Abstract
Two new bracelet cyclotides from roots of Pombalia calceolaria with potential anticancer activity have been characterized in this work. The cyclotides Poca A and B (1 and 2) and the previously known CyO4 (3) were de novo sequenced by MALDI-TOF/TOF mass spectrometry (MS). The MS2 spectra were examined and the amino acid sequences were determined. The purified peptides were tested for their cytotoxicity and effects on cell migration of MDA-MB-231, a triple-negative breast cancer cell line. The isolated cyclotides reduced the number of cancer cells by more than 80% at 20 μM, and the concentration-related cytotoxic responses were observed with IC50 values of 1.8, 2.7, and 9.8 μM for Poca A (1), Poca B (2), and CyO4 (3), respectively. Additionally, the inhibition of cell migration (wound-healing assay) exhibited that CyO4 (3) presents an interesting activity profile, in being able to inhibit cell migration (50%) at a subtoxic concentration (2 μM). The distribution of these cyclotides in the roots was analyzed by MALDI imaging, demonstrating that all three compounds are present in the phloem and cortical parenchyma regions.
Pombalia calceolaria (L.) Paula-Souza (synonyms:1 ≡ Hybanthus calceolaria (L.) Oken, ≡ Viola calceolaria L., ≡ Hybanthus ipecacuanha (L.) Oken., ≡ Pombalia ipecacuanha Vand.), commonly known as “papaconha” and “ipepacuanha”, is a herbaceous plant of the Violaceae family that is found in northeastern Brazil.2 Until 2014, this species was catalogued as Hybanthus calceolaria (L.) Oken, but more recent molecular-based phylogeny and morphological studies helped to re-establish Pombalia as a distinct segregate genus from Hybanthus; hence H. calceolaria should be termed Pombalia calceolaria.1
P. calceolaria is implicated by farmers as the cause of neurological symptoms during livestock grazing.2 Several poisoning outbreaks associated with the ingestion of this plant were observed in cattle, and the principal manifestation were ataxia, recumbency, and myokymia.2 In traditional medicine, its roots are used as decoction, infusion, and syrup as purgatives (amebicides) and expectorants.3
Phytochemically, this plant is still under investigation, and only the compound inulin, a naturally occurring polysaccharide, has been described to date as a constituent.3 However, it is thought that all violet species (Violaceae) contain small cyclic cystine-knot proteins, named cyclotides.4 These peptides are characterized by a head-to-tail cyclized backbone, usually composed of 28–37 amino acids, and a unique knotted disulfide topology involving six conserved cysteine residues termed the cyclic cystine-knot (CCK) motif.5,6 The CCK motif is responsible for their exceptional resistance to chemical, enzymatic, and thermal degradation.7 CCK proteins occur naturally in plants and have a wide range of bioactivities that make them interesting for biological or potential therapeutic applications. This includes their anti-HIV,8−10 anthelmintic,11−13 insecticidal,14−16 molluscicidal,17 antimicrobial,18,19 uterotonic,20−23 cytotoxic,24−26 hemolytic,27,28 trypsin inhibitory,29 and immunosuppressive activities.30−32
Cyclotides have been classified into two subfamilies based on topological differences. The Möbius subfamily contains a cis-proline bond in loop 5, resulting in a 180° twist of the peptide backbone, while the bracelet subfamily has all the backbone amide bonds in the trans configuration.33 Besides the presence of the cis-Pro bond, Möbius and bracelet cyclotides differ in size and amino acid composition. Moreover, a typical bracelet cyclotide contains a higher number of cationic residues as compared to their Möbius counterparts.34
It is known that the expression and the regulation of cyclotides differ among plant organs.35,36 Despite this difference, in the current literature there are only two published studies describing the distribution of the cyclotides in plant tissues by MALDI imaging37 and immunolocalization.38
In this study, an analytical characterization was carried out on three cyclotides from P. calceolaria, namely, Poca A (1), Poca B (2), and the previously known CyO45,999 (3). We investigated their effects on breast cancer cell migration and cytotoxicity. On the basis of MALDI imaging experiments, it was demonstrated that the cyclotides accumulated in the phloem and cortical parenchyma of the plant roots, with higher levels of theses peptides to be found in primary cell walls.
Results and Discussion
Cyclotide Identification from P. calceolaria
The peptide-rich fractions from the root extract (C18_80%) of P. calceolaria (L.) Oken were subjected to MALDI-TOF MS to identify the compounds’ molecular weight signals in the range between 2500 and 4500 Da. The presence of many mass signals indicated a great diversity of possible cyclotides in the C18_80% root extract, with signals in the region between 2900 and 3400 Da (Figure S1A, Supporting Information). To analyze the number of disulfide bonds of these putative peptides, the fractions were subjected to S-reduction and S-carbamidomethylation modifications. Each S-alkylated cysteine residue increased the mass of a peptide by 58 Da. All peptide signals displayed a mass shift of 348 Da, indicating the presence of six cysteine residues (Figure S1B, Supporting Information).
Cyclotide Isolation and de Novo Sequencing
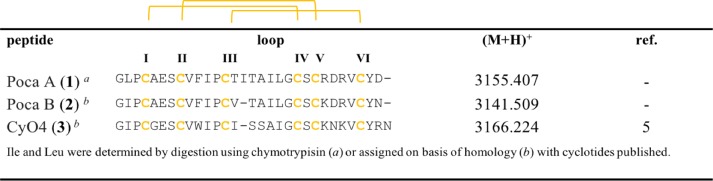
Aiming at the purification of these cyclotides, successive elution using preparative analysis was employed to the root C18_80% extract (464 mg), and the purities and the monoisotopic masses of compounds were verified by analytical RP-HPLC and MALDI-TOF MS. From this fraction, it was possible to isolate three cyclotides, namely, Poca A (1) at m/z 3155, Poca B (2) at m/z 3141, and the known CyO4 (3) at m/z 3166 (Figure S2B, Supporting Information). Other cyclotides occurred in the root C18_80% extract, and their masses and retention times were measured (Table S1, Supporting Information).
Reduction and alkylation of the peptides resulted in a mass gain of 348 Da, which confirmed the presence of six cysteine residues (to form three disulfide bonds). To sequence cyclotides by MS/MS, the peptides were hydrolyzed enzymatically by targeting the conserved Glu residue in loop 1 using endoproteinase GluC (Endo-GluC), which commonly results in a single (linearized cyclotide) fragment and also allowed the presence of a cyclic backbone to be inferred, with a mass increase of 18 Da (in total 366 Da compared to the native form). To achieve faster, more readily interpretable peptide fragments, other enzymes such as trypsin or chymotrypsin were used. The resultant mass spectra were carefully examined for each peptide, and the sequence was determined based on the presence of both the b- and y-ion series (N- and C-terminal fragments).
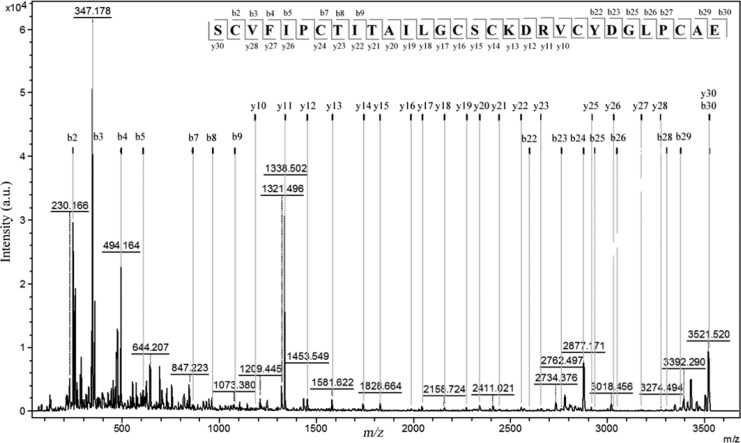
The enzymatic digestion of Poca A (1) using Endo-GluC provided an ion at m/z 3521, indicating the ring-opening of the cyclotide backbone. This ion was fragmented by MS/MS, allowing the sequence of the y-ion series to be determined as SCVFI/LPCTI/LTAI/LI/LGCSCKDRV and YDGI/LPCAE as b-ion series (Figure 1). The tryptic digestion of Poca A (1) showed the presence of peaks at m/z 3145 and 2067. The analysis of these ions suggested that the m/z 2067 ion is a part of the m/z 3145 ion. All fragments generated from both these ions (tryptic digestion) yielded a complementary sequence (Table S2, Supporting Information).
Figure 1.
MALDI-TOF/TOF spectra of Poca A (1) cyclotide isolated from the roots of P. calceolaria. Tandem MS analysis of the fragment m/z 3521.5 obtained by digestion with Endo-GluC.
There is an intrinsic problem in distinguishing between isobaric residues, i.e., Leu and Ile, using MS/MS. Therefore, we used chymotrypsin to attempt to solve this question, knowing that it hydrolyzes amino acid sequences after Leu but not Ile. If, however, Leu is followed by a Pro, cleavage is impossible, and hence the results are difficult to interpret. Thus, the chymotrypsin digestion of Poca A (1) yielded two fragments, at m/z 3503 and 1023. The m/z 1023 ion was generated from digestion of the aromatic amino acids Phe10 and Leu19 present in the chain. This analysis allowed only three out of five isobaric Leu/Ile residues of Poca A (1), i.e., Ile15, Ile18, and Leu19 residues (loop 3), to be distinguished. The other two isobaric residues, i.e., Leu2 and Ile11, were determined by homology and sequence comparison to known cyclotides (http://www.cybase.org.au), and in this case, the amino acid analysis was not performed. The complete sets of b- and y-ions observed for all enzymes allowed proposing an amino acid sequence of the bracelet cyclotide, Poca A (1), to be cyclo-GLPCAESCVFIPCTITAILGCSCKDRVCYD.
The characterization of both Poca B (2) and CyO4 (3) was accomplished using reduction and alkylation and the Endo-GluC, trypsin, and chymotrypsin enzymes, as described above, with the sequences determined accordingly (Figure 2). The observed fragments are available in the Supporting Information (Table S2).
Figure 2.
Structures and sequences of identified cyclotides. Sequence alignment and monoisotopic masses (M+H)+ of new cyclotides (1 and 2) and a known cyclotide (3) isolated from P. calceolaria.
Cancer Cell Cytotoxicity of Cyclotides
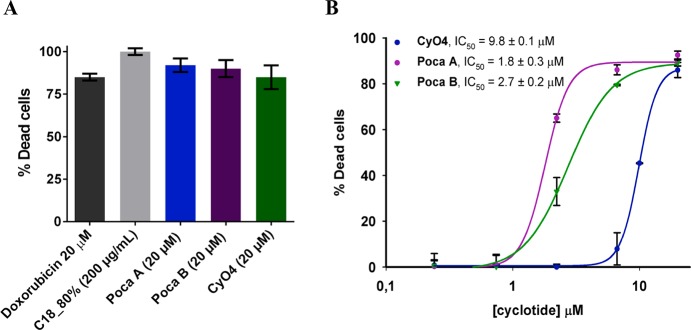
In recent years, several cyclotides have been reported as cytotoxic or antitumor agents with a mechanism of action associated with membrane-disrupting properties.39,24 Their remarkable stability has led to further investigations of their therapeutic potential.24 Owing to its high cyclotide content, the root C18_80% fraction was evaluated for its cytotoxic activity against MDA-MB-231 breast cancer cells at 200 μg/mL (Figure 3A). This cyclotide mixture showed a high cytotoxic efficiency and markedly reduced the viability of the cells at this concentration (100% dead cells). The activities of the isolated cyclotides Poca A (1), Poca B (2), and CyO4 (3) were evaluated to verify their cytotoxic effects. The cyclotides led to more than 80% of the cancer cell death at 20 μM (Figure 3A), yielding a concentration-related cytotoxic response with IC50 values determined to be 1.8, 2.7, and 9.8 μM, respectively, for Poca A (1), Poca B (2), and CyO4 (3) (Figure 3B). The most potent cyclotide, Poca A (1), exhibited an IC50 value similar to that of the positive control substance doxorubicin (IC50 of 0.9 μM), an effective antitumor drug used in chemotherapy.40 The bioactivity of these novel cyclotides against the MDA-MB-231 cell line is supportive of further studies using their scaffold as the basis for the design of new analogues to be studied as cytotoxic agents.
Figure 3.
Cytotoxicity of the plant extract and isolated cyclotides. (A) Cytotoxicity at a single concentration. (B) Concentration–response curves. Data are presented as the means ± SD of triplicate measurements.
Inhibition of Cell Migration of Cyclotides
The cell migration inhibitory activities of these compounds were evaluated, taking into account that cell migration of tumor cells is crucial to metastasis (the cause of 90% of human cancer deaths).41 Although a large number of studies have been conducted on the structure–activity of many classes of compounds, only a few studies have described the effects of cyclotides on cancer cell migration.42 Thus, to evaluate these effects, a wound-healing assay was conducted, using the MDA-MB-231 breast cancer cell line (Table 1). The cyclotides were tested at four different concentrations (1, 2, 5, and 10 μM), and the experiments demonstrated that Poca A (1) and Poca B (2) were inactive at 1 μM and toxic at higher concentrations, which is in agreement with their cytotoxic potency observed (Figure 4B). However, CyO4 (3) showed an interesting activity profile, in being capable of inhibiting cell migration at subtoxic concentrations (Table 1). Thus, this cyclotide inhibited 50% of wound closures while exhibiting no toxicity at a concentration of 2 μM.
Table 1. Effects of Cyclotides (1–3) on Cell Migration.
| wound
closure inhibitiona (%) |
||||
|---|---|---|---|---|
| compound | 1 μM | 2 μM | 5 μM | 10 μM |
| Poca A (1) | 0% | b | b | b |
| Poca B (2) | 0% | b | b | b |
| CyO4 (3) | 15 ± 1 | 50 ± 4 | 60 ± 3 | b |
| colchicine | 75 ± 2 | n.d. | n.d. | 85 ± 7 |
Results are presented as the means of two independent experiments ± SD; n.d. not determined.
Toxic (toxicity was visually assessed: cells detached and presented morphological alterations such as volume reduction and membrane disintegration).
Figure 4.

Transverse sections of the root tissues from P. calceolaria and MALDI-MS images reconstructed with ions at m/z 3166.3807 [M + H]+ (C), m/z 3155.3866 [M + H]+ (D), and m/z 3141.3062 [M + H]+ (E). CP, cortical parenchyma; Pe, peridermis; Ph, phloem; Xy, xylem.
Anatomical and Imaging Studies of P. calceolaria Roots
Plant roots are organs at the beginning of secondary growth. They are composed of a short peridermis, with two to three layers of phellem, a remaining cortical parenchyma, and a vascular cylinder with an inner xylem and outer phloem. The phloem is formed by companion cells, a sieve tube, and rays of parenchyma cells. The xylem is formed of fibers, wide xylem vessels, and rays of fibers and parenchymatic thick-walled cells. The xylem occupies the whole center of the root.
The peptides previously isolated from P. calceolaria root showed ions at m/z 3155.3866 [M + H]+ Poca A (1), m/z 3141.3062 [M + H]+ Poca B (2), and m/z 3166.3870 [M + H]+ CyO4 (3), by MALDI-MS analysis. The identity and distribution of these peptides in root tissue were confirmed by MALDI imaging. None of the ions were localized in xylem tissue, and all were detected in the region of phloem and cortical parenchyma as demonstrated by higher accumulation of these peptides in primary cell wall (Figure 4). Prior immunolocalization studies of the roots of Viola odorata and Viola uliginosa determined that cyclotides are present in the vascular bundles, specifically in the phloem, collenchyma, and xylem parenchyma, but not in the xylem vessels.38 Thus, the present anatomical studies using MALDI imaging reinforce that the distribution of cyclotides in the plant roots supports their proposed role in defense.38
Experimental Section
General Experimental Procedures
Analytical HPLC analysis was carried out on a Dionex Ultimate 3000 instrument (Dionex, Switzerland) and a Shimadzu Prominence LC-20A instrument (Tokyo, Japan), using a LC-6AD pump, a DAD SPD-M20A detector, a SIL-10AF autosampler, a CBM-20A controller, and a DGU 20-A5R degassing unit, with a Phenomenex C18 column (4.6 × 250 mm, 100 Å pore size, 5 μm particle size, 1 mL min–1) (Torrance, CA, USA). Preparative purifications were performed on the Shimadzu Prominence LC-20A instrument, with an LC-20AT pump, a DAD SPD-M20A detector, a SIL-20A autosampler, a CBM-20A controller, a DGU 20-A3 degassing unit, and a CTO-20A oven, with a Phenomenex Luna C18 column (21.2 × 250 mm, 100 Å pore size, 5 μm particle size, 10 mL min–1). For all analytical procedures, two detection wavelengths were selected: 220 and 280 nm. The solvents consisted of buffer A (0.1% aqueous trifluoroacetic acid) and B (90% CH3CN, 0.08% trifluoroacetic acid).
Plant Material
The roots P. calceolaria were collected in Chico Mendes Natural Park, Recreio dos Bandeirantes, Rio de Janeiro, Brazil, during February 2013. The plant specimen were identified by one of the authors (M.T.), and a voucher has been deposited at Rio de Janeiro Botanical Garden with the collection number MT576 (JBRJ).
Extraction and Isolation
Dried and pulverized roots (22.3 g) of P. calceolaria were extracted with 400 mL of CH3OH–H2O (6:4, v/v) at room temperature four times over a period of 24 h. The extract was partitioned with CH2Cl2–CH3OH–H2O (1:1:1, v/v/v) (four times), and the aqueous phases were separated and concentrated on a rotary evaporator prior to freeze-drying, yielding an aqueous extract (1.17 g). The root aqueous extract was dissolved in CH3CN–H2O (1:9; v/v) and immediately used for solid-phase extraction (SPE). A C18 SPE cartridge (Strata-Phenomenex C18 55 μm, 70 Å, 500 mg) was activated with CH3OH and subsequently equilibrated with aqueous 0.1% trifluoroacetic acid. After application of the extract, the cartridge was washed with mixtures of buffer A (0.1% aqueous trifluoroacetic acid) in B (90% CH3CN, 0.08% trifluoroacetic acid) in the proportions 8:2 (v/v) and 2:8 (v/v), respectively. The fraction eluted in buffer B 80%, named roots C18_80%, was considered a peptide-rich fraction (464.4 mg). A 106.5 mg aliquot of this fraction was submitted to preparative HPLC using a linear gradient from 30% to 60% buffer B in buffer A over 60 min, yielding three cyclotides: Poca A (1) (1.7 mg), Poca B (2) (6.0 mg) and CyO4 (3), (0.8 mg).
Sequencing of Pombalia Cyclotides
These cyclotides were characterized by de novo peptide sequencing using enzymatic digestion, MALDI-TOF/TOF MS, and sequence comparison using CyBase tools (www.cybase.org.au). The amino acid sequences of Poca A and B (1 and 2) and CyO4 (3) were elucidated by MS using a Bruker Daltonics Ultraflex MALDI TOF/TOF mass spectrometer (Bremen, Germany) and a MALDI-TOF/TOF 4800 proteomics analyzer (Concord, Ontario, Canada). The reflector positive-ion mode was used, and 1000–2000 laser shots were acquired per spectrum. The calibration was performed using Bruker’s Peptide Calibration Standard II (Pepcal II). For acquisition of unmodified samples the dried compounds (1–3) were dissolved in 0.1% trifluoroacetic acid and mixed at a ratio of 1:3 (v/v) with a matrix solution of saturated α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO, USA) in ddH2O–CH3CN–trifluoroacetic acid, 50:50:0.1% (v/v/v). An aliquot (2 μL) of the sample/matrix mixture was directly spotted onto the MALDI target plate and allowed to air-dry. Mass spectra were obtained over the spectral range m/z 2500 to 4500. For sequencing, the peptides were reduced and alkylated according to a previously established protocol described by Gruber et al.43 The reduced and alkylated cyclotides were digested using individual endoGlu-C, trypsin, and chymotrypsin enzymes, as described previously.43 The peptides obtained from enzymatic digestion were desalted using C18 ZipTips (Millipore, Billerica, MA, USA) and reconstituted in 10 μL of buffer B (90% CH3CN, 0.08% trifluoroacetic acid) in buffer A (0.1% aqueous trifluoroacetic acid) (8:2) (v/v). MS/MS spectra were recorded from enzymatic fragments. Each cyclotide amino acid sequence was obtained by manual assignment of the N-terminal b-ion and C-terminal y-ion series using the Bruker FlexAnalysis 3.3 software and Data Explorer software version 4.3.
Cell Culture
The human cell lines were obtained from the Rio de Janeiro Cell Bank (BCRJ, Rio de Janeiro, Brazil). The MDA-MB-231 (metastatic breast cancer) cells were maintained in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% fetal bovine serum (FBS). The cultures were maintained at 37 °C in a 5% CO2 humidified atmosphere. Cell viability was assessed using the Trypan blue exclusion test before the experiments.
Wound-Healing Assay
The influence of the substances in directional cell migration was assessed qualitatively by a wound-healing assay.44,45 Using this procedure, 1 × 105 MDA-MB-231 cells/well were seeded in 24-well culture plates (TPP) and allowed to grow a confluent monolayer. The monolayers were scratched off using a sterile 100 μL pipet tip, and fresh medium without FBS was added to clean the debris. Finally, the cells were incubated for 22 h at 37 °C with supplemented medium (10% FBS) and different concentrations of test compounds (10, 5, 2, and 1 μM). Photographs of the wounds at 0 and 22 h were taken under a 4× objective (Optiphase). The areas of the scratches were analyzed using ImageJ software, and two independent experiments were conducted in triplicate. Colchicine was used as a positive control.
MTS Cytotoxicity Assay
The cells were plated in 96-well culture plates (TPP) (100 μL of DMEM medium per well) at a density of 4 × 103 cells/well for MDA-MB-231. After 24 h, seven concentrations (20–0.03 μM) of the test substances and doxorubicin (as control) were added in triplicate to the wells. After 24 h of incubation at 37 °C in a 5% CO2 humidified atmosphere, 20 μL of the MTS reagent46 (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega) was added to the wells, and the cells were incubated for 4 h at 37 °C. Absorbance was measured at 490 nm using a spectrophotometer (SpectraMax Plus384, Molecular Devices), and the percentage of viable cells was determined in relation to the control wells. At least two independent experiments in triplicate were carried out for each test compound. The IC50 values were calculated using nonlinear regression analysis using Sigmaplot 13.0 (Systat Software).
Histological Analysis of P. calceolaria Roots
A flesh root was sectioned in transverse plan (30 μm) using a rotatory microtome (Leica RM 2255), stained with safrablau (safranine/astra blue), mounted on a slide using water, and observed under a light microscope. Photomicrographs were obtained using a Leica DM 4500 B light microscope coupled to a Leica DFC 320 digital camera.
MALDI Imaging Analysis
Analysis was performed using the MALDI-TOF/TOF UltrafleXtreme (Bruker Daltonics) equipped with a Smartbeam-II laser system and operated in the reflector positive-ion mode, with the instrument controlled using FleXcontrol v.3.3 (Bruker Daltonics), employing the AutoXecute mode. MSI data were analyzed and normalized using SCiLS Lab MVS 2018a. The settings of the instrument were as follows: positive-ion reflector mode; ion source 1, 25.00 kV; ion source 2, 22.40 kV; lens, 8.30 kV; pulsed ion extraction, 120 ns; matrix suppression mass cutoff, m/z 1000. The spectra were recorded across a mass range of m/z 2–4 kDa by accumulating 2000 shots per spectrum, using a laser spot diameter of 40 μm. A peptide calibration standard II (Bruker Daltonics) was used for external calibration of the mass spectrometer. The average mass deviation was below 10 ppm. The transverse sections of roots were adhered with double-sided carbon tape to an indium tin oxide-coated conductive slide (Bruker Daltonics) for MALDI analysis. The matrix α-cyano-4-hydroxycinnamic acid solution (7 mg/mL) was prepared in CH3CN–H2O (1:1, v/v) with 0.2% trifluoroacetic acid. The matrix was applied to the tissue by an ImagePrep apparatus using N2 flux during the spray process, totaling 60 cycles of spray, then incubated and dried.
Acknowledgments
The authors are thankful for grant 2017/17098-4, São Paulo Research Foundation (FAPESP), and grant 162855/2015-0, National Council of Technological and Scientific Development (CNPq) for a scholarship awarded to M.E.F.P. SISBIOTA-CNPq-FAPESP grant 2010/52327-5, CEPID-FAPESP grant 2013/07600-3, INCT-CNPq grant 465637/2014-0, L.P. Lopes FAPESP grant 2014/50265-3, A.F. Bobey CNPq grant 150244/2014-2, and CAPES are acknowledged for scholarships and other financial support. The research of C.W.G. was supported by the Austrian Science Fund (FWF) through project I3243.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.7b00969.
Mass spectra of root extract before and after S-reduction and S-alkylation (Figure S1); RP-HPLC analytical profile of root extract and mass spectra of isolated peptides (Figure S2); molecular weights and retention times of cyclotides identified in roots (Table S1); fragments and sequences from Poca A, B (1, 2) and CyO4 (3 obtained by enzymatic digestion and de novo sequencing (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- de Paula-Souza J.; Ballard J. H. E. Phytotaxa 2014, 183, 1–15. 10.11646/phytotaxa.183.1.1. [DOI] [Google Scholar]
- Carvalho F. K. L.; Nascimento E. M.; Rocha B. P.; Mendonça F. S.; Veschi J. L. A.; Silva S. M. M. S.; Medeiros R. M. T.; Riet-Correa F. J. Vet. Diagn. Invest. 2014, 26, 674–677. 10.1177/1040638714544685. [DOI] [PubMed] [Google Scholar]
- Pontes A. G. O.; Silva K. L.; Fonseca S. G. d. C.; Soares A. A.; Feitosa J. P. d. A.; Braz-Filho R.; Romero N. R.; Bandeira M. A. M. Carbohydr. Polym. 2016, 149, 391–398. 10.1016/j.carbpol.2016.04.108. [DOI] [PubMed] [Google Scholar]
- Burman R.; Yeshak M. Y.; Larsson S.; Craik D. J.; Rosengren K. J.; Göransson U. Front. Plant Sci. 2015, 6, 855. 10.3389/fpls.2015.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J.; Daly N.; Bond T.; Waine C. J. Mol. Biol. 1999, 294, 1327–1336. 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- Craik D. J. Toxicon 2001, 39, 1809–1813. 10.1016/S0041-0101(01)00129-5. [DOI] [PubMed] [Google Scholar]
- Colgrave M. L.; Craik D. J. Biochemistry 2004, 43, 5965–5975. 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- Gustafson K. R.; Sowder R. C.; Henderson L. E.; Parsons I. C.; Kashman Y.; Cardellina J. H. II; McMahon J. B.; Buckheit R. W.; Pannell L. K.; Boyd M. R. J. Am. Chem. Soc. 1994, 116, 9337–9338. 10.1021/ja00099a064. [DOI] [Google Scholar]
- Daly N.; Koltay A.; Gustafson K.; Boyd M.; Casas-Finet J.; Craik D. J. J. Mol. Biol. 1999, 285, 333–345. 10.1006/jmbi.1998.2276. [DOI] [PubMed] [Google Scholar]
- Hallock Y. F.; Sowder R. C.; Pannell L. K.; Hughes C. B.; Johnson D. G.; Gulakowski R.; Cardellina J. H. II; Boyd M. R. J. Org. Chem. 2000, 65, 124–128. 10.1021/jo990952r. [DOI] [PubMed] [Google Scholar]
- Colgrave M. L.; Kotze A. C.; Huang Y. H.; O’Grady J.; Simonsen S. M.; Craik D. J. Biochemistry 2008, 47, 5581–5589. 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- Colgrave M. L.; Kotze A. C.; Ireland D. C.; Wang C. K.; Craik D. J. ChemBioChem 2008, 9, 1939–1945. 10.1002/cbic.200800174. [DOI] [PubMed] [Google Scholar]
- Colgrave M. L.; Kotze A. C.; Kopp S.; McCarthy J. S.; Coleman G. T.; Craik D. J. Acta Trop. 2009, 109, 163–166. 10.1016/j.actatropica.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Jennings C.; West J.; Waine C.; Craik D. J.; Anderson M. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 10614–10619. 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings C. V.; Rosengren K. J.; Daly N. L.; Plan M.; Stevens J.; Scanlon M. J.; Waine C.; Norman D. G.; Anderson M. A.; Craik D. J. Biochemistry 2005, 44, 851–860. 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- Barbeta B. L.; Marshall A. T.; Gillon A. D.; Craik D. J.; Anderson M. A. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 1221–1225. 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plan M. R. R.; Saska I.; Cagauan A. G.; Craik D. J. J. Agric. Food Chem. 2008, 56, 5237–5241. 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- Tam J. P.; Lu Y.-A.; Yang J.-L.; Chiu K.-W. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 8913–8918. 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pränting M.; Lööv C.; Burman R.; Göransson U.; Andersson D. I. J. Antimicrob. Chemother. 2010, 65, 1964–1971. 10.1093/jac/dkq220. [DOI] [PubMed] [Google Scholar]
- Gran L. Acta Pharmacol. Toxicol. 1973, 33, 400–408. 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Gran L. Lloydia 1973, 6, 174–178. [PubMed] [Google Scholar]
- Saether O.; Craik D. J.; Campbell I. D.; Sletten K.; Juul J.; Norman D. J. Biochemistry 1995, 34, 4147–4158. 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- Koehbach J.; O’Brien M.; Muttenthaler M.; Miazzo M.; Akcan M.; Elliott A. G.; Daly N. L.; Harvey P. J.; Arrowsmith S.; Gunasekera S.; Smith T. J.; Wray S.; Göransson U.; Dawson P. E.; Craik D. J.; Freissmuth M.; Gruber C. W. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 21183–21188. 10.1073/pnas.1311183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm P.; Göransson U.; Johansson S.; Claeson P.; Gullbo J.; Larsson R.; Bohlin L.; Backlund A. Mol. Cancer Ther. 2002, 1, 365–369. [PubMed] [Google Scholar]
- Svangård E.; Göransson U.; Hocaoglu Z.; Gullbo J.; Larsson R.; Claeson P.; Bohlin L. J. Nat. Prod. 2004, 67, 144–147. 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- Herrmann A.; Burman R.; Mylne J. S.; Karlsson G.; Gullbo J.; Craik D. J.; Clark R. J.; Göransson U. Phytochemistry 2008, 69, 939–952. 10.1016/j.phytochem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Schöpke T.; Hasan A. M. I.; Kraft R.; Otto A.; Hiller K. Sci. Pharm. 1993, 61, 145–153. [Google Scholar]
- Chen B.; Colgrave M. L.; Wang C.; Craik D. J. J. Nat. Prod. 2006, 69, 23–28. 10.1021/np050317i. [DOI] [PubMed] [Google Scholar]
- Hernandez J. F.; Gagnon J.; Chiche L.; Nguyen T. M.; Andrieu J.-P.; Heitz A.; Hong T. T.; Chau P. T. T.; Le-Nguyen D. Biochemistry 2000, 39, 5722–5730. 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- Thell K.; Hellinger R.; Sahin E.; Michenthaler P.; Gold-Binder M.; Haider T.; Kuttke M.; Liutkevičiu̅tė Z.; Göransson U.; Gründemann C.; Schabbauer G.; Gruber C. W. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 3960–3965. 10.1073/pnas.1519960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinger R.; Koehbach J.; Fedchuk H.; Sauer B.; Huber R.; Gruber C. W.; Gründemann C. J. Ethnopharmacol. 2014, 151, 299–306. 10.1016/j.jep.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründemann C.; Koehbach J.; Huber R.; Gruber C. W. J. Nat. Prod. 2012, 75, 167–174. 10.1021/np200722w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J.; Daly N. L. Mol. BioSyst. 2007, 3, 257–265. 10.1039/b616856f. [DOI] [PubMed] [Google Scholar]
- Ireland D. C.; Colgrave M. L.; Craik D. J. Biochem. J. 2006, 400, 1–12. 10.1042/BJ20060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson U.; Broussalis A. M.; Claeson P. Anal. Biochem. 2003, 318, 107–117. 10.1016/S0003-2697(03)00114-3. [DOI] [PubMed] [Google Scholar]
- Trabi M.; Craik D. J. Plant Cell 2004, 16, 2204–2216. 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth A. G.; Mylne J. S.; Grassl J.; Lyons R. E.; Millar H.; Colgrave M. L.; Craik D. J. J. Biol. Chem. 2012, 287, 27033–27046. 10.1074/jbc.M112.370841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slazak B.; Kapusta M.; Malik S.; Bohdanowicz J.; Kuta E.; Malec P.; Göransson U. Planta 2016, 244, 1029–1040. 10.1007/s00425-016-2562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardino K.; Pinto M. E. F.; Bolzani V. S.; de Moura A. F.; Batista J. M. Jr. Chem. Commun. 2017, 53, 7337–7340. 10.1039/C7CC02333B. [DOI] [PubMed] [Google Scholar]
- Svangård E.; Burman R.; Gunasekera S.; Lövborg H.; Gullbo J.; Göransson U. J. Nat. Prod. 2007, 70, 643–647. 10.1021/np070007v. [DOI] [PubMed] [Google Scholar]
- Kim H. S.; Lee Y. S.; Kim D. K. Pharmacology 2009, 84, 300–309. 10.1159/000245937. [DOI] [PubMed] [Google Scholar]
- Wells A.; Grahovac J.; Wheeler S.; Ma B.; Lauffenburger D. Trends Pharmacol. Sci. 2013, 34, 283–289. 10.1016/j.tips.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E.; Wang D.; Chen J.; Tao X. Int. J. Clin. Exp. Med. 2015, 8, 4059–4065. [PMC free article] [PubMed] [Google Scholar]
- Gruber C. W.; Elliott A. G.; Ireland D. C.; Delprete P. G.; Dessein S.; Göransson U.; Trabi M.; Wang C. K. B.; Robbrecht E.; Craik D. Plant Cell 2008, 20, 2471–2483. 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürk R. R. Proc. Natl. Acad. Sci. U. S. A. 1973, 70, 369–372. 10.1073/pnas.70.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. C.; Park A. Y.; Guan J. L. Nat. Protoc. 2007, 2, 329–333. 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Barltrop J. A. Bioorg. Med. Chem. Lett. 1991, 1, 611–614. 10.1016/S0960-894X(01)81162-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.