Abstract
Advanced reproductive age is unequivocally associated with increased aneuploidy in human oocytes, which contributes to infertility, miscarriages, and birth defects. The frequency of meiotic chromosome segregation errors in oocytes derived from reproductively aged mice appears to be similar to that observed in humans, but a limitation of this important model system is our inability to accurately identify chromosome-specific aneuploidy. Here we report the validation and application of a new low-pass whole-genome sequencing approach to comprehensively screen chromosome aneuploidy in individual mouse oocytes and blastocysts. First, we validated this approach by using single mouse embryonic fibroblasts engineered to have stable trisomy 16. We further validated this method by identifying reciprocal chromosome segregation errors in the products of meiosis I (gamete and polar body) in oocytes from reproductively aged mice. Finally, we applied this technology to investigate the incidence of aneuploidy in blastocysts derived from in vitro- and in vivo-matured oocytes in both young and reproductively aged mice. Using this next generation sequencing approach, we quantitatively assessed meiotic and mitotic segregation errors at the single chromosome level, distinguished between errors due to premature separation of sister chromatids and classical nondisjunction of homologous chromosomes, and quantified mitochondrial DNA (mtDNA) segregation in individual cells. This whole-genome sequencing technique, therefore, greatly improves the utility of the mouse model system for the study of aneuploidy and is a powerful quantitative tool with which to examine the molecular underpinnings of mammalian gamete and early embryo chromosome segregation in the context of reproductive aging and beyond.
Keywords: aging, aneuploidy, embryo, oocyte, whole-genome sequencing
Introduction
Female gamete quality is dictated by acquisition of both cytoplasmic and meiotic competence [1]. Cytoplasmic competence refers to the ability of the oocyte to accumulate critical stores of maternally derived proteins, mRNAs, and organelles during oogenesis, which are essential for supporting meiotic maturation, fertilization, and early embryo development [2]. Meiotic competence refers to the ability of the oocyte to resume meiosis and undergo two sequential rounds of cell division without an intervening round of DNA replication to ultimately produce a haploid gamete [3]. It is well established that advanced reproductive age is associated with increased chromosome segregation errors in the oocyte during meiosis [4]. For example, the incidence of trisomy increases from 2%–3% to >35% as women age from their third to fifth decades of life [5]. Postfertilization mitotic errors are also present in human preimplantation embryos but occur at a lower incidence in mice [6]. This age-associated increase in aneuploidy contributes to adverse reproductive outcomes including miscarriages, infertility, and birth defects [7].
Understanding the origins of aneuploidy is essential as more women globally are delaying child bearing [8, 9]. In the United States, for example, 40% of live births are from women >30 years old. When women reach their mid-30s, they experience a decline in both gamete quality and quantity that can contribute to subfertility or infertility [10]. Evidence accumulated from clinical assisted reproductive technology suggests that the maternal age-associated decline in fertility is attributable primarily to deterioration in oocyte quality [3, 11, 12]. More specifically, when women of advanced reproductive age conceive using donor oocytes from reproductively young individuals, implantation and live birth outcomes reflect the age of the oocyte donor [13].
The most physiologically relevant method for studying the underlying mechanisms of reproductive aging is to analyze gametes from reproductively old animals. The physiologic aging mouse is an increasingly well-accepted model for reproductive aging, and recent studies have demonstrated that, across mouse strains, the mouse exhibits several phenotypes that occur in humans [14-26]. As in humans, reproductively aged mice have significantly fewer ovarian follicles than young controls, and this diminished ovarian reserve translates into altered hormone production, disrupted estrous cyclicity, and reduced fertility [27, 28]. In addition to endocrine alterations and reduced gamete quantity, reproductively aged mice, like humans, have reduced gamete quality. Oocytes isolated from reproductively old mice exhibit 30%–50% aneuploidy depending on strain, mirroring the incidence of common age-associated trisomies in human oocytes [14, 16, 19, 21-23, 26]. Use of the mouse model system has produced key insights into the cause of age-associated increases in aneuploidy. Although maternal age effects are likely multifactorial, leading mechanisms include alterations in recombination sites that generate chromosomes susceptible to nondisjunction, insensitive spindle assembly checkpoints, improper microtubule-kinetochore attachments, deterioration of chromosome cohesion, and changes in the micromechanical properties of meiotic chromosomes [23, 29-31].
A limitation of existing mouse studies is that they are unable to distinguish whether the proposed aneuploidy mechanisms affect all chromosomes equally or whether there is chromosomal specificity. This is in large part due to the lack of established methodologies for comprehensive, affordable, and chromosome-specific evaluation of aneuploidy in individual mouse cells. Routine methods for evaluating aneuploidy in mouse gametes and early embryos include chromosome spreads and fluorescence in situ hybridization [21]. These methods are technically challenging, and the number of chromosomes that can be reliably and efficiently detected in individual cells is low. An in situ chromosome spreading technique has been developed, which allows for accurate chromosome numbers to be determined in the context of intact oocytes [16]. Although this technique has been widely used in reproductive aging studies, it does not provide information about aneuploidy at the resolution of the individual chromosome [14, 19, 32].
In contrast to the mouse model, methods of comprehensive chromosome screening (CCS) are well established and widely used to assess aneuploidy in human oocytes and preimplantation embryos in the setting of clinical assisted reproductive technology. Aneuploidy assessment during in vitro fertilization (IVF) now involves a variety of methods which allow for analysis of all 24 chromosomes in parallel. Some methods in clinical use involve whole-genome amplification (WGA) with subsequent analysis by array comparative genomic hybridization, single-nucleotide polymorphism arrays, or next generation sequencing (NGS) [33]. One advantage of using WGA and NGS is the ability to amplify and analyze DNA from any species using a published genome sequence. Recently a powerful protocol for genome-wide copy number analysis from single cells has been established using WGA and NGS [34]. Here we report for the first time the validation and application of such methodologies to comprehensively screen aneuploidy and mitochondrial DNA (mtDNA) copy number in individual mouse oocytes and preimplantation embryos. This strategy provides the fields of reproductive science and medicine with a new research tool to improve the basic understanding of mechanisms contributing to maternal age-associated aneuploidy in mammals.
Materials and Methods
Cell Lines
Cell lines included previously characterized mouse embryonic fibroblasts (MEFs) with trisomy 16 female and normal male karyotypes (a generous gift from A. Amon, Massachusetts Institute of Technology, Cambridge, MA) [35]. DNA from large quantities of cells was purified using Qiagen columns as recommended for cell cultures by the supplier (Qiagen, Inc, Valencia, CA). Single cells were obtained using a dissecting microscope as previously described and prepared for cell lysis and WGA as described below [36].
Animals
Extensive studies in several mouse strains from different laboratories have detailed a reproductive age-associated deterioration in oocyte quality, including an increase in egg aneuploidy [14-26]. Because our research goal was methodological and because of the limited availability of aged mice, we performed experiments using two outbred strains (CD-1 and CF1), both of which exhibit prominent reproductive aging phenotypes [17, 19, 24, 26]. We selected the ages of our reproductively young (6–9 wk old) and old cohorts (13.5–19 mo old) based on existing publications to keep consistent with the field [14-26].
In the validation portion of our study examining reciprocal chromosome segregation errors in the oocyte and polar body, oocyte and polar body sample collection was performed at Northwestern University. Gametes were collected from a total of 8 CD-1 female mice (Harlan Laboratories, Indianapolis, IN) of advanced reproductive age (16-19 mo old). Mice were housed in a controlled-barrier facility at Northwestern University's Center of Comparative Medicine under constant temperature, humidity, and light (12L:12D). Food and water were provided ad libitum. In the application portion, the study determining the incidence of aneuploidy in in vitro maturation (IVM)- and IVF-derived blastocysts, sample collection was performed at the National Foundation for Fertility Research. CF-1 mice were obtained from Harlan Laboratories and maintained on a 14L:10D cycle with access to food and water ad libitum. We used a total of 14 females 6–9 wk old (designated young) and 65 females 13.5 mo old (designated aged) for the described experiments. All mice used in this study were retired breeders.
Ethical Approval
All animal experiments were approved by the Institutional Animal Care and Use Committee (Northwestern University) or the National Foundation for Fertility Research Ethics in Research Committee and were carried out in accordance with National Institutes of Health Guidelines and the Society for the Study of Reproduction's specific guidelines and standards.
In Vitro Maturation and Oocyte and Polar Body Processing
To validate WGA NGS-based CCS for accurate detection of aneuploidy in an unbiased manner, we examined mouse polar bodies and oocytes after spontaneous meiotic maturation as is routinely done in studies investigating molecular mechanisms of age-associated increases in egg aneuploidy [14, 16, 25, 26]. Meiosis I (MI) results in an asymmetric reductive division in which homologous chromosomes are segregated between the oocyte and the first polar body. Thus, chromosomes that are lost or gained in the oocyte are reciprocally gained or lost in the first polar body. Thus, NGS of both the oocyte and the polar body represents a useful strategy for additional validation of the CCS methodology.
Ovaries were harvested from each animal and cumulus-oocyte-complexes (COC) were released into Leibovitz L-15 medium (Life Technologies, Foster City, CA) containing 3 mg/ml polyvinylpyrrolidone (Sigma-Aldrich Inc, St. Louis, MO) by puncturing antral follicles with insulin needles. Oocytes were mechanically stripped of cumulus cells and cultured in α-MEM-Glutamax (Life Technologies) medium containing 3 mg/ml bovine serum albumin (MP Biomedicals, Santa Ana, CA) at 37°C in a humidified atmosphere of 5% CO2 in air to induce spontaneous meiotic maturation. Cells were cultured for a total of 14–17 h, and meiotic progression was scored by morphology as assessed by light microscopy. Cells that had extruded the first polar body were arrested at metaphase of MII and were thereby considered mature oocytes. Only mature MII oocytes were used for subsequent processing, and oocytes from individual animals were analyzed separately. Images were obtained using an inverted model DM IRB microscope with 4× and 40× objectives (Leica Microsystems, Buffalo Grove, IL).
To obtain matched oocyte and polar body samples for analysis, we incubated intact oocytes briefly in acidified Tyrode solution (EMD Millipore, Billerica, MA) to remove the zona pellucida. The polar body was gently aspirated from the oocyte, and matched polar body and oocyte samples were washed in hypotonic wash buffer (HWB) and snap frozen individually in a sterile PCR tube containing 1 μl of HWB on a dry ice and ethanol bath. HWB alone served as the negative control, and HWB remaining from a wash drop served as background. All samples were shipped to Reproductive Medicine Associates of New Jersey, where further processing, sequencing, and analysis were performed.
Blastocyst Production and Processing
To apply WGA NGS technology to the investigation of aneuploidy in mice using a more physiologic model, we generated embryos by IVM or in vivo ovulation (IVO) of oocytes followed by IVF using young and aged females. Our objective was to determine whether NGS-based CCS would identify differences between these distinct age groups (young vs. aged) and oocyte maturation methods (IVM vs. IVO). To obtain COC for IVM, mice were stimulated with 5 IU of pregnant mare's serum gonadotropin (PMSG; Calbiochem, Billerica, MA) administered by i.p. injection. Ovaries were harvested 46 to 48 h after PMSG injection, and COCs were recovered (composition of maturation and culture media were described previously [24]); all incubation steps were performed at 37°C in 7.5% CO2 and 6.5% O2 [37-39]. From the COC recovered, only those with several layers of compact cumulus cells were selected and subsequently matured in vitro (10 COC per 50-μl drop under oil immersion [Ovoil; Vitrolife, Gothenburg, Sweden) for 18 h and then used for IVF. To obtain in vivo-matured oocytes, 46 to 48 h after PMSG ovulation was stimulated with 5 IU of human chorionic gonadotrophin (Calbiochem); fully-expanded COC were collected from the oviduct 16 h later. Mature oocytes were fertilized (10 COC per 50-μl drop under oil immersion) using spermatozoa from B6D2F1 males (≥8 wk old) at a final concentration of 1 × 106 sperm/ml. Gametes were co-incubated for 6 h. After IVF, presumptive zygotes (10 per 20-μl drop under oil immersion) were placed in the first step of a sequential culture medium. Cleavage was evaluated 24 h post IVF, and embryos that had not cleaved and/or had fragmented were removed from culture. Embryos were moved into step 2 medium after 48 h of culture, and blastocysts were collected following 96–116 h of culture. Zona pellucidae were removed using acidified Tyrode solution. Single whole blastocysts were washed in Dulbecco phosphate-buffered saline (Life Technologies) plus 0.01% polyvinylpyrrolidone (Sigma-Aldrich) and then transferred to HWB. Individual blastocysts were picked up and placed into sterile PCR tubes in 1 μl of HWB. Samples were stored at 2–8°C and shipped to Reproductive Medicine Associates of New Jersey where further processing, sequencing, and analysis were performed. Differences in the incidence of aneuploidy between the type of oocyte maturation within age were analyzed using the Fisher exact test. A P value of <0.05 was considered statistically different.
Whole-Genome Amplification
Single fibroblast cells, first polar bodies, oocytes, and blastocysts were lysed in alkaline lysis buffer as previously described [40] and processed using the GenomePlex WGA4 kit as recommended by the supplier (Sigma-Aldrich). WGA DNA was purified using GenElute PCR cleanup columns (Sigma-Aldrich) and quantified using a Nanodrop 8000 spectrophotometer (Fisher Scientific Inc., Waltham, MA).
Next Generation Sequencing
WGA DNA was normalized to 200 ng in a total volume of 35 μl of molecular biological grade water (Lonza, Rockland, ME). Ion Plus fragment library kit, Ion Xpress Plus fragment library kit, and Ion Xpress barcode adapters 1-96 kit were used to construct the WGA library as recommended by the supplier (Life Technologies). WGA DNA was fragmented with Ion Shear Plus reagent for 20 min to generate 150 to 250 base pair fragments. Fragmented DNA was then purified with Agencourt AMPure XP reagent beads as recommended (Beckman Coulter Inc., Brea, CA). Barcoded adapter ligation and nick repair were performed, followed by another Agencourt AMPure XP reagent bead purification. A peak size of 270 base pairs was selected with an E-Gel SizeSelect agarose gel (Life Technologies). Size-selected DNA was amplified with 8 cycles, using Platinum PCR SuperMix High Fidelity (Life Technologies). After Agencourt AMPure XP Reagent bead purification, 1 μl of amplified library was assessed with D1k ScreenTape (Agilent Technologies Inc., Wilmington, DE). Individual libraries were diluted to 100 picomolars with low-Tris-EDTA buffer (Life Technologies). Equal amounts taken from each of 24 samples (including 4 normal male control samples) were pooled for one Ion PI Chip V2 (Life Technologies). Ion Sphere particles containing clonally amplified DNA were prepared with Ion PI template OT2 200 kit version 3, and the template-positive Ion Sphere particles were then enriched with the Ion OneTouch ES (Life Technologies). The enriched template-positive Ion Sphere particles were then sequenced with Ion PI Chip V2 and Ion PI sequencing 200 Kit v3 using the Ion Proton instrument (Life Technologies).
Data Processing and Mapping
Ten bases were trimmed from the 5′ and 3′ ends of each reading, and trimmed reads were aligned to the reference Mus musculus genome (mm10) using Bowtie2 version 2.1.0 software (Bowtie; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) with the “sensitive-local” preset mode. The number of reads mapping to each chromosome were counted using SAMtools version 0.1.19 software (http://samtools.sourceforge.net), ignoring alignments with MAPQ <20. The number of mapped reads averaged 2.0 ± 1.2 million per sample by using this protocol. Average depth of coverage was 0.10 ± 0.06 X. The mean read length ranged from 63 to 125 base pairs.
Calculation of Chromosomal Copy Numbers
For each sample, the read count for each chromosome was divided by the average read count across all autosomes for that sample, that is, the normalized read count for chromosome i in sample j, denoted normcountij, is given by normcountij =
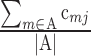 , where cmj are the raw read counts for sample j, and A is the set of autosomes. Next, for each chromosome i in sample j, the normalized read count was divided by the mean of the normalized read counts for the chromosome across all euploid male reference samples. To obtain the copy number, this result was multiplied by 2 for autosomes or multiplied by 1 for sex chromosomes:
, where cmj are the raw read counts for sample j, and A is the set of autosomes. Next, for each chromosome i in sample j, the normalized read count was divided by the mean of the normalized read counts for the chromosome across all euploid male reference samples. To obtain the copy number, this result was multiplied by 2 for autosomes or multiplied by 1 for sex chromosomes:
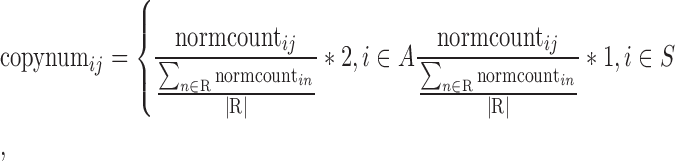 |
where R is the set of euploid reference samples, and S is the set of sex chromosomes. To establish a threshold for an aneuploid versus a euploid chromosome, we plotted the chromosomal copy number values obtained from samples with confirmed aneuploidies including the trisomy 16 MEF samples and reciprocal aneuploidy polar body and oocyte samples (chromosomes 11 and 12 from animal A, chromosome 15 from animal E, and chromosome 7 from animal G) (Table 1). We generated distributions for monosomy, disomy, and trisomy and obtained the midpoint between the minimum disomy and maximum monosomy for monsomy cutoff designation, and the maximum disomy and minimum trisomy for the trisomy cutoff. Chromosomes with copy number values <1.415 (monosomy) and >2.625 (trisomy) were considered aneuploid (Supplemental Fig. S1; supplemental data are available online at www.biolreprod.org).
Table 1.
Summary of egg aneuploidy.
| Animal ID | No. of aneuploid eggs/total (%) | Egg aneuploidy | Reciprocal PB aneuploidy† |
|---|---|---|---|
| A | 1/5 (20%) | +11, −12 | −11, +12 |
| B | 1/5 (20%) | −9 | ++9 |
| C | 1/5 (20%) | Chaotic | −5 |
| D | 0/5 (0%) | NA* | NA |
| E | 1/8 (13%) | +15 | −15 |
| F | 0/2 (0%) | NA | NA |
| G | 1/5 (20%) | +7 | −7 |
| H | 1/5 (20%) | Chaotic | +2 |
NA = not applicable.
PB = polar body.
Relative Mitochondrial DNA Assessment
Mitochondrial genome read counts were sufficient to make predictions of relative copy numbers in all cases. The number of reads, which aligned to the mitochondrial genome, was normalized by dividing it by the total sum of reads aligning to the autosomes from the same sample. Because the first polar body and remaining oocyte contained the same amount of chromosomal DNA following meiosis I, the normalized values were directly comparable to one another. Therefore, in order to determine the percentage of mitochondrial DNA which segregated to the first polar body relative to remaining oocyte, the normalized polar body mitochondrial DNA read counts were divided by the same value obtained from the remaining paired oocyte.
Results
NGS-Based CCS Successfully Identifies Aneuploidy in Single Mouse Cells
To validate WGA and NGS in individual mouse cells, we evaluated the technique by sequencing genomic DNA isolated from pooled cells known to possess trisomy 16 (35]. Using these cells, we were able to detect the presence of the extra copy of chromosome 16 and zero copies of chromosome Y, as expected (Fig. 1A). We next extended this technique to single cells from the same cell line and a known normal male cell line by incorporating WGA. We were able to amplify DNA from single cells (291.2 ± 26.4 ng/μl of DNA/fibroblast cell) and obtain accurate chromosome-specific copy number assignments for both of these cell types (Fig. 1, B and D). As expected for the aneuploid female MEFs, the copy number for all autosomes (1–19) was 2, except for chromosome 16, which had a copy number of 3, reflecting the stable trisomy (Fig. 1C). In addition, given the sex of the cell line, there was a copy number of 2 for the X chromosome and zero for the Y chromosome (Fig. 1C). For the euploid male MEFs, the copy number for all autosomes was 2, but the sex chromosomes (X and Y) each had a copy number of 1 (Fig. 1D). Taken together, these findings validate the application of CCS using WGA and NGS to individual mouse somatic cells.
Fig. 1.
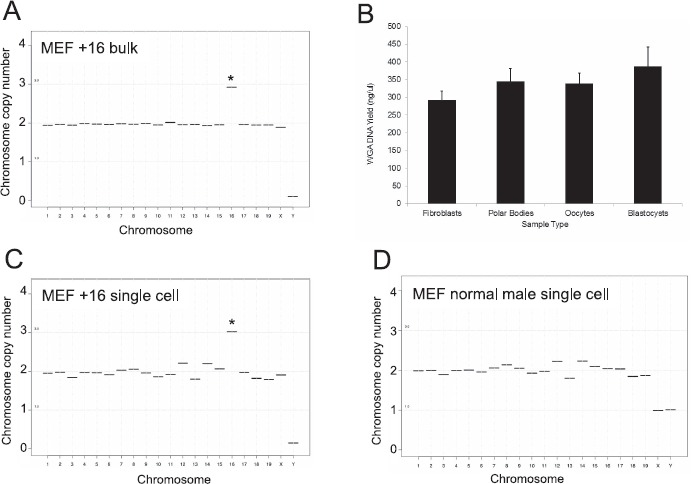
NGS can be used to identify the karyotype of mouse cells of known ploidy. A) Sequencing results are shown of genomic DNA isolated from pooled mouse embryonic fibroblasts engineered to have stable Trisomy 16 (copy number of 3). This cell line is of female origin. B) Average yield of DNA isolated from individual fibroblasts, polar bodies, oocytes, or blastocysts following WGA. Representative sequencing plots are shown from a single Trisomy 16 female MEF (C) and a single euploid male MEF (D) following WGA and NGS. *Autosomes that have copy numbers that deviate from 2, which indicates aneuploidy.
NGS-Based CCS Successfully Detects Reciprocal Errors Due to Premature Separation of Sister Chromatids and Homologous Chromosome Nondisjunction in Polar Bodies and Oocytes
To perform reciprocal oocyte and polar body sequencing, we isolated fully grown germinal vesicle-intact oocytes from COC and obtained mature MII oocytes following IVM (Supplemental Fig. S2, A and B). Gametes from reproductively aged mice were used for this validation study because this population is known to have increased aneuploidy [31]. We used oocytes derived from spontaneous IVM rather than in vivo ovulation because polar bodies following IVM tend to be larger and more stable than those obtained following ovulation [41]. In addition, IVM has been routinely used as a model for studying chromosome segregation errors during meiosis [14, 16, 17]. Using this approach, we collected an average of 15 ± 3 oocytes from 8 individual CD-1 mice ranging in age from 15-19 months. Following IVM, approximately 80% (range 57%-96%) of oocytes from these reproductively aged mice resumed meiosis and reached MII (Supplemental Fig. S2, B and C).
Using these mature oocytes, we separated the oocyte from the first polar body and then performed WGA and NGS using these matched cells (42]. We amplified 345.1 ± 36.7 ng/μl of DNA/polar body and 338 ± 30.2 ng/μl of DNA/oocyte (Fig. 1B). We performed sequencing using a total of 40 matched first polar body and oocyte samples, representing a subset of 2–8 oocytes per mouse. We obtained interpretable sequencing data from 37 of 40 oocytes and 35 of 40 polar bodies (Table 2). The remaining sequencing data from the oocytes (3 of 40) and polar bodies (5 of 40) were considered chaotic due to the noise level in the observed copy number assignment patterns. Such patterns could potentially reflect complex aneuploidy, biological mosaicism, sample degradation, or methodological artifacts (Table 2 and Supplemental Fig. S3). Because we were unable to distinguish among these possibilities, we considered these data nonresults, unless we were able to obtain clear sequencing results from the reciprocal polar body or gamete (Table 1). As expected, euploid mouse oocytes and their matched polar bodies had a copy number of 2 for all nineteen autosomes and the X chromosome but a copy number of zero for the Y chromosome (Fig. 2, C and D). Any copy number pattern that differed from this was considered aneuploid. In the subset of 40 oocytes that were sequenced, we observed an overall aneuploidy incidence of 15% (6 of 40 oocytes) and a euploidy incidence of 85% (34 of 40 oocytes) (Table 1). From an individual animal standpoint, 6 of 8 mice had aneuploid oocytes, with an incidence ranging from 13% to 20%, whereas 2 mice did not (Table 1).
Table 2.
Summary of chaotic egg and polar body (PB) reads.
| Animal ID | No. of chaotic eggs/total (%) | No. of chaotic PB/total (%) |
|---|---|---|
| A | 0/5 (0%) | 1/5 (20%) |
| B | 0/5 (0%) | 1/5 (20%) |
| C | 2/5 (40%) | 0/5 (0%) |
| D | 0/5 (0%) | 0/5 (0%) |
| E | 0/8 (0%) | 2/8 (25%) |
| F | 0/2 (0%) | 0/2 (0%) |
| G | 0/5 (0%) | 0/5 (0%) |
| H | 1/5 (20%) | 1/5 (20%) |
| Total | 3/40 (8%) | 5/40 (13%) |
Fig. 2.
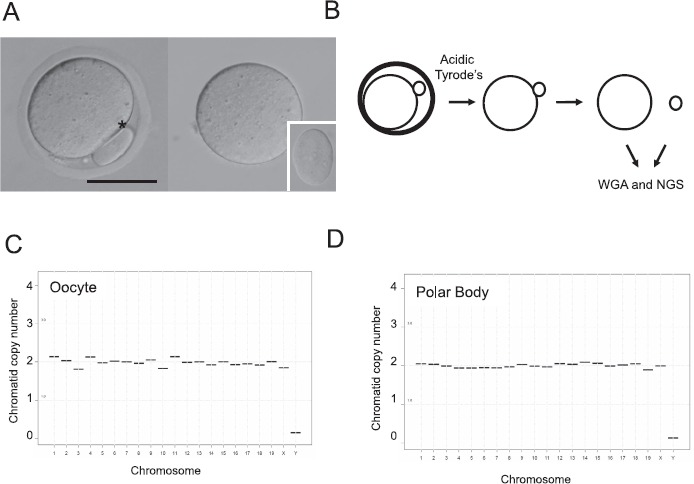
Use of WGA and NGS for chromosome assessment in individual matched polar bodies and chromosomes is shown. A) MII oocytes with visible polar bodies were used for sequencing analysis. A representative image is shown in the left panel (*polar body). The oocyte and polar body were separated from each other following removal of the zona pellucida. The matched oocyte and polar body (inset) are shown in the right panel following separation. Bar = 50 μm. B) Schematic shows the experimental workflow. The zona pellucida was removed from each MII oocyte using acidic Tyrode solution, the matched polar body and oocyte were gently separated by aspiration and WGA, and NGS was performed in each cell. Representative sequencing plots of a euploid oocyte (C) and its matched polar body (D) are shown. Note, all autosomes and the X chromosome have a copy number of 2, whereas the Y chromosome has a copy number of 0.
In the 6 aneuploid oocytes, we identified chromosome segregation errors in seven different autosomes: 2, 5, 7, 9, 11, 12, and 15 (Table 1 and Figs. 3 and 4). In five of the oocytes, the aneuploidy was attributed to premature separation of sister chromatids (PSSC). This was evident as a reciprocal gain or loss of a single chromatid, or a copy number of three or one for a particular chromosome(s), in the respective matched oocyte and polar body samples (Table 1 and Fig. 3, A and B, chromosomes 2, 5, 7, 11, 12, and 15). In one oocyte, we identified nondisjunction of chromosome 9 homologs that resulted in a copy number of 4 in the polar body and a reciprocal copy number of zero in the oocyte (Table 1 and Fig. 3, C and D). In the majority of the aneuploid oocytes (5 of 6), we detected the mis-segregation of a single chromosome/chromatid. However, in one oocyte, we identified PSSC of two chromosomes, 11 and 12 (Table 1 and Fig. 3, E and F). Taken together, these results demonstrate that NGS-based CCS is a reliable and quantitative tool with which to identify chromosome-specific aneuploidy and the type of mis-segregation in the mouse oocyte.
Fig. 3.
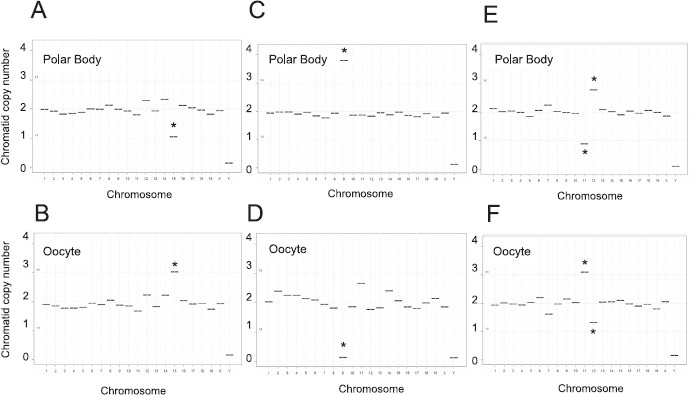
Detection of reciprocal aneuploidies in metaphase II oocytes and matched polar bodies. Sequencing plots from matched polar bodies (A, C, and E) and oocytes (B, D, and F) that reveal reciprocal aneuploidies, including premature separation of sister chromatids (loss/gain of 1 copy of chromosome 15 [A and B]) non-disjunction of homologs (loss/gain of 2 copies of chromosome 9 [C and D]), and complex aneuploidy (loss/gain of 1 copy of chromosomes 11 and 12 [E and F]). *Chromosomes with an abnormal copy number indicative of aneuploidy.
Fig. 4.
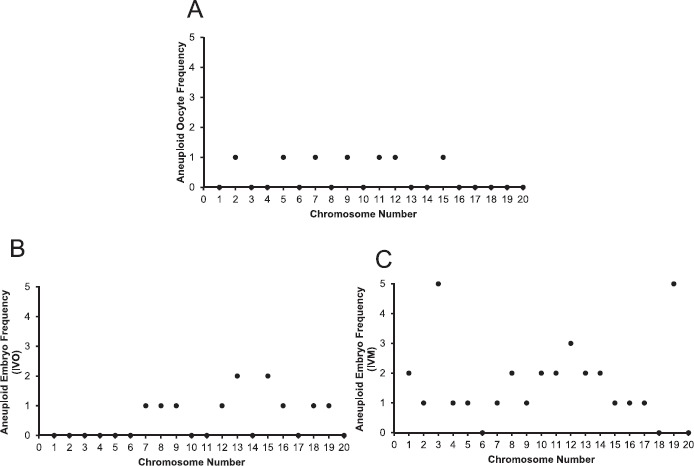
Frequency of chromosome-specific aneuploidies in oocytes and embryos from reproductively aged mice. Histograms show frequency of segregation errors for each specific chromosome observed in aneuploid oocytes (A), IVO-derived embryos (B), and IVM-derived embryos (C).
NGS can be used to Evaluate Relative Quantities of Mitochondrial DNA
In addition to evaluation of the chromosome complement of individual oocytes and polar bodies, WGA and NGS can be used to assess mtDNA (Table 3). Specifically, the percentage of mtDNA in the polar body relative to that in the matched oocyte samples was evaluated. The first polar body contained between 0.1% and 8.8% of the total mtDNA relative to that in the oocyte. This relatively small amount of mtDNA in the polar body is expected, as meiosis results in highly asymmetric cell divisions to produce small polar bodies, such that the bulk cytoplasm remains in the oocyte to support further development.
Table 3.
Summary of relative mtDNA segregation.
| Animal ID | Percentage of mtDNA in PB relative to egg* |
|---|---|
| A | 0.7, 0.9,1.5, 2.6, 8.8 |
| B | 0.6, 1.4, 1.9, 1.9, 2.3 |
| C | 0.2, 0.5, 1.2, 3.8, 4.7 |
| D | 0.5, 0.8, 1.1, 1.0, 3.0 |
| E | 0.4, 0.6, 0.7, 0.7, 1.0, 1.2, 1.3, 1.5 |
| F | 0.7, 1.9 |
| G | 0.1, 0.3, 0.2, 0.2, 0.5 |
| H | 0.3, 0.7, 1.2, 1.5, 1.9 |
Data from aneuploid oocytes are shown in boldface. PB = polar body.
NGS Based CCS Successfully Identifies Aneuploidy in Blastocysts
We next applied our NGS-based CCS technology for aneuploidy detection in individual preimplantation mouse blastocysts produced by IVM or IVO from reproductively young and aged females (Fig. 5A). Importantly, our analysis was done using intact blastocysts, which precludes detection of mitotic errors. Thus, the aneuploidy we determined with this particular approach is limited to meiotic errors and may underestimate the total aneuploidy incidence in the preimplantation embryo. We amplified 387 ± 55.6 ng/μl of DNA/blastocyst (Fig. 1B). In the reproductively young cohort, a total of 89 morphologically healthy COC were selected for IVM from 9 mice (∼10 COC/female), and 199 COC were isolated from the oviducts of 5 mice (∼40 COC/female) after IVO. Of fertilized IVM eggs from young females, 56.1% developed to the blastocyst stage. A subset of 33 blastocysts was analyzed by CCS; WGA was successful in 32 of them. Forty-six percent of the embryos were male. Aneuploidy in three blastocysts was detected (9.4%) (Table 4), with mis-segregation of single chromosomes (chromosomes 2 and 13) (Table 5). Of fertilized IVO eggs from young animals, 56.4% developed to the blastocyst stage. WGA was successful in 27 of 29 embryos examined; 15% of the embryos were male. Only one blastocyst was aneuploid (3.7%, Table 4), with a segregation error in chromosome 8 (Table 5).
Fig. 5.
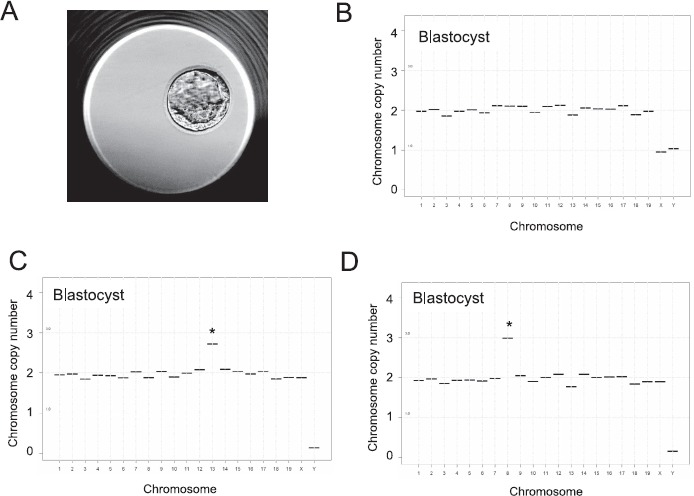
Use of WGA and NGS for chromosome assessment in individual blastocysts originating from young and reproductively aged female. A) Blastocysts derived after IVM and IVF were used for sequencing. A representative blastocyst is shown. Representative sequencing plots of euploid (B) and aneuploid blastocysts (C and D) are shown. Aneuploidy is due to a gain of 1 copy of chromosome 13 (C) and a gain of 1 copy of chromosome 8 (D). *Chromosomes with an abnormal copy number indicative of aneuploidy.
Table 4.
Summary of embryo aneuploidy.
| Age group | Maturation | Aneuploid | Euploid | Total | No result | % Aneuploidy |
|---|---|---|---|---|---|---|
| Young | IVO | 1 | 26 | 27 | 2 | 3.7a |
| IVM | 3 | 29 | 32 | 1 | 9.4a | |
| Aged | IVO | 6 | 41 | 47 | 3 | 12.8a |
| IVM | 20 | 45 | 65 | 2 | 30.8b |
Groups with different superscripts are statistically different from each other (P < 0.05).
Table 5.
Description of embryo aneuploidy.
| Group | Maturation | Embryo ID | Aneuploidy | Sex |
|---|---|---|---|---|
| Aged | IVM | 1 | +1, +5, +8, +13 | Female |
| Aged | IVM | 2 | +3, −14, −17, +19 | Male |
| Aged | IVM | 3 | +16 | Female |
| Aged | IVM | 4 | +19 | Male |
| Aged | IVM | 5 | −12 | Female |
| Aged | IVM | 6 | +2 | Male |
| Aged | IVM | 7 | +8 | Female |
| Aged | IVM | 8 | +3 | Male |
| Aged | IVM | 9 | +3, +10 | Female |
| Aged | IVM | 10 | +13 | Female |
| Aged | IVM | 11 | +15 | Male |
| Aged | IVM | 12 | −19 | Male |
| Aged | IVM | 13 | +10, +12 | Male |
| Aged | IVM | 14 | +7, +11 | Male |
| Aged | IVM | 15 | −9, +19 | Female |
| Aged | IVM | 16 | −3 | Female |
| Aged | IVM | 17 | −4 | Female |
| Aged | IVM | 18 | −3, +11, +14, +19 | Male |
| Aged | IVM | 19 | +12 | Female |
| Aged | IVM | 20 | +1 | Male |
| Aged | IVO | 21 | +13 | Male |
| Aged | IVO | 22 | +8 | Female |
| Aged | IVO | 23 | +19 | Male |
| Aged | IVO | 24 | +15 | Male |
| Aged | IVO | 25 | +7, −9, +12, +15, +16, +18 | Female |
| Aged | IVO | 26 | −13 | Female |
| Young | IVM | 27 | +2 | Female |
| Young | IVM | 28 | +13 | Male |
| Young | IVM | 29 | +13 | Female |
| Young | IVO | 30 | +8 | Female |
In contrast to reproductively young mice, aged mice had fewer morphologically healthy COCs to be used for IVM, fewer ovulated COCs following IVO, and an increased incidence of aneuploidy compared to younger counterparts, despite successful fertilization and blastocyst development. Specifically, for aged IVM oocytes, a total of 55 CF1 mice 13.5 months of age were used to obtain 223 morphologically healthy COC (∼4 COC/female). Following IVF and embryo culture, 75.8% of the fertilized eggs reached the blastocyst stage (160 of 211). For aged IVO oocytes, 10 CF1 females 13.5 months of age were used to obtain 106 COC following superovulation (∼11 COC/female), and 72.7% of the fertilized eggs following IVF reached blastocyst stage (64 of 88 eggs).
To assess aneuploidy in these two aged cohorts, a subset of 67 IVM blastocysts was submitted for chromosomal analysis, and WGA was successful in 65 cases (Fig. 1B). Twenty of the 65 embryos (30.8%) were aneuploid (Table 4), with segregation errors involving multiple chromosomes (Table 4 and Figs. 4 and 5). Thirty-two of 65 embryos (45.7%) were male based on copy number states of the X and Y chromosomes (Table 4). Thirteen of the 20 aneuploid embryos possessed aneuploidy of a single chromosome, whereas the remaining embryos each had 2, 4, or 6 different chromosomes that contributed to aneuploidy (Table 5). For the IVO group, a subset of 50 blastocysts was analyzed, and WGA was successful in 47 cases. Six of 47 blastocysts were aneuploid (12.8%) (Table 4), with segregation errors involving chromosomes 7, 8, 9, 12, 13, 15, 16, 18, and 19 (Fig. 4 and Table 4). Twenty-eight of 47 embryos were male (59.6%); 5 of the 6 aneuploid embryos had aneuploidy of a single chromosome (Table 5).
Discussion
In these studies, we validated and applied a sophisticated comprehensive chromosome screening method to further inform our fundamental understanding of aneuploidy in mouse oocytes and preimplantation embryos. We used physiological mouse models of reproductive aging to develop and optimize this technique, as it is well established that there is an age-associated increase in the incidence of oocyte aneuploidy [29, 31]. Using WGA, we were able to reliably amplify DNA from single gametes (polar bodies and oocytes) and blastocysts and perform NGS to obtain a comprehensive and quantitative analysis of the ploidy status of each sample. We validated our approach using several controls including MEFs of known trisomy, the lack of the Y chromosome in oocytes, and the identification of reciprocal aneuploidies in matched oocyte and polar body pairs. In addition, we applied this technology to investigate aneuploidy in blastocysts originating from IVO and IVM oocytes from young and aged females. Using the combined approach of WGA and NGS, we noted that the incidence of aneuploidy in IVM oocytes and preimplantation embryos from aged females ranges between 15% and 31%. This degree of aneuploidy is higher than that reported in individual mouse somatic cells, in which the prevalence of aneuploidy in skin, liver, and brain cells was determined to be less than 5% [43]. Such differences are likely attributable to age-associated changes in aneuploidy, as the incidence of aneuploidy in young IVO-derived blastocysts was more similar to somatic cells at only 3.7%.
Although we documented aneuploidy in oocytes from mice of advanced reproductive age (0–20% per female), this percentage is lower than that described previously [14, 16, 19, 21]. For example, the incidence of aneuploidy in B6D2F1 mice of advanced reproductive age was 35% [16]. Using the CD1 mouse strain, Merriman et al. [19] reported the incidence of aneuploidy to be 37.5% at 12 months and 60% at 15 months. There are several explanations for this apparent discrepancy. First, these previously published results are based on an in situ chromosome spreading technique, which is more subjective and therefore potentially more error-prone than the quantitative technique of NGS [44]. NGS is also not able to distinguish sister chromatids that have prematurely separated at metaphase II and that would then mis-segregate and contribute to aneuploidy upon completion of MII following fertilization. This would result in an underestimation of the aneuploidy incidence because premature dyad separation in MII may be a major origin of segregation error in mouse oocytes [26]. To fully elucidate these mechanisms, future studies performing CCS in the oocyte, first and second polar bodies are warranted to distinguish reproductive age-associated aneuploidies specifically due to MI and MII errors. Interestingly, the frequency of aneuploidy observed in IVM blastocysts from aged mice was markedly higher than that in IVM oocytes in this study. This may be due to a number of factors including the additional contribution of MII errors, the use of ovarian stimulation to obtain eggs for blastocyst production, the potential strain-specific differences in susceptibility to age related aneuploidy, and the difference in medium and protocols used for obtaining oocytes and embryos. All these factors represent areas of great interest for future investigation using this technology and model system.
With this technology, we were able to obtain quantitative copy number information, providing insight into the type of meiotic errors that occur—either nondisjunction of homologous chromosomes or PSSC. Reciprocal gains or losses of individual chromatids were the most commonly observed mis-segregation events in oocytes from reproductively old mice. Although the dataset we generated as part of this proof-of-concept study is small, these findings are consistent with previous mouse studies and clinical data from human assisted reproductive technology in which most aneuploidies are due to PSSC rather than nondisjunction [45, 46]. In fact, age-associated loss of centromeric cohesion has been identified as a prime mechanism underlying PSSC in mouse and human [14, 32]. Our findings, therefore, add to the evidence that the aging mouse may provide a relevant model for the mechanisms of aneuploidy development in the human oocyte.
Using NGS-based CCS, we were able to identify the specific chromosome mis-segregation events that resulted in aneuploid oocytes and embryos. We found that, in most cases, complex aneuploidies were relatively rare (13% of all detected embryo aneuploidies, for example), and instead, aneuploidy was attributable to the mis-segregation of 1 or 2 chromosomes within a given oocyte or embryo. These results are consistent with those of previous mouse studies, which indicate that although advanced reproductive age is associated with general alterations in factors such as chromosome micromechanical properties and cohesion, these alterations do not translate into massive global aneuploidy of every chromosome [14, 16, 17, 19, 23, 26]. Interestingly, there did not appear to be an obvious pattern of chromosome-specific mis-segregation in the subset of oocytes and embryos we analyzed (Fig. 4). For example, all aneuploidies did not appear due to a single chromosome or only small or large chromosomes. These findings are intriguing, and their significance will require further analysis.
An additional strength of this NGS strategy is that it can be used to simultaneously evaluate aneuploidy and mtDNA copy number within individual cells. Mitochondrial number, which can be inferred by mtDNA copy number, may be an important marker of gamete quality [47, 48]. Oocyte mitochondria provide critical energy stores to support the events of meiosis, fertilization, and early embryo development. Published data suggest that there is an association between oocyte mitochondria and aneuploidy in the human embryo, and this sequencing approach provides a new methodology to address this question [49, 50]. In our study, in half of the animals that had aneuploid oocytes, the aneuploid oocyte had the highest percentage of mtDNA in the polar body compared to the euploid oocytes from that individual animal (Table 3, animals A, B, G). These results suggest that relative mtDNA quantities may be associated with aneuploidy during MI in mouse oocytes. However, additional studies with a larger sample size are warranted to examine this possibility. Should this association be confirmed, there are promising therapeutic strategies to reverse mitochondrial loss with ER stress inhibitors or chaperone inducers [51].
When analyzing blastocysts, CCS using WGA and NGS confirmed the lower incidence of aneuploidy in blastocysts originating from young females compared to aged females. Thus, even though similar percentages of fertilized eggs from the two age cohorts progressed to the blastocyst stage, the higher incidence of aneuploidy in the aged females underscores significant reproductive age-associated differences in egg and embryo quality. When comparing the type of oocyte maturation used, there was no difference in blastocyst aneuploidy in young females, but blastocysts from females of advanced maternal age produced after IVM had more aneuploidy compared to IVO blastocysts. Data regarding aneuploidy in primate embryos produced after IVM are contradictory. Some reports have shown that there is not an increase in oocyte chromosomal abnormalities after IVM in humans [52, 53]. However, IVM can induce meiotic errors in oocytes from aged macaque females [54], suggesting the susceptibility of older females to chromosome errors after IVM, similar to our report. Generally, oocytes have a higher frequency of abnormal meiotic spindles and chromosomal mis-alignment after IVM [55]. Meiotic aneuploidies are the major contributor to the increased rate of embryo aneuploidy with maternal age in humans, and chromosomal instability is common [56]. Our results suggest this deleterious effect may be accentuated in females of advanced reproductive age. The additional contribution of mitotic errors and mosaicism to chromosomal errors in the preimplantation embryo from reproductively aged females will require follow up studies using individual cells from trophectoderm biopsies or defining the sensitivity and specificity of identifying mosaicism within a multi-cell biopsy using cell line mixtures as a model.
Taken together, our results demonstrate that CCS using WGA and NGS is a robust technique to evaluate chromosome segregation errors that can be applied to gametes and embryos of any species with a published and annotated genome. Moreover, this method is highly cost effective, making it accessible for use in most laboratories [49]. Because this technology identifies aneuploidy in a chromosome-specific and quantitative manner, it provides the field of reproductive science and medicine with a new tool for evaluating how age, environmental factors, and disease conditions influence aneuploidy in the gamete and preimplantation embryo. In addition, it could be used in future research to assess the effectiveness of therapeutic interventions to lower the incidence of oocyte and embryo aneuploidy in aged females.
Supplementary Material
Acknowledgment
We thank Professor Angelika Amon and Sarah Pfau (MIT) for providing the Trisomy 16 MEFs. We also acknowledge Dr. Jessica Hornick for technical assistance in processing the mouse egg and polar body samples.
References
- 1. Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev 1996; 8:485–489. [DOI] [PubMed] [Google Scholar]
- 2. Gosden R, Lee B.. Portrait of an oocyte: our obscure origin. J Clin Invest 2010; 120:973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hunt PA, Hassold TJ.. Human female meiosis: what makes a good egg go bad? Trends Genet 2008; 24:86–93. [DOI] [PubMed] [Google Scholar]
- 4. Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT Jr. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril 2014; 101:656–663.e651. [DOI] [PubMed] [Google Scholar]
- 5. Hassold T, Chiu D.. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 1985; 70:11–17. [DOI] [PubMed] [Google Scholar]
- 6. Mantikou E, Wong KM, Repping S, Mastenbroek S.. Molecular origin of mitotic aneuploidies in preimplantation embryos. Biochim Biophys Acta 2012; 1822:1921–1930. [DOI] [PubMed] [Google Scholar]
- 7. Hassold T, Hunt P.. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2:280–291. [DOI] [PubMed] [Google Scholar]
- 8. Johnson JA, Tough S;. Society of Obstetricians and Gynaecologists of Canada. Delayed child-bearing. J Obstet Gynaecol Can 2012; 34:80–93. [DOI] [PubMed] [Google Scholar]
- 9. Mathews TJH. Hamilton BE. First births to older women continue to rise. NCHS Data Brief 2014; 152:1–8. [PubMed] [Google Scholar]
- 10. Broekmans FJ, Soules MR, Fauser BC.. Ovarian aging: mechanisms and clinical consequences. Endocr Rev 2009; 30:465–493. [DOI] [PubMed] [Google Scholar]
- 11. Hassold T, Hunt P.. Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr Opin Pediatr 2009; 21:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keefe D, Kumar M, Kalmbach K.. Oocyte competency is the key to embryo potential. Fertil Steril 2015; 103:317–322. [DOI] [PubMed] [Google Scholar]
- 13. Check JH, Jamison T, Check D, Choe JK, Brasile D, Cohen R.. Live delivery and implantation rates of donor oocyte recipients in their late forties are similar to younger recipients. J Reprod Med 2011; 56:149–152. [PubMed] [Google Scholar]
- 14. Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA.. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 2010; 20:1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chiang T, Schultz RM, Lampson MA.. Age-dependent susceptibility of chromosome cohesion to premature separase activation in mouse oocytes. Biol Reprod 2011; 85:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan FE, Chiang T, Schultz RM, Lampson MA.. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod 2009; 81:768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hornick JE, Duncan FE, Sun M, Kawamura R, Marko JF, Woodruff TK.. Age-associated alterations in the micromechanical properties of chromosomes in the mammalian egg. J Assist Reprod Genet 2015; 32:765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, Jessberger R, Kirkwood TB et al. . Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 2010; 20:1511–1521. [DOI] [PubMed] [Google Scholar]
- 19. Merriman JA, Jennings PC, McLaughlin EA, Jones KT.. Effect of aging on superovulation efficiency, aneuploidy rates, and sister chromatid cohesion in mice aged up to 15 months. Biol Reprod 2012; 86:49. [DOI] [PubMed] [Google Scholar]
- 20. Paczkowski M, Schoolcraft WB, Krisher RL.. Dysregulation of methylation and expression of imprinted genes in oocytes and reproductive tissues in mice of advanced maternal age. J Assist Reprod Genet 2015; 32:713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan H, Ma P, Zhu W, Schultz RM.. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol 2008; 316:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL.. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc Natl Acad Sci U S A 2011; 108:12319–12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shomper M, Lappa C, FitzHarris G.. Kinetochore microtubule establishment is defective in oocytes from aged mice. Cell Cycle 2014; 13:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva E, Greene AF, Strauss K, Herrick JR, Schoolcraft WB, Krisher RL.. Antioxidant supplementation during in vitro culture improves mitochondrial function and development of embryos from aged female mice. Reprod Fertil Dev (in press)Published online ahead of print 5 March 2015 as DOI: 10.1071/RD14474. [DOI] [PubMed] [Google Scholar]
- 25. Yun Y, Holt JE, Lane SI, McLaughlin EA, Merriman JA, Jones KT.. Reduced ability to recover from spindle disruption and loss of kinetochore spindle assembly checkpoint proteins in oocytes from aged mice. Cell Cycle 2014; 13:1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yun Y, Lane SI, Jones KT.. Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development 2014; 141:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirshfeld-Cytron JE, Duncan FE, Xu M, Jozefik JK, Shea LD, Woodruff TK.. Animal age, weight and estrus cycle stage impact the quality of in vitro grown follicles. Hum Reprod 2011; 26:2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eppig JJ, O'Brien M.. In vitro maturation and fertilization of oocytes isolated from aged mice: a strategy to rescue valuable genetic resources. J Assist Reprod Genet 1995; 12:269–273. [DOI] [PubMed] [Google Scholar]
- 29. Chiang T, Schultz RM, Lampson MA.. Meiotic origins of maternal age-related aneuploidy. Biol Reprod 2012; 86:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones KT, Lane SI.. Chromosomal, metabolic, environmental, and hormonal origins of aneuploidy in mammalian oocytes. Exp Cell Res 2012; 318:1394–1399. [DOI] [PubMed] [Google Scholar]
- 31. Jones KT, Lane SI.. Molecular causes of aneuploidy in mammalian eggs. Development 2013; 140:3719–3730. [DOI] [PubMed] [Google Scholar]
- 32. Duncan FE, Hornick JE, Lampson MA, Schultz RM, Shea LD, Woodruff TK.. Chromosome cohesion decreases in human eggs with advanced maternal age. Aging Cell 2012; 11:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Handyside AH. 24 chromosome copy number analysis: a comparison of available technologies. Fertil Steril 2013; 100:595–602. [DOI] [PubMed] [Google Scholar]
- 34. Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, Wigler M, Navin N et al. . Genome-wide copy number analysis of single cells. Nat Protoc 2012; 7:1024–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang YC, Williams BR, Siegel JJ, Amon A.. Identification of aneuploidy-selective antiproliferation compounds. Cell 2011; 144:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Treff NR, Su J, Tao X, Levy B, Scott RT Jr. Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril 2010; 94:2017–2021. [DOI] [PubMed] [Google Scholar]
- 37. Herrick JR, Strauss KJ, Schneiderman A, Rawlins M, Stevens J, Schoolcraft WB, Krisher RL.. The beneficial effects of reduced magnesium during the oocyte-to-embryo transition are conserved in mice, domestic cats and humans. Reprod Fertil Dev 2015; 27:323–331. [DOI] [PubMed] [Google Scholar]
- 38. Paczkowski M, Schoolcraft WB, Krisher RL.. Fatty acid metabolism during maturation affects glucose uptake and is essential to oocyte competence. Reproduction 2014; 148:429–439. [DOI] [PubMed] [Google Scholar]
- 39. Paczkowski M, Silva E, Schoolcraft WB, Krisher RL.. Comparative importance of fatty acid beta-oxidation to nuclear maturation, gene expression, and glucose metabolism in mouse, bovine, and porcine cumulus oocyte complexes. Biol Reprod 2013; 88:111. [DOI] [PubMed] [Google Scholar]
- 40. Cui XF, Li HH, Goradia TM, Lange K, Kazazian HH Jr, Galas D, Arnheim N.. Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci U S A 1989; 86:9389–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ibanez E, Sanfins A, Combelles CM, Overstrom EW, Albertini DF.. Genetic strain variations in the metaphase-II phenotype of mouse oocytes matured in vivo or in vitro. Reproduction 2005; 130:845–855. [DOI] [PubMed] [Google Scholar]
- 42. Jiao ZX, Xu M, Woodruff TK.. Age-associated alteration of oocyte-specific gene expression in polar bodies: potential markers of oocyte competence. Fertil Steril 2012; 98:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knouse KA, Wu J, Whittaker CA, Amon A.. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A 2014; 111:13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stein P, Schindler K.. Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp 2011; (53):e2851,DOI:10.3791/2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Handyside AH, Montag M, Magli MC, Repping S, Harper J, Schmutzler A, Vesela K, Gianaroli L, Geraedts J.. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur J Hum Genet 2012; 20:742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fragouli E, Alfarawati S, Goodall NN, Sanchez-Garcia JF, Colls P, Wells D.. The cytogenetics of polar bodies: insights into female meiosis and the diagnosis of aneuploidy. Mol Hum Reprod 2011; 17:286–295. [DOI] [PubMed] [Google Scholar]
- 47. Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA.. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010; 83:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shoubridge EA, Wai T, Mitochondrial DNA.. and the mammalian oocyte. Curr Top Dev Biol 2007; 77:87–111. [DOI] [PubMed] [Google Scholar]
- 49. Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, Munne S.. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet 2014; 51:553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Su J, Tao X, Baglione G, Treff NR, Scott RT Jr.. Mitochondrial DNA Is Significantly increased in aneuploid human embryos. Fertil Steril 2010; 94:S88–S89. [Google Scholar]
- 51. Wu LL, Russell DL, Wong SL, Chen M, Tsai TS, St John JC, Norman RJ, Febbraio MA, Carroll J, Robker RL.. Mitochondrial dysfunction in oocytes of obese mothers: transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 2015; 142:681–691. [DOI] [PubMed] [Google Scholar]
- 52. Edwards RGIVF. IVM, natural cycle IVF, minimal stimulation IVF - time for a rethink. Reprod Biomed Online 2007; 15:106–119. [DOI] [PubMed] [Google Scholar]
- 53. Requena A, Bronet F, Guillen A, Agudo D, Bou C, Garcia-Velasco JA.. The impact of in-vitro maturation of oocytes on aneuploidy rate. Reprod Biomed Online 2009; 18:777–783. [DOI] [PubMed] [Google Scholar]
- 54. Nichols SM, Gierbolini L, Gonzalez-Martinez JA, Bavister BD.. Effects of in vitro maturation and age on oocyte quality in the rhesus macaque Macaca mulatta. Fertil Steril 2010; 93:1591–1600. [DOI] [PubMed] [Google Scholar]
- 55. Li Y, Feng HL, Cao YJ, Zheng GJ, Yang Y, Mullen S, Critser JK, Chen ZJ.. Confocal microscopic analysis of the spindle and chromosome configurations of human oocytes matured in vitro. Fertil Steril 2006; 85:827–832. [DOI] [PubMed] [Google Scholar]
- 56. Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, Fryns JP, Verbeke G et al. . Chromosome instability is common in human cleavage-stage embryos. Nat Med 2009; 15:577–583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


