Abstract
The bacterial membrane protein SecDF enhances protein translocation across the membrane driven by the complex of SecA ATPase and SecYEG. Many newly synthesized proteins in the cytoplasm are programmed to be translocated to the periplasm via the narrow channel that is formed in the center of SecYEG. During the protein-translocation process, SecDF is proposed to undergo repeated conformational transitions to pull out the precursor protein from the SecYEG channel into the periplasm. Once SecDF captures the precursor protein on the periplasmic surface, SecDF can complete protein translocation even if SecA function is inactivated by ATP depletion, implying that SecDF is a protein-translocation motor that works independent of SecA. Structural and functional analyses of SecDF in 2011 suggested that SecDF utilizes the proton gradient and interacts with precursor protein in the flexible periplasmic region. The crystal structures of SecDF in different states at more than 3Å resolution were reported in 2017 and 2018, which further improved our understanding of the dynamic molecular mechanisms of SecDF. This review summarizes recent structural studies of SecDF.
Keywords: protein translocation, SecDF, SecYEG, crystal structure, proton driven
Crystal structures and subsequent functional analyses have elucidated the dynamics of proton-driven SecDF, which is involved in mediating protein translocation across the membrane.
INTRODUCTION
One of the essential biological phenomena conserved in all organisms is protein translocation across the membrane. More than 30% of proteins that are newly synthesized by ribosomes are translocated via a protein-conducting channel called the Sec translocon, which is composed of membrane proteins SecY, SecE and SecG in bacteria (Chatzi et al. 2014; Tsirigotaki et al. 2017), corresponding to Sec61α, Sec61γ and Sec61β, respectively, in eukaryotes (Rapoport, Li and Park 2017). The Sec translocon provides a pathway for precursors via its structural changes. After SecY was first identified as a component of the protein-translocation machinery (Ito et al. 1983), many genetic, biochemical and structural studies have been performed, among which, the first report of the Sec translocon crystal structure is particularly important (van den Berg et al. 2004). The structure revealed that transmembrane (TM) helices of SecY form a penetrated hourglass-like pore, in the center of which a constricted ring prevents leakage of water, ions and substrates in the resting state. A series of structure-based functional analyses of the Sec translocon machinery have provided mechanistic clues regarding how proteins are transported across the membrane. A common mechanism supported by the crystal structures of the Sec translocon (Tsukazaki et al. 2008; Zimmer, Nam and Rapoport 2008; Egea and Stroud 2010; Tanaka et al. 2015; Li et al. 2016) is that SecY allows unfolded precursor proteins to pass through the membrane, following appropriate expansion of the pore size and constriction of the ring. The pore is closed by the plug domain on the periplasmic side in the resting state, whereas the loop of SecG covers the cytoplasmic side to completely seal the translocation pathway. The Sec translocon provides not only the vertically oriented pore for protein translocation, but also a lateral gate opening for membrane protein insertion. During integration of membrane proteins, bacterial YidC (Samuelson et al. 2000; Kumazaki et al. 2014a,b; Xin et al. 2018) functions as a chaperon in the membrane in concert with SecYEG (Hennon et al. 2015).
The precursor proteins in unfolded state are translocated through the Sec translocon and then folded into mature proteins in the periplasm in bacteria (Fig. 1A) or in the endoplasmic reticulum in eukaryotes. There are two types of protein-translocation mechanisms that occur via the Sec translocon, viz., co-translational and post-translational translocation. In co-translational translocation, the Sec translocon is directly linked to the ribosome, and protein export from the cytosol occurs simultaneously with polypeptide elongation. Recent cryo-electron microscopy studies have illustrated the intermediate states of the ribosome-nascent chain complex at medium resolution (Bischoff et al. 2014; Gogala et al. 2014; Park et al. 2014; Voorhees et al. 2014; Pfeffer et al. 2015; Jomaa et al. 2016), providing insights into the dynamics of the Sec translocon, including lateral gate opening, expansion of pore size and dislocation of the plug. In post-translational translocation in bacteria, the synthesized proteins in the cytoplasm are retained in the unfolded state by chaperons such as SecB, and targeted to the membrane by information provided from the signal sequence. Membrane-associated SecA, which has affinity for SecY and is involved in targeting of precursors to the membrane, repeatedly pushes the precursor protein into SecYEG using the energy from ATP hydrolysis to complete the translocation. Several attractive molecular mechanisms of SecA-driven protein translocation have been proposed, although they are still controversial (Zimmer, Nam and Rapoport 2008; Chatzi et al. 2014; Allen et al. 2016; Hsieh et al. 2017).
Figure 1.
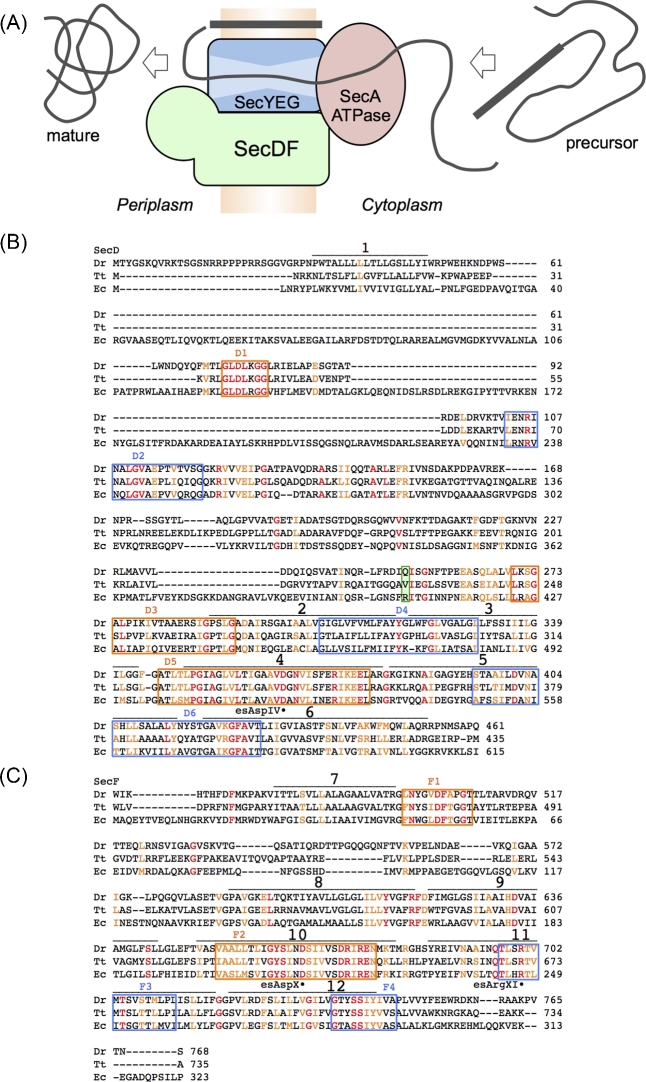
(A) Bacterial protein translocation across the membrane. SecA and SecDF drive protein translocation via the SecYEG complex, a protein conducting channel. (B) Sequence alignment of SecD from Deinococcus radiodurans (Dr), Thermus thermophilus (Tt) and Escherichia coli (Ec). (C) Sequence alignments of SecF from Dr, Tt and Ec. The TM numbers are shown. Perfectly and highly conserved residues (Furukawa et al. 2017) are colored red and orange, respectively. Essential residues Asp in TM4 (esAspIV), Asp in TM10 (esAspX) and Arg in TM11 (esArgXI) are indicated by solid circles. The conserved regions D1-6 and F1-4 (Eichler 2003) are highlighted by colored squares. The green box indicates EcSecD R407 and its corresponding residues (see also Fig. 5).
SecDF is a protein-translocation factor, which is different from the ATP-driven SecA motor. It functions at the periplasmic side independent of SecA (Tsukazaki et al. 2011). However, some reports have suggested that SecDF is related to SecA or SecG functions (Economou et al. 1995; Duong and Wickner 1997). In addition, the interaction between SecDF and YidC is involved in the integration of membrane proteins (Nouwen and Driessen 2002; Chen et al. 2005). Although a previous study indicated that SecDF is associated with a late step of protein translocation at the periplasmic side (Matsuyama, Fujita and Mizushima 1993), the detailed mechanism of SecDF remained unclear. In 2011, the structural and functional analyses of SecDF proposed that SecDF is a protein-translocation motor that pulls precursor proteins from the SecYEG channel to the periplasmic space using the energy of a proton gradient across the membrane (Tsukazaki et al. 2011). Furthermore, two recent reports regarding the crystal structures of SecDF at more than 3 Å resolution have allowed us to discuss the detailed molecular mechanisms of the complex (Furukawa et al. 2017, 2018).
In this review, I have summarized structural information regarding SecDF and proposed a working model of SecDF-assisted protein translocation based on the structural biology analyses of SecDF.
CHARACTERIZATION OF SecDF
Genes secD and secF are involved in protein translocation (Gardel et al. 1987, 1990) and have been shown to be conserved in bacteria and archaea (Eichler 2003). Most species, including Escherichia coli (Ec) and Vibrio alginolyticus (Va) have consecutive secD and secF. The membrane proteins, SecD and SecF, which are encoded by these genes (Pogliano and Beckwith 1994a,b), form a stable heterodimer called SecDF. Most of the SecDFs that have been characterized in vivo are derived from Ec and Va. The amino acid sequences of SecDFs are shown in Fig. 1B and C. SecDF-depleted Ec strains showed decreased efficiency of protein translocation, resulting in cell growth inhibition, particularly at lower temperatures (Pogliano and Beckwith 1994a; Nouwen and Driessen 2005; Hand et al. 2006). The antibody that recognizes the periplasmic region of SecDF inhibits protein translocation, suggesting that SecDF may be involved in the release of precursor protein from the membrane to the periplasmic space (Matsuyama, Fujita and Mizushima 1993). Certain SecDFs are expressed as a single membrane protein possessing 12 TM helices, such as those from Thermus thermophilus (Tt) or Deinococcus radiodurans (Dr), the crystal structures of which have been reported (Tsukazaki et al. 2011; Furukawa et al. 2017, 2018). Based on its amino acid sequence, SecDF is classified as a member of the resistance-nodulation-division (RND) superfamily of proteins, which includes 12 TM helices (Tseng et al. 1999); however, the homology between SecDFs and other proteins of the RND superfamily is low. Importantly, the size of the periplasmic region of SecDF is completely different from that of other members of the RND superfamily. In addition, unlike MexB, AcrB, CusA and ZneA, which exist as homotrimers (Pak et al. 2013; Yamaguchi, Nakashima and Sakurai 2015) and are involved in the export of specific ions and small molecules, SecDF exists as a monomer and forms a holo-translocon complex with SecYEG and YidC to export proteins (Botte et al. 2016). Some conserved, essential residues in the TM region of AcrB transporters are not conserved in SecDFs. These differences between SecDF and other members of the RND superfamily imply that the working mechanism of SecDF is likely to be different from those of the other members. In Ec, SecDF forms a stable complex with a membrane protein, YajC, which might be involved in protein translocation; however, the details of this mechanism are unclear (Pogliano and Beckwith 1994a,b). Although YajC may function to stabilize the SecDF complex, yajC, located just upstream of secD, is not an essential factor. YajC possesses one TM helix that can form a complex with TM segments 2, 11, and 12 of AcrB; however, the importance of these interactions is not known (Tornroth-Horsefield et al. 2007). YajC may peripherally interact with the TM2, 11 and 12 of SecDF in the same manner as indicated in the crystal structure of the YajC-AcrB complex. Certain marine bacteria, including Va, utilize two sets of SecDF proteins for efficient protein translocation; the first is sodium ion-driven and the other is proton-driven (Ishii et al. 2015). Sodium ions, which are abundant in the ocean, are primarily used for SecDF function as an alternative to protons. In Va, the expression level of proton-driven SecDF is elevated when the efficiency of protein translocation decreases. This mechanism is regulated by the biogenesis of the Vibrioprotein export monitoring polypeptide (VemP) (Ishii et al. 2015; Su et al. 2017; Mori et al. 2018). Although it was previously not known how the proton-motive force was related to Sec protein translocation (Arkowitz and Wickner 1994), it is now clear that SecDF uses the proton gradient for its function (Tsukazaki et al. 2011).
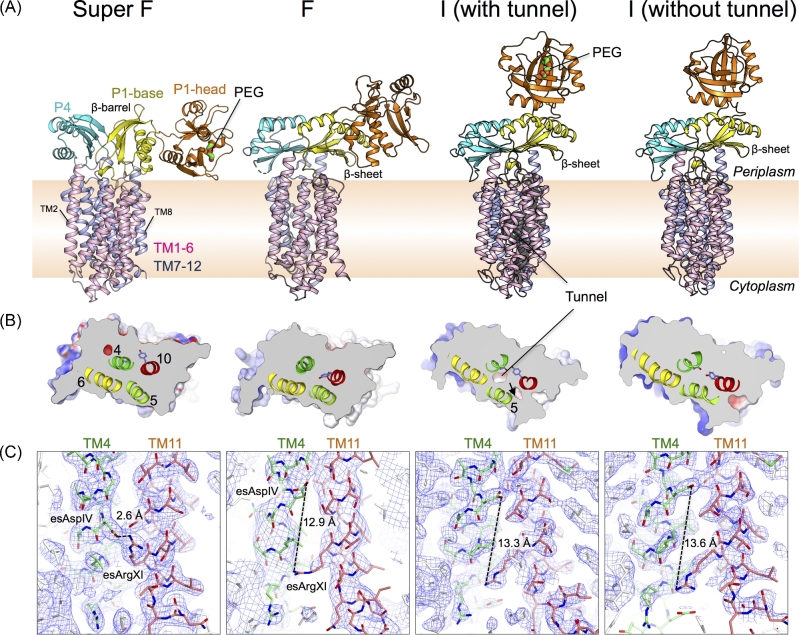
Structural determination of SecDF in different forms has advanced our understanding of the molecular mechanism of SecDF (Tsukazaki et al. 2011; Furukawa et al. 2017, 2018). The TM region of SecDF is composed of 12 helices, as predicted from its amino acid sequence. The periplasmic region consists of three domains, P1-head, P1-base and P4 (Fig. 2). The first report of the crystal structure of SecDF and its functional analyses revealed that the TM region conducts protons, whereas the flexible periplasmic region interacts with an unfolded protein mimicking a precursor protein (Tsukazaki et al. 2011). This study proposed a model in which structural transitions in the periplasmic region are crucial for the protein-translocation activity of SecDF. Because the proton-transporting region of SecDF, which is in the membrane, is distant from the substrate interaction area in the periplasmic domain of SecDF, there must be a coupling mechanism to transmit structural changes from the TM to the periplasmic region (Yamaguchi, Nakashima and Sakurai 2015). Currently, the available crystal structures of SecDF represent the super membrane facing (Super F), membrane facing (F) and intermediate (I) forms (Figs 2A and 3A). The major differences in architecture among these forms are in the orientation of the P1-head domain. In the I form structures, the P1-head is located on the P1 base domain. In contrast, in the Super F and F form structures, the P1-head is close to the membrane surface. Moreover, the P1-base and P4 in the Super F form a β-barrel architecture instead of the β-sheet observed in the I and F forms (described in detail later). Comparison of the P1 domains in the three forms shows dramatic conformational changes. SecDF is likely to undergo these structural transitions during its function.
Figure 2.
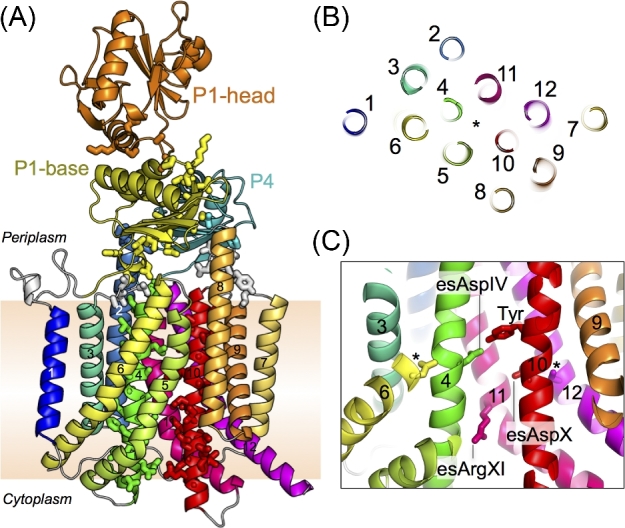
(A) The crystal structure of SecDF in the I form (PDB ID: 5XAP). SecDF consists of 12 TMs (TM1, blue; TM2, pale blue; TM3, turquoise; TM4, green; TM5, greenish yellow; TM6, pale greenish yellow; TM7, yellow; TM8, pale orange; TM9, orange; TM10, red; TM11, deep pink; and TM12, magenta), P1-base (yellow), P1-head (orange), and P4 (cyan) domains. Side chains of the highly conserved regions D1, D3, D5, F1 and F2 are depicted as stick representations. (B) TM region of SecDF cross-sectioned at the middle of the membrane and viewed from the periplasm. The asterisk shows the pseudosymmetrical center. The TM numbers are shown. (C) The important area in the center of the TM region. The essential, conserved residues, esAspIV, esAspX and esArgXI, are shown as stick representations. The important Tyr is located near the esAspIV. The corresponding positions of the inactive mutants of EcSecDF (Nouwen and Driessen 2005) in TM6 and TM12 are also shown as stick representations and indicated by asterisks.
Figure 3.
Crystal structures of SecDF. (A) The overall structures of the Super F, F, I (with tunnel) and I (without tunnel) forms (PDB ID: 3AQP, 5YHF, 5XAN-Mol B, and 5XAN-Mol A, respectively). PEG molecules in the periplasmic cavity are shown by stick representations. (B) Cut-away models of the surface representations of the middle TM regions viewed from the periplasm. Displacement of TM5 creates a tunnel. The important Tyr and esAspIV are shown by stick representations. (C) Electron density maps of SecDF. The 2Fo-Fc electron density maps in the Super F form (contoured at 1.0 σ), F form (contoured at 1.5 σ), and I form (contoured at 1.5 σ). The esAspIV and esArgXI residues are labeled. Distance between the OD2 of esAspIV and NH2 of esArgXI are shown.
TM REGION OF SecDF
Analysis of the crystal structures of SecDF has revealed that the overall arrangement of the TM helices is similar to that of the monomer of other RND superfamily proteins. TM helices TM 1–6 and TM7–12 are assembled in a pseudosymmetrical manner (Fig. 2A, B) and correspond to SecD and SecF regions of some bacteria, such as Ec and Va. The periplasmic regions, consisting of P1 (P1-base and P1-head) and P4 domains, exist between TM1 and TM2, and between TM7 and TM8. The P1 region is much larger than the P4 region. SecDF has conserved regions called D1–D6 and F1–F4 (Figs 1B and 2A) (Eichler 2003). TM4 and TM10, corresponding to the highly conserved regions D5 and F2, are positioned at the center of the TM bundle, and interact with each other. This interaction appears to stabilize the TM region and plays an important role in SecDF function. Moreover, TM4 and TM10 are surrounded by other TM helices (Fig. 2B), in which TM2, TM5, TM6, TM8, TM11 and TM12 are longer than the thickness of the membrane. TM helices TM5, TM6, TM11 and TM12 are tilted and curved at the middle of the membrane region. On the periplasmic side, both TM2 and TM8 have helices that extend approximately 10 Å from the membrane plane. The conserved, essential residues D340, D637 and R671 of Tt in TM4, TM10 and TM11 are termed esAspIV, esAspX and esArgXI, respectively, in this review (Figs 1B, C and 2C). Mutations at these three residues cannot complement the growth deficiency that results from the reduction of protein-translocation activity in SecDF-depleted conditions (Tsukazaki et al. 2011; Furukawa et al. 2017). Moreover, mutations in esAspIV and esArgXI show a loss of proton conducting activity. The functional importance of esAspIV and esArgXI was proposed in 2017 and 2018 (Furukawa et al. 2017, 2018), which will be discussed later. Notably, although esAspX plays an important role in protein translocation, the mechanism has not yet been elucidated (Tsukazaki et al. 2011). In addition, strains harboring mutations A593 and S288 in Ec SecD and SecF, respectively, showed cold-sensitive phenotype and abolished protein-translocation activity of SecDF in vitro; however, the functional importance of this loss in activity is still unknown (Nouwen and Driessen 2005).
As shown in Fig. 3A and B, the TM regions of three of the four displayed crystal structures are completely sealed, preventing the transport of small molecules and ions across the membrane. In contrast, one of the I form structures shows a tunnel architecture formed by TM4, TM5, TM6 and TM10, which penetrates the cytosol and the periplasm. Compared to other crystal structures, TM5 is 5 Å-dislocated to the outside, although the other TMs are placed in similar positions, which generates a tunnel structure. In the tunnel undefined ambiguous electron densities, presumably due to water molecules or small molecules, are present. Interestingly, esAspIV is positioned at the center of the tunnel. The pKa of esAspIV is approximately 7, based on the crystal structure, implying that there is a transition between protonation and deprotonation of esAspIV. Molecular dynamics (MD) simulations of SecDF using the highest resolution structure of the I form without the tunnel architecture (PDB ID: 5XAP) in a state of deprotonated esAspIV temporarily showed water molecules in a row across the TM region of SecDF (Fig. 4). The water molecule queue was essentially consistent with the tunnel position of the I form with the tunnel (Fig. 3A). In contrast, the MD simulation in a state of protonated esAspIV did not show invasion of water molecules into the membrane region. It is conceivable that the proton flow occurs through a network of hydrogen bonds; therefore, this tunnel may conduct protons. A series of observations showed that esAspIV significantly contributes to the formation of the tunnel architecture (Furukawa et al. 2017).
Figure 4.
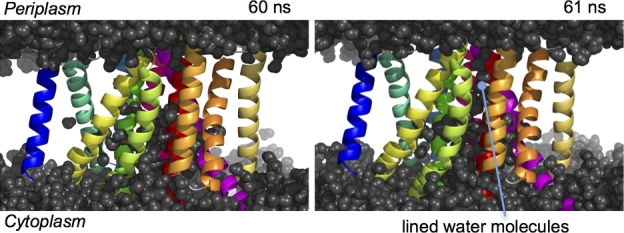
Snapshots of the MD simulation at 60 and 61 ns with dehydration of esArpIV. The water molecules queue from the cytoplasm to the periplasm through the TM region at 61 ns.
As described above, sodium ion-driven SecDF exists in certain marine species. Even in such cases, the size of the tunnel architecture could adequately accommodate dehydrated sodium ions as well as water molecules. When such a penetrating tunnel is formed, a large amount of protons can flow rapidly inside the cell, which is consistent with the high proton transport activity demonstrated by patch clamp and proton influx experiments (Furukawa et al. 2017, 2018). The importance of conserved Tyr residues in the vicinity of esAspIV (Figs. 2C and 3B) was also shown (Furukawa et al. 2017). Substitutions with Ala, Asn and Gln abolish the protein-translocation activity of SecDF, whereas Phe substitution does not. The side chain of Tyr is oriented toward the tunnel and may be involved in tunnel formation and regulation of proton transport. Despite these extensive studies, the functional importance of the large proton influx associated with SecDF (Furukawa et al. 2017) remains unknown as it appears to produce excessive energy.
IMPORTANCE OF THE FLEXIBLE PERIPLASMIC REGION
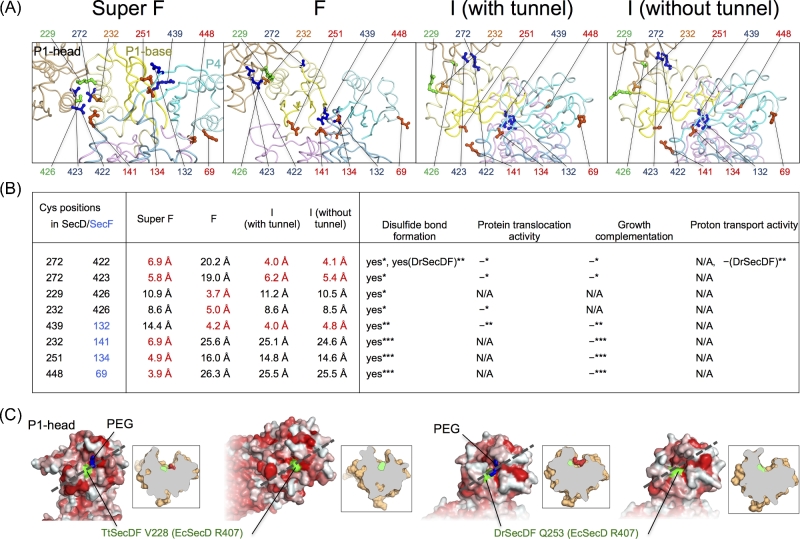
As shown in Figs. 3A and 5A, each P1-head domain of the Super F, F and I forms is differently positioned, indicating that the P1 region is inherently flexible. To investigate the importance of the flexibility of this region for SecDF function, several double cysteine mutants were constructed based on the structural information to immobilize the periplasmic domain in place (Tsukazaki et al. 2011; Furukawa et al. 2017, 2018) (Fig. 5A and B). The distance between each Cys-substituted position is close in at least one form and distant in at least one other form. All the double cysteine mutants formed disulfide bonds in vivo, which were easily recognized, suggesting that the cysteine residues could temporarily interact with each other in several formations and that the P1 region may continually fluctuate in vivo. In fact, the P1 region can take on numerous different forms as demonstrated by the crystal structures. The formation of the disulfide bonds restricts the movement of the P1 region, resulting in loss of growth complementation of SecDF-depleted Ec cells. As previously reported (Tsukazaki et al. 2011; Furukawa et al. 2017), the formation of disulfide bonds decreases protein-translocation and proton-transport activities. Moreover, the effects of disulfide bond formation on proton transport activity demonstrate the correlation between structural changes in the periplasmic region and proton transport in the membrane region.
Figure 5.
Flexibility of the periplasmic region. (A) Close up views of the periplasmic domain of SecDF (color scheme same as in Fig. 3A). The substituted corresponding positions of double cysteine mutants of EcSecDF are shown by the ball and stick model. (B) Summary of previous functional analyses of the EcSecDF mutants. The distance between the Cβ atoms of the substituted positions of SecD (black) and SecF (blue), protein translocation activity, growth complementation, and proton transport activity of the double cysteine mutants in disulfide bond form are summarized *(Tsukazaki et al. 2011); **(Furukawa et al. 2017); ***(Furukawa et al. 2018). –: inactive, N/A: not available. C, P1-head cavities. Surface representation (left) colored according to hydrophobicity, from white (hydrophilic) to red (hydrophobic). The corresponding residue of EcSecD R407 interacting with precursor is colored green and indicated in Fig. 1. PEG molecules in the cavity are shown by stick representations. Cut-away models (right) of the surface representations along the dotted line.
All the P1-head domains in the crystal structures contain an amphiphilic cavity (Fig. 5C). Although the amino acid residues in the cavity are not well conserved, the overall shapes of the P1-head cavity are similar among the crystal structures. In the SecDF structures in Super F and I forms (with channel), electron densities in the cavities can be visualized and are thought to represent molecules of polyethylene glycol (PEG), which is used as a precipitant for crystallization. The interaction between the cavity of the P1-head and precursor proteins was proposed and confirmed by site-specific photocrosslinking using p-benzoyl-L-phenylalanine mutant of SecDF. (colored green in Fig. 5C) (Furukawa et al. 2017). Therefore, the cavity of the P1-head would be a contact site of precursor proteins. Comparison of the P1-head domain structures shows that both the orientation and size of the cavity are variable (Figs. 3A and 5C). This flexible feature of the P1-head may be important for capturing and releasing various regions of precursor proteins by promiscuous recognition. This flexibility presumably allows appropriate interactions with various precursor proteins through such structural changes. The crosslinking experiment showed that only one residue interacted with the precursor proteins; however, it is possible that other sites in the periplasmic region also interact with substrate proteins.
REMOTE-COUPLED STRUCTURAL CHANGES
A recent report demonstrated that structural transitions in the TM domain induced dramatic structural changes in the periplasmic region (Furukawa et al. 2018). Notable structures include β-strands in the P1-base and P4 domains (Fig. 3A). The eight β-strands in the F and I forms reported in 2011 and 2017 form a β-sheet architecture, whereas in the Super F form reported in 2018; the eight β-strands make up a β-barrel structure. The side chain orientations of esArgXI in the Super F form is different from that in the other structures; esArgXI interacts with esAspIV via a hydrogen bond (Fig. 3C), resulting in a structural transition, which induces proximity between TM4 and TM10. Structural changes in the TM region are transmitted to the periplasmic side via the D1, D3, D5, F1 and F2 conserved regions (Fig. 2C) and can drive the dramatic transition from the β-sheet to the β-barrel structure. At the same time, structural changes from α-helical to unfolded occur in protruding parts of the helices at the periplasmic sides of TM2 and TM8 (Fig. 3A). Intramolecular disulfide bond formation, as described above, supports the existence of the Super F form in vivo. Furthermore, disulfide bonds were not formed in esAspIV and esARgXI mutants, suggesting that esAspIV and esARgXI are critical for the formation of the Super F form. These findings provide insight into the coupling of structural transitions on the periplasmic side with those in the membrane region. However, these results still cannot explain the mechanism underlying the structural transitions between the F form and I form (without tunnel) because the TM regions of these forms are similar. Presumably, some other unknown factors contribute to the swinging motion of the P1-head domain.
WORKING MODEL OF SecDF
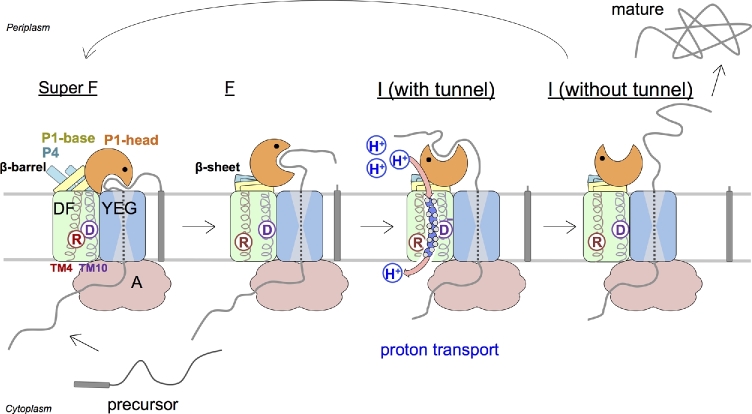
Based on the four available structures introduced here (Fig. 3A), I propose the following power stroke-based model (Fig. 6), although the ratchet mechanism of SecDF-dependent protein translocation, which includes capturing of the precursor protein at the periplasmic side for enhancing net forward movement, cannot be excluded. Because the cavity of the P1-head domain may interact with precursor proteins, the cavity may capture proteins emerging from the SecYEG channel. For this process to be efficient, the SecYEG complex must be located in the vicinity of SecDF, where the P1-head domain may incline toward SecYEG. If SecYEG and SecDF are positioned as shown in Fig. 6, the interacting cavity of the Super F form at the periplasmic side would continue seamlessly from the exit of the SecYEG translocon. In this case, the precursor proteins emerging from the SecYEG channel could interact with the P1 cavity without delay. Because SecDF is proposed to be functionally related to SecA and SecG (Economou et al. 1995; Duong and Wickner 1997), SecDF may directly interact with them in the Sec complex and regulate their activity. The Super F form, which exhibits the greatest incline in the P1-head toward the membrane, may be a resting state. Although the order of the structural changes in the periplasmic region after the first interaction with a precursor protein may not be correct, I arrayed them for the purpose of this review to account for the inclination in each P1-head domain; the underlying concept is that transitions may occur from the Super F form to the I form via the F form. The first interaction between a substrate and SecDF in the Super F form may trigger conformational changes. In this model, SecDF in the Super F form initially interacts with a precursor protein, followed by structural changes to the I form. The P1-head stands up holding the precursor protein. In this way binding of the precursor may allow protons to move through the membrane. The I form could transport protons via the tunnel, which may induce the release of the precursor protein. After that, the TM domain would close the proton channel. Subsequently, the conformation of SecDF reverts to the Super F form, and the protein interacts again with another region of the precursor protein. SecDF repeatedly undergoes these structural changes to complete protein translocation. The Super F form may represent the most stable configuration of SecDF. Thus, based on our current knowledge, SecDF may be postulated to act as a proton-driven protein-translocation motor by undergoing the presented conformational transitions.
Figure 6.
Working model of SecDF based on structural and functional analyses. The TM, P1-head, P1-base and P4 regions of SecDF are colored pale green, yellow, cyan, and orange, respectively. SecYEG and SecA ATPase are shown in pale blue and pink, respectively. Precursor protein (gray) is captured and translocated along with repeating SecDF transitions. The esAspIV and esArgXI are shown as circled D and R, respectively. The I form forms the tunnel architecture for proton transport.
PERSPECTIVES
Various crystal structures of SecDF have been reported (Tsukazaki et al. 2011; Furukawa et al. 2017, 2018), suggesting that SecDF undergoes more dynamic structural changes during protein translocation than expected. Although some of the mechanisms through which SecDF is remotely coupled between the periplasmic and TM regions have been elucidated based on a comparison of the Super F form (with the β-barrel structure) and the F form (with the β-sheet structure) at the periplasmic region, it has not been possible to elucidate the structural changes in the P1 domain underlying the Super F, F and I forms. Further structural and functional analyses are required to completely elucidate the molecular mechanisms of SecDF. Importantly, interactions with an unfolded protein, which mimics a precursor protein, enhance the proton-transport activity of SecDF (Tsukazaki et al. 2011). To elucidate these mechanisms, interaction analyses between substrate proteins and SecDF using nuclear magnetic resonance may provide meaningful results. I propose that structural changes may occur in the order shown in Fig. 6; however, because the crystal structures show snapshots of SecDF, the mechanism underlying the repeated conformational changes of the P1-head remains unclear. For example, does the P1-head show repetitive power stroke motion, such as that observed for SecA ATPase or that observed for the proton-driven FoF1ATP synthase? To resolve the dynamics of P1 motion, methods such as single-unit observations using high-speed atomic force microscopy or real-time single-molecule fluorescence may be suitable.
Although SecDF, which interacts with YidC and SecYEG, is proposed to be a component of the Sec holo-translocon (Botte et al. 2016), the structural details of the holo-complex are still unclear. Using the structural information of SecYEG, SecDF and YidC reported at maximum resolutions of 2.7, 2.6 and 2.4 Å, respectively (Kumazaki et al. 2014b; Tanaka et al. 2015; Furukawa et al. 2017), we can perform site-specific disulfide bond crosslinking and photocrosslinking experiments to uncover the interactions among these proteins. Furthermore, using a system with a strong ultraviolet source, it is possible to track the time-dependent interactions and structural changes in Sec and precursor proteins (Miyazaki et al. 2018). Further developments in Sec protein research are expected in the near future.
Eukaryotic SecDF homologs present in the endoplasmic reticulum have not yet been identified, although a YidC homolog has been found in the endoplasmic reticulum (Anghel et al. 2017). The Bip chaperone, an essential component of the endoplasmic reticulum, interacts with precursor proteins and is involved in their translocation (Dudek et al. 2015). Therefore, Bip may be functionally similar to SecDF. Electron microscopic analysis of the complex of the Sec translocon and ribosome from the endoplasmic reticulum showed a relatively large soluble domain derived from the translocon-associated protein (TRAP) complex (Pfeffer et al. 2017; Braunger et al. 2018). This domain is located at the endoplasmic reticulum side of the Sec translocon, similar to the location of the periplasmic domain of SecDF. Although the molecular mechanisms of action of the TRAP complex and SecDF may differ significantly, the soluble domain of the TRAP complex, similar to SecDF, appears to interact with substrate proteins emerging from the Sec translocon and enhance protein translocation. Since SecDF is proposed to be one of the smallest proteins that is a proton-driven motor, the molecular mechanism of SecDF involves fundamental and essential principles that underlie many proton-driven biological processes. Therefore, understanding the fundamental characteristics of SecDF will provide important insights. Moreover, as the number of multidrug-resistant bacteria is increasing, new types of antibiotics targeting SecDF may be developed after understanding the molecular mechanisms of SecDF function (Yan and Wu 2016).
Acknowledgements
We thank C. Suzuki for administrative assistance and S. Nakayama and A. Furukawa for support with graphics.
FUNDING
This work was financially supported by the JSPS/MEXT KAKENHI (grant nos. JP26119007, JP26291023, and JP16K14713), Mitsubishi Foundation, Noguchi Institute, Naito Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and PRESTO, JST (grant no. JPMJPR12L3).
Conflict of interest. None declared.
REFERENCES
- Allen WJ, Corey RA, Oatley P et al. Two-way communication between SecY and SecA suggests a Brownian ratchet mechanism for protein translocation. Elife 2016;5:e15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anghel SA, McGilvray PT, Hegde RS et al. Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep 2017;21:3708–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz RA, Wickner W. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. Embo J 1994;13:954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff L, Wickles S, Berninghausen O et al. Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nat Commun 2014;5:4103. [DOI] [PubMed] [Google Scholar]
- Botte M, Zaccai NR, Nijeholt JL et al. A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci Rep 2016;6:38399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger K, Pfeffer S, Shrimal S et al. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science. 2018;360:215–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi KE, Sardis MF, Economou A et al. SecA-mediated targeting and translocation of secretory proteins. Biochim Biophys Acta 2014;1843:1466–74. [DOI] [PubMed] [Google Scholar]
- Chen M, Xie K, Yuan J et al. Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry 2005;44:10741–9. [DOI] [PubMed] [Google Scholar]
- Dudek J, Pfeffer S, Lee PH et al. Protein transport into the human endoplasmic reticulum. J Mol Biol 2015;427:1159–75. [DOI] [PubMed] [Google Scholar]
- Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. Embo J 1997;16:4871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A, Pogliano JA, Beckwith J et al. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 1995;83:1171–81. [DOI] [PubMed] [Google Scholar]
- Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci 2010;107:17182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. Evolution of the prokaryotic protein translocation complex: a comparison of archaeal and bacterial versions of SecDF. Mol Phylogenet Evol 2003;27:504–9. [DOI] [PubMed] [Google Scholar]
- Furukawa A, Nakayama S, Yoshikaie K et al. Remote coupled drastic beta-barrel to beta-sheet transition of the protein translocation motor. Structure 2018;26:485–489.e2. [DOI] [PubMed] [Google Scholar]
- Furukawa A, Yoshikaie K, Mori T et al. Tunnel formation inferred from the I -Form structures of the proton-driven protein secretion motor SecDF. Cell Rep 2017;19:895–901. [DOI] [PubMed] [Google Scholar]
- Gardel C, Johnson K, Jacq A et al. The secD locus of E. coli codes for two membrane proteins required for protein export. Embo J 1990;9:4205–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel C, Benson S, Hunt J et al. SecD, a new gene involved in protein export in Escherichia coli. J Bacteriol 1987;169:1286–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogala M, Becker T, Beatrix B et al. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 2014;506:107–10. [DOI] [PubMed] [Google Scholar]
- Hand NJ, Klein R, Laskewitz A et al. Archaeal and bacterial SecD and SecF homologs exhibit striking structural and functional conservation. J Bacteriol 2006;188:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L et al. YidC/Alb3/Oxa1 family of insertases. J Biol Chem 2015;290:14866–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YH, Huang YJ, Zhang H et al. Dissecting structures and functions of SecA-only protein-conducting channels: ATPase, pore structure, ion channel activity, protein translocation, and interaction with SecYEG/SecDF• YajC. PLoS One 2017;12:e0178307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii E, Chiba S, Hashimoto N et al. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci USA 2015;112:E5513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Wittekind M, Nomura M et al. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell 1983;32:789–97. [DOI] [PubMed] [Google Scholar]
- Jomaa A, Boehringer D, Leibundgut M et al. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat Commun 2016;7:10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki K, Kishimoto T, Furukawa A et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep 2014a;4:7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazaki K, Chiba S, Takemoto M et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 2014b;509:516–20. [DOI] [PubMed] [Google Scholar]
- Li L, Park E, Ling J et al. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 2016;531:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. Embo J 1993;12:265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R, Myougo N, Mori H et al. A photo-cross-linking approach to monitor folding and assembly of newly synthesized proteins in a living cell. J Biol Chem 2018;293:677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Sakashita S, Ito J et al. Identification and characterization of a translation arrest motif in VemP by systematic mutational analysis. J Biol Chem. 2018;293:2915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N, Driessen AJ. SecDFyajC forms a heterotetrameric complex with YidC. Mol Microbiol 2002;44:1397–405. [DOI] [PubMed] [Google Scholar]
- Nouwen N, Driessen AJ. Inactivation of protein translocation by cold-sensitive mutations in the yajC-secDF operon. J Bacteriol 2005;187:6852–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JE, Ekende EN, Kifle EG et al. Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter. Proc Natl Acad Sci 2013;110:18484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Menetret JF, Gumbart JC et al. Structure of the SecY channel during initiation of protein translocation. Nature 2014;506:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Burbaum L, Unverdorben P et al. Structure of the native Sec61 protein-conducting channel. Nat Commun 2015;6:8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Schaffer M et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat Commen 2017;8:14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. Embo J 1994a;13:554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano KJ, Beckwith J. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol 1994b;176:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Li L, Park E. Structural and mechanistic insights into protein translocation. Annu Rev Cell Dev Biol. 2017;33:369–90. [DOI] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F et al. YidC mediates membrane protein insertion in bacteria. Nature 2000;406:637–41. [DOI] [PubMed] [Google Scholar]
- Su T, Cheng J, Sohmen D et al. The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling. Elife 2017;6: e25642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sugano Y, Takemoto M et al. Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep 2015;13:1561–8. [DOI] [PubMed] [Google Scholar]
- Tornroth-Horsefield S, Gourdon P, Horsefield R et al. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 2007;15:1663–73. [DOI] [PubMed] [Google Scholar]
- Tseng TT, Gratwick KS, Kollman J et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol 1999;1:107–25. [PubMed] [Google Scholar]
- Tsirigotaki A, De Geyter J, Sostaric N et al. Protein export through the bacterial Sec pathway. Nat Rev Micro 2017;15:21–36. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Fukai S et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 2008;455:988–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Echizen Y et al. Structure and function of a membrane component SecDF that enhances protein export. Nature 2011;474:235–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM Jr, Collinson I et al. X-ray structure of a protein-conducting channel. Nature 2004;427:36–44. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH et al. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 2014;157:1632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y, Zhao Y, Zheng J et al. Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion. FASEB J. 2018;32:2411–21. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol 2015;6:2411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wu G. Evolutionary evidence on suitability of SecD as a target for development of antibacterial agents against Staphylococcus aureus. Ecol Evol 2016;6:1393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 2008;455:936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]