Abstract
Background
Predicting death in a cohort of clinically diverse, multicondition hospitalized patients is difficult. Prognostic models that use electronic medical record (EMR) data to determine 1-year death risk can improve end-of-life planning and risk adjustment for research.
Objective
Determine if the final set of demographic, vital sign, and laboratory data from a hospitalization can be used to accurately quantify 1-year mortality risk.
Design
A retrospective study using electronic medical record data linked with the state death registry.
Participants
A total of 59,848 hospitalized patients within a six-hospital network over a 4-year period.
Main Measures
The last set of vital signs, complete blood count, basic and complete metabolic panel, demographic information, and ICD codes. The outcome of interest was death within 1 year.
Key results
Model performance was measured on the validation data set. Random forests (RF) outperformed logisitic regression (LR) models in discriminative ability. An RF model that used the final set of demographic, vitals, and laboratory data from the final 48 h of hospitalization had an AUC of 0.86 (0.85–0.87) for predicting death within a year. Age, blood urea nitrogen, platelet count, hemoglobin, and creatinine were the most important variables in the RF model. Models that used comorbidity variables alone had the lowest AUC. In groups of patients with a high probability of death, RF models underestimated the probability by less than 10%.
Conclusion
The last set of EMR data from a hospitalization can be used to accurately estimate the risk of 1-year mortality within a cohort of multicondition hospitalized patients.
Electronic supplementary material
The online version of this article (10.1007/s11606-018-4316-y) contains supplementary material, which is available to authorized users.
KEY WORDS: machine learning, hospital outcomes, predictive models, data mining
INTRODUCTION
During hospitalizations, seriously ill patients are frequently exposed to unwanted interventions.1 Surveys of seriously ill hospitalized patients have found that communication about end-of-life planning (EOLp) is an area of potential improvement for hospitals.2 An informed EOLp discussion is based on an accurate estimation of a patient’s likelihood of death. Estimates of the probability of patient survival are also needed to adjust for the risk of death as a confounder. This is useful in comparing outcomes between hospitals.3 Some commonly used prognostic models are applicable to specific diseases and subpopulations (e.g., the MELD-Na score in end-stage liver disease). Some studies have used Electronic Health Record (EHR) data to estimate inpatient mortality risk.4,5 However, there is a lack of prognostic models in diverse, multicondition, hospitalized patients for estimating longer term outcomes, such as mortality at 1 year.6
Estimating the risk of 1-year mortality in a clinically heterogeneous cohort of hospitalized patients is difficult.3 Existing prognostic models in this area frequently rely on variables that may not be available to clinicians.6,7 Some examples of such variables are comorbidity scores based on billing data, Activities of Daily Living assessments, and the number of hospitalizations prior to the index hospitalization.6 As an example, the widely used Charlson score relies on billing data and has been shown to have poor reproducibility.6 It is frequently not available for weeks after the hospitalization ends and needs manual abstraction by chart review. Reliance on such variables has limited the broad automation, utilization, and deployment of prognostic models at the bedside. Ideally, a prognostic model would rely solely on clinical EMR data. Clinical EMR data (i.e., vitals signs, common laboratory values, clinical and demographic information) are ubiquitously available at point of care and amenable to automated abstraction. Such an approach will facilitate the deployment of automated, EHR-interfaced models for decision support.8
During an emergent hospitalization, patients are typically admitted with an acute alteration in physiology.9,10 It is reasonable to expect that patients, who have significant abnormalities in vital signs and biochemistry at discharge will have worse outcomes. Instability in vital signs at discharge is associated with a higher 30-day readmission rate.11,12 Nguyen et al. found that models utilizing granular EHR data from a hospitalization exhibited modest performance in predicting readmission.11 Their study did not model 1-year mortality. A predictive model that uses commonly available EHR data to accurately prognosticate 1-year mortality in diverse, multicondition, hospitalized patients remains elusive.3,6 In this study, we aim to develop models that estimate the 1-year mortality risk based on EHR data available at the end of a hospitalization.
Machine learning (ML) is a field of computer science that develops techniques to learn and extract knowledge from data. ML algorithms can deal with complex multivariate data and model non-linear relationships and are able to handle missing or noisy data.13 ML is very promising from the standpoint of handling EHR data, which is frequently “dirty.”14 Our proof-of-concept study aims to construct models that utilize EHR data to prognosticate 1-year mortality in a large, diverse cohort of multicondition hospitalizations. Our study also answers the following questions: (1) Do ML approaches outperform regression-based models? (2) Which variables are most important for prognostication?
METHODS
Inclusion Criterion
After obtaining institutional review board approval, we used our institution’s clinical data warehouse to create an EMR-derived data set of 98,643 emergent hospital admissions for 59,848 patients within a six-hospital network in the Twin Cities area, Minnesota. The encounters spanned a 4-year period ranging from 2012 to 2016. The hospital system consists of one 450-bed university tertiary care center and 5 community hospitals ranging from 100 to 450 beds in capacity. Patients were excluded if they were non-emergent admissions, less than 18 years of age, did not consent to their medical record being used for research purposes, or had less than a year of follow-up mortality data. We included hospitalizations to all units and services as long as they met the above criterion. Vital status and death dates were obtained from the state death registry. Our database had the complete death record issued from 2011 onwards for deceased individuals who were born in Minnesota, had died in Minnesota, or have ever had a permanent address in the state.
Model Variables
Our data set included four broad classes of variables (features) that were very commonly available in the EMR from most hospitalizations and were clinically relevant. (1) Demographic variables: age, length of stay, sex. (2) Physiologic variables: systolic blood pressure, diastolic blood pressure, pulse, respiratory rate, temperature, pulse-oximetry, and body mass index. (3) Biochemical variables: Serum sodium, potassium, chloride, bicarbonate, creatinine, urea nitrogen, ALT, AST, total bilirubin, albumin, white blood cell count, hematocrit, hemoglobin, platelet count, and mean corpuscular volume. (4) Clinical comorbidity variables: We created a comorbidity profile for each patient across the 30 classes of diseases in the AHRQ comorbidity category index from ICD codes billed during an encounter.15
All laboratory and physiologic data were time-stamped and obtained within 48 h of the end of the hospitalization. For each data element, the last available measurement during an encounter was used in the model. The primary outcome of interest was predicting whether death occurred within a year of the end of the hospitalization.
Missing Data
We tested two imputation strategies to deal with missing data. The first was the k-nearest neighbor approach, which replaced missing data in an encounter with the values of its nearest neighbor based on a distance measure.13 The second was the median imputation approach where missing values for a variable were replaced with median values for the variable.13,16 The two approaches did not change model performance significantly. 16 Due to its simplicity and fast computation time, the median imputation was used.
Data Set Partitioning
The data set was partitioned into a derivation and a validation data set with encounters selected randomly at a ratio of 0.80/0.20.
Modeling
We compared the performance of logistic regression (LR) to a class of ML models known as random forest models (RF).13,17 A more detailed explanation of RFs can be obtained by referring to existing reviews on this methodology.18 RFs are known for their superior “out of box” performance, are able to handle non-linear data, and are less prone to over-fitting. 13,19 RFs are based on decision trees. Decision tree algorithms formulate decision rules to fit the underlying data. However, decision trees are frequently “unstable” and are sensitive to minor alterations in the data. RFs aggregate the results of many different decision trees to eliminate this instability. 13,19,20 RFs utilize two basic strategies to achieve this objective. (1) The algorithm utilizes a random subset of the training data to build each new tree in the ensemble. (2) A random subset of features is utilized for constructing each decision rule in a tree. This approach avoids introducing an inordinate degree of bias in the classification, stemming from a few influential observations.13,19 Variable importance is interpreted in RFs by using an importance measure known as the ‘mean decrease in the Gini Index’ (MDGini). MDGini measures a variable’s performance by randomly permuting it and measuring the resultant change in classification error.13,21 For each RF classifier, 501 trees were used in the ensemble in our analysis. The mtry parameter, which is the number of variables randomly sampled as candidates at each split, was sqrt(p) where p is number of variables in the model. We used the RF implementation from the ‘randomForest’ package in R for our analysis.
Statistical Tests
For non-normal variables, median values with interquartile range (IQR) were reported. Mean with standard deviation (SD) was reported for normal variables. The significance of comparisons between two non-normal continuous variables was tested using the Wilcoxon test. For comparisons between two categorical variables the Fisher Test was used. The Hosmer-Lemeshow test was used to assess the goodness of fit.
Model Validation and Testing
The discriminative performance of the models was measured by constructing receiver-operator curves (ROC) and calculating the area under the curve (AUC) on the validation data set.13 In clinical studies, the AUC gives the probability that a randomly selected patient who experienced an event (e.g., a disease or condition) had a higher risk score than a patient who had not experienced the event. It is equal to the area under the receiver-operating characteristic (ROC) curve and ranges from 0.5 to 1.13 The 95% confidence intervals around the AUC estimates were estimated using the DeLong method, which is implemented in the pROC package in R.22 To evaluate whether the predicted probability of 1-year mortality from the random forest model reflected the observed probabilities, we constructed model calibration plots using the PresenceAbsence package in R. In a perfectly calibrated model, all the points would fall along the diagonal straight line.
RESULTS
Characteristics of the Cohort
The demographic, physiologic, and laboratory characteristics of the encounters are shown in Table 1. In 15.1% percent of the hospitalizations, death occurred within a year of the last day of hospitalization (Table 1). The median age, body mass index (BMI), length of stay, creatinine, blood urea nitrogen (BUN), mean corpuscular volume (MCV), white blood count (WBC), respiratory rate, and pulse were significantly higher in hospitalizations that were followed by death within a year (Table 1). The albumin, hemoglobin, and pulse-oximetry readings were significantly lower at the end of the hospitalizations in which patients died within a year (Table 1). The distribution of comorbidities in the cohort is shown in online supplementary Table 1.
Table 1.
Demographic, Laboratory, and Physiologic Characteristics of the Cohort
| Stratified by death within 1 year | False | True | p Value |
|---|---|---|---|
| Number of hospitalizations | 83,652 | 14,991 | |
| Age (years, median [IQR]) | 59.85 [43.56, 75.07] | 73.32 [60.49, 84.63] | < 0.001 |
| BMI (kg/m2, median [IQR]) | 27.19 [23.24, 32.31] | 25.51 [22.02, 30.05] | < 0.001 |
| Sex = male (number, %) | 39,610 (47.4) | 7474 (49.9) | < 0.001 |
| Race = white (number, %) | 69,924 (83.6) | 13,165 (87.8) | 0.028 |
| AST (U/l, median [IQR]) | 28.00 [22.00, 47.00] | 37.00 [23.00, 74.00] | < 0.001 |
| ALT (U/l, median [IQR]) | 31.00 [20.00, 44.00] | 33.00 [22.00, 56.00] | < 0.001 |
| Bilirubin.total (mg/dl, median [IQR]) | 0.60 [0.40, 0.90] | 0.70 [0.40, 1.60] | < 0.001 |
| Albumin (g/dl, median [IQR]) | 3.50 [3.00, 4.00] | 2.80 [2.30, 3.20] | < 0.001 |
| Carbon dioxide (mEq/l, median [IQR]) | 26.00 [24.00, 28.00] | 26.00 [23.00, 29.00] | 0.519 |
| Chloride (mEq/l, median [IQR]) | 105.00 [102.00, 108.00] | 104.00 [100.00, 108.00] | < 0.001 |
| Creatinine (mg/dl, median [IQR]) | 0.82 [0.67, 1.05] | 1.00 [0.71, 1.56] | < 0.001 |
| Glucose (mg/dl, median [IQR]) | 101.00 [90.00, 122.00] | 107.00 [91.00, 135.00] | < 0.001 |
| Potassium (3.5–5.1 mEq/l, median [IQR]) | 4.00 [3.70, 4.20] | 4.00 [3.70, 4.40] | < 0.001 |
| Sodium (mEq/l, median [IQR]) | 139.00 [137.00, 141.00] | 138.00 [135.00, 141.00] | < 0.001 |
| Urea nitrogen (mg/dl,median [IQR]) | 14.00 [10.00, 20.00] | 22.00 [14.00, 36.00] | < 0.001 |
| Hemoglobin (mmol/l, median [IQR]) | 11.80 [10.10, 13.40] | 9.90 [8.70, 11.40] | < 0.001 |
| MCV (femtoliters/cell, median [IQR]) | 90.00 [87.00, 94.00] | 92.00 [88.00, 97.00] | < 0.001 |
| Platelet count (× 109/l, median [IQR]) | 209.00 [162.00, 265.00] | 177.00 [114.00, 249.00] | < 0.001 |
| WBC (× 109 per liter (l), median [IQR]) | 7.50 [5.70, 9.70] | 8.20 [5.80, 11.80] | < 0.001 |
| Length of stay (days, median [IQR]) | 3.00 [2.00, 4.00] | 4.00 [2.00, 7.00] | < 0.001 |
| Discharge disposition (%) | < 0.001 | ||
| Acute rehab facility | 1132 (1.4) | 160 (1.1) | |
| Expired in the hospital | 26 (0.0) | 2713 (18.1) | |
| Home IV drug therapy | 857 (1.0) | 158 (1.1) | |
| Home or self care | 54,623 (65.3) | 3877 (25.9) | |
| Home-health care svc | 8674 (10.4) | 2369 (15.8) | |
| Hospice/home | 356 (0.4) | 852 (5.7) | |
| Left against medical advice | 922 (1.1) | 48 (0.3) | |
| Psychiatric hospital | 1054 (1.3) | 23 (0.2) | |
| Short-term hospital | 1878 (2.2) | 363 (2.4) | |
| Skilled nursing facility | 11,964 (14.3) | 3693 (24.6) | |
| Other | 2166 (2.6) | 735 (4.9) | |
| Systolic blood pressure, mmHg (median [IQR]) | 126.00 [114.00, 140.00] | 122.00 [107.00, 138.00] | < 0.001 |
| Diastolic blood pressure, mmHg (median [IQR]) | 72.00 [63.00, 81.00] | 67.00 [57.00, 77.00] | < 0.001 |
| Temperature (Fahrenheit, median [IQR]) | 98.00 [97.50, 98.40] | 97.90 [97.40, 98.40] | 0.007 |
| Pulse (beats per minute, median [IQR]) | 78.00 [68.00, 89.00] | 83.00 [71.00, 96.00] | < 0.001 |
| Pulse oxymetry (%, mean [SD]) | 96.14 (2.87) | 93.51 (10.44) | < 0.001 |
| Respiratory rate (breaths per minute, median [IQR]) | 16.00 [16.00, 18.00] | 18.00 [16.00, 20.00] | < 0.001 |
Cohort characteristics. The table is stratified by whether death occurred within a year of the end of the hospitalization. Interquartile ranges are listed in the parentheses. Median values are reported for non-normal variables. The Wilcox test is used for comparisons between the non-normal continuous variables, and the Fisher test is used for comparisons between categorical variables. IQR: Interquartile range. SD: Standard deviation
Predictive Performance of Models for Death Within 1-Year of the End of a Hospitalization
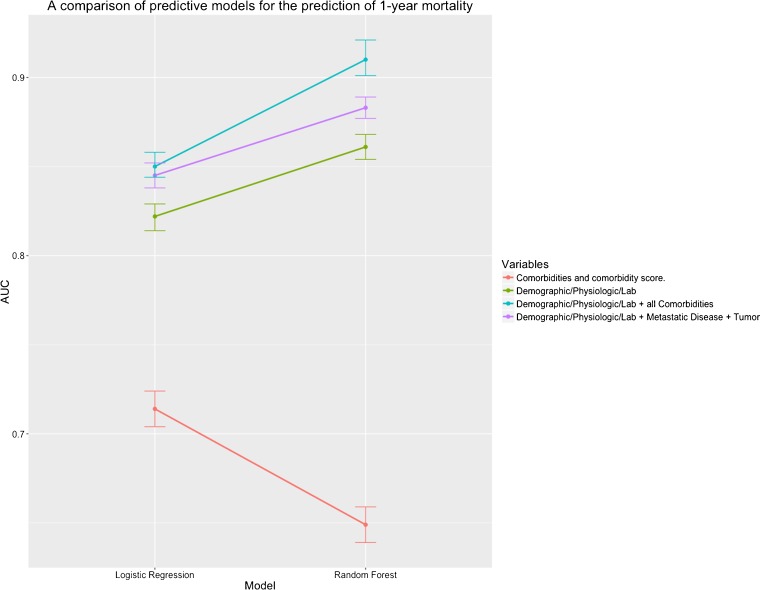
Models with all four classes of variables had the highest AUC (blue line, Fig. 1 and online supplementary Table 2). Models with clinical-comorbidity variables alone had the worst performance (red line, Fig. 1 and online supplementary Table 2). RF models generally outperformed the LR models (Fig. 1 and online supplementary Table 2). The AUC of models when inpatient death was excluded was slightly lower (online supplementary Table 2). Metastatic disease and tumors increased the model AUC slightly but significantly when added to the Demographic/Physiologic/Lab model (Fig. 1, purple and green lines, online supplementary Table 2).
Figure 1.
Line chart of model AUCs for predicting 1-year mortality. The AUC of each model on the validation data set is plotted on the vertical (y-axis) and the model type is indicated on the horizontal (x-axis). The variables incorporated into each model are listed in the color-coded legend on the right hand side of the figure. The vertical error bars show the 95% confidence intervals around each AUC estimate. Each point represents the AUC of one model.
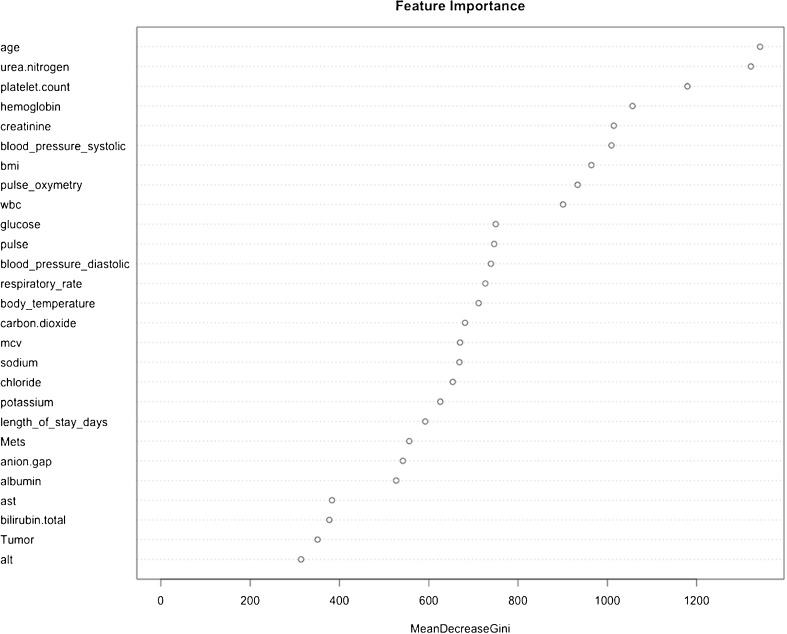
Variable Importance in the Random Forest Variables
The highest ranking 27 features are shown in Fig. 1. Twenty-five of the top 27 features are physiologic, laboratory, and demographic variables (Fig. 2). Among the comorbidity variables, metastatic disease and tumor were the highest ranking (Fig. 2).
Figure 2.
Feature importance in the random forest models. The features are ranked by importance as measured by the mean decrease in the Gini Index.
Model Calibration
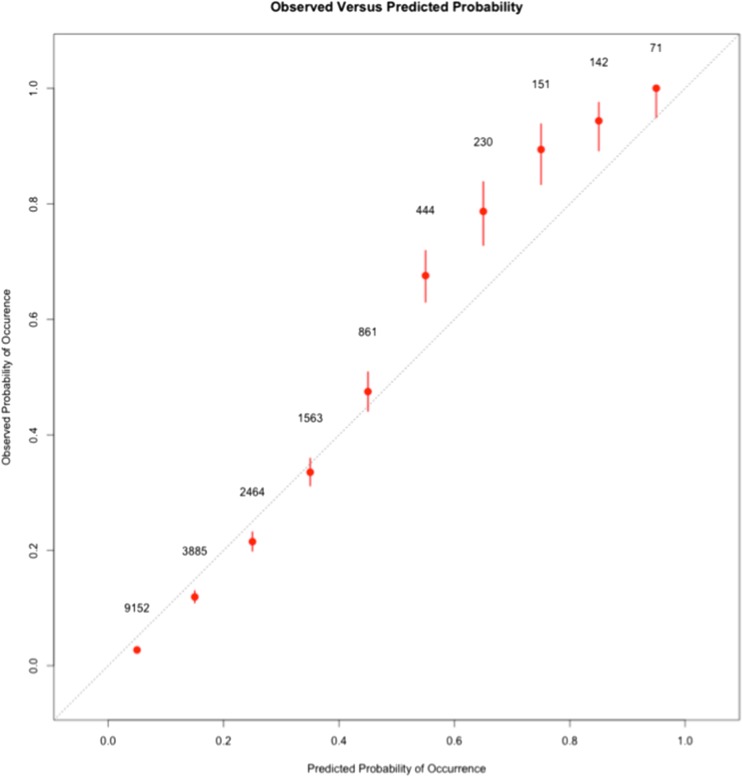
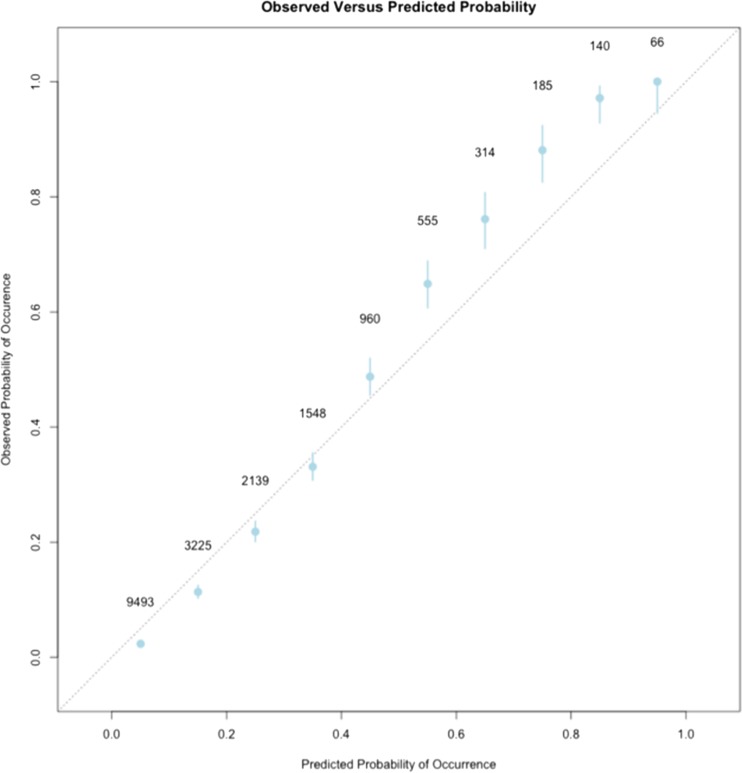
The Hosmer-Lemeshow statistic for the Demographic/Physiologic/Lab model was significant (p < 0.001, 10 bins) indicating that the predicted probabilities deviated from the observed probabilities within certain probability ranges. The calibration of a model is a measure of how well the probabilities estimated by the model reflect the observed probabilities. A calibration plot of the RF models revealed that when the probability of death was greater than 45%, the predicted probability was slightly lower than the observed probability (Figs. 3 and 4, online supplementary Tables 3 and 4). Similarly, at low probabilities of death, the model slightly overestimated the probability of death (Figs. 3 and 4, online supplementary Tables 3 and 4). The difference between predicted probability and the actual probability of death was always less than 10% (Figs. 3 and 4, online supplementary Tables 3 and 4.
Figure 3.
Calibration plots of the Demographic/Physiologic/Lab RF model. The observed rate of death at 1 year within each one of the ten probability bins is plotted on the y-axis. The predicted probability from the RF model is indicated on the x-axis. The dotted diagonal line represents points along a perfectly calibrated model. Each point on the graph represents one of the ten bins of probability. The number beside each point represents the total number of hospitalizations that fall within the particular bin. The bars delineate the 95% confidence intervals around the observed probability.
Figure 4.
Calibration plot of the Demographic/Physiologic/Lab + Metastasis + Tumor RF model. The observed rate of death at 1 year within each one of the ten probability bins is plotted on the y-axis. The predicted probability from the RF model is indicated on the x-axis. The dotted diagonal line represents points along a perfectly calibrated model. Each point on the graph represents one of the ten bins of probability. The number beside each point represents the total number of hospitalizations that fall within the particular bin. The bars delineate the 95% confidence intervals around the observed probability.
Addressing Potential Selection Bias
We repeated the model development and testing using one of two approaches: Approach 1: In this approach, each distinct hospitalization was treated as a unit of analysis. We used the last set of data from each available hospitalization for each patient. Approach 2: In this approach, each unique patient was treated as a unit of analysis. The data set was sampled and one hospitalization for each patient was randomly selected for inclusion analysis (i.e., random admission model). This was done to test the effect of potential selection bias that could be theoretically introduced by using multiple data points from the same patient.23 Both these strategies yielded models with nearly identical AUCs and predictive performance.
DISCUSSION
In our proof-of-concept study, we demonstrate that the last set of EMR data from the end of a hospitalization (vital signs, laboratory tests, and demographic information) can accurately estimate the probability of death within a year. EMR data are “dirty” with a significant amount of missing and erroneous data. This makes it challenging to develop accurate models.8,24 Our approach relies on a relatively simple imputation strategy to deal with missing data. We use an ML algorithm (i.e., RF) capable of handling “noisy” data. Our results highlight the effectiveness of this approach for utilizing EMR data in prognostic models.
We achieve an excellent discriminative performance for predicting 1-year mortality by constructing an RF model that utilizes Demographic/Laboratory/Physiologic variables. The AUC of the model is 0.86 on the validation data set (Fig. 1, online supplementary Table 2). This is one of the highest AUCs described in multicondition, diverse, hospitalized patients for predicting 1-year mortality.6 Adding the two highest ranking comorbidity variables (i.e., tumor and metastatic disease) to this model increases the AUC by a small amount (Fig. 1, online supplementary Table 2). However, the calibration curves for both these RF models are similar, yielding identical probabilities of death (Figs. 3 and 4, online supplementary Tables 3 and 4). At high probabilities of death, both the RF models underestimate the risk of death at 1 year by less than 10%. Compared to existing models in multicondition hospitalized patients, these are well-calibrated models.6 The AUC of the RF models is slightly lower when in-patient deaths are excluded from the outcome. However, this degradation in discriminative performance is not large enough to be clinically meaningful (online supplementary Table 2). The Demographic/Laboratory/Physiologic/Tumor/Metastasis model is web-deployed at https://niceguy.shinyapps.io/shiny_model2/ for purposes of demonstration.
Age, BUN, platelet count, hemoglobin, creatinine, systolic blood pressure, BMI, and pulse oxymetry readings are the most important variables in the RF models (MD Gini score of greater than 800). The majority of the predictive performance of the RF model is derived from physiologic, biochemical, and demographic variables. The clinical comorbidity variables are less important (Figs. 1 and 2). This is not surprising, as the billing data-based comorbidity index does not capture the granularity of the physiologic/biochemical aspects of disease. Although the Demographic/Laboratory/Physiologic RF model has a very good discrimination and calibration, the highest AUC is achieved when all the comorbidity variables are added to this model (AUC 0.91, Fig. 1 and online supplementary Table 2). It is worth highlighting the superior discriminative performance of RF models compared to LR models (Fig. 1, online supplementary Table 2). Unlike LRs, RFs are able to capture non-linear relationships and deal with “noisy” data.20 However, one drawback of using RF models is that the interpretation of variable importance is lost (i.e., there are no odds ratios to interpret for variables).
Our models were developed and validated on a demographically, economically, and clinically diverse cohort. Our data set includes data from a large multihospital health system. The system encompasses a university tertiary care center and urban, suburban and semi-rural hospitals. Ultimately, our model needs to be validated in other settings to demonstrate geographic and temporal portability.6,7 We used state death registry data for ascertaining the date of death (for out-of-hospital deaths). If a death were not reported to the Minnesota state registry, then it would not be captured in our data set. To minimize the impact of this issue, we included only emergent hospitalizations in our analysis. This may result in our data reflecting a “sicker” subset of patients than if elective admissions were also included. It is likely that if we had included non-emergent hospitalizations, the discriminative performance would have been better (since our model performs very well at identifying low-risk subgroups).
To the best of our knowledge, our study is the first and largest study attempting to develop models for 1-year mortality by using granular EHR data at the end of a hospitalization.6,25 Prior attempts at developing a 1-year mortality model in diverse, multicondition hospitalized patients have suffered from a number of drawbacks.6 They have relied on administrative, non-clinical data that frequently need to be manually abstracted.3 Model development has been limited to specific subpopulations of patients.6 These issues have limited model use in clinical settings.6 We utilize variables directly available to clinicians from the EHR. Compared to previous studies of multicondition hospitalized patients, our cohort is significantly larger and clinically diverse.6 Prior models have reported AUCs in the 0.7–0.8 range for 1-year mortality.6,3 Our models demonstrate excellent discrimination and calibration in comparison. Models that are based on Demographic/Laboratory/Physiologic variables can be interfaced with the EHR using common messaging standards such as HL7 to “pull” structured data elements. This can facilitate automated deployment for decision support.14
Our work demonstrates that ML approaches such as RF can utilize ubiquitously available granular EMR elements from the end of a hospitalization to accurately estimate the risk of 1-year mortality in a large heterogeneous multicondition cohort of hospitalized patients. Future work is needed to validate such models in other settings to test whether this approach is widely portable.
Electronic supplementary material
(DOCX 494 kb)
Acknowledgements
We thank Zohara Cohen, MS, and Justin Dale for help with the technical aspects of data management. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- AST
Aspartate amino transferase
- ALT
Alanine amino transferase
- AHRQ
Agency for Health Care Research and Quality
- EOL
End of life
- EOLp
End of life planning
- EMR
Electronic medical record
- MCV
Mean corpuscular volume
- WBC
White blood cell count
- CMP
Complete metabolic panel
- CBC
Complete blood count
- BMP
Basic metabolic panel
- RF
Random forest
- LR
Likelihood ratio
- LR
Logistic regression
- SD
Standard deviation
- AUC
Area under the curve
- ROC
Receiver-operator curve
- OOB
Out of bag
- ICD9-CM
International Classification of Diseases 9-Clinical Modification
- ICD10
International Classification of Diseases 10
- MD-Gini
Mean decrease in Gini Index
- ML
Machine learning
Compliance with Ethical Standards
Conflicts of Interest
The authors report no conflicts of interest in this work.
References
- 1.Frost DW, Cook DJ, Heyland DK, Fowler RA. Patient and healthcare professional factors influencing end-of-life decision-making during critical illness: a systematic review*. Crit Care Med. 2011;39(5):1174–1189. doi: 10.1097/CCM.0b013e31820eacf2. [DOI] [PubMed] [Google Scholar]
- 2.You JJ, Downar J, Fowler RA, et al. Barriers to goals of care discussions with seriously ill hospitalized patients and their families: a multicenter survey of clinicians. JAMA Intern Med. 2015;175(4):549–556. doi: 10.1001/jamainternmed.2014.7732. [DOI] [PubMed] [Google Scholar]
- 3.Van Walraven C, McAlister FA, Bakal JA, Hawken S, Donzé J. External validation of the Hospital-patient One-year Mortality Risk (HOMR) model for predicting death within 1 year after hospital admission. CMAJ. 2015;187(10):725–733. doi: 10.1503/cmaj.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabak YP, Sun X, Nunez CM, Johannes RS. Using electronic health record data to develop inpatient mortality predictive model: Acute Laboratory Risk of Mortality Score (ALaRMS) J Am Med Informatics Assoc. 2014;21(3):455–463. doi: 10.1136/amiajnl-2013-001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escobar GJ, Greene JD, Scheirer P, Gardner MN, Draper D, Kipnis P. Risk-Adjusting Hospital Inpatient Mortality Using Automated Inpatient, Outpatient, and Laboratory Databases. Med Care. 2008;46(3):232–239. doi: 10.1097/MLR.0b013e3181589bb6. [DOI] [PubMed] [Google Scholar]
- 6.Yourman LC, Lee SJ, Schonberg MA, Widera EW, Smith AK. Prognostic indices for older adults: a systematic review. JAMA. 2012;307(2):182–192. doi: 10.1001/jama.2011.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Walraven C. The Hospital-patient One-year Mortality Risk score accurately predicted long-term death risk in hospitalized patients. J Clin Epidemiol. 2014;67(9):1025–1034. doi: 10.1016/j.jclinepi.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Hripcsak G, Bloomrosen M, FlatelyBrennan P, et al. Health data use, stewardship, and governance: ongoing gaps and challenges: a report from AMIA’s 2012 Health Policy Meeting. J Am Med Inform Assoc. 2013;21(2):204–211. doi: 10.1136/amiajnl-2013-002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fialho AS, Cismondi F, Vieira SM, Reti SR, Sousa JMC, Finkelstein SN. Data mining using clinical physiology at discharge to predict ICU readmissions. Expert Syst Appl. 2012;39(18):13158–13165. doi: 10.1016/j.eswa.2012.05.086. [DOI] [Google Scholar]
- 10.Rothman MJ, Rothman SI, Beals J. Development and validation of a continuous measure of patient condition using the Electronic Medical Record. J Biomed Inform. 2013;46(5):837–848. doi: 10.1016/j.jbi.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen OK, Makam AN, Clark C, et al. Predicting all-cause 30-day hospital readmissions using electronic health record data over the course of hospitalization: Model derivation, validation and comparison. J Gen Intern Med. 2015;30(0):S231. 10.1002/jhm.2568.
- 12.Nguyen OK, Makam AN, Clark C, et al. Vital Signs Are Still Vital: Instability on Discharge and the Risk of Post-Discharge Adverse Outcomes. J Gen Intern Med. 2017;32(1):42–48. doi: 10.1007/s11606-016-3826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. Vol 103.; 2013. 10.1007/978-1-4614-7138-7.
- 14.Hripcsak G, Albers DJ. Correlating electronic health record concepts with healthcare process events. J Am Med Inform Assoc. 2013;20(e2):e311–8. doi: 10.1136/amiajnl-2013-001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43(11):1130–1139. doi: 10.1097/01.mlr.0000182534.19832.83. [DOI] [PubMed] [Google Scholar]
- 16.Hu Z, Melton GB, Arsoniadis EG, Wang Y, Kwaan MR, Simon GJ. Strategies for handling missing clinical data for automated surgical site infection detection from the electronic health record. J Biomed Inform. 2017;68:112–120. doi: 10.1016/j.jbi.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L. Random forest. Mach Learn. 1999;45(5):1–35. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 18.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition (Springer Series in Statistics) (9780387848570): Trevor Hastie, Robert Tibshirani, Jerome Friedman: Books. In: The Elements of Statistical Learning: Dta Mining, Inference, and Prediction. ; 2011:501–520. http://www.amazon.com/Elements-Statistical-Learning-Prediction-Statistics/dp/0387848576/ref=sr_1_14?ie=UTF8&qid=1429565346&sr=8-14&keywords=machine+learning.
- 19.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 20.Shi T, Horvath S. Unsupervised Learning With Random Forest Predictors. J Comput Graph Stat. 2006;15(1):118–138. doi: 10.1198/106186006X94072. [DOI] [Google Scholar]
- 21.Grömping U. Variable Importance Assessment in Regression: Linear Regression versus Random Forest. Am Stat. 2009;63(4):308–319. doi: 10.1198/tast.2009.08199. [DOI] [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 23.Krumholz HM, Wang Y, Mattera JA, et al. An administrative claims model suitable for profiling hospital performance based on 30-day mortality rates among patients with heart failure. Circulation. 2006;113(13):1693–1701. doi: 10.1161/CIRCULATIONAHA.105.611194. [DOI] [PubMed] [Google Scholar]
- 24.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen OK, Makam AN, Clark C, et al. Predicting all-cause readmissions using electronic health record data from the entire hospitalization: Model development and comparison. J Hosp Med. 2016;11(7):473–480. doi: 10.1002/jhm.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 494 kb)