Abstract
Mounting evidence indicates that microRNAs play important roles in the development of various cancers. Aberrant expression of microRNA-199a-5p has been frequently reported in cancer studies; however, the mechanistic details of the role of microRNA-199a-5p in colorectal cancer still remain unclear. Our study aimed to explore the role of microRNA-199a-5p in colorectal cancer cells by targeting Rho-associated coiled coil-containing protein kinase 1. Here, we showed that microRNA-199a-5p was significantly downregulated in colorectal cancer cell lines and tissue samples and was associated with a poor prognostic phenotype. MicroRNA-199a-5p suppressed colorectal cancer cell proliferation, migration, and invasion and induced cell apoptosis. Moreover, we identified Rho-associated coiled coil-containing protein kinase 1 as the direct target of microRNA-199a-5p using luciferase and Western blot assays. Importantly, Rho-associated coiled coil-containing protein kinase 1 overexpression rescued the microRNA-199a-5p-induced suppression of proliferation, migration, and invasion of colorectal cancer cells. Furthermore, the overexpression of microRNA-199a-5p inhibited tumor growth and metastasis by inactivating the phosphoinositide 3-kinase/AKT and Janus kinase 1/signal transducing activator of transcription signaling pathways through downregulation of Rho-associated coiled coil-containing protein kinase 1. Altogether, microRNA-199a-5p/Rho-associated coiled coil-containing protein kinase 1 may be a potential therapeutic target for colorectal cancer therapy.
Keywords: colorectal cancer, miR-199a-5p, ROCK1, progression, cell apoptosis
Introduction
Colorectal cancer (CRC) is the fourth most common cancer in the world, with the highest rate of cancer-related morbidity and mortality.1,2 There are more than 1 million new cases diagnosed every year.1 Surgery, adjuvant chemotherapy, and radiation therapy are known to be the approaches to the treatment of CRC.3 In spite of advances in surgery and adjuvant chemotherapies, which remain the standard treatment options, the prognosis for patients with CRC still remains poor.4 Therefore, new diagnostic markers or therapeutic targets for CRC are critical.
MicroRNAs (miRNAs) are a type of single-stranded RNAs which composed of 19 to 25 nucleotides and work as the negative posttranscriptional regulators of their target gene.5,6 Researches had revealed that miRNAs correlated with the development and progression processes in many different cancer types.7-10 MicroRNA-199a (miR-199a) has been reported to be downregulated and represses tumor progression in prostate adenocarcinoma,11 chondrosarcoma,12 hepatocellular carcinoma,13 and ovarian cancer.14 It has been reported recently that miR-199a-5p is abnormally expressed in CRCs.15-18 For example, Hu et al 19 reported that miR-199a-5p inhibited the migration and invasion of CRC cells by targeting discoidin domain receptor 1 (DDR1). In addition, miR-199a decreased CRC proliferation, migration, and invasion through the Hypoxia-inducible factor 1-alpha/vascular endothelial growth factor (HIF-1α/VEGF) pathway.20 These findings suggest that miR-199a-5p has a tumor suppressive role in CRC. The underlying mechanism of miR-199a in regulating the development and progression of CRC cells still remains unclear. In addition, as miR has various target genes,21 other targets of miR-199a-5p may also be involved in the effect of miR-199a-5p on the tumor initiation and progression of CRC. In this study, we concentrated on the investigation of the mechanism of miR-199a-5p in CRC progression and development.
Materials and Methods
Cell Lines and CRC Clinical Samples
Six CRC cell lines (SW480, HT29, LS174T, LoVo, SW620, and HCT116) and a normal colonic cell line (NCM460) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and cultured in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% of fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 mg/mL) at 37°C. Colorectal cancers and adjacent samples of normal tissues were obtained after the approval of the protocol from research ethics. The samples were collected from the First Affiliated Hospital of Wenzhou Medical University, Wenzhou City, Zhejiang, China. The medical ethics committee of the First Affiliated Hospital of Wenzhou Medical University approved the protocol, and written informed consent was obtained from all participants.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, California). To detect miR-199a-5p expression, SYBR Premix Ex Taq (Takara, Dalian, China) was used for reverse transcription process and quantitative real-time polymerase chain reaction (qRT-PCR) process. Primers used were: miR-199a-5p forward: 5′-TCAAGAGCAATAACGAAAAATGT-3′, reverse: 5′-GCTGTCAACGATACGCTACGT-3′; U6 small nuclear RNA (snRNA) used as internal control; Rho-associated coiled coil-containing protein kinase 1 forward: 5′-AGGAAGGCGGACATATTAGTCCCT-3′, reverse: 5′-AGACGATAGTTGGGTCCCGGC-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward: 5′-AACGTGTCAGTGGTGGACCTG-3′, reverse: 5′-AGTGGGTGTCGCTGTTGAAGT-3′. Data were normalized with either U6 or GAPDH; 2−ΔΔCt was used to calculate the relatively expressed miRNA or messenger RNA (mRNA) expressions.
Lentivirus Infection
We purchased pLV-has-miR-199a plasmid and pLV-miRNA-vector control plasmid from Biosettia (San Diego, California). Viruses were packaged (LV-miR-199a, LV-Ctrl) and infected following protocol of the company.
Luciferase Reporter Assay and Plasmid Construction
To perform luciferase reporter assays, we first inserted the 3′-untranslated region (UTR) of miR-199a-5p into ROCK1, then the DNA fragment was packed into pGL3-control vector (Promega, Madison, Wisconsin). Mutation of specific binding sites of miR-199a-5p within ROCK1 3′-UTR was achieved by using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, California). After that, LV-miR-199a and LV-Ctrl were transfected with either wild-type (WT) or Mut plasmids into target cells. Dual-Luciferase Reporter Assay System (Promega) was used to measure the luciferase level of each group 48 hours after the transfection. For the construction of ROCK1 expression vector, we purchased the plasmid which contains ROCK1 complementary DNA (cDNA) without 3′-UTR from GeneCopoeia (Rockville, Maryland); pcDNA.3 (Invitrogen) expression vector was used to transfect the purchased plasmid.
3-(4, 5-Dimethylthiazol-2-yl)-,2,5-Diphenyltetrazolium Bromide, Colony Formation, Cell Migration, and Invasion Assays
For cell proliferation assay, we used 3-(4, 5-dimethylthiazol-2-yl)-,2,5-diphenyltetrazolium bromide (MTT) assay. First, 4 × 103 cells per well were seeded in 96-well plates. Cells were then incubated in reagent of MTT. After that, supernatants of each well were removed and we added dimethyl sulfoxide into each well. For data analysis, absorbance values (OD) from each well was analyzed at 490 nm.
For colony formation, 24 hours after infection, 300 infected cells were placed in a fresh 6-well plate and maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet in 20% methanol for 15 minutes.
Cell migration was determined using a wound healing assay. Cells were seeded in 6-well plates and incubated for 24 hours in a humidified chamber with 5% CO2 at 37°C. At the end of the incubation, 3 scratch wounds were made across each well using a P-200 pipette tip. Fresh RPMI-1640 medium containing 5% FBS was added. The cells were continuously incubated, and the wound-closing procedure was observed for 48 hours. Photographs were taken at 0 and 48 hours.
For cell invasion assay, Matrigel invasion chamber (BD Biosciences, San Jose, CA, USA) was used. Cells at 1 × 105 per well for 24-well plates were added into the insert part of the chamber. Serum-free medium was used for the chamber that added cells and 10% FBS medium for the lower chamber. Plates were then incubated at 37°C for 8 hours. Then cells were washed with PBS and top chamber was removed. Crystal violet was used to stain the remaining cells in the lower chamber.
Apoptosis Assay
Annexin V/fluorescein isothiocyanate apoptosis detection kit (Keygen, Nanjing, China) were used for the apoptosis analysis. All processes were performed following the manufacturer’s instructions. Becton Dickinson (Franklin Lakes, New Jersey) FACSCalibur and CellQuest software were used to acquire and analyze data.
Western Blot Analysis
Radio immunoprecipitation assay (RIPA) reagent was used for cell lysis with proteinase inhibitor. After this, protein was separated by performing gel electrophoresis. Then, protein was transferred to Polyvinylidene Fluoride (PVDF) membranes. After blocking with 5% nonfat milk, the membranes were incubated with primary antibodies against ROCK1, p-PI3K (Tyr458), PI3K, p-AKT (Ser473), AKT, p-Janus kinase 1 (JAK1), JAK1, p-signal transducing activator of transcription (STAT) 3, STAT3, and GAPDH (1:1000; Santa Cruz Biotechnology, Santa Cruz, California) at 4°C overnight. Then, the membranes were incubated with secondary antibodies for 1 hour at room temperature. At last, enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Little Chalfont, UK) were used for chemiluminescence detection.
Immunohistochemistry
Immunohistochemistry and evaluation of ROCK1 (1:100; Santa Cruz Biotechnology Inc) and Ki-67 (1:100; Boster, Wuhan, China) staining were performed with tumor tissue sections prepared from in vivo experiments according to previously described methods.22
In Vivo Tumorigenesis
All animal procedures were performed in accordance with institutional guidelines. SW480 cells 4 × 106 were transplanted into 4 nude mice for each group at the flanks. Tumor volume was calculated by using the formula: Volume (mm3) = L × W × W/2 (length L, mm; width W, mm).
Statistical Analysis
All values are presented as the mean ± standard error of the mean, and 2-tailed Student t test or 1-way analysis of variance was used to compare the differences among different groups. Survival analysis was performed by using Kaplan-Meier method. For statistical analysis of all data, SPSS 16.0 (SPSS Inc) was used and P < .05 represented the significant difference between different groups.
Results
Decreased miR-199a-5p Are Related to Poor Prognostic in CRCs
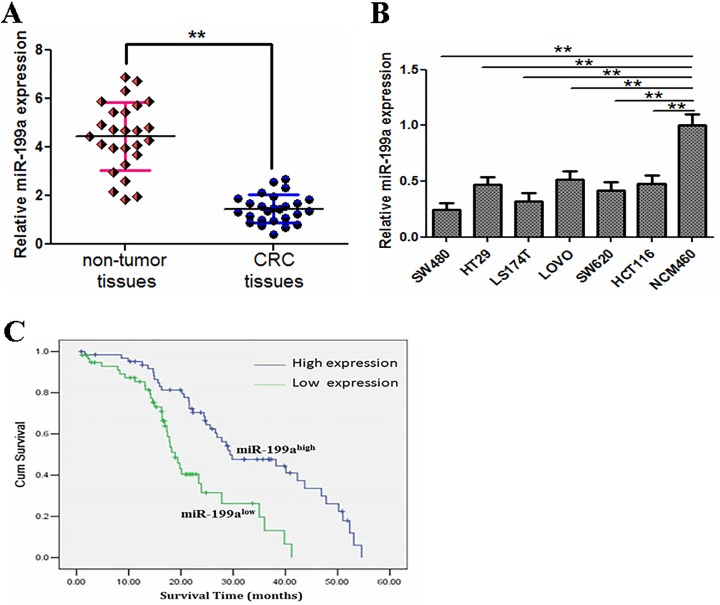
The level of miR-199a-5p was detected in either CRC tissues or adjacent normal tissues in 30 patients by using qRT-PCR. Data indicated that miR-199a-5p was significantly decreased in CRC tissues when compared to adjacent normal tissues (Figure 1A; P < .01). Moreover, in 6 CRC cell lines (SW480, HT29, LS174T, LoVo, SW620, and HCT116), miR-199a-5p expression was significantly downregulated compared with the normal colonic cell line (NCM460; Figure 1B, P < .01). Furthermore, the relationship between miR-199a-5p expression and overall survival was determined through Kaplan-Meier survival analysis in 102 patients with CRC. Data indicated that lower expression of miR-199a-5p was related to the poorer survival rates in patients with CRC when compared with those groups who had relatively higher miR-199a-5p levels (Figure 1C; P < .05).
Figure 1.
Lower levels of miR-199a are frequently detected in CRC tissues and cell lines and are associated with a poor prognostic phenotype. A, The expression of miR-199a is significantly downregulated in CRC compared with adjacent nontumor tissues (n = 30). B, The miR-199a expression is markedly downregulated in 6 human CRC cell lines compared to a normal colonic cell line. C, Patients with CRC with lower median levels of miR-199a expression have poorer survival in contrast to those with higher median levels of miR-199a expression. **P < .01. CRC indicates colorectal cancer; miR-199a, microRNA-199a.
Rho-Associated Coiled Coil-Containing Protein Kinase 1 Is the Target of miR-199a-5p in CRC Cells
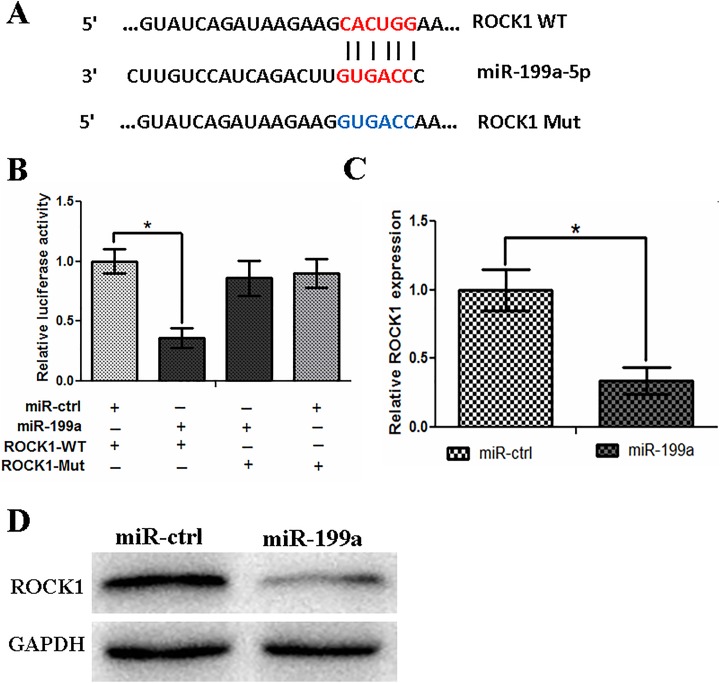
MicroRNAs were known as the small RNA which can regulate downstream gene functions by binding to their 3′-UTRs. To analyze the function of miR-199a-5p, we used TargetScan and miRanda to predict the potential target gene of it. Results indicated that ROCK1 was one of those target genes. We found the potential binding sites on ROCK1 (Figure 2A) and then subcloned both WT and mutant 3′-UTR (MUT) plasmids. A luciferase reporter vector was used to insert these 2 plasmids. Then, we transfected these 2 vectors into SW480 cells with the cotransfection of miR-199a-5p or a scrambled control. Results showed that miR-199a-5p and ROCK1 WT cotransfection reduced the luciferase level when compared with MUT group (Figure 2B; P < .05). Furthermore, miR-199a overexpression significantly decreased both ROCK1 mRNA and protein expressions in SW480 cells (Figure 2C and D).
Figure 2.
Rho-associated coiled coil-containing protein kinase 1 is a direct target of miR-199a in SW480 cells. A, Sequence alignment of miR-199a and the ROCK1 3′-UTR, which contains one predicted miR-199a binding site. The seed-recognizing sites in the ROCK1 3′-UTR are indicated in red, while the sites in the mutant ROCK1 3′-UTR are indicated in blue. B, Luciferase assay in SW480 cells cotransfected with miR-199a and a luciferase reporter containing the ROCK1 3′-UTR (WT) or a mutant (Mut). C, The miR-199a transfection affects ROCK1 mRNA levels. D, The miR-199a transfection affects ROCK1 protein levels. *P < .05. mRNA indicates messenger RNA; miR-199a, microRNA-199a; ROCK1, Rho-associated coiled coil-containing protein kinase 1; 3-UTR, 3-untranslated region; WT, wild type.
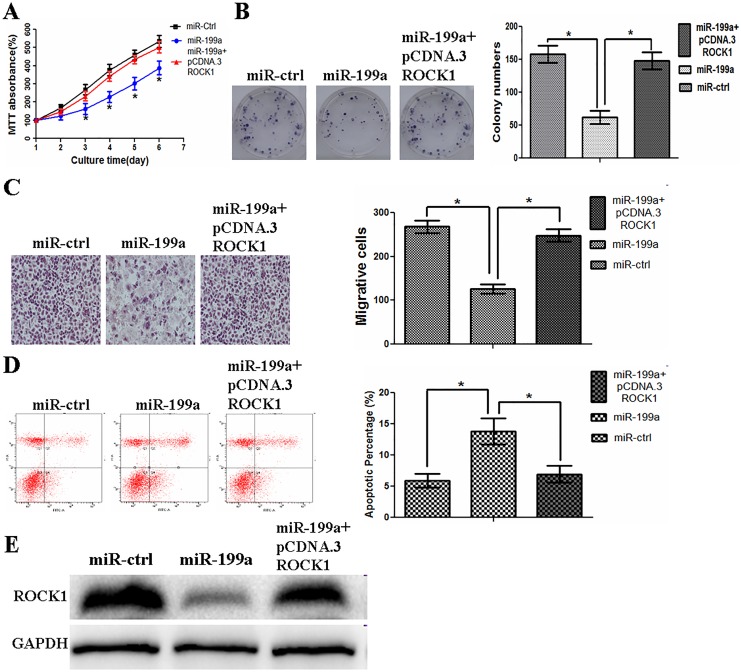
The Overexpression of MiR-199a-5p Inhibits CRC Cell Proliferation, Migration, and Invasion But Induces Cell Apoptosis
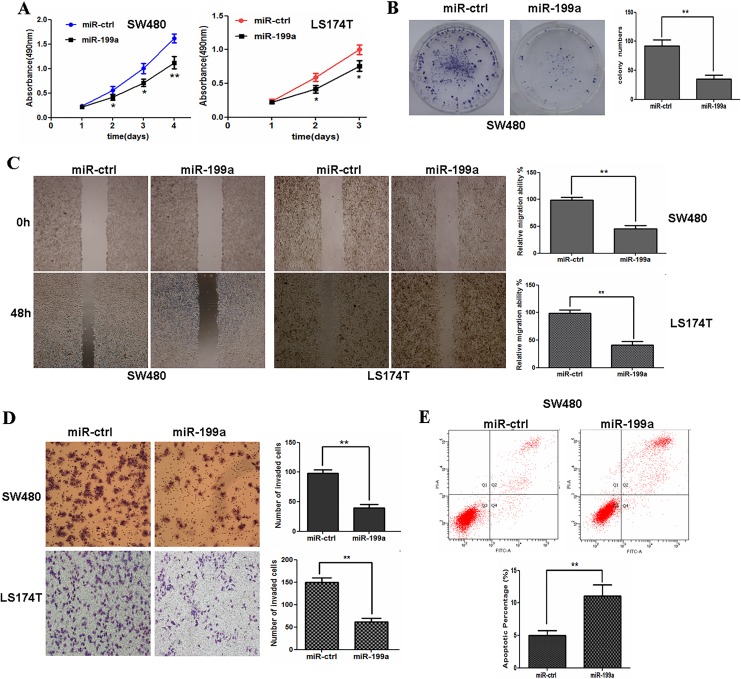
Rho-associated coiled coil-containing protein kinase 1 was reported to be involved in the key process of many cells.23 We investigated whether miR-199a-5p expression was involved in cell proliferation, migration, and invasion abilities in CRC cells. Data in Figure 3A and B showed that miR-199a-5p overexpression inhibited CRC cell proliferation significantly. Next, the effect of miR-199a-5p on the migrative and invasive capacity of CRC cells was characterized using wound-healing assays and Matrigel invasion assays. The results demonstrated that exogenetic overexpression of miR-199a-5p markedly reduced the migration and invasiveness of CRC cells (Figure 3C and D). We then investigated whether miR-199a-5p may affect cell apoptosis in CRC cells. Flow cytometric analysis was used to analyze apoptosis level of SW480 cells. In Figure 3E, the overexpression of miR-199a-5p induced SW480 cell apoptosis.
Figure 3.
Exogenetic overexpression of miR-199a inhibits cell proliferation, migration, and invasion but induces apoptosis in CRC cells. A, Restoration of miR-199a inhibits SW480 or LS174T cell proliferation. B, Overexpression of miR-199a impedes the colony formation ability of SW480 cells. C, Exogenetic overexpression of miR-199a markedly reduces the migration of SW480 or LS174T cells. D, Exogenetic overexpression of miR-199a significantly reduces the invasiveness of SW480 or LS174T cells. E, miR-199a overexpression induces apoptosis in SW480 cells. **P < .01. CRC indicates colorectal cancer; miR-199a, microRNA-199a.
MicroRNA-199a-5p Suppresses Tumor Progression In Vivo
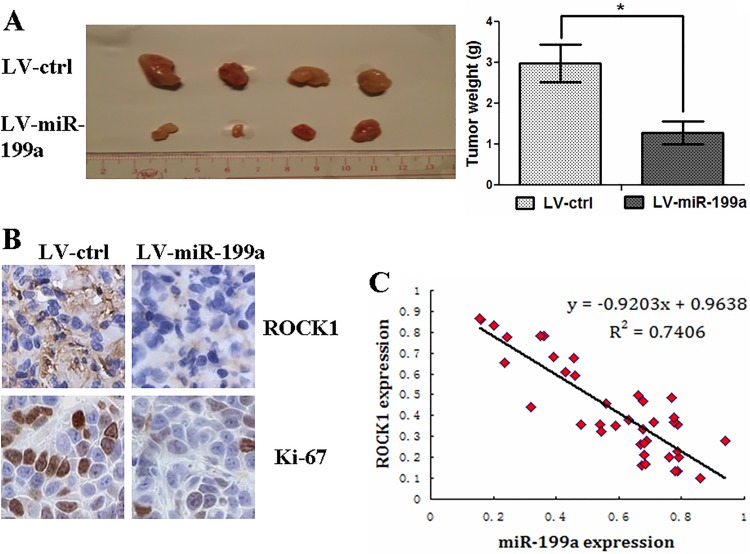
For in vivo analysis of miR-199a-5p on tumor progression, we infected SW480 cells with LV-miR-199a or LV-Ctrl. Decreased tumor weight was observed in mice subcutaneously inoculated with SW480 cells that transfected with LV-miR-199a when compared with empty vector control inoculation group (Figure 4A). Immunohistochemical staining (IHC) data indicated that tumors originated from LV-miR-199a transfection groups had a decreased Ki-67 and ROCK1 level when compared with LV-Ctrl originated tumors (Figure 4B). This part of data indicated that miR-199a-5p inhibits tumor progression in vivo.
Figure 4.
MicroRNA-199a inhibits tumor growth in vivo. A, Stable transfection of miR-199a into SW480 cells results in decreased growth and tumor weight. B, immunohistochemical staining demonstrates that the tumors originating from LV-miR-199a cells have decreased expression of Ki-67 and ROCK1 compared with that from LV-Ctrl cells. C, The inverse correlation between the levels of miR-199a and mRNA expression of ROCK1 in CRC tissues (n = 40). *P < .05. CRC indicates colorectal cancer; mRNA, messenger RNA; miR-199a, microRNA-199a; ROCK1, Rho-associated coiled coil-containing protein kinase 1
MicroRNA-199a-5p Inversely Correlated With ROCK1 in CRC Tissues at the mRNA Level
By analyzing ROCK1 mRNA expressions in 40 CRC tissue samples, we found that miR-199a-5p was reversely correlated with ROCK1 mRNA expression (Figure 4C, P < .05).
Overexpression of ROCK1 Reverses the Tumor Suppressive Effect of miR-199a-5p in CRC Cells
To evaluate whether ROCK1 is responsible for the functional effects of miR-199a in CRC cells, we generated a ROCK1-overexpressing vector (pcDNA3-ROCK1) and used it to transfect SW480 cells overexpressing miR-199a-5p. We found that the overexpression of ROCK1 partially rescued the miR-199a-5p-induced suppression of cell proliferation ability in SW480 cells (Figure 5A and B). Restoration of ROCK1 partially attenuated the suppression of cell migration and invasion induced by miR-199a-5p (Figure 5C). Furthermore, upregulated ROCK1 blocked the effect of miR-199a-5p on apoptosis (Figure 5D). We also found that the overexpression of ROCK1 restored expression of ROCK1 protein that had been suppressed by the overexpression of miR-199a-5p (Figure 5E). These data indicated that miR-199a-5p exerted its suppressive role in CRC cells by targeting ROCK1.
Figure 5.
Overexpression of ROCK1 reverses the tumor suppressive effect of miR-199a in CRC cells. A, Exogenetic overexpression of ROCK1 partially rescues the miR-199a-induced suppression of cell proliferation in SW480 cells. B, Restoration of ROCK1 partially rescues the miR-199a-induced suppression of colony formation ability in SW480 cells. C, Restoration of ROCK1 partially attenuates the suppression of cell migration and invasion induced by miR-199a. D, Exogenetic overexpression of ROCK1 blocks the effect of miR-199a on apoptosis. E, Exogenetic overexpression of ROCK1 restores the miR-199a-induced suppression of expression of ROCK1 protein in SW480 cells. *P < .05. CRC indicates colorectal cancer; miR-199a, microRNA-199a; ROCK1, Rho-associated coiled coil-containing protein kinase 1.
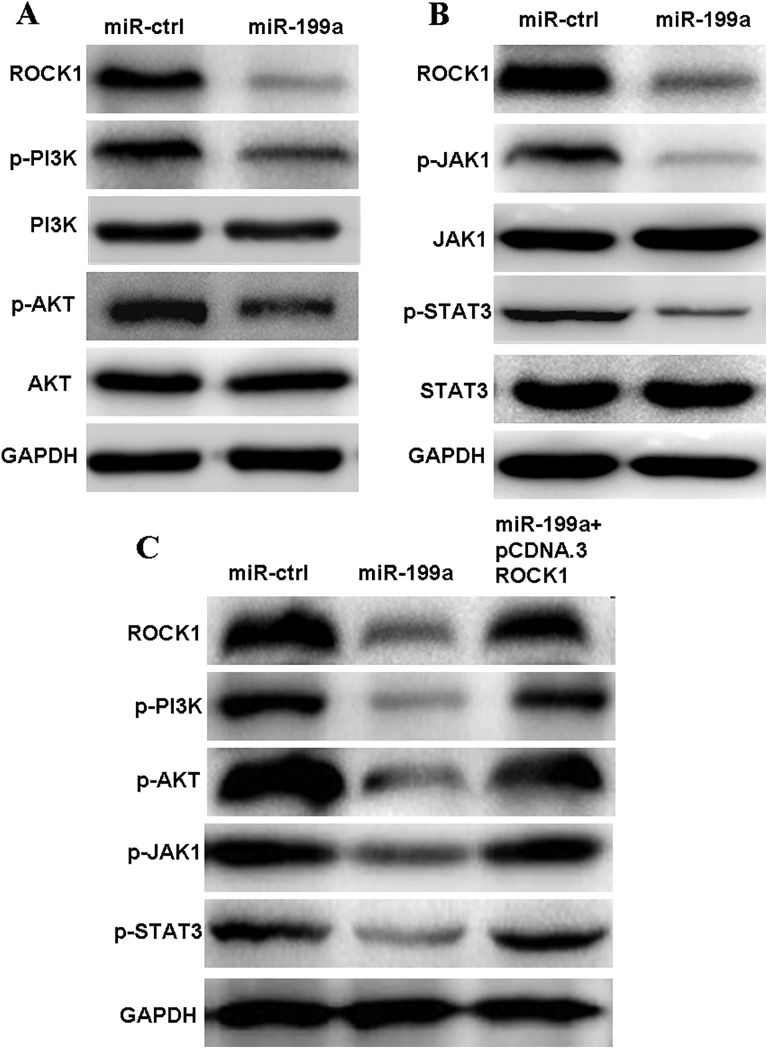
MicroRNA-199a-5p Modulates the Progression of CRC Cells Via the PI3K/Akt and JAK/STAT Signaling Pathways
To clarify the signaling pathways involved in the functional effects of SW480 cells mediated by the interaction between miR-199a-5p and ROCK1, we also examined the effect of miR-199a-5p on the PI3K/Akt and JAK/STAT signaling pathways. We found that miR-199a-5p overexpression reduced the expression of p-PI3K, p-AKT, p-JAK1, and p-STAT3, whereas that of PI3K, Akt, JAK1, and STAT3 remained unchanged (Figure 6A and B). Furthermore, we also found that inactivation of PI3K/AKT and JAK/STAT signaling pathways could be rescued at least partly by the overexpression of ROCK1 in miR-199a-5p-overexpressed cells (Figure 6C).
Figure 6.
MicroRNA-199a modulates the progression of CRC cells via the PI3K/AKT and JAK/STAT signaling pathways. A, The miR-199a overexpression reduces PI3K/AKT signaling through ROCK1 downregulation. B, Overexpression of miR-199a suppresses JAK/STAT signaling through ROCK1 downregulation. C, Exogenetic overexpression of ROCK1 partially restores the miR-199a-induced inactivation of PI3K/AKT and JAK/signal transducing activator of transcription signaling pathways. CRC indicates colorectal cancer; JAK1, Janus kinase 1; miR-199a, microRNA-199a; PI3K, phosphoinositide 3-kinase; ROCK1, Rho-associated coiled coil-containing protein kinase 1; STAT, signal transducing activator of transcription.
Discussion
In the present study, we investigated the function of miR-199a-5p in CRC cell progression. Data indicated that miR-199a-5p expression was downregulated in CRC tissues when compared to adjacent normal tissues. Besides, miR-199a-5p plays the role of tumor suppressor both in vitro and in vivo.
MicroRNAs were reported to widely contribute to many key cell processes and play important roles in many human cancers.24 The deregulation of different miRNAs is associated with many different diseases, including cancers.25 According to the importance of miRNAs in human cancers, the therapeutic method which targets miRNAs is becoming widely investigated.26 The roles of miR-199a-5p in CRC have been discussed in many previous works. Hu et al demonstrated that miR-199a-5p was significantly downregulated in CRC tissues and cells and inhibited the migration and invasion of CRC cells by targeting DDR1.19 Ye et al found that miR-199a reduced the proliferation, migration, and invasion and that miR-199a might potentially act through the HIF-1α/VEGF pathway.20 Kim et al suggested that miR-199a-5p acts as a tumor suppressor via inhibition of FZD6 expression, resulting in reduced invasiveness.15 In conclusion, miR-199a-5p played an important role in the suppression of cell proliferation and invasion in CRC cells. We further confirmed that ROCK1 was a direct target of miR-199a using a dual-luciferase reporter assay and Western blot analysis.
Rho-associated coiled coil-containing protein kinase 1, one of the Rho family of GTPase proteins, is a serine/threonine kinase which facilitates the reorganization of the actin cytoskeleton and plays a key role in cell migration and invasion.27 Rho-associated coiled coil-containing protein kinase 1 is overexpressed in various cancers, including CRC.23,28 Rho-associated coiled coil-containing protein kinase 1 is important in many pathways which regulate cell apoptosis, proliferation, and invasion.29,30 Our data indicated that miR-199a exerts its tumor suppressor function partially through ROCK1 in CRC cells. Consistent with a previous study in which miR-199a/b-5p inhibits hepatocellular carcinoma progression by posttranscriptionally suppressing ROCK1,31 we reached similar conclusions in CRC. Moreover, we proved miR-199a-5p was correlated with ROCK1 reversely by regulating ROCK1 mRNA expression in CRC tissues.
The PI3K/AKT signaling pathway is a classical signaling pathway in tumorigenesis.32,33 Several studies have demonstrated that the PI3K/Akt pathway is the most frequently activated in CRC, which contributes to tumor cell invasion and metastasis, uncontrolled cell proliferation, angiogenesis, and inhibition of apoptosis.34,35 In our investigation, we found that the overexpression of miR-199a decreased the expression of pPI3K, pAkt, p-JAK1, and p-STAT3. This result suggests that miR-199a suppresses the growth and metastasis of CRC cells at least in part through inactivating the PI3K/AKT and JAK/STAT signaling pathways via downregulation of ROCK1. Similarly, JAK/STAT was also inactivated by miR-199a.
In conclusion, our study identified miR-199a as a tumor suppressor in CRC cells both in vivo and in vitro. MicroRNA-199a-5p had proved to be suppressed in human CRCs and the reintroduction of it could be a potential method of suppressing the proliferation and progression of CRCs. Our finding may provide a potential method in CRC treatment in the future.
Abbreviations
- CRC
colorectal cancer
- DDR1
discoidin domain receptor 1
- FBS
fetal bovine serum
- GAPDH, glyceraldehyde-3-phosphate dehydrogenase
- JAK1
Janus kinase 1
- miRNAs
microRNAs; miR-199a-5p, microRNA-199a-5p
- MTT
3-(4,5-dimethylthiazol-2-yl)-,2,5-diphenyltetrazolium bromide
- PI3K
phosphoinositide 3-kinase
- ROCK1
Rho-associated coiled coil-containing protein kinase 1
- RT-PCR
real-time polymerase chain reaction
- STAT
signal transducing activator of transcription
- UTRs
untranslated regions
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from Natural Science Foundation of Zhejiang Province, china (NO.LY15H030015/NO.LY18H160056)
ORCID iD: Fang Wu  http://orcid.org/0000-0003-2321-7643
http://orcid.org/0000-0003-2321-7643
References
- 1. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2. Lucas C, Barnich N, Nguyen HTT. Microbiota, inflammation and colorectal cancer. Int J Mol Sci. 2017;18(6):E1310 doi:10.3390/ijms18061310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203. doi:10.1016/j.clcc.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 4. Camp ER, Patterson LD, Kester M, Voelkel-Johnson C. Therapeutic implications of bioactive sphingolipids: a focus on colorectal cancer. Cancer Biol Ther. 2017;18(9):640–650. doi:10.1080/15384047.2017.1345396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The long tail of mRNA regulation. Cell. 2017;168(3):335–338. doi:10.1016/j.cell.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 6. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi:10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 7. Chi Y, Zhou D. MicroRNAs in colorectal carcinoma—from pathogenesis to therapy. J Exp Clin Cancer Res. 2016;35:43 doi:10.1186/s13046-016-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi:10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Amato NC, Howe EN, Richer J.K. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013;341(1):46–55. doi:10.1016/j.canlet.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 10. Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331(6017):550–553. doi:10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhong J, Huang R, Su Z, et al. Downregulation of miR-199a-5p promotes prostate adeno-carcinoma progression through loss of its inhibition of HIF-1α. Oncotarget. 2017;8(48):83523–83538. doi:10.18632/oncotarget.18315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu GT, Huang YL, Tzeng HE, Tsai CH, Wang SW, Tang CH. CCL5 promotes vascular endothelial growth factor expression and induces angiogenesis by down-regulating miR-199a in human chondrosarcoma cells. Cancer Lett. 2015;357(2):476–487. doi:10.1016/j.canlet.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 13. Ghosh A, Dasgupta D, Ghosh A, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8(3):e2706 doi:10.1038/cddis.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gadducci A, Sergiampietri C, Lanfredini N, Guiggi I. Micro-RNAs and ovarian cancer: the state of art and perspectives of clinical research. Gynecol Endocrinol. 2014;30(4):266–271. doi:10.3109/09513590.2013.871525. [DOI] [PubMed] [Google Scholar]
- 15. Kim BK, Yoo HI, Kim I, Park J, Kim Yoon S. FZD6 expression is negatively regulated by miR-199a-5p in human colorectal cancer. BMB Rep. 2015;48(6):360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han Y, Kuang Y, Xue X, et al. NLK, a novel target of miR-199a-3p, functions as a tumor suppressor in colorectal cancer. Biomed Pharmacother. 2014;68(5):497–505. doi:10.1016/j.biopha.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 17. Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30(25):2888–2899. doi:10.1038/onc.2011.13. [DOI] [PubMed] [Google Scholar]
- 18. Kong Y, Bai PS, Sun H, Nan KJ, Chen NZ, Qi XG. The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int J Biochem Cell Biol. 2012;44(12):2321–2332. doi:10.1016/j.biocel.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 19. Hu Y, Liu J, Jiang B, et al. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig Dis Sci. 2014;59(9):2163–2172. doi:10.1007/s10620-014-3136-0. [DOI] [PubMed] [Google Scholar]
- 20. Ye H, Pang L, Wu Q, et al. A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn Pathol. 2015;10:65 doi:10.1186/s13000-015-0260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2(11):e363 doi:10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen T, Wang C, Wu F, et al. Altered localization of p120 catenin in the cytoplasm rather than the membrane correlates with poor prognosis in esophageal squamous cell carcinoma. PLoS One. 2015;10(3):e0118645 doi:10.1371/journal.pone.0118645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schofield AV, Bernard O. Rho-associated coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem Mol Biol. 2013;48(4):301–316. doi:10.3109/10409238.2013.786671. [DOI] [PubMed] [Google Scholar]
- 24. Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One.2010;5(9):e13067 doi:10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60 doi:10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi:10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 27. Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28(1-2):65–76. doi:10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 28. Xi ZW, Xin SY, Zhou LQ, Yuan HX, Wang Q, Chen KX. Downregulation of rho-associated protein kinase 1 by miR-124 in colorectal cancer. World J Gastroenterol. 2015;21(18):5454–5464. doi:10.3748/wjg.v21.i18.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vemula S, Shi J, Mali RS, et al. ROCK1 functions as a critical regulator of stress erythropoiesis and survival by regulating p53. Blood. 2012;120(14):2868–2878. doi:10.1182/blood-2011-10-384172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kümper S, Mardakheh FK, McCarthy A, et al. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. Elife. 2016;5:e12994 doi:10.7554/eLife.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhan Y, Zheng N, Teng F, et al. MiR-199a/b-5p inhibits hepatocellular carcinoma progression by post-transcriptionally suppressing ROCK1. Oncotarget. 2017;8(40):67169–67180. doi:10.18632/oncotarget.18052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28. doi:10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 33. Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell Adh Migr. 2015;9(4):317–324. doi:10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu YZ, Wu K, Huang J, et al. The PTEN/PI3K/Akt and Wnt/β-catenin signaling pathways are involved in the inhibitory effect of resveratrol on human colon cancer cell proliferation. Int J Oncol. 2014;45(1):104–112. doi:10.3892/ijo.2014.2392. [DOI] [PubMed] [Google Scholar]
- 35. Johnson SM, Gulhati P, Rampy BA, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210(5):767–776, 776–778. doi:10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]