Abstract
The protein ataxin-3 contains a polyglutamine region; increasing the number of glutamines beyond 55 in this region gives rise to the neurodegenerative disease spinocerebellar ataxia type 3. This disease and other polyglutamine expansion diseases are characterized by large intranuclear protein aggregates (nuclear inclusions). By using full-length human ataxin-3, we have investigated the changes in secondary structure, aggregation behavior, and fibril formation associated with an increase from the normal length of 27 glutamines (Q27 ataxin-3) to a pathogenic length of 78 glutamines (Q78 ataxin-3). Q78 ataxin-3 aggregates strongly and could be purified only when expressed with a solubility-enhancing fusion-protein partner. A marked decrease in α-helical secondary structure accompanies expansion of the polyglutamine tract, suggesting destabilization of the native protein. Proteolytic removal of the fusion partner in the Q78 protein, but not in the Q27 protein, leads to the formation of SDS-resistant aggregates and Congo-red reactive fibrils. Infrared spectroscopy of fibrils reveals a high β-sheet content and suggests a parallel, rather than an antiparallel, sheet conformation. We present a model for a polar zipper composed of parallel polyglutamine β-sheets. Our data show that intact ataxin-3 is fully competent to form aggregates, and posttranslational cleavage or other processing is not necessary to generate a misfolding event. The data also suggest that the protein aggregation phenotype associated with glutamine expansion may derive from two effects: destabilization of the native protein structure and an inherent propensity for β-fibril formation on the part of glutamine homopolymers.
Spinocerebellar ataxia type-3 (SCA3) is an autosomal-dominant neurodegenerative disorder caused by the expansion of a CAG repeat in the coding region of the MJD1 gene. Like Huntington's disease, SCA3 belongs to a superfamily of trinucleotide-repeat disorders in which expanded CAG repeats direct the formation of large polyglutamine tracts in proteins. This condition leads to a toxic gain of function associated with intranuclear protein aggregation and altered protein interactions, which becomes evident once the number of glutamines surpasses a pathogenic threshold (1). Ataxin-3, the product of the MJD1 gene, is a 42-kDa protein of unknown function that is widely expressed in the brain. Normally, ataxin-3 contains a tract of 12–40 glutamines near its C terminus, but in the pathogenic form, this domain increases to 55–84 glutamines (2).
Wild-type ataxin-3 is present in both the nucleus and cytoplasm (3). Pathogenic forms of ataxin-3 containing expanded glutamine tracts aggregate into dense nuclear inclusions, much like those seen in other polyglutamine diseases (4). Expression of expanded polyglutamine forms of ataxin-3 in cultured cells or transgenic animals gives rise to nuclear inclusions and cell death (5, 6). Nuclear inclusions containing expanded polyglutamine ataxin-3 are ubiquitinated and associated with the proteasome. In certain cases, inhibition of the ubiquitin/proteasome pathway increases polyglutamine-mediated cell death (7), whereas in other cases, polyglutamine-containing nuclear inclusions themselves seem to act as inhibitors of the ubiquitin/proteasome system (8). In Drosophila, coexpression of chaperonins Hsp40 or Hsp70 with expanded repeat forms of ataxin-3 or huntingtin reduces toxicity and increases cell survival (9, 10), suggesting a role for protein misfolding in the pathogenesis of polyglutamine expansion diseases.
Many diseases are characterized by aberrant protein folding, which typically leads to the formation of stable β -structures and the deposition of amyloid material. The presence of β-structure has been demonstrated in Aβ-amyloid fibrils derived from both Alzheimer's patients and synthetic peptides (11, 12). Many other proteins can be induced to form β-fibrils, either through mutations or exposure to harsh environments, and it has been argued that the ability to form β-fibrils might be an inherent property of polypeptides (13–15). However, homopolymeric glutamine repeats may be uniquely predisposed toward the formation of stable, extended β-structures. Synthetic poly-l-glutamine has been shown to aggregate into “polar zippers” composed of β pleated sheets stabilized by hydrogen bonds between the glutamine side chains (16). As β-fibrils are intimately associated with the pathology of Alzheimer's disease, related polyglutamine fibrils may play similar roles in spinocerebellar ataxia type-3 and other neurodegenerative diseases (1, 17).
Truncated N-terminal fragments of huntingtin that contain the polyglutamine tract show a strong propensity to aggregate into insoluble β-structures, with the aggregation being sharply dependent on the length of the glutamine region (18). However, the protein context in which a polyglutamine region occurs might exert a strong effect on its behavior. Unfortunately, no data have been available on the aggregation behavior of full-length, intact polyglutamine proteins. In this study, we show that full-length human ataxin-3 containing a 78-residue glutamine repeat forms fibrils in vitro under near-physiological conditions. Fibril formation is accompanied by a dramatic increase in the amount of β-sheet. Under the same conditions, a nonpathogenic form of ataxin-3 containing 27 glutamines neither fibrillizes nor acquires increased β-structure.
Materials and Methods
Protein Expression and Purification.
Human ataxin-3 cDNAs containing 27-residue and 78-residue glutamine repeats (obtained from H. Paulson and R. Pittman, University of Pennsylvania, Philadelphia) were subcloned into pProEX-HTb (Life Technologies, Rockville, MD) to produce N-terminal hexahistidine-tagged proteins, parallel maltose-binding protein (MBP) vectors, and MBP/ataxin-3 fusion proteins (19). Proteins expressed in Escherichia coli were purified by nickel chelate or amylose-affinity chromatography followed by anion exchange. Final purity was at least 90–95%, as assessed by SDS/PAGE.
Solution Characterization.
Gel-permeation chromatography was performed with a TSK-SW3000 column (TosoHaas, Montgomeryville, PA) on a BioSys 2000 HPLC (Beckman Coulter). The column was equilibrated with PBS (pH 7.4) plus 5 mM mercaptoethanol; samples were eluted at 1.0 ml/min at room temperature. Calibration was performed with proteins of known Stokes radius (20). Analytical ultracentrifugation experiments were performed at 10°C with an XL-I analytical ultracentrifuge (Beckman Coulter). Sedimentation equilibrium experiments were performed at 20,000 RPM, and velocity sedimentation experiments were performed at 40,000 RPM. Data were analyzed with sedinterp (21).
Electrophoresis and Western Blot Analysis.
Polyclonal anti-ataxin-3 was a gift from H. Paulson and R. Pittman. Polyclonal anti-MBP was purchased from New England Biolabs. Protein (100 ng per well) in SDS sample buffer was electrophoresed in SDS/12% PAGE gels and transferred to poly(vinylidene difluoride) membranes. Membranes were blocked for 2 hr in 5% dry milk and 0.2% Tween-20 in Tris-buffered saline, incubated for 2 hr with primary antibody at a dilution of 1:15,000, washed five times in blocking solution, and incubated for 1 hr with peroxidase-conjugated goat anti-rabbit antisera (1:5,000). Bands were visualized by using enhanced chemiluminescence (Amersham Pharmacia). For Coomassie staining (see Fig. 2D), 3 μg of protein were loaded per lane.
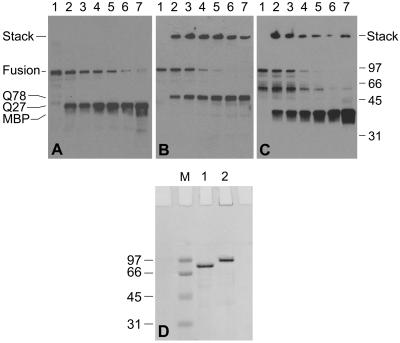
Figure 2.
(A–C) Western blots showing the time course of TEV protease digestion of MBP/ataxin-3 fusion proteins. (A) Digestion of MBP/Q27 ataxin-3. (B and C) Digestion of MBP/Q78 ataxin-3. A and B are probed with polyclonal anti-ataxin-3; C is probed with anti-MBP. Lane 1: time = 0; lane 2: 15 min; lane 3: 30 min; lane 4: 1 hr; lane 5: 2 hr; lane 6: 3 hr; lane 7: 4 hr. The high-molecular-weight species seen in B and C have failed to enter the resolving gel and remain in the stacking gel. (D) Coomassie-stained SDS/PAGE gel of MBP/ataxin-3 fusion proteins. Lane 1: MBP/Q27 ataxin-3; lane 2: MBP/Q78 ataxin-3; lane M: molecular-weight markers.
Infrared Spectroscopy.
For the transmission studies, protein solutions (3–5 mg/ml in 20 mM Hepes plus 2 mM mercaptoethanol in D2O, pD 8.0) were placed between CaF2 windows with a 0.012 mm Teflon spacer at room temperature. For the internal reflection studies, samples were evaporated onto a 50 × 10 × 2 mm germanium crystal configured as a 45° internal reflection element. For both types of studies, single-beam spectra (1,024 coadded interferograms) were collected at 2 cm−1 resolution with triangular apodization and one level of zero filling on an FTS 6000 Fourier transform infrared spectrometer (Bio-Rad) that was purged with dry air and equipped with a liquid-nitrogen-cooled mercury–cadium–tellurium (MCT) detector. No deconvolution, baseline correction, smoothing, or water vapor subtraction were performed on the internal reflection spectra, but water vapor subtraction was performed on the transmission spectra.
Fibrillogenesis.
Fibril samples or controls were obtained by incubating MBP fusions of Q27 or Q78 ataxin-3 (2 mg/ml) with 25 units of tobacco etch virus (TEV) protease per mg of fusion at 37°C with shaking at 225 rpm. Fibril formation typically required at least 5 days. Fibril formation was assayed with the Congo red (CR) spectroscopic assay (22). Fibril formation sample (100 μg) was added to 5 μM CR in 10 mM sodium phosphate (pH 7.27), mixed thoroughly, and allowed to incubate for 30 min at room temperature. Absorbance spectra then were measured with a Cary Model 400 spectrophotometer.
Circular Dichroism (CD).
Proteins were dialyzed into 20 mM sodium phosphate plus 2 mM mercaptoethanol and spectra were collected with a 0.1-cm quartz cuvette (Hellma, Forest Hills, NY) in a 62DS CD spectrometer (Aviv Associates, Lakewood, NJ). Protein concentrations were determined by a modified Edelhoch analysis (23); concentrations used were 7.5 μM for the hexahistidine-labeled Q27 ataxin-3 and 4 μM for the MBP fusions with Q27 and Q78 ataxin-3. Spectra were obtained by taking measurements every 1 nm with an averaging time of 5 s at 20°C; five spectra were averaged and corrected for buffer absorbance.
Electron Microscopy.
Proteins were dialyzed into 5 mM Tris/2 mM mercaptoethanol (pH 8.0) and fibrillized where appropriate. Samples were diluted 5-fold with H2O and applied to colloidal/carbon-coated grids that were glow-discharged just before use. After negative staining with 2% aqueous uranyl acetate, samples were imaged with a Philips CM-100 transmission electron microscope at 80 kV.
Results
Protein Expression.
It was possible to produce milligram quantities of both wild-type and expanded repeat forms of full-length ataxin-3 as recombinant proteins in E. coli. The wild type (Q27 ataxin-3), containing 27 glutamines in the polyglutamine tract, was well behaved and could be purified as either a hexahistidine or MBP fusion; proteolytic removal of the fusion partner did not alter the solubility of the protein. In contrast, all attempts to express the expanded repeat version (Q78 ataxin-3; 78 glutamines in the polyglutamine tract) as a hexahistidine fusion yielded aggregated, insoluble protein. However, the use of MBP as an N-terminal fusion partner greatly enhanced the solubility of Q78 ataxin-3 (24). Whereas the Q78 ataxin-3 fusion with MBP was considerably more labile than any of the forms of Q27 ataxin-3 studied, it was still possible to obtain >90% pure fusion protein, albeit in low yield. Proteolytic removal of the MBP fusion partner from Q78 ataxin-3 resulted in protein aggregation, which necessitated performing many experiments with the fusion proteins.
Biophysical Characterization.
Size-exclusion chromatography of Q27 ataxin-3 showed a single species with a 4.2-nm Stokes radius. Equilibrium analytical ultracentrifugation revealed the protein to be monomeric at concentrations up to 30 μM. Velocity sedimentation yielded a sedimentation coefficient of 1.77 S; Teller's method (implemented in the program sedinterp; ref. 21) predicts an elongated shape for the molecule, which is consistent with a prolate ellipsoid of Stokes radius 4.3 nm and a long/short axis ratio of 11.3.
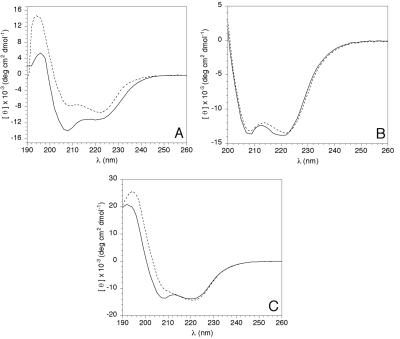
The CD spectrum of Q27 ataxin-3 indicates a high α-helical content (Fig. 1A); spectral analysis by the method of Scholtz (25) yields an estimate of 35% α-helix. Because it was not possible to purify Q78 ataxin-3 in stable form, secondary structure comparisons were performed with the MBP fusion forms of Q27 and Q78 ataxin-3. These two proteins contain 755 and 806 amino acids, respectively, and differ only in the length of the homopolymeric glutamine region. Comparison of the spectra of the MBP fusions of Q27 and Q78 ataxin-3 reveals a significant difference in secondary structure between the two fusion proteins (Fig. 1C). A reduction in the minimum at 208 nm for MBP/Q78 ataxin-3 suggests a loss of helical structure in the expanded repeat protein. The α-helical content was calculated to be 32% for MBP/Q27 ataxin-3 and 20% for MBP/Q78 ataxin-3. The MBP fusion partner has a negligible effect on the secondary structure of ataxin-3, because the calculated spectrum obtained by summing the spectra of MBP alone and Q27 ataxin-3 alone is nearly identical to the CD spectrum of the MBP/Q27 ataxin-3 fusion (Fig. 1B). Additionally, the presence of ataxin-3 does not perturb the MBP structure, as MBP binding to amylose resin is unaffected in the fusion proteins. Therefore, differences in the spectra for the Q27 and Q78 fusion proteins reflect changes in the ataxin-3 structure.
Figure 1.
CD spectra of various ataxin-3 species. (A) Solid line, hexahistidine-Q27 ataxin-3; dotted line, MBP. (B) Solid line, MBP/Q27 ataxin-3 fusion protein; dotted line, calculated spectrum obtained from a mass-weighted average of the MBP and Q27 ataxin-3 spectra found in A. (C) Solid line, MBP/Q27 ataxin-3 fusion; dotted line, MBP/Q78 ataxin-3 fusion.
Aggregation of Q78 Ataxin-3.
The strong tendency of Q78 ataxin-3 to aggregate precluded isolation of pure protein. However, the MBP fusion construct allowed for the purification of the protein (Fig. 2D) and subsequent removal of the MBP fusion partner by proteolytic cleavage. This result made it possible to examine the time dependence of the aggregation phenomenon. When the fusion proteins are cleaved with TEV protease to release ataxin-3, the Q27 form produces only monomeric ataxin-3 at the appropriate molecular weight (Fig. 2A); other Q27 constructs lacking the MBP partner also show no aggregation (data not shown). In contrast, the Q78 protein produces two species after cleavage, a 48-kDa monomer and a high-molecular-weight SDS-resistant aggregate, with the aggregate appearing only minutes after cleavage is initiated (Fig. 2B). Interestingly, the high-molecular-weight aggregate can be labeled with an anti-MBP antibody, suggesting the aggregates are able to recruit either MBP or MBP/Q78 ataxin-3 molecules that do not themselves form aggregates (Fig. 2C). Although demonstrating a high molecular weight on SDS/PAGE, these aggregates did not form visible precipitates, as would be expected for the nonspecific aggregation associated with protein denaturation.
Fibril Formation and Characterization.
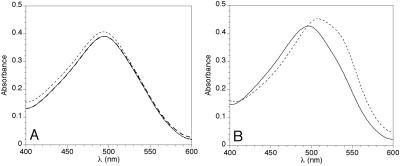
The propensity of Q78 ataxin-3 to aggregate led us to examine the ability of the protein to form β-fibrils. CR-binding assays invariably revealed the characteristic red shift and intensity increase (22) in the Q78 ataxin-3 that was released from its MBP fusion partner and incubated at 37°C for at least 5 days (Fig. 3B). Q78 ataxin-3 fibril stained with CR and dried onto microscope slides exhibited birefringence when viewed with polarized light (data not shown). Uncleaved MBP/Q78 ataxin-3 fusion protein would sometimes form CR-positive material, but with greatly reduced CR binding. Q27 ataxin-3, either alone or with MBP, never formed CR-positive fibrils under the conditions of the experiment (Fig. 3A). No CR binding was seen for three control proteins subjected to the same conditions (BSA, TEV protease, and MBP; data not shown). Alkylation of the cysteines of the Q78 ataxin-3 protein had no effect on its fibril-forming behavior, excluding the possibility that fibrils or aggregates form by disulfide cross-linking.
Figure 3.
CR binding by MBP/ataxin-3 fusion proteins, before and after exposure to fibril-growth conditions. TEV protease was added at day 0, and fusion proteins were incubated at 37°C, as described under Materials and Methods. (A) MBP/Q27 ataxin-3 fusion. CR only (no protein added), thick dashed line; solid line, day 0; dotted line, day 5. No significant dye-binding by the protein is observed at either day 0 or day 5. In fact, the figure shows that the spectra for CR alone and for protein at day 0 are virtually identical. (B) MBP/Q78 ataxin-3 fusion. Solid line, day 0; dotted line, day 5. A marked increase in dye-binding is seen to accompany fibril formation at day 5.
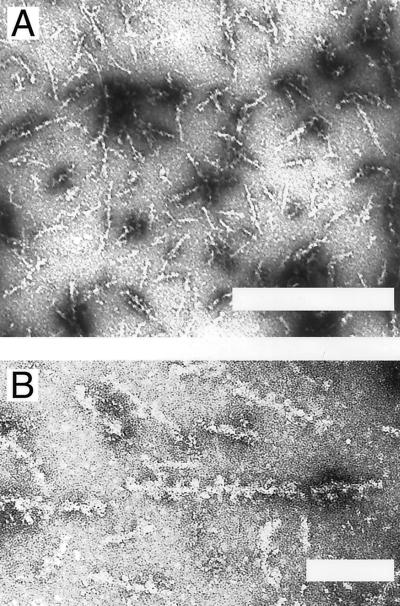
Electron micrographs of negatively stained CR-reactive Q78 ataxin-3 samples show unbranched fibrils of ≈200-nm length and 10-nm width (Fig. 4 A and B). The appearance of these fibrils is similar to that of other amyloid fibrils (18, 26). High magnification images (Fig. 4B) of well-stained populations reveal small beads ≈5 nm in diameter occurring along the entire length of the fibrils, with a periodicity of ≈12 nm. Control micrographs of other samples, including Q27 ataxin-3 that was subjected to fibrillizing conditions, showed only amorphous aggregate and no fibrils.
Figure 4.
Electron micrographs of negatively stained Q78/ataxin-3 fibrils at (A) 33,000× (Bar = 500 nm) and (B) 100,000× magnification (Bar = 100 nm).
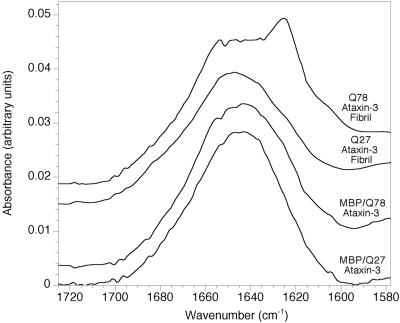
The secondary structure of both the soluble fusion proteins and the fibrils was examined with Fourier transform infrared spectroscopy (Fig. 5). MBP/Q27 ataxin-3 has an absorbance maximum at 1,643 cm−1. After fusion protein cleavage and incubation for 5 days at 37°C, this maximum shifts slightly to 1,647 cm−1, and a minor shoulder becomes apparent between 1,620 and 1,630 cm−1. However, neither of these spectra yielded specific information about conformation. The spectrum of the MBP/Q78 ataxin-3 fusion protein is similar to that of MBP/Q27 ataxin-3, although there is a suggestion of increased absorbance in the region of 1,620 and 1,630 cm−1. When Q78 ataxin-3 is cleaved from its fusion partner and permitted to form fibrils, however, there is a major new absorption at 1,625 cm−1, indicating that Q78 has significantly more secondary β-structure than Q27. The lack of any apparent shoulder in the 1,680–1,690 cm−1 region suggests that this β-structure may be parallel rather than anti-parallel.
Figure 5.
Infrared spectra of intact MBP/ataxin-3 fusion proteins and of cleaved fusion proteins exposed to fibril-forming conditions. Transmission spectra are shown for intact fusions, and internal reflection spectra are shown for cleaved and fibrillized fusions.
Discussion
The link between polyglutamine-mediated protein aggregation and neuronal pathology has kindled considerable interest in the biophysical and biochemical properties of proteins and peptides containing homopolymeric glutamine tracts. Because many of the proteins containing disease-related expanded glutamine repeats are large and difficult to express and purify, much work has focused on synthetic peptides and small fragments of larger proteins. In this way, for example, it has been possible to demonstrate that synthetic peptides containing stretches of 15 glutamines form cross β-fibrils (16), and that N-terminal fragments of huntingtin that contain expanded polyglutamine tracts form aggregates in a concentration and time-dependent manner (13). However, although this work has provided considerable insight into the behavior of polyglutamine tracts themselves, it has been unable to address the effects of large glutamine repeats on intact proteins. In addition, many studies have revealed toxic consequences associated with the expression of protein fragments containing polyglutamine tracts (7). It has not been clear whether this toxicity is unique to the fragment, implying some posttranslational cleavage event may be required to create the toxic species, or whether intact proteins containing expanded polyglutamine regions might exert similar toxic effects. By using full-length human ataxin-3 containing either 27 or 78 glutamines, we are able to compare directly the behavior of the normal and pathogenic forms of an intact human disease protein.
The Q27 and Q78 forms of ataxin-3 behave differently during expression, aggregation, and fibrillization. Q27 ataxin-3 can be purified readily; Q78 ataxin-3 aggregates strongly, and can be purified only by using the highly soluble MBP fusion partner. The Q27 and Q78 fusion proteins differ in their CD spectra, suggesting that the addition of glutamines is accompanied by a loss of α-helix and a gain of random coil. Hence, even before aggregation takes place, the expanded glutamine repeat perturbs the native structure of the ataxin-3 protein.
Once Q78 ataxin-3 is released from its fusion partner, it almost immediately forms SDS-resistant aggregates. These aggregates do not behave like aggregates of denatured proteins, in that they do not precipitate or strongly scatter light. They are not fibrils, because many days are necessary to acquire CR reactivity and form fibrils that can be readily identified by electron microscopy. The initial aggregates might represent protofibrils or fibril nuclei, as has been observed with other amyloid proteins (12); however, this possibility is difficult to reconcile with the kinetics—as with crystallization, nucleation is expected to be slow, and fibril growth more rapid. Hence, the initial aggregates might actually lie off the fibrillization pathway. Another possibility is that the initial aggregates represent polar zipper formation by only the polyglutamine regions; over time, transient unfolding of the remainder of the protein could allow it to be recruited into the β-structure as well. Initial SDS-resistant aggregates might, therefore, correspond to limited β-structures involving only the polyglutamine region, whereas the full-fledged fibrils would represent more extensive β-structures comprising the bulk of the protein.
Electron micrographs reveal 5-nm beads occurring at regular 12-nm spacings along the fibril, suggesting a higher-order structure; however, the precise nature of this structure is unclear, as yet. The 12-nm spacings would correspond to roughly 25 strands of cross β-structure; if one assumes that the 10-nm width of the fibril corresponds to an elongated strand of ≈40 residues, then a 12-nm repeat would require at least two molecules (40 × 25 = 800; there are 412 residues in Q78 ataxin-3).
Liu et al. (27) have recently suggested that the phenomenon of amyloid-fibril formation by proteins might, in many cases, reflect domain swapping. Thus, when the interactions between a peripheral domain and the core domain of a protein are weakened, the peripheral domain can be released from the central core and bind to the core of another protein molecule; successive domain-swapping events lead to oligomer formation. In some instances, the linker connecting the two domains might contribute to oligomerization by interacting with linker domains from other molecules.
In the context of this domain-swapping model, our data suggest that expanded glutamine tracts might contribute to oligomerization in two separate ways. First, we show that an expanded glutamine repeat alters and presumably destabilizes the native state of ataxin-3. When inserted between two domains of a protein, expanded glutamine repeats may promote domain swapping by partially destabilizing the interactions between those domains. Indeed, it has been shown that the insertion of a small glutamine repeat into chymotrypsin inhibitor 2 causes dimerization by means of domain swapping (28). Second, the propensity of isolated polyglutamine stretches to assemble into hydrogen-bonded polar zippers is well documented (29). Therefore, if two protein domains are connected by a polyglutamine linker, once the domains are separated, the linker can become extended, bind to linker regions from other molecules, and assemble into a polymeric β-structure. This dual contribution of expanded glutamine regions to oligomerization may explain why polyglutamine proteins are associated so strongly with an aggregatory phenotype and disease pathology.
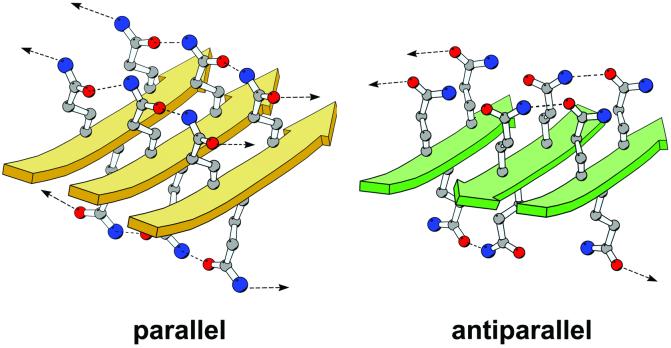
It is clear that polyglutamine polar zippers adopt β-pleated sheet structures (30), but it is less clear whether these sheets favor either a parallel or antiparallel topology. Models for antiparallel β-structures of polyglutamine have been described as examples of possible polar zipper architecture (16). However, our infrared data hint that fibrils composed of Q78 ataxin-3 might possess significant parallel β-structure. Interpretation of infrared spectra is not straightforward, particularly for entire proteins (31), and the presence of β-sheet twist can confound the distinction between parallel and antiparallel structures (32). Nonetheless, because a parallel organization has recently been suggested for a variety of different amyloid fibrils, including those generated from insulin (33), from the yeast prion protein Sup35 (34), and the Aβ-fibrils associated with Alzheimer's disease (35, 36), we constructed a model for an extended β-sheet composed of parallel strands of polyglutamine (Fig. 6), with the side-chain torsion angles for each residue approximately χ1 = −0°, χ2 = 180°, and χ3 = −0°. After energy minimization, the quality of this parallel model, as assessed by standard stereochemical criteria, is comparable to that of an antiparallel β model constructed by similar methods. One important difference between the two models lies in the hydrogen-bonding pattern adopted by the glutamine side chains: in the parallel model, each side chain hydrogen-bonds to two other side chains, one from each of the two adjoining strands, whereas in the antiparallel model, each side chain hydrogen-bonds to only one other side chain. Hence, in the parallel model, the strands are stabilized by continuous chains of side-chain–side-chain hydrogen bonds, and the hydrogen-bonding capacity of both the carbonyl oxygen and the amide nitrogen of each side chain is satisfied. In contrast, in the antiparallel model, adjoining strands are joined by pairwise hydrogen bonds, so for a given side chain, either the oxygen or the nitrogen participates in a hydrogen bond, but not both. Thus, parallel polyglutamine β-sheets may prove to be more energetically favorable than antiparallel β-sheets. In either case, oligomers containing small numbers of molecules are unlikely to be stable, because the hydrogen-bonding potentials at the edges of the nascent β-sheet would not be satisfied.
Figure 6.
Molecular models of homopolymeric glutamine peptides forming either parallel β-sheets (gold) or antiparallel β-sheets (green). Models containing four 11-residue strands were built in QUANTA (Accelrys, San Diego) and energy-minimized with CHARMM (Harvard Univ., Cambridge, MA; ref. 37); only portions of the models are shown for the sake of clarity. The parallel model was constructed starting from a canonical parallel β-sheet architecture followed by a search for optimal side-chain torsion angles. The antiparallel model is based upon figure 1 in ref. 16. The strands of the β-sheet are represented by solid arrows, and the glutamine side chains are shown in a ball-and-stick rendering, with carbon atoms colored in gray, nitrogen atoms in blue, and oxygen atoms in red. Dashed lines represent hydrogen bonds between side chains. Arrows represent hydrogen bonds to adjacent strands that are not included in the figure. The figure was made with the program MOLSCRIPT (38).
Parallel and antiparallel strand arrangements have different topological consequences for the fibril architecture. At least two types of antiparallel structures are possible: extended structures (in which the entire polyglutamine region of one molecule forms a single strand that associates with strands from adjacent molecules), and hairpin structures (in which the polyglutamine region forms one or more antiparallel β-hairpins, which can then associate with hairpins from neighboring molecules). Extended antiparallel structures would lead to the N and C termini of any given ataxin-3 molecule lying on opposite sides of the cross β-fibril; each edge of the fibril would be lined with alternating N and C termini from adjacent molecules. Antiparallel hairpins could associate either in a completely antiparallel manner, which would place the N and C termini from all molecules along one edge of the fibril, or in a mixed parallel/antiparallel fashion, which would place the N and C termini from one molecule together on one edge of the fibril, the termini from the adjacent molecule on the opposite edge, and so on in an alternating manner. In contrast to the antiparallel arrangements, parallel β-structures would not form hairpins, but would be found only as extended structures in which all N termini lay on one edge of the fibril and all C termini on the opposite edge. Given that the disposition of the nonpolyglutamine portions of the protein within the fibril depends strongly on the packing arrangement that is adopted, it seems likely that the protein context will determine whether a parallel, antiparallel, or mixed architecture results.
Acknowledgments
We thank Paul Axelsen, Jim Lear, Hank Paulson, Randy Pittman, and Carmen San Martin for technical assistance and invaluable discussions. This work was supported by National Institutes of Health Grant GM-55171.
Abbreviations
- CR
Congo red
- MBP
maltose binding protein
- TEV
tobacco etch virus
- CD
circular dichroism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Cummings C J, Zoghbi H Y. Hum Mol Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 3.Paulson H L, Das S S, Crino P B, Perez M K, Patel S C, Gotsdiner D, Fischbeck K H, Pittman R N. Ann Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- 4.Paulson H L. Brain Pathol. 2000;10:293–299. doi: 10.1111/j.1750-3639.2000.tb00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoghbi H Y, Orr H T. Curr Opin Neurobiol. 1999;9:566–570. doi: 10.1016/S0959-4388(99)00013-6. [DOI] [PubMed] [Google Scholar]
- 6.Lin X, Cummings C J, Zoghbi H Y. Neuron. 1999;24:499–502. doi: 10.1016/s0896-6273(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 7.Wyttenbach A, Carmichael J, Swartz J, Furlong R A, Narain Y, Rankin J, Rubinsztein D C. Proc Natl Acad Sci USA. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 9.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 10.Chan H Y, Warrick J M, Gray-Board G L, Paulson H L, Bonini N M. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 11.Harper J D, Lansbury P T., Jr Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 12.Harper J D, Wong S S, Lieber C M, Lansbury P T., Jr Biochemistry. 1999;38:8972–8980. doi: 10.1021/bi9904149. [DOI] [PubMed] [Google Scholar]
- 13.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobson C M. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 15.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perutz M F, Windle A H. Nature (London) 2001;412:143–144. doi: 10.1038/35084141. [DOI] [PubMed] [Google Scholar]
- 18.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 19.Sheffield P, Garrard S, Derewenda Z. Protein Expression Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 20.Harlan J E, Picot D, Loll P J, Garavito R M. Anal Biochem. 1995;224:557–563. doi: 10.1006/abio.1995.1087. [DOI] [PubMed] [Google Scholar]
- 21.Laue T, Shaw B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 22.Klunk W E, Jacob R F, Mason R P. Methods Enzymol. 1999;309:285–305. doi: 10.1016/s0076-6879(99)09021-7. [DOI] [PubMed] [Google Scholar]
- 23.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapust R B, Waugh D S. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholtz J M, Qian H, York E J, Stewart J M, Baldwin R L. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 26.Guijarro J I, Sunde M, Jones J A, Campbell I D, Dobson C M. Proc Natl Acad Sci USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Gotte G, Libonati M, Eisenberg D. Nat Struct Biol. 2001;8:211–214. doi: 10.1038/84941. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y W, Stott K, Perutz M F. Proc Natl Acad Sci USA. 1999;96:1257–1261. doi: 10.1073/pnas.96.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perutz M. Protein Sci. 1994;3:129–1637. doi: 10.1002/pro.5560031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perutz M F. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 31.Surewicz W K, Mantsch H H, Chapman D. Biochemistry. 1993;32:389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- 32.Kubelka J, Keiderling T A. J Am Chem Soc. 2001;123:6142–6150. doi: 10.1021/ja010270x. [DOI] [PubMed] [Google Scholar]
- 33.Bouchard M, Zurdo J, Nettleton E J, Dobson C M, Robinson C V. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balbirnie M, Grothe R, Eisenberg D S. Proc Natl Acad Sci USA. 2001;98:2375–2380. doi: 10.1073/pnas.041617698. . (First Published February 20, 2001; 10.1073/pnas.041617698). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antzutkin O N, Balbach J J, Leapman R D, Rizzo N W, Reed J, Tycko R. Proc Natl Acad Sci USA. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. . (First Published November 7, 2000; 10.1073/pnas.230315097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benzinger T L, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Biochemistry. 2000;39:3491–3499. doi: 10.1021/bi991527v. [DOI] [PubMed] [Google Scholar]
- 37.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 38.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]