SUMMARY
BCR/ABL1 -like acute lymphoblastic leukaemia (ALL) is a subgroup of B-lineage acute lymphoblastic leukaemia that occurs within cases without recurrent molecular rearrangements. Gene expression profiling (GEP) can identify these cases but it is expensive and not widely available. Using GEP, we identified 10 genes specifically overexpressed by BCR/ABL1-like ALL cases and used their expression values - assessed by quantitative real time-polymerase chain reaction (Q-RT-PCR) in 26 BCR/ABL1-like and 26 non-BCR/ABL1-like cases to build a statistical “BCR/ABL1-like predictor”, for the identification of BCR/ABL1-like cases. By screening 142 B-lineage ALL patients with the “BCR/ABL1-like predictor”, we identified 28/142 BCR/ABL1-like patients (19.7%). Overall, BCR/ABL1-like cases were enriched in JAK/STAT mutations (p<0.001), IKZF1 deletions (p<0.001) and rearrangements involving cytokine receptors and tyrosine kinases (p=0.001), thus corroborating the validity of the prediction.
Clinically, the BCR/ABL1-like cases identified by the BCR/ABL1-like predictor achieved a lower rate of complete remission (p=0.014) and a worse event-free survival (p=0.0009) compared to non-BCR/ABL1-like ALL. Consistently, primary cells from BCR/ABL1-like cases responded in vitro to ponatinib.
We propose a simple tool based on Q-RT-PCR and a statistical model that is capable of easily, quickly and reliably identifying BCR/ABL1-like ALL cases at diagnosis.
Keywords: Acute lymphoblastic leukaemia, BCR/ABL1-like, adults, prognosis, tyrosine kinase inhibitors
INTRODUCTION
Among B-lineage acute lymphoblastic leukaemia (ALL), BCR/ABL1-like ALL is one of the most clinically relevant subsets because it is characterized by a poor outcome and could potentially benefit from the use of tyrosine kinase inhibitor (TKI) therapy (Ofran & Izraeli, 2017). Mullighan et al (2009a) identified a subgroup of paediatric B-lineage ALL (B-ALL) with a gene expression profile similar to that of BCR/ABL1-positive patients, frequent IKZF1 deletions and poor outcome. Simultaneously, Den Boer et al (2009) performed gene expression profiling (GEP) analysis on a cohort of paediatric B-ALL cases and termed this subgroup BCR/ABL1-like; it represented 15–20% of B-ALL cases, showed an unfavourable outcome and was associated with IKZF1, TCF3, EBF1, PAX5 and VPREB1 deletions, and upregulation of CRLF2.
Subsequently, the BCR/ABL1-like ALL subgroup has been extensively evaluated, particularly in paediatric cohorts (Mullighan et al, 2009b; Mullighan et al, 2009c; Harvey et al, 2010a; Harvey et al, 2010b; Yoda et al, 2010; Chen et al, 2012; van der Veer et al, 2013; Asai et al, 2013) and more recently in adults (Tokunaga et al, 2013; Boer et al, 2015a; Herold et al, 2017; Roberts et al, 2017; Jain et al, 2017a). In addition to the association with IKZF1 deletions and CRLF2 deregulation, Roberts et al (2012) unveiled that kinase activating alterations characterize the majority (91%) of paediatric BCR/ABL1-like ALL cases. The most frequent alterations involve ABL1, JAK2, PDGFRB, CRLF2 and EPOR, activating mutations of IL7R, FLT3-internal tandem duplication mutations and deletion of SH2B3 (Roberts et al, 2012; Roberts et al, 2014a). Several of these alterations are targeted by TKIs, suggesting a potential role for tailored treatment (Roberts et al, 2012; Roberts et al, 2014a). Furthermore, all authors confirmed an association with poor outcome, while the relationship with minimal residual disease (MRD) is still debated (Roberts et al, 2014b; Heatley et al, 2017).
Despite this in-depth genetic characterization, the identification of BCR/ABL1-like patients is still challenging: it relies on GEP and/or a multistep approach, such as a combination of next generation sequencing (NGS) and fluorescence in situ hybridisation (FISH) (Roberts et al, 2014a). However, GEP is largely cohort/experiment-dependent and a consensus diagnostic signature has not been agreed upon (Boer et al, 2015b). Similarly, NGS is costly and requires bioinformatic skills. Alternatively, these cases can be recognized with Low Density Assay (LDA) of selected genes, but the full methodology has not been published (Harvey et al, 2013). Thus, this study aimed to: i) produce an easy, rapid and reproducible assay, based on quantitative real time-polymerase chain reaction (Q-RT-PCR) analysis, for the recognition of BCR/ABL1-like ALL cases; ii) define the molecular background, clinico-biological features and outcome of the cases thus identified; and iii) verify the in vitro response of primary BCR/ABL1-like ALL cells to the pan-TKI, ponatinib. The final goal is to provide a rapid, user-friendly and economically-viable diagnostic tool that can recognize these cases at presentation, a step towards a refined prognostic and therapeutic management of these poor prognosis patients.
METHODS
Identification of BCR/ABL1-like cases and genes
To identify the core BCR/ABL1-like cases and BCR/ABL1-like specific genes (Figure S1), we used two in house GEP cohorts: GEP1, run on HG-U133 plus2 (Affymetrix, Santa Clara, CA, USA) - comprising 148 B-ALL cases (70 BCR/ABL1-positive and 78 B-NEG, i.e. negative for BCR/ABL1, ETV6/RUNX1, TCF3/PBX1 and KMT2A rearrangements) (Haferlach et al, 2010; Messina et al, 2010) - and GEP2 - including 79 cases (37 BCR/ABL1-positive and 42 B-NEG) (Chiaretti et al, 2005) - evaluated with an older version of the array (HG-U95 Av2, Affymetrix). In both GEP1 and GEP2, a t-test between BCR/ABL1-positive and B-NEG cases was performed to recognize the BCR/ABL1-like ALL cases. B-NEG cases clustering within the BCR/ABL1-positive cluster were regarded as the “core” BCR/ABL1-like samples.
To select the BCR/ABL1-like genes (Figure S2), the 16 “core” BCR/ABL1-like ALL cases of GEP1 were compared with the remaining 62 B-NEG cases by t-test. The genes selected by this approach were compared with the literature (Harvey et al, 2010b) and the overlapping genes, together with CRLF2 (Yoda et al, 2010; van der Veer et al, 2013; Herold et al, 2017; Chiaretti et al, 2016), were used to build the Q-RT-PCR-based BCR/ABL1-like predictor. GEP analyses are detailed in Supplementary material.
Development and validation of the “BCR/ABL1-like predictor”
To generate the BCR/ABL1-like predictor, we validated the expression levels of the selected genes in the discovery cohort, including 26 core BCR/ABL1-like samples (16 from GEP1 and 10 from GEP2) and 26 non-BCR/ABL1-like selected from GEP1. The quantification of transcript levels was performed by Q-RT-PCR (Supplementary material and Table S1) and computed as 2−ΔCt.
Q-RT-PCR results were used to build the “BCR/ABL1-like predictor”, extensively described in the Supplementary material. Briefly, expression values were shrunk into principal components (PCs) and a logistic regression model was used to examine the association among the PCs and BCR/ABL1-like ALL cases. Subsequently, a score on PCs was built and used to classify 142 additional B-NEG ALL cases, representing the screening panel.
The analysis of the genetic and clinical features was performed on a total of 194 B-NEG ALL, 52 belonging to the discovery and 142 to the screening panels (Table S2 and Supplementary material). Patients were enrolled in Gruppo Italiano Malattie EMatologiche dell’Adulto (GIMEMA) and Associazione Italiana di Ematologia ed Oncologia Pediatrica (AIEOP) protocols (Table S3).
The study was approved by the local IRB, in accordance with the Helsinki Declaration.
Analysis of genetic features
Recurrently mutated JAK/STAT and RAS pathways genes (Messina et al, 2016) were sequenced in 182/194 samples (Table S4). Copy number aberrations (Messina et al, 2017) were assessed by multiplex ligation-dependent probe amplification (MLPA) in 111/194 samples. RNA-sequencing was performed in 54 samples by the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA). RNA libraries were paired-end sequenced (2×100 bp) using the Illumina HiSeq2500 platform. Identification of fusion transcripts was performed by STAR-fusion (STAR-Fusion_v0.5.1) and Fusioncatcher (v0.99.3e) (Nicorici et al, 2014).
Statistical analysis of clinical features and event-free survival (EFS)
Patients’ characteristics were compared by chi-squared or Fisher’s exact test for categorical variables and by Wilcoxon test for continuous data. Event-free survival (EFS) and overall survival (OS) was estimated by Kaplan-Meier: EFS was estimated from the time of diagnosis to the occurrence of refractoriness, relapse or death, while OS was estimated from the time of diagnosis to death. Multivariate analysis was performed using the Cox proportional model to adjust the effect of BCR/ABL1-like ALL cases to white blood cell (WBC) count, age and CRLF2 on EFS. All tests were two-sided and P values <0.05 were considered statistically significant. Analyses were performed using the SAS software (release 9.4; SAS Institute, Cary, NC, USA).
In vitro experiments
To assess the sensitivity to ponatinib, the annexin V/7-aminoactinomycin D (7AAD) apoptotic test (BD Bioscience, San Josè, CA) and 3H-thymidine (Perkin Elmer, Waltham, MA) proliferation assays were performed on primary cells from 7 BCR/ABL1-like, 6 non-BCR/ABL1-like and 4 BCR/ABL1-positive cases. Ponatinib (Selleck Chemicals, Houston, TX) at 0 μM (dimethyl sulfoxide only) or at increasing doses (0.01–10 μM) was added at time 0 and viability was measured after 48 and 72 h (Supplementary data).
RESULTS
Identification and quantification of BCR/ABL1-like predictor genes in the discovery cohort
The comparison of BCR/ABL1-positive and B-NEG cases of GEP1 (Haferlach et al, 2010; Messina et al, 2010) using a t-test recognized 16 B-NEG cases that clustered with the BCR/ABL1-positive cases: these 16 misclustered B-NEG cases were regarded as the “core” BCR/ABL1-like samples (Figure S2).
To select the BCR/ABL1-like genes (Figure S2) the “core” BCR/ABL1-like ALL cases were compared with the remaining B-NEG cases by t-test, that resulted in the identification of 285 genes (Table S5), of which 9 had been previously reported as part of the BCR/ABL1-like signature (Harvey et al, 2010b). These 9 genes (SOCS2, IFITM1, CD99, TP53INP1, IFITM2, JCHAIN, NUDT4, ADGRE5 and SEMA6A) together with CRLF2 (Yoda et al, 2010; van der Veer et al, 2013; Herold et al, 2017) were used to build the Q-RT-PCR-based BCR/ABL1-like predictor.
By analysing GEP2 (Chiaretti et al, 2005) we selected 10 additional BCR/ABL1-like ALLs, leading to a total of 26 BCR/ABL1-like cases that were included in the discovery panel of this study.
Next, the expression levels of these 10 genes were quantified by Q-RT-PCR in the discovery cohort (n=52), comprising 26 BCR/ABL1-like and 26 non-BCR/ABL1-like ALLs, classified according to GEP (Chiaretti et al, 2005; Haferlach et al, 2010; Messina et al, 2010). All genes were significantly overexpressed in BCR/ABL1-like ALL samples (Figure S3).
Development of the “BCR/ABL1-like predictor”
The Q-RT-PCR expression values of the 10 genes were used to build the “BCR/ABL1-like ALL predictor”. First, we verified that in univariate analysis, all genes were risk factors for a higher BCR/ABL1-like ALL probability (Table S6). As a high correlation was detected among the expression levels of the 10 genes (Figure S4), it was possible to summarize the variability by means of PC analysis; by definition, the PCs thus identified are uncorrelated. In detail, expression values were shrunk into 3 PCs (accounting for >80% of the variability): the contributions of each gene to each component are expressed by factor loadings, a measure of their relationship (Table S7). Each component is mainly explained by genes with the highest loadings: PC1 is explained by the expression values of NUDT4, SEMA6A, ADGRE5, SOCS2 and JCHAIN, PC2 by CRLF2, TP53INP1, CD99 and PC3 by IFITM1 and IFITM2.
Second, a logistic regression model was used to estimate the probability of a case being BCR/ABL1-like using the first 3 PCs; all components were statistically significant in multivariate analysis (Table S8).
Finally, to generate a predictive model, a score was computed by means of a linear combination of the PCs, as a result of the above mentioned multivariate logistic regression model. Figure S5 illustrates the generation of the score. The optimal cut-off was set at −0.30: cases with a score ≥−0.30 were defined as BCR/ABL1-like. This cut-off provides the best distinction between BCR/ABL1-like and non-BCR/ABL1-like cases and ensures the optimal compromise between sensitivity and specificity (88.5% and 100%, respectively). The mathematical equation is provided in Supplementary material.
An online BCR/ABL1-like score calculator was implemented into the web-based application GIMEMA REDCap system (Harris et al 2009).
Identification of the BCR/ABL1-like ALL cases and genomic characterization
The predictive model was then validated in a screening cohort of 142 B-NEG ALL patients: 28 cases (19.7%) were classified as BCR/ABL1-like (min score −0.279, max score 2.176) and 114 as non-BCR/ABL1-like ALL (min score −1.810, max score −0.353). The comparison of the clinico-biological features of BCR/ABL1-like and non-BCR/ABL1-like ALL cases at diagnosis, carried out in the whole cohort, showed that BCR/ABL1-like ALLs were associated with a significantly higher WBC count at diagnosis (22.9 vs 12.6 x109/L, p=0.013) while no differences were observed for the other parameters (Table I).
Table I.
Comparison between BCR/ABL1-like and non-BCR/ABL1-like clinico-biological features.
| BCR/ABL1-like n=54 | non-BCR/ABL1-like n=140 | p-value | |
|---|---|---|---|
| Gender (male/female) | 36/18 | 78/62 | p=ns |
| Median age (range), years | 32 (6–72) | 28 (0–78) | p=ns |
| Age cohort 0–15 years (n=21) | 2 (9.5%) | 19 (90.5%) | |
| Age cohort 15–35 years (n=98) | 29 (29.5%) | 69 (70.4%) | p=ns |
| Age cohort >35 years (n=75) | 23 (30.6%) | 52 (69.3%) | |
| Median (range) WBC count, x109/l | 22.6 (1.89–239) | 12.4 (0.6–425) | p=0.023 |
| Median (range) platelet count, x109/l | 47 (0.15–283) | 47 (1–308) | p=ns |
| Median (range) Hb g/l | 97 (41–153) | 89 (37–158) | p=ns |
ns, not significant; WBC: white blood cell.
Recurrently mutated genes were investigated in 182/194 cases. Mutations of the JAK/STAT pathway (i.e. JAK1/2, CRLF2 and IL7R) were detected in 44.2% (23/52) of BCR/ABL1-like cases and only in 7.7% (10/130) of the non-BCR/ABL1-like cases (p<0.001), RAS pathway mutations were more frequent in non-BCR/ABL1-like (n=38/130, 29.2%) than in BCR/ABL1-like cases (n=9/52, 17.3%, p=0.068). Details are provided in Table II.
Table II.
Comparison between BCR/ABL1-like and non-BCR/ABL1-like genetic features.
| BCR/ABL1-like | non-BCR/ABL1-like | p-value | |
|---|---|---|---|
| JAK/STAT pathway members mutated cases | 23/52 (44.2%)^ | 10/130 (7.7%) | p<0.001 |
|
| |||
| JAK1/2 mutations | 14/52 (26.9%) | 4/130 (3.1%) | p<0.001 |
| CRLF2 mutations | 6/52 (11.5%) | 2/130 (1.5%) | p=0.007 |
| IL7R mutations | 6/52 (11.5%) | 4/130 (3.1%) | p=0.033 |
|
| |||
| RAS pathway members mutated cases | 9/52 (17.3%) | 38/130 (29.2%)* | p=0.068 |
|
| |||
| FLT3 mutations | 0/52 (0%) | 10/130 (7.7 %) | p=0.031 |
| KRAS/NRAS mutations | 9/52 (17.3%) | 30/130 (23.1%) | p=ns |
| IKZF1 deletions | 29/35 (82.8%) | 31/76 (40.8%) | p<0.001 |
| EBF1 deletions | 14/35 (40%) | 5/76 (6.6%) | p<0.001 |
| BTG1 deletions | 10/35 (28.5%) | 5/76 (6.6%) | p=0.003 |
|
| |||
| CRLF2 overexpressing cases$ | 33/54 (61.1%) | 25/140 (17.8%) | p<0.001 |
| CRLF2 expression levels | 6.7 (0.6–16.6) | 10.9 (3.2–17.9) | p<0.001 |
|
| |||
| TK/cytokine receptor fusions |
EBF1/PDGFRB (N=3) JAK2-fusions (N=3) RCSD1/ABL1 (N=1) TRIM24/FGFR1 (N=1) |
P2RY8/CRLF2 (N=1) | |
|
P2RY8/CRLF2 (N=2) TSLP-fusions (N=2) 12/28 (42.8%) |
1/26 (3.8%) | p=0.001 | |
Three cases carried 2 concomitant JAK/STAT pathway mutations: 2 cases harboured JAK2 and CRLF2 mutations, 1 case harboured IL7R and CRLF2 mutations.
Two cases carried 2 concomitant RAS pathway mutations: 1 case harboured NRAS and KRAS mutations, 1 case FLT3 and NRAS mutations.
Overexpression was defined at ΔCt<8 as previously described by Chiaretti et al (2016).
We also examined copy number aberrations in 111 cases: IKZF1 deletions were significantly more frequent in BCR/ABL1-like than non-BCR/ABL1-like cases (82.8% vs 40.8%, p<0.001). We found that EBF1 and BTG1 deletions were significantly more frequent in BCR/ABL1-like cases (Table II).
Finally, CRLF2 levels were significantly higher (p≤0.001) in BCR/ABL1-like (median ΔCt = 6.7, range 0.6–16.6) than in non-BCR/ABL1-like cases (median ΔCt = 11.3, range 3.2–17.9). Complete results are detailed in Tables S9 and S10. Comparable results were obtained when separating the discovery (Table S11) and screening cohorts (Table III).
Table III.
Comparison between BCR/ABL1-like and non-BCR/ABL1-like ALL cases included in the screening panel.
| BCR/ABL1-like | non-BCR/ABL1-like | p-value | |
|---|---|---|---|
| JAK/STAT pathway members mutated cases | 12/27 (44.4%)^ | 10/107 (9.3%) | p<0.001 |
|
| |||
| JAK1/2 mutations | 7/27 (25.9%) | 4/107 (3.7%) | p=0.001 |
| CRLF2 mutations | 2/27 (7.4%) | 2/107 (1.9%) | ns |
| IL7R mutations | 4/27 (14.8%) | 4/107 (3.7%) | p=0.05 |
|
| |||
| RAS pathway members mutated cases | 6/27 (22.2%) | 32/107 (29.9%)* | ns |
|
| |||
| FLT3 mutations | 0 | 9/107 (8.4%) | ns |
| KRAS/NRAS mutations | 6/27 (22.2%) | 24/107 (22.4%) | ns |
| IKZF1 deletions | 14/18 (77.7%) | 23/62 (37.1%) | p=0.029 |
| EBF1 deletions | 6/18 (33.3%) | 4/62 (6.5%) | p=0.007 |
| BTG1 deletions | 4/18 (22.2%) | 4/62 (6.5%) | p=0.071 |
|
| |||
| CRLF2 overexpressing cases | 16/28 (57.1%) | 21/114 (18.4%) | p<0.001 |
| CRLF2 median expression levels (range) | 7.6 (2–16.6) | 11.3 (3.2–17.5) | p<0.001 |
|
| |||
| TK/cytokine receptor fusions |
JAK2-fusions (N=1) TRIM24/FGFR1 (N=1) TSLP-fusions (N=2) 4/13 (30.7%) |
P2RY8/CRLF2 (N=1) 1/21 (4.7%) |
p=0.037 |
ns, not significant
One case harboured IL7R and CRLF2 mutations
One 1 case harboured NRAS and FLT3 mutations
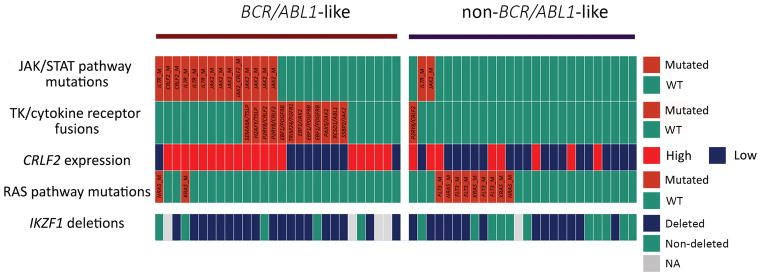
RNA-sequencing, performed in 54 samples (28 BCR/ABL1-like and 26 non-BCR/ABL1-like), identified 13 fusion transcripts targeting tyrosine kinases (TKs)/cytokine receptors, of which 12 were detected in BCR/ABL1-like cases (12/28 vs 1/26 p=0.001). The most recurrent fusion was EBF1/PDGFRB found in 3 cases; JAK2 rearrangements with different partners (i.e. PAX5, EBF1, SSBP2) were detected in 3other cases, P2RY8/CRLF2 and TSLP-fusion genes in 2 samples; finally, RCSD1/ABL1 and TRIM24/FGFR1 were found in 1 case each. Within non-BCR/ABL1-like cases, 1 case harboured P2RY8/CRLF2 while no fusion genes targeting TKs were documented. Figure 1 shows that 27/28 (96.4%) had at least one lesion typical of the BCR/ABL1-like profile and suggests that the BCR/ABL1-like profile is sustained by at least 2 different mechanisms: one represented by TK-rearrangements only and the other by CRLF2 overexpression plus JAK/STAT mutations.
Figure 1.
Distribution of BCR/ABL1-like specific genetic lesions in the samples with a complete molecular characterization. Legend: green boxes for JAK/STAT pathway mutations: wild-type; red boxes: mutation detected. The mutated gene name is provided in the figure; green boxes for RAS pathway mutations: wild-type; red boxes: mutation detected; the mutated gene name is provided in the figure; CRLF2 expression; red boxes: overexpression; TK/cytokine fusions: green boxes: no rearrangement detected; red boxes: rearrangement detected. The fusion gene is specified in the figure; IKZF1 deletions: green boxes: no deletions; blue boxes: presence of deletions; grey boxes: sample not evaluated.
Outcome of the BCR/ABL1-like ALL samples
We analysed the complete remission (CR) rate and survival in the adolescents and adults with clinical data available (n=142, Table S3)
In the whole cohort, the CR rate was lower in BCR/ABL1-like than in non-BCR/ABL1-like cases (77.8% vs 89.6%, p=0.06).
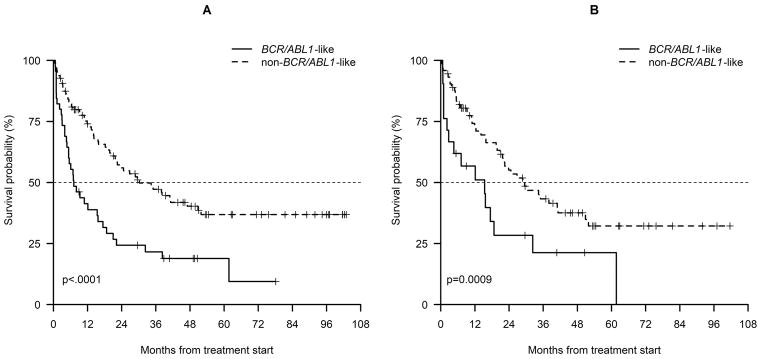
Consistently, EFS at 36 months was significantly inferior for BCR/ABL1-like cases compared to non-BCR/ABL1-like cases (21.6% vs 47.2%, p<0.0001; Figure 2A).
Figure 2.
Event-free survival at 36 months of adolescents and adults classified as BCR/ABL1-like and non-BCR/ABL1-like belonging to the whole cohort (A) and the screening panel only (B).
When we considered the screening cohort only (n=95), we confirmed that the CR rate was significantly lower in BCR/ABL1-like than in non-BCR/ABL1-like cases (71.4% vs 91.8%, p=0.014). Similarly, EFS at 36 months was significantly inferior for BCR/ABL1-like cases compared to non-BCR/ABL1-like cases (21.3% vs 43.3%, p=0.0009; Figure 2B).
In line with EFS estimates, OS at 36 months was significantly inferior for BCR/ABL1-like cases compared to non-BCR/ABL1-like cases (37.3% vs 60.7%, p=0.05; Figure S6A) in the whole cohort and a similar trend was observed in the screening cohort only (Figure S6B). The impact of BCR/ABL1-like prediction retained statistical significance on EFS in multivariate analysis (Hazard ratio: 2.12, 95% confidence interval: 1.18–3.82, p=0.01) in a model adjusted for age and WBC count. We also evaluated the interaction on EFS between the BCR/ABL1-like signature and CRLF2 overexpression: BCR/ABL1-like prediction retained statistical significance on EFS (p=0.05) in the bivariate model adjusted by CRLF2 overexpression.
In vitro sensitivity to ponatinib
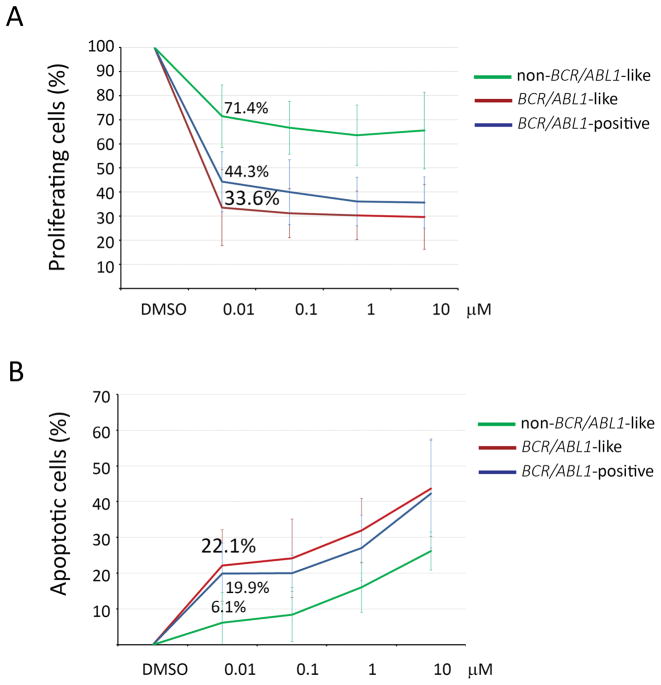
After 72 h of incubation with ponatinib (0.01 μM), a 3H-thymidine uptake assay showed that the proliferation rate of primary cells from 7 BCR/ABL1-like cases (2 EBF1/PDGFRB-positive, 1 JAK2-mutated and P2RY8/CRLF2-positive, 1 RCSD1/ABL1, 3 wild type WT for JAK/STAT and RAS mutations) decreased to 33.6%±15%, comparable to the sensitivity observed in 4 BCR-ABL1-positive cases (44.3%±12.4%). Contrarily, in the non-BCR-ABL1-like (non-BCR-ABL1-positive) samples (n=6) - all WT for JAK/STAT and RAS hotspot mutations - the proliferation rate upon ponatinib treatment decreased only to 71.5%±28.7% (p=0.0007) (Figure 3A). Ponatinib (0.01 μM) also increased the apoptotic rate in both BCR/ABL1-like and BCR/ABL1-positive primary ALL cells (22.1%±10%, 19.9%±8.5%, respectively), while the apoptotic response in non-BCR/ABL1-like/non-BCR/ABL1-positive ALL was significantly inferior (6.1%±8.4%, p=0.023, Figure 3B)
Figure 3.
In vitro response to ponatinib in BCR/ABL1-like and non-BCR/ABL1-like primary cells; BCR/ABL1+ cells were used as control. A) Average values of 3H-thymidine incorporation of 16 primary B-ALL cells samples after 72 h of treatment with ponatinib (0.01 μM); B) Average values of Annexin V positive primary B-ALL cells samples after 72 h of treatment with ponatinib (0.01 μM). Samples are grouped according to BCR/ABL1-like (n=7), BCR/ABL1-positive (n=4) and non-BCR/ABL1-like (n=6).
Discussion
Although the recognition of the BCR/ABL1-like ALL subset dates back to 2009, a consensus on a BCR/ABL1-like signature has not been reached (Mullighan et al, 2009a; Den Boer et al, 2009) and a standardized tool to identify these cases is currently not available (Ofran & Izraeli, 2017). Roberts et al (2014a) and Fasan et al (2015) proposed a combination of different methods: analysis of CRLF2 expression, FISH targeting ABL1 and JAK activating rearrangements, fusion-specific RT-PCR for the identification of the ABL and JAK partners and MRD monitoring. However, this approach relies on multiple techniques and can only recognise cases carrying already known fusion transcripts.
Simultaneously, Harvey et al (2013) developed a method based on the quantification of 15 transcripts by LDA and several groups adopted this method (Heatley et al, 2017; Reshmi et al, 2017). However, the mathematical equations were not provided. Our approach took advantage of previously reported GEP data (Messina et al, 2010; Chiaretti et al, 2005) to identify a narrow list of 10 transcripts (CRLF2, SOCS2, IFITM1, CD99, TP53INP1, IFITM2, JCHAIN, NUDT4, ADGRE5, SEMA6A), capable of accurately identifying the BCR/ABL1-like ALLs.
The genes chosen to build the predictive model are also in common with other algorithms recently used to identify BCR/ABL1-like cases (Roberts et al, 2012; Harvey et al, 2013).
Using Q-RT-PCR of these 10 transcripts, we built an algorithm capable of identifying BCR/ABL1-like cases with a high sensitivity and specificity. Furthermore, we generated a user-friendly tool, which requires only the upload of gene expression values to assess whether a sample is BCR/ABL1-like or non-BCR/ABL1-like.
Next, the screening of 142 B-NEG ALL samples by the BCR/ABL1-like predictor assigned 28 cases (19.7%) to the BCR/ABL1-like ALL subset, in line with the reported incidence (Ofran & Izraeli, 2017).
The prediction accuracy was indirectly corroborated by the analysis of the genetic features of BCR/ABL1-like ALL cases. As reported in both paediatric and adult cohorts, BCR/ABL1-like ALL is associated with IKZF1 deletions, CRLF2 deregulation/rearrangements, JAK1/2 mutations, rearrangements of genes coding for TKs and cytokine receptors (Mullighan et al, 2009b; Mullighan et al, 2009c; Harvey et al, 2010a; Harvey et al, 2010b; Yoda et al, 2010; Chen et al, 2012; van der Veer et al, 2013; Asai et al, 2013; Tokunaga et al, 2013; Boer et al, 2015a; Ge et al, 2016; Roberts et al, 2017; Jain et al, 2017a; Herold et al, 2017). Consistently, our BCR/ABL1-like cases frequently carried JAK/STAT pathway mutations, the most recurrent targeting JAK2, followed by CRLF2 and IL7R. In addition, BCR/ABL1-like cases were enriched in IKZF1 deletions, detected in 80% of cases, in line with Herold and colleagues (Herold et al, 2017).
More importantly, RNA-sequencing - performed in 28 BCR/ABL1-like cases - revealed that 7 carried TK rearrangements, 2 P2RY8/CRLF2 and 2 TSLP-rearrangements. TK-activating fusion genes are specific to BCR/ABL1-like cases and may be targeted by TKIs, as shown in pre-clinical models and sporadic case reports (Roberts et al, 2012; Tasian et al, 2012; Maude et al, 2012; Weston et al, 2013; Lengline et al, 2013; Roberts et al, 2014a; Roberts et al, 2014c; Shi et al, 2014; Shi et al, 2015; Francis et al, 2016).
The integration of all the molecular features, feasible in 28 BCR/ABL1-like, demonstrated that 96.4% had at least one lesion typical of the BCR/ABL1-like profile and suggested that the BCR/ABL1-like profile is sustained by at least two mechanisms, either a TK-activating fusion or CRLF2 overexpression with a concomitant JAK/STAT mutation; at variance, CRLF2 overexpression alone seems insufficient to induce a BCR/ABL1-like profile.
From a clinical standpoint, the features of the BCR-ABL1-like cases hereby identified were in line with a BCR-ABL1-like profile: they displayed a significantly higher WBC count at diagnosis, a lower CR rate and a significantly worse EFS than non-BCR-ABL1-like patients; we observed no differences related to gender, in agreement with other reports (Boer et al, 2015b; Herold et al, 2017). An association with MRD levels was not feasible since this parameter was not available for a large set of patients. Furthermore, the incidence in paediatric cases was lower than in adolescents and adults (9.5% vs 29.5% and 30.6%, respectively) in our cohort.
Finally, in vitro experiments showed that the pan-TKI, ponatinib, the most potent inhibitor in BCR/ABL1-positive ALL (Jabbour et al, 2015), was able to reduce the proliferative rate in BCR-ABL1-like samples and to increase apoptosis, similarly to that observed in BCR-ABL1-positive ALL. Notably, the BCR-ABL1-like samples analysed by in vitro experiments comprised ABL class lesions, JAK/STAT mutated and WT cases, indicating that ponatinib is active in all cases regardless of the underlying lesion and may represent an alternative to ruxolitinib whose clinical activity remains to be determined (Jain et al, 2017b).
In conclusion, we hereby describe a Q-RT-PCR based assay capable of singling out BCR-ABL1-like patients from the B-NEG ALL cohort. This approach has many advantages: first, it requires minimal amounts of diagnostic RNA; second, it is simple and cost-effective, being based on Q-RT-PCR; third, it is rapid, because the screening can be completed within a few days. This is essential, considering the high rate of refractory cases that - if promptly recognized - could benefit from an alternative approach, contemplating the use of the pan-TKI, ponatinib. These advantages make this tool suitable to be introduced in the diagnostic workflow and appears applicable to many haematology centres, followed by further genomic screens that comprise RNA-sequencing characterization to fully elucidate the underlying molecular lesion in the patients thus identified. Finally, because this assay is not based on a specific target identification, it allows the potential recognition of all BCR/ABL1-like cases, including those carrying novel genetic lesions.
Supplementary Material
Acknowledgments
The authors wish to thank Associazione Italiana per la Ricerca sul Cancro (AIRC), Special Program Molecular Clinical Oncology-Extension program, 5 × 1000 (10007), Milan (Italy) to RF; Finanziamento per l’avvio alla ricerca 2015 (Sapienza University of Rome) to MM; Finanziamento Medi Progetti Universitari 2015 to SC (Sapienza University of Rome); Fondazione Le Molinette Onlus, Turin (Italy); MM was partly supported by Associazione Cristina Bassi Onlus (Genova). DMW is supported by NCI 5R01CA151898 and 5R01 CA17238.
SC designed research, analysed data and wrote the manuscript; MM performed experiments, analysed data and wrote the manuscript; SG performed experiments and analysed data; AP designed the BCR/ABL1-like predictor model and performed statistical analyses; ALF, VG, AL, NP performed experiments; FDG performed RNA sequencing experiments; MV performed statistical analyses; MPM, VA, AV and CS provided samples and clinico-biological data; OE and RB analysed RNA-sequencing data; LSL and DW provided clinical samples; GI analysed data and critically revised the manuscript; AG and RF designed the study, analysed data and critically revised the manuscript.
References
- Asai D, Imamura T, Suenobu S, Saito A, Hasegawa D, Deguchi T, Hashii Y, Matsumoto K, Kawasaki H, Hori H, Iguchi A, Kosaka Y, Kato K, Horibe K, Yumura-Yagi K, Hara J, Oda M. IKZF1 deletion is associated with a poor outcome in pediatric B-cell precursor acute lymphoblastic leukemia in Japan. Cancer Med. 2013;2:412–419. doi: 10.1002/cam4.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer JM, Koenders JE, van der Holt B, Exalto C, Sanders MA, Cornelissen JJ, Valk PJ, den Boer ML, Rijneveld AW. Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. Haematologica. 2015a;100:e261–264. doi: 10.3324/haematol.2014.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer JM, Marchante JR, Evans WE, Horstmann MA, Escherich G, Pieters R, Den Boer ML. BCR-ABL1-like cases in pediatric acute lymphoblastic leukemia: a comparison between DCOG/Erasmus MC and COG/St. Jude signatures. Haematologica. 2015b;100:e354–357. doi: 10.3324/haematol.2015.124941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H, Borowitz MJ, Camitta BM, Carroll AJ, Devidas M, Pullen DJ, Payne-Turner D, Tasian SK, Reshmi S, Cottrell CE, Reaman GH, Bowman WP, Carroll WL, Loh ML, Winick NJ, Hunger SP, Willman CL. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: A Children’s Oncology Group study. Blood. 2012;119:3512–3522. doi: 10.1182/blood-2011-11-394221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaretti S, Li X, Gentleman R, Vitale A, Wang KS, Mandelli F, Foà R, Ritz J. Gene expression profiles of B-lineage adult acute lymphocytic leukemia reveal genetic patterns that identify lineage derivation and distinct mechanisms of transformation. Clin Cancer Res. 2005;11:7209–7219. doi: 10.1158/1078-0432.CCR-04-2165. [DOI] [PubMed] [Google Scholar]
- Chiaretti S, Brugnoletti F, Messina M, Paoloni F, Fedullo AL, Piciocchi A, Elia L, Vitale A, Mauro E, Ferrara F, De Fabritiis P, Luppi M, Ronco F, De Propris MS, Raponi S, Kronnie GT, Vignetti M, Guarini A, Foà R. CRLF2 overexpression identifies an unfavourable subgroup of adult B-cell precursor acute lymphoblastic leukemia lacking recurrent genetic abnormalities. Leuk Res. 2016;41:36–42. doi: 10.1016/j.leukres.2015.11.018. [DOI] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasan A, Kern W, Nadarajah N, Weber S, Schindela S, Schlenther N, Schnittger S, Haferlach T, Haferlach C. Three steps to the diagnosis of adult Ph-like ALL. Blood; 57th ASH meeting; Orlando. December 5–8, 2015; 2015. p. 2610. [Google Scholar]
- Francis OL, Milford TA, Martinez SR, Baez I, Coats JS, Mayagoitia K, Concepcion KR, Ginelli E, Beldiman C, Benitez A, Weldon AJ, Arogyaswamy K, Shiraz P, Fisher R, Morris CL, Zhang XB, Filippov V, Van Handel B, Ge Z, Song C, Dovat S, Su RJ, Payne KJ. A novel xenograft model to study the role of TSLP-induced CRLF2 signals in normal and malignant human B lymphopoiesis. Haematologica. 2016;101:417–426. doi: 10.3324/haematol.2015.125336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Gu Y, Zhao G, Li J, Chen B, Han Q, Guo X, Liu J, Li H, Yu MD, Olson J, Steffens S, Payne KJ, Song C, Dovat S. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget. 2016;7:49722–49732. doi: 10.18632/oncotarget.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK, Mills KI, Gilkes A, Chiaretti S, Shurtleff SA, Kipps TJ, Rassenti LZ, Yeoh AE, Papenhausen PR, Liu WM, Williams PM, Foà R. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, Carroll WL, Devidas M, Bowman WP, Camitta BM, Reaman GH, Hunger SP, Downing JR, Willman CL. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010a;115:5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, Wilson CS, Wharton W, Murphy M, Devidas M, Carroll AJ, Borowitz MJ, Bowman WP, Downing JR, Relling M, Yang J, Bhojwani D, Carroll WL, Camitta B, Reaman GH, Smith M, Hunger SP, Willman CL. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010b;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Kang H, Roberts KG, Atlas SR, Bedrick EJ, Gastier-Foster JM, Zhang J, Gerhard DS, Smith MA, Larsen EC, Raetz EA, Winick NJ, Carroll WL, Stonerock E, Heerema NA, Carroll AJ, Chen S, Song G, Becksfort J, Rusch M, Li Y, Ma J, Ell D, Reshmi SC, Loh ML, Davidas M, Hunger SP, Mullighan CG, Willman CL. Development and Validation Of a Highly Sensitive and Specific Gene Expression Classifier To Prospectively Screen and Identify B-Precursor Acute Lymphoblastic Leukemia (ALL) Patients With a Philadelphia Chromosome-Like (“Ph-like” or “BCR-ABL1-Like”) Signature For Therapeutic Targeting and Clinical Intervention. Blood; 55th ASH meeting; New Orleans. December 7–10, 2013; 2013. p. 826. [Google Scholar]
- Heatley SL, Sadras T, Kok CH, Nievergall E, Quek K, Dang P, McClure B, Venn N, Moore S, Suttle J, Law T, Ng A, Muskovic W, Norris MD, Revesz T, Osborn M, Moore AS, Suppiah R, Fraser C, Alvaro F, Hughes TP, Mullighan CG, Marshall GM, Pozza LD, Yeung DT, Sutton R, White DL. High prevalence of relapse in children with Philadelphia-like acute lymphoblastic leukemia despite risk-adapted treatment. Haematologica. 2017;102:e490–e493. doi: 10.3324/haematol.2016.162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold T, Schneider S, Metzeler K, Neumann M, Hartmann L, Roberts KG, Konstandin NP, Greif PA, Bräundl K, Ksienzyk B, Huk N, Schneider I, Zellmeier E, Jurinovic V, Mansmann U, Hiddemann W, Mullighan CG, Bohlander SK, Spiekermann K, Hoelzer D, Brüggemann M, Baldus CD, Dreyling M, Gökbuget N. Philadelphia chromosome-like acute lymphoblastic leukemia in adults have frequent IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102:130–138. doi: 10.3324/haematol.2015.136366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E, Kantarjian H, Ravandi F, Thomas D, Huang X, Faderl S, Pemmaraju N, Daver N, Garcia-Manero G, Sasaki K, Cortes J, Garris R, Yin CC, Khoury JD, Jorgensen J, Estrov Z, Bohannan Z, Konopleva M, Kadia T, Jain N, DiNardo C, Wierda W, Jeanis V, O’Brien S. Combination of hyper-CVAD with ponatinib as first-line therapy for patients with Philadelphia chromosome-positive acute lymphoblastic leukaemia: a single-centre, phase 2 study. Lancet Oncol. 2015;16:1547–1555. doi: 10.1016/S1470-2045(15)00207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Roberts KG, Jabbour E, Patel K, Eterovic AK, Chen K, Zweidler-McKay P, Lu X, Fawcett G, Wang SA, Konoplev S, Harvey RC, Chen IM, Payne-Turner D, Valentine M, Thomas D, Garcia-Manero G, Ravandi F, Cortes J, Kornblau S, O’Brien S, Pierce S, Jorgensen J, Shaw KR, Willman CL, Mullighan CG, Kantarjian H, Konopleva M. Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood. 2017a;129:572–581. doi: 10.1182/blood-2016-07-726588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Jabbour E, Zweidler-McKay P, Ravandi F, Takahashi K, Kadia T, Wierda WG, Rytting ME, Nunez C, Patel K, Lu X, Tang G, Konoplev S, Wang SA, Han L, Thakral B, Deshmukh A, Garris R, Jorgensen JL, Cavazos A, Verstovsek S, Garcia-Manero G, O’Brien S, Cortes J, Mullighan CG, Kantarjian H, Konopleva M. Ruxolitinib or Dasatinib in Combination with Chemotherapy for Patients with Relapsed/Refractory Philadelphia (Ph)-like Acute Lymphoblastic Leukemia: A Phase I-II Trial. Blood. 2017b;130:1322. [Google Scholar]
- Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E. Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica. 2013;98:e146–e148. doi: 10.3324/haematol.2013.095372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, Mullighan CG, Hunger SP, Harvey RC, Willman CL, Fridman JS, Loh ML, Grupp SA, Teachey DT. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina M, Chiaretti S, Tavolaro S, Peragine N, Vitale A, Elia L, Sica S, Levis A, Guarini A, Foà R. Protein kinase gene expression profiling and in vitro functional experiments identify novel potential therapeutic targets in adult acute lymphoblastic leukemia. Cancer. 2010;116:3426–3437. doi: 10.1002/cncr.25113. [DOI] [PubMed] [Google Scholar]
- Messina M, Chiaretti S, Wang J, Fedullo AL, Peragine N, Gianfelici V, Piciocchi A, Brugnoletti F, Di Giacomo F, Pauselli S, Holmes AB, Puzzolo MC, Ceglie G, Apicella V, Mancini M, Te Kronnie G, Testi AM, Vitale A, Vignetti M, Guarini A, Rabadan R, Foà R. Prognostic and therapeutic role of targetable lesions in B-lineage acute lymphoblastic leukemia without recurrent fusion genes. Oncotarget. 2016;7:13886–138901. doi: 10.18632/oncotarget.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina M, Chiaretti S, Fedullo AL, Piciocchi A, Puzzolo MC, Lauretti A, Gianfelici V, Apicella V, Fazi P, Te Kronnie G, Testi AM, Vitale A, Guarini A, Foà R. Clinical significance of recurrent copy number aberrations in B-lineage acute lymphoblastic leukaemia without recurrent fusion genes across age cohorts. Br J Haematol. 2017;178:583–587. doi: 10.1111/bjh.14721. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR Children’s Oncology Group. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009a;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, Mikhail FM, Meyer J, Carroll AJ, Williams RT, Cheng J, Heerema NA, Basso G, Pession A, Pui CH, Raimondi SC, Hunger SP, Downing JR, Carroll WL, Rabin KR. Rearrangement of CRLF2 in B-progenitor and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009b;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, Devidas M, Atlas SR, Chen IM, Clifford RJ, Gerhard DS, Carroll WL, Reaman GH, Smith M, Downing JR, Hunger SP, Willman CL. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2009c;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kilkku O. FusionCatcher - a Tool for Finding Somatic Fusion Genes in Paired-End RNA-Sequencing Data. bioRxiv. 2014:011650. doi: https://doi.org/10.1101/011650.
- Ofran Y, Izraeli S. BCR-ABL (Ph)-like acute leukemia-Pathogenesis, diagnosis and therapeutic options. Blood Rev. 2017;31:11–16. doi: 10.1016/j.blre.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, Chen IM, Valentine M, Liu Y, Li Y, Shao Y, Easton J, Payne-Turner D, Gu Z, Tran TH, Nguyen JV, Devidas M, Dai Y, Heerema NA, Carroll AJ, 3rd, Raetz EA, Borowitz MJ, Wood BL, Angiolillo AL, Burke MJ, Salzer WL, Zweidler-McKay PA, Rabin KR, Carroll WL, Zhang J, Loh ML, Mullighan CG, Willman CL, Gastier-Foster JM, Hunger SP. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood. 2017;129:3352–3361. doi: 10.1182/blood-2016-12-758979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, Chen X, Kasap C, Yan C, Becksfort J, Finney RP, Teachey DT, Maude SL, Tse K, Moore R, Jones S, Mungall K, Birol I, Edmonson MN, Hu Y, Buetow KE, Chen IM, Carroll WL, Wei L, Ma J, Kleppe M, Levine RL, Garcia-Manero G, Larsen E, Shah NP, Devidas M, Reaman G, Smith M, Paugh SW, Evans WE, Grupp SA, Jeha S, Pui CH, Gerhard DS, Downing JR, Willman CL, Loh M, Hunger SP, Marra MA, Mullighan CG. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen SC, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrózek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014a;371:1005–1115. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Pei D, Campana D, Payne-Turner D, Li Y, Cheng C, Sandlund JT, Jeha S, Easton J, Becksfort J, Zhang J, Coustan-Smith E, Raimondi SC, Leung WH, Relling MV, Evans WE, Downing JR, Mullighan CG, Pui CH. Outcomes of children with BCR-ABL1–like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014b;32:3012–3020. doi: 10.1200/JCO.2014.55.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Yang Y, Payne-Turner D, Harvey RC, Chen IM, Reshmi SC, Gastier-Foster J, Loh ML, Willman CL, Hunger SP, Mullighan CG. Functional analysis of kinase-activating fusions in Ph-like acute lymphoblastic leukemia. Blood. 2014c;124:786. [Google Scholar]
- Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, Pei D, Iacobucci I, Valentine M, Pounds SB, Shi L, Li Y, Zhang J, Cheng C, Rambaldi A, Tosi M, Spinelli O, Radich JP, Minden MD, Rowe JM, Luger S, Litzow MR, Tallman MS, Wiernik PH, Bhatia R, Aldoss I, Kohlschmidt J, Mrózek K, Marcucci G, Bloomfield CD, Stock W, Kornblau S, Kantarjian HM, Konopleva M, Paietta E, Willman CL, Mullighan CG. High Frequency and Poor Outcome of Philadelphia Chromosome-Like Acute Lymphoblastic Leukemia in Adults. J Clin Oncol. 2017;35:394–401. doi: 10.1200/JCO.2016.69.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Han L, Tabe Y, Hong Mu, Wu S, Zhou J, Zeng Z, Fruman DA, Tasian SK, Weinstock DM, Konopleva M. Dual targeting of JAK2 signaling with a type II JAK2 inhibitor and of mTOR with a TOR kinase inhibitor induces apoptosis in CRLF2-rearranged Ph-like acute lymphoblastic leukemia. Blood. 2014;124:3706. [Google Scholar]
- Shi C, Han L, Zhang Q, Roberts KG, Park E, Tabe Y, Jacamo RO, Mu H, Wu S, Zhou J, Ma H, Zeng Z, Jain N, Jabbour EJ, Muschen M, Tasian SK, Mullighan CG, Weinstock DM, Fruman D, Konopleva M. Combined targeting of JAK2 with a type II JAK2 inhibitor and mTOR with a TOR kinase inhibitor constitutes synthetic activity in JAK2-driven Ph-like acute lymphoblastic leukemia. Blood. 2015;126:2529. [Google Scholar]
- Tasian SK, Doral MY, Borowitz MJ, Wood BL, Chen IM, Harvey RC, Gastier-Foster JM, Willman CL, Hunger SP, Mullighan CG, Loh ML. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–842. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K, Yamaguchi S, Iwanaga E, Nanri T, Shimomura T, Suzushima H, Mitsuya H, Asou N. High frequency of IKZF1 genetic alterations in adult patients with B-cell acute lymphoblastic leukemia. Eur J Haematol. 2013;91:201–208. doi: 10.1111/ejh.12155. [DOI] [PubMed] [Google Scholar]
- van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, Harrison CJ, Evans WE, van der Velden VH, Hoogerbrugge PM, Van Leeuwen F, Escherich G, Horstmann MA, Mohammadi Khankahdani L, Rizopoulos D, De Groot-Kruseman HA, Sonneveld E, Kuiper RP, Den Boer ML. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–2629. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston BW, Hayden MA, Roberts KG, Bowyer S, Hsu J, Fedoriw G, Rao KW, Mullighan CG. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31:e413–e416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, West N, Xiao Y, Brown JR, Mitsiades C, Sattler M, Kutok JL, DeAngelo DJ, Wadleigh M, Piciocchi A, Dal Cin P, Bradner JE, Griffin JD, Anderson KC, Stone RM, Ritz J, Foà R, Aster JC, Frank DA, Weinstock DM. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2010;107:252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.