelium by CD45+ immune cells, and the enhanced production of IFN-γ, as demonstrated by increased epithelial IRF-1 levels. Equally interesting was the response of our human OTC esophageal epithelium to the acute inflammatory microenvironment. Specifically, there was a significant thickening of the epithelium because of enhanced basal cell proliferation and suprabasal hyperplasia. In addition, we also demonstrated elevated epithelial oxidative stress, DNA damage, and epithelial cell apoptosis associated with this microenvironment. These responses only occurred in the OTC setting of human PBMCs treated with the proinflammatory ILs (IL-2, -7, and -15), implicating the acute inflammatory environment as a driver of these epithelial responses.
One benefit of our OTC approach was our ability to specifically establish the specific role played by the acute inflammatory environment on these epithelial cell responses. It has been reported that the reflux of acid and bile can increase epithelial cell proliferation, oxidative stress, DNA damage, and apoptosis using both in vivo and in vitro models.39, 40, 41, 42 Although the infiltration of the acute inflammatory immune cells in response to the reflux injury has long been thought to further elevate local ROS production and to exacerbate epithelial cell oxidative stress,43, 44 until the OTC/PBMC culture system there was no way to separate the oxidative stress caused by the refluxate and that caused by the inflammatory cells. Our modifications of the OTC system permit this isolation and testing of inflammation as an individual driver of ROS production and oxidative stress in the setting of esophagitis.
Our inflammatory organotypic system confirmed that an acute inflammatory microenvironment contributes significantly to elevated ROS levels and DNA damage in overlying esophageal epithelial cells. This implies the inflammation is an important driver of epithelial responses and not simply a bystander reaction to the epithelial injury.29, 39, 40 Moreover, these in vitro findings support the conclusions from a recent in vivo study in human patients with severe reflux disease. In this study of 12 patients with severe gastroesophageal reflux disease who had their antireflux medication withdrawn, the acute epithelial response to the refluxate included basal cell hyperplasia and an influx of T cells but not loss of surface epithelial cells from the caustic injury.29 This work suggested that much of the epithelial injury in response to the reflux was caused by local cytokine production and the resulting immune response and not to the refluxate directly, placing greater importance on understanding the role of the immune response in esophageal reflux disease. Our findings in the OTCs with human PBMCs and proinflammatory cytokines substantiate their findings and suggest a change in the acid reflux paradigm might be in order, one placing greater significance on the immune response and its critical role in promoting disease pathogenesis.
Equally instructive are the pathologic features of reflux esophagitis our OTC system did not manifest. Specifically, we did not observe dilated intercellular spaces, elongated papillae, or ballooning degeneration of epithelial cells25 in the OTCs. It might be these elements are the specific products of the caustic chemical injury. This is a testable hypothesis, one we may explore in future studies. However, of greater interest to us is the potential for OTC and other advanced human 3D culture systems to model normal human physiology and disease pathophysiology.13, 36 A number of laboratories have begun using this platform to model complex processes such as metastasis in esophageal squamous carcinogenesis.37, 45, 46, 47 We anticipate future efforts using this platform to model complex disorders such as gastroesophageal reflux disease and eosinophilic esophagitis. In these future studies, epithelial cells can be engineered to express key cytokines involved in gastroesophageal reflux disease pathogenesis such as IL-1β or IL-6, or IL-5 and eotaxin-3 as in eosinophilic esophagitis. It is also entirely feasible in the future to model the effects of microbial components or microbes themselves on these complex processes. We plan in future studies to use enhanced OTCs to model effects of the inflammatory microenvironment on two key steps in the pathogenesis and progression of BE to esophageal adenocarcinoma. Specifically, we will explore the early and late stages of this pathway by examining the effects of a chronic inflammatory environment on esophageal squamous cell responses and genomic integrity, and on the progression of nondysplastic BE to dysplasia and cancer.
Conclusions
We provide evidence that OTC is a useful platform to model esophageal inflammation because it can recapitulate the characteristics of esophagitis in a well-controlled in vitro setting. Future studies will be directed toward dissecting the molecular mechanisms by which inflammation induces oxidative stress and DNA damage, and to continue to enhance the OTC approach to better model esophagitis and BE, such as by the addition of a microbial component. In conclusion, our results further highlight the importance of immune-cell–mediated esophageal injury in esophagitis and support the use of this innovative platform to study human epithelial cell responses to esophagitis.
Acknowledgments
We thank the University of Pennsylvania's Morphology and Pathology Imaging Core and Flow Cytometry and Cell Sorting facility for technical expertise and assistance.
Footnotes
Supported by National Cancer Institute Barrett's Esophagus Translational Research Network program grant CA163004 (A.K.R., G.G.G., G.W.F., and J.P.L.), the Landenberger Family Foundation (G.W.F.), a Rosztóczy Foundation Fellowship (D.L.), a NIH career development award OD 012097 (J.P.L.), and Center for Molecular Studies in Digestive and Liver Disease at the University of Pennsylvania grants P30-DK050306 (A.K.R.) and PO1 CA098101 (A.K.R.).
Disclosures: None declared.
References
- 1.Kulig M., Leodolter A., Vieth M., Schulte E., Jaspersen D., Labenz J., Lind T., Meyer-Sabellek W., Malfertheiner P., Stolte M., Willich S.N. Quality of life in relation to symptoms in patients with gastro-oesophageal reflux disease—an analysis based on the ProGERD initiative. Aliment Pharmacol Ther. 2003;18:767–776. doi: 10.1046/j.1365-2036.2003.01770.x. [DOI] [PubMed] [Google Scholar]
- 2.Spechler S.J., Souza R.F. Barrett's esophagus. N Engl J Med. 2014;371:836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 3.Jankowski J.A., Wright N.A., Meltzer S.J., Triadafilopoulos G., Geboes K., Casson A.G., Kerr D., Young L.S. Molecular evolution of the metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Pathol. 1999;154:965–973. doi: 10.1016/S0002-9440(10)65346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rustgi A., El-Serag H.B. Esophageal carcinoma. N Engl J Med. 2015;372:1472–1473. doi: 10.1056/NEJMc1500692. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa H., Whelan K., Lynch J.P. Mechanisms of Barrett's oesophagus: intestinal differentiation, stem cells, and tissue models. Best Pract Res Clin Gastroenterol. 2015;29:3–16. doi: 10.1016/j.bpg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quante M., Bhagat G., Abrams J.A., Marache F., Good P., Lee M.D., Lee Y., Friedman R., Asfaha S., Dubeykovskaya Z., Mahmood U., Figueiredo J.L., Kitajewski J., Shawber C., Lightdale C.J., Rustgi A.K., Wang T.C. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D.H., Tiwari A., Kim M.E., Clemons N.J., Regmi N.L., Hodges W.A., Berman D.M., Montgomery E.A., Watkins D.N., Zhang X., Zhang Q., Jie C., Spechler S.J., Souza R.F. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett's metaplasia. J Clin Invest. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seto Y., Kobori O. Role of reflux oesophagitis and acid in the development of columnar epithelium in the rat oesophagus. Br J Surg. 1993;80:467–470. doi: 10.1002/bjs.1800800420. [DOI] [PubMed] [Google Scholar]
- 9.Kalabis J., Wong G.S., Vega M.E., Natsuizaka M., Robertson E.S., Herlyn M., Nakagawa H., Rustgi A.K. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc. 2012;7:235–246. doi: 10.1038/nprot.2011.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong J., Crissey M.A., Stairs D.B., Sepulveda A.R., Lynch J.P. Cox2 and beta-catenin/T-cell factor signaling intestinalize human esophageal keratinocytes when cultured under organotypic conditions. Neoplasia. 2011;13:792–805. doi: 10.1593/neo.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong J., Nakagawa H., Isariyawongse B.K., Funakoshi S., Silberg D.G., Rustgi A.K., Lynch J.P. Induction of intestinalization in human esophageal keratinocytes is a multistep process. Carcinogenesis. 2009;30:122–130. doi: 10.1093/carcin/bgn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosoff R.E., Gardiner K.L., Merlo L.M., Pavlov K., Rustgi A.K., Maley C.C. Development and characterization of an organotypic model of Barrett's esophagus. J Cell Physiol. 2012;227:2654–2659. doi: 10.1002/jcp.23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman K.G., Bortner J.D., Falk G.W., Yu J., Martín M.G., Rustgi A.K., Lynch J.P. Modeling inflammation and oxidative stress in gastrointestinal disease development using novel organotypic culture systems. Stem Cell Res Ther. 2013;4(Suppl 1):S5. doi: 10.1186/scrt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemons N.J., McColl K.E., Fitzgerald R.C. Nitric oxide and acid induce double-strand DNA breaks in Barrett's esophagus carcinogenesis via distinct mechanisms. Gastroenterology. 2007;133:1198–1209. doi: 10.1053/j.gastro.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 15.Olive P.L., Banath J.P. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 16.Gyori B.M., Venkatachalam G., Thiagarajan P.S., Hsu D., Clement M.V. OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2014;2:457–465. doi: 10.1016/j.redox.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heufler C., Topar G., Grasseger A., Stanzl U., Koch F., Romani N., Namen A.E., Schuler G. Interleukin 7 is produced by murine and human keratinocytes. J Exp Med. 1993;178:1109–1114. doi: 10.1084/jem.178.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry T.J., Mackall C.L. Interleukin-7: from bench to clinic. Blood. 2002;99:3892–3904. doi: 10.1182/blood.v99.11.3892. [DOI] [PubMed] [Google Scholar]
- 19.Anderson D.M., Kumaki S., Ahdieh M., Bertles J., Tometsko M., Loomis A., Giri J., Copeland N.G., Gilbert D.J., Jenkins N.A., Valentine V., Shapiro D.N., Morris S.W., Park L.S., Cosman D. Functional characterization of the human interleukin-15 receptor alpha chain and close linkage of IL15RA and IL2RA genes. J Biol Chem. 1995;270:29862–29869. doi: 10.1074/jbc.270.50.29862. [DOI] [PubMed] [Google Scholar]
- 20.Steel J.C., Waldmann T.A., Morris J.C. Interleukin-15 biology and its therapeutic implications in cancer. Trends Pharmacol Sci. 2012;33:35–41. doi: 10.1016/j.tips.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dooms H. Interleukin-7: fuel for the autoimmune attack. J Autoimmun. 2013;45:40–48. doi: 10.1016/j.jaut.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Boehm U., Klamp T., Groot M., Howard J.C. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 23.Kong J., Crissey M.A., Sepulveda A.R., Lynch J.P. Math1/Atoh1 contributes to intestinalization of esophageal keratinocytes by inducing the expression of Muc2 and Keratin-20. Dig Dis Sci. 2012;57:845–857. doi: 10.1007/s10620-011-1998-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D.H., Souza R.F. Biology of Barrett's esophagus and esophageal adenocarcinoma. Gastrointest Endosc Clin N Am. 2011;21:25–38. doi: 10.1016/j.giec.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takubo K., Honma N., Aryal G., Sawabe M., Arai T., Tanaka Y., Mafune K., Iwakiri K. Is there a set of histologic changes that are invariably reflux associated? Arch Pathol Lab Med. 2005;129:159–163. doi: 10.5858/2005-129-159-ITASOH. [DOI] [PubMed] [Google Scholar]
- 26.Jolly A.J., Wild C.P., Hardie L.J. Acid and bile salts induce DNA damage in human oesophageal cell lines. Mutagenesis. 2004;19:319–324. doi: 10.1093/mutage/geh035. [DOI] [PubMed] [Google Scholar]
- 27.Hong J., Chen Z., Peng D., Zaika A., Revetta F., Washington M.K., Belkhiri A., El-Rifai W. APE1-mediated DNA damage repair provides survival advantage for esophageal adenocarcinoma cells in response to acidic bile salts. Oncotarget. 2016;7:16688–16702. doi: 10.18632/oncotarget.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandner A., Illert J., Koitzsch S., Unverzagt S., Schon I. Reflux induces DNA strand breaks and expression changes of MMP1+9+14 in a human miniorgan culture model. Exp Cell Res. 2013;319:2905–2915. doi: 10.1016/j.yexcr.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Dunbar K.B., Agoston A.T., Odze R.D., Huo X., Pham T.H., Cipher D.J., Castell D.O., Genta R.M., Souza R.F., Spechler S.J. Association of acute gastroesophageal reflux disease with esophageal histologic changes. JAMA. 2016;315:2104–2112. doi: 10.1001/jama.2016.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feagins L.A., Zhang H.Y., Zhang X., Hormi-Carver K., Thomas T., Terada L.S., Spechler S.J., Souza R.F. Mechanisms of oxidant production in esophageal squamous cell and Barrett's cell lines. Am J Physiol Gastrointest Liver Physiol. 2008;294:G411–G417. doi: 10.1152/ajpgi.00373.2007. [DOI] [PubMed] [Google Scholar]
- 31.Federico A., Morgillo F., Tuccillo C., Ciardiello F., Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381–2386. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- 32.Abdel-Latif M.M., Duggan S., Reynolds J.V., Kelleher D. Inflammation and esophageal carcinogenesis. Curr Opin Pharmacol. 2009;9:396–404. doi: 10.1016/j.coph.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Turinetto V., Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 35.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 36.Hartman K.G., Bortner J.D., Jr., Falk G.W., Ginsberg G.G., Jhala N., Yu J., Martin M.G., Rustgi A.K., Lynch J.P. Modeling human gastrointestinal inflammatory diseases using microphysiological culture systems. Exp Biol Med (Maywood) 2014;239:1108–1123. doi: 10.1177/1535370214529388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega M.E., Giroux V., Natsuizaka M., Liu M., Klein-Szanto A.J., Stairs D.B., Nakagawa H., Wang K.K., Wang T.C., Lynch J.P., Rustgi A.K. Inhibition of Notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett's-like metaplasia via KLF4. Cell Cycle. 2014;13:3857–3866. doi: 10.4161/15384101.2014.972875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stairs D.B., Nakagawa H., Klein-Szanto A., Mitchell S.D., Silberg D.G., Tobias J.W., Lynch J.P., Rustgi A.K. Cdx1 and c-Myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward Barrett's esophagus. PLoS One. 2008;3:e3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvorak K., Payne C.M., Chavarria M., Ramsey L., Dvorakova B., Bernstein H., Holubec H., Sampliner R.E., Guy N., Condon A., Bernstein C., Green S.B., Prasad A., Garewal H.S. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett's oesophagus. Gut. 2007;56:763–771. doi: 10.1136/gut.2006.103697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong J., Whelan K.A., Laczko D., Dang B., Caro Monroig A., Soroush A., Falcone J., Amaravadi R.K., Rustgi A.K., Ginsberg G.G., Falk G.W., Nakagawa H., Lynch J.P. Autophagy levels are elevated in barrett's esophagus and promote cell survival from acid and oxidative stress. Mol Carcinog. 2016;55:1526–1541. doi: 10.1002/mc.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wetscher G.J., Hinder R.A., Bagchi D., Hinder P.R., Bagchi M., Perdikis G., McGinn T. Reflux esophagitis in humans is mediated by oxygen-derived free radicals. Am J Surg. 1995;170:552–556. doi: 10.1016/s0002-9610(99)80014-2. discussion 556–557. [DOI] [PubMed] [Google Scholar]
- 42.McQuaid K.R., Laine L., Fennerty M.B., Souza R., Spechler S.J. Systematic review: the role of bile acids in the pathogenesis of gastro-oesophageal reflux disease and related neoplasia. Aliment Pharmacol Ther. 2011;34:146–165. doi: 10.1111/j.1365-2036.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Ding Y.W., Yang G., Bondoc F., Lee M.J., Yang C.S. Oxidative damage in an esophageal adenocarcinoma model with rats. Carcinogenesis. 2000;21:257–263. doi: 10.1093/carcin/21.2.257. [DOI] [PubMed] [Google Scholar]
- 44.Olliver J.R., Hardie L.J., Dexter S., Chalmers D., Wild C.P. DNA damage levels are raised in Barrett's oesophageal mucosa relative to the squamous epithelium of the oesophagus. Biomarkers. 2003;8:509–521. doi: 10.1080/13547500310001644961. [DOI] [PubMed] [Google Scholar]
- 45.Basu D., Bewley A.F., Sperry S.M., Montone K.T., Gimotty P.A., Rasanen K., Facompre N.D., Weinstein G.S., Nakagawa H., Diehl J.A., Rustgi A.K., Herlyn M. EGFR inhibition promotes an aggressive invasion pattern mediated by mesenchymal-like tumor cells within squamous cell carcinomas. Mol Cancer Ther. 2013;12:2176–2186. doi: 10.1158/1535-7163.MCT-12-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long A., Giroux V., Whelan K.A., Hamilton K.E., Tetreault M.P., Tanaka K., Lee J.S., Klein-Szanto A.J., Nakagawa H., Rustgi A.K. WNT10A promotes an invasive and self-renewing phenotype in esophageal squamous cell carcinoma. Carcinogenesis. 2015;36:598–606. doi: 10.1093/carcin/bgv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natsuizaka M., Kinugasa H., Kagawa S., Whelan K.A., Naganuma S., Subramanian H., Chang S., Nakagawa K.J., Rustgi N.L., Kita Y., Natsugoe S., Basu D., Gimotty P.A., Klein-Szanto A.J., Diehl J.A., Nakagawa H. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res. 2014;4:29–41. [PMC free article] [PubMed] [Google Scholar]