Abstract
Polyploidy in animals is much less common than in plants, where it is thought to be pervasive in all higher plant lineages. Recent studies have highlighted the impact of polyploidization and the associated process of diploidy restoration on the evolution and speciation of selected taxonomic groups in the animal kingdom: from vertebrates represented by salmonid fishes and African clawed frogs to invertebrates represented by parasitic root-knot nematodes and bdelloid rotifers. In this review, we focus on the unique and diverse roles that transposable elements may play in these processes, from marking and diversifying subgenome-specific chromosome sets prior to hybridization, to influencing genome restructuring during rediploidization, to affecting subgenome-specific regulatory evolution, and occasionally providing opportunities for domestication and gene amplification to restore and improve functionality. There is still much to be learned from the future comparative genomic studies of chromosome-sized and haplotype-aware assemblies, and from post-genomic studies elucidating genetic and epigenetic regulatory phenomena across short and long evolutionary distances in the metazoan tree of life.
Introduction
Polyploidy in animals has long been thought to be quite rare [1,2], in contrast to plant polyploidy, which is now recognized as being present in almost all lineages of higher plants [3,4]. Most of the polyploidization events investigated in the animal kingdom were initially focused on the more easily discernible paleopolyploidy, noted in several rounds of whole-genome duplications (WGD) at the base of vertebrate evolution [5], rather than neopolyploidy, which is much harder to disentangle experimentally and bioinformatically. In recent years, we have witnessed much progress in genome sequencing technologies, which fortunately shows no signs of slowing down. With genome size and complexity no longer being the limiting factor, comparative genomics has finally drifted away from sequencing the genomes of haploids or highly inbred diploids. It is now rapidly moving towards resolving haplotypes in heterozygous diploids and towards analyzing complex genomes with several sets of chromosomes co-inhabiting the nuclei of polyploid plants and animals, including recent polyploids.
While ploidy changes can account, to a certain extent, for the huge variation in genome size that can be observed between different species, another major contribution to such variation is often provided by the expansion and contraction of the so-called “fluid component” of eukaryotic genomes [6], which is represented largely by various types of transposable elements (TEs) (Fig. 1a). Furthermore, the interplay of these two major contributing factors can result not simply in an increase or decrease in the genome size, but can bring about more profound genetic and epigenetic changes that could further define the evolutionary trajectories of individual species and larger taxonomic groups.
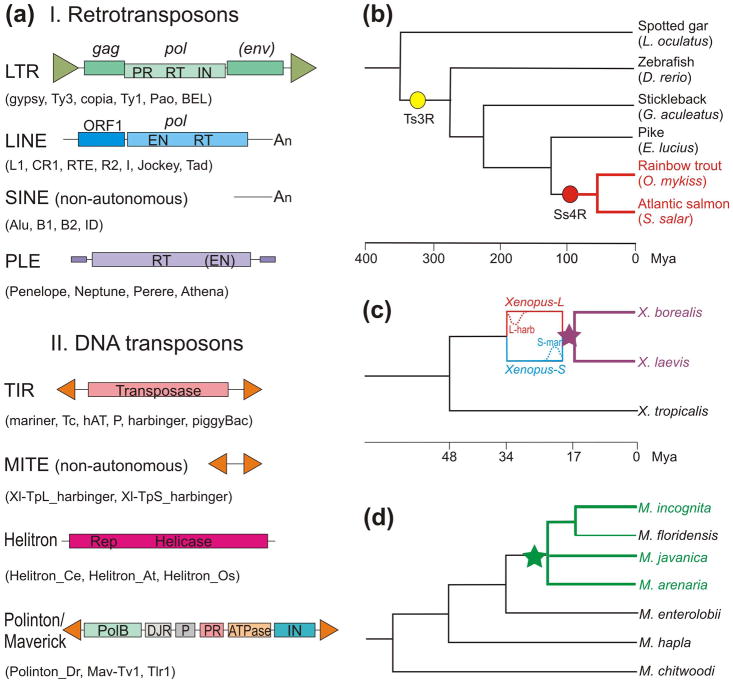
Figure 1.
Transposable elements and representative species discussed in the text. (a) A compilation of major structural features for class I TE (retrotransposons) and class II TE (DNA transposons). Shown are the characteristic ORFs with functional domains (RT, reverse transcriptase; IN, integrase; PR, protease; EN, endonuclease; PolB, family B DNA polymerase; DJR, double jelly-roll capsid-like protein) and direct or inverted repeats or poly-A stretches at the termini. Selected representatives, including those mentioned in the text and figures, are listed in parentheses. Not to scale. (b) Phylogenetic relationships in teleost fish and the ancestral spotted gar, with salmonids shown in red (after ref. [15]). Yellow and red circles represent the teleost-specific whole genome duplication (Ts3R) and the salmonid-specific whole genome duplication (Ss4R), respectively. (c) Phylogeny of Xenopus frogs according to [22]. Hybridization between the progenitor Xenopus-L and Xenopus-S species is denoted by purple star. Estimated peaks of L-harbinger and S-mariner activity are shown at 33–34 Mya and 18 Mya, respectively. (d) A consensus phylogram illustrating relationships between root-knot nematodes discussed in the text, based on [24–26]. Species with elevated ploidy are shown in green; the green star denotes the presumed recent hybridization(s), the precise time estimate for which was not reported. M. javanica is shown as a polytomy, since its phylogenetic placement differs in [24] and [26]. In (b-d), the cases of ploidy increase are shown by colored lines with double thickness.
With several recent reviews summarizing the impact of TE proliferation and cyclical ploidy changes on the evolution of genome architecture in plants, both on the genetic and the epigenetic side [7–11], the timing is right to revisit the subject of TE-associated genetic and epigenetic changes in hybrid and polyploid animals, which we reviewed a few years ago [12], focusing specifically on those polyploid genomes for which TE analysis has been performed. The recently sequenced genomes of polyploid animals, from vertebrates such as fish and amphibians, to invertebrates including ecdysozoan and lophotrochozoan taxa, reveal some interesting parallels with plant genomes, but also suggest certain differences related to the less frequent occurrence of polyploidy in animals.
Fishes: Transposons and rediploidization in salmonids
Vertebrate genomes were initially shaped by two rounds of ancient WGD events, which in fishes were followed by a teleost-specific round of WGD (Ts3R, also called TGD for teleost genome duplication), and additionally by a salmonid-specific round (Ss4R) [13] (Fig. 1). For this reason, aided by economic interest, the salmonids have attracted special attention from comparative genomicists seeking to uncover gene fates after this relatively “recent” WGD, when the genome is still in the process of returning to the diploid state and is undergoing “genome fractionation”, as the differential loss of homeologs is often called in plants [14].
The genome of the Atlantic salmon (Salmo salar), produced by an international collaborative effort, provided new insights into re-diploidization pathways after Ss4R, which they dated at 80 Mya [15]. In contrast to most other known WGD events, this one likely reflects autopolyploidy, i.e. had resulted from the actual whole-genome duplication event rather than from successful interspecific hybridization (allopolyploidy), with the chromosomes still capable of forming multivalents. In this study, Lien et al. concluded that large genomic reorganizations, coinciding with bursts of TE-mediated repeat expansions, were crucial for the post-Ss4R rediploidization process. The TE content in S. salar is one of the highest reported in vertebrates (Fig. 2), and is dominated by members of the Tc/mariner superfamily of DNA TEs, which make up 13% of the genome and have experienced several waves of proliferation [15,16] (Fig. 3a). Ancient bursts of Tc1 expansion may have coincided with the initiation of rediploidization well after Ss4R (at 85–87% sequence similarity), which may suggest that Ss4R eventually led to TE expansion by compromising regulatory processes responsible for control of TE mobility in the course of gene loss [15,16]. This, in turn, may have further driven the genome towards a diploid state through chromosome rearrangements, which may have been caused by ectopic recombination and TE-induced chromosomal breakage. A detailed inspection for TE remnants at the breakpoints could further strengthen this case.
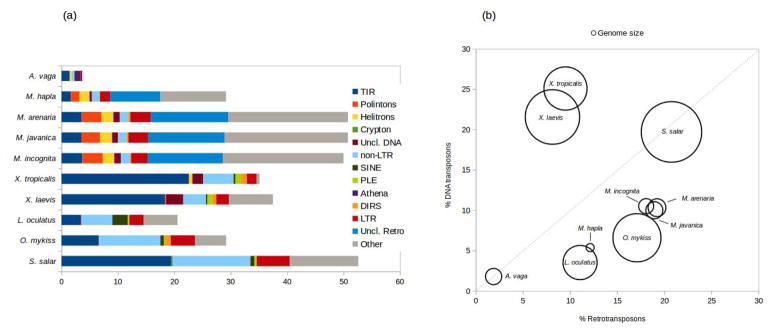
Figure 2.
TE class abundance and diversity in selected polyploid animal species A. vaga, M. arenaria, M. incognita, M. javanica, X. laevis, O. mykiss and S. salar, discussed in the text. For comparison, the diploid species M. hapla, L. oculatus and X. tropicalis are included. (a) Histogram of TE content profile. The X-axis shows the percentage of the genome assembly occupied by each TE class/order, as specified in the legend. “Other” denotes unclassified repeats. (b) Relationship between TE content and genome size. Content is given as the percentage of coverage of the genome assembly by retrotransposons (X-axis) and DNA transposons (Y-axis). The area of each bubble representing a species is proportional to its genome size (Mb): M. hapla (53.6), M. incognita (183.5), A. vaga (213.8), M. javanica (235.8), M. arenaria (258), L. oculatus (945), X. tropicalis (1513), O. mykiss (1877.5), X. laevis (2408.8) and S. salar (2970). Data sources: [15,17,18,22,24,29,44].
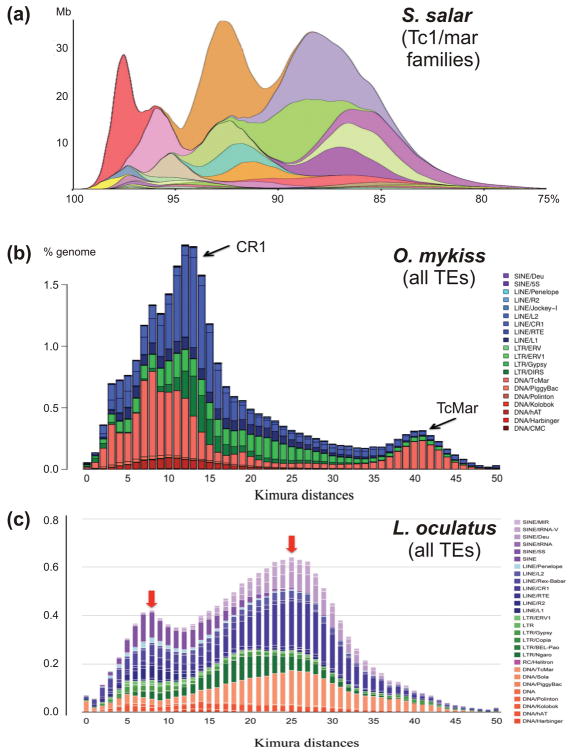
Figure 3.
A compilation of TE divergence plots over the evolutionary time scales in selected fish genomes discussed in the text. The S. salar plot (a) shows Tc1-like DNA TEs, with the X-axis showing per cent similarity to the consensus for each family and the Y-axis showing its genome abundance in Mb [15]. The O. mykiss (b) [17] and L. oculatus (c) [18] panels show the canonical RepeatMasker [45] TE landscape divergence plots with Kimura distances on the X-axis and per cent of the genome occupied by each TE superfamily on the Y-axis. The scales on the X-axes differ for (a) vs (b) and (c), thus the S. salar plot was mirrored and stretched to extend the X scale to the limit of detection beyond which no data can be plotted (25% nucleotide divergence, or Kimura distances of 50). Arrows indicate major TE expansions.
These results do not entirely parallel the picture observed in the first sequenced salmonid genome, that of the rainbow trout (Oncorhynchus mykiss) [17]. With the reported 38% TE content, the trout genome also experienced several waves of TE expansion, however rediploidization appears to have been slow and stepwise, with gene loss mostly occurring through pseudogenization, i.e. with no visible shrinkage of intergenic regions between adjacent pairs of ohnologs (homeologs), and the singleton genes between homeologs can still be paired with a corresponding non-functional region for 66% of the singletons. The Ss4R event was dated by Berthelot et al. at 90–102 Mya [17], after which no pervasive chromosome rearrangement was observed. A possible explanation of the different deletion rates in homeologous chromosomes of salmon and trout may lie in the fact that the trout genome has experienced post-Ss4R proliferation of CR1 non-LTR retrotransposons, which may be not as prone to generation of double-strand breaks (DSB) or rearrangements during transposition as are Tc/mariner-like DNA TEs. Nevertheless, a relatively recent peak of proliferation of mariner-like TEs was also observed (Fig. 3b).
For comparison with these post-Ts3R and post-Ss4R genomes, the genome of the spotted gar (Lepisosteus oculatus) provided an overview of a genome without these two duplications, forming a sister group to teleosts [18] (Fig. 1b). This genome was found to be slow-evolving, however two peaks of TE activity were observed involving mostly Tc/mariner and LINE/SINE elements (Fig. 3c). Chalopin and Volff [19] estimated the overall TE content in gar at 20% of the genome, with family diversity matching that in other teleost genomes, and concluded that most of the TE superfamilies were present in the last common ancestor of gars and teleosts, and no major TE diversification has occurred since then. Nevertheless, horizontal acquisitions of different families could not be ruled out. Notably, in most of the inspected fish genomes the current levels of TE activity appear to be much lower than in the past, with the lowest level of recent activity visible in rainbow trout (Fig. 3b). In sum, TEs appear to have actively participated in re-diploidization of the salmon genome, but much less so in the rainbow trout. However, as evident from the analysis of the spotted gar genome, TE de-activation and re-activation may always occur for other reasons unrelated to ploidy changes.
In addition to autopolyploidy, comparable challenges may be experienced by genomes during allopolyploidization and hybrid formation. For instance, in crosses between recently diverged dwarf and normal populations of lake whitefish, which show clear signs of emerging reproductive isolation, hybrid formation was shown to result in a “transcriptomic shock” manifested in extensive transcriptome-wide deregulation of gene expression and massive TE reactivation in malformed backcrosses [20]. Similarly, during formation of allopolyploids, challenges resulting from the collision of two different genomes may often involve TE derepression in a way reminiscent of a “genomic shock” described by McClintock [21], and would need to be overcome to form a stable polyploid lineage.
Amphibians: Marking subgenomes with transposons
The genome of the allotetraploid African clawed frog (Xenopus laevis) was shown to have diverged from its diploid relative X. tropicalis 48 Mya, and the two now-extinct X. laevis progenitor species diverged 34 Mya, fusing their genomes into the allopolyploid X. laevis 17 Mya [22] (Fig. 1c). The time interval since allopolyploidization was short enough to preserve the species-specific DNA TEs which have independently amplified in each of the two progenitor subgenomes, providing a unique molecular fossil record for the autonomous existence of each subgenome prior to the interspecific hybridization event. Most excitingly, these TE families (two of which were classified as MITEs, i.e. non-autonomous derivatives of the PIF/harbinger DNA TEs; Fig. 1a) were abundant enough to mark the entire chromosome subset belonging to each subgenome, one of which is called L (large) and the other S (small). The DNA TE called Xl-TpL_harbinger was amplified in the L subgenome 33–34 Mya, while Xl-TpS_mariner, with modest contribution from Xl-TpS_harbinger, dominated the amplification landscape in the S subgenome 17–18 Mya, right before Ss4R, painting exclusively the S chromosomes. The existence of this molecular fossil record in the absence of extant progenitor species made it possible to unequivocally establish the allopolyploid origin by separating the two subgenomes, and to uncover the asymmetry of evolution of each subgenome post-hybridization. Specifically, all the S chromosomes, marked by TpS-mar and TpS-harb, are appreciably shorter due to extensive gene loss from the S subgenome. The observed lack of recombination between subgenomes testifies to the absence of inter-homeolog meiotic pairing (homeologs are on average 6% divergent, as opposed to 0.4% divergence between alleles). The gene-wise deletions mostly affected the S subgenome, with 31.5% of genes lost, while the L subgenome lost only 8.3% of genes. Session et al. [22] hypothesized that the asymmetry between subgenomes, also observed in plants and yeast [14], could have stemmed either from intrinsic difference between progenitors, or from a ‘genomic shock’ resulting from post-merger TE activation, with TpS_mariner activity possibly putting the S-progenitor at a disadvantage in the immediate aftermath of tetraploidization.
A correlated asymmetry was identified between the two subgenomes at the regulatory element level [23]. The coactivator protein p300 (EP300), which can acetylate histones, is in part recruited by active enhancers that are modified with mono-methylated H3K4 (H3K4me1). The gene regulation markers H3k4me3, H3K36me3, RNA polymerase II and the transcription factor p300, all of which are associated with gene activation, were studied in X. laevis early gastrula embryos by ChIP-Seq profiling. No overall difference was observed between the two subgenomes in the number of regulatory elements per gene, as the same deletion rate applies to genes and to regulatory elements within each subgenome. However, the two subgenomes have evolved differently with regard to gene content and regulatory elements. Differential distribution of H3K4me3 and p300 regulatory elements is observed at many L and S loci, with enrichment for TEs that is proportional to a more profound loss of homeologous genes from the S subgenome. Altogether, this scenario suggests that the initial TE activation after WGD may have contributed to evolution of subgenome-specific enhancer elements, in addition to deletions and higher mutation rates in the S chromosomes. Furthermore, consistent TE reactivation was detected in artificial hybrids of X. tropicalis x X. laevis. Promoter methylation showed similar H3K4 patterns in hybrids and normal embryos, while recruitment of p300 was increased or lost at specific X. tropicalis loci in hybrid embryos where TEs were present at many locations, possibly recapitulating the processes in the L and S subgenomes following hybridization.
Nematodes: Increased transposon content in hybrids
Blanc-Mathieu et al. [24] sought to understand the genomic basis for the unusually high degree of evolutionary success in three obligatory asexual (apomictic) parasitic root-knot nematodes from the genus Meloidogyne (M. incognita, M. javanica and M. arenaria) in comparison with their sexual relatives. Intriguingly, these asexual species turned out to be polyploids, and the increase in ploidy was seemingly non-uniform between species, estimated as 3, 3–4, and 4–5 for M. incognita, M. javanica and M. arenaria, respectively. For comparison, the authors used a congeneric facultatively sexual M. hapla, which is capable of meiosis and sexual reproduction and reproduces by automictic parthenogenesis [25]. The origin of allopolyploidy in M. incognita, M. javanica and M. arenaria was estimated to be quite recent, as follows from the near-identity of mitochondrial genes (with interspecific divergence averaging 0.17%), and the authors hypothesized at least two successive hybridization events with the same or closely related maternal donor lineage and different paternal donors to explain the observed patterns. Nucleotide divergence between homeologous nuclear gene copies within each species averaged 8%, from 4.5–6% in the coding regions to 10–11% in introns, apparently reflecting divergence of ancestral copies prior to species hybridization. No substantial bias in gene fractionation towards any of the subgenomes was observed, however the proportion of collinear genes lost in duplicated genomic blocks was very low. Furthermore, over 60% of homeologous gene pairs displayed diverged expression patterns, and 22% showed evidence of positive selection.
Interestingly, the fraction of the genomes occupied by TEs was significantly higher in each of the three polyploids than in the diploid sister species, covering ~50% of each polyploid genome vs ~30% in the homozygous diploid M. hapla (Fig. 2). Whether the increase in TE content may have resulted from ploidy changes or from abandonment of sexual reproduction remains to be investigated, however the authors note that in all cases the TE load, which has increased substantially, may have contributed to increased genome plasticity, especially in the absence of sex. It is plausible that multiple TE insertions may have been associated with pervasive changes in gene regulation observed in homeologous pairs.
Szitenberg et al. independently sequenced multiple isolates of the same three apomictic root-knot nematodes, supplemented by the automictic M. floridensis, and by an outgroup M. enterolobii which is also apomictic [26]. They similarly observed an increase in ploidy, but after applying several filters to remove presumably unreliable duplicates, they concluded that most apomictic genomes in this genus are hypotriploids, i.e. a subset of genes from one of the subgenomes is present in a second copy, forming 2:1 A–B ortholog groups. This arrangement would be best explained by a hybridization event involving unreduced (2n) and reduced (n) gametes from the A and B genomes. Interestingly, the automictic M. floridensis displayed a high degree of homozygosity throughout the entire genome, having reverted to the diploid state. These authors also placed the interspecific hybridization event in the common ancestor of M. incognita, M. javanica, M. arenaria and M. floridensis, with the latter subsequently losing most of its homeologous pairs. Although they also observed higher TE content in apomicts compared to M. hapla and M. chitwoodi, the difference was not correlated with the reproductive mode, as the automictic M. floridensis also shows elevated TE content [26,27]. This is not too surprising, as the post-hybridization transition to automixis in M. floridensis is not expected to have resulted in a drop in TE content, since recombination in the absence of outcrossing would not exert the same effect on TE removal as it would in interbreeding populations. To reconcile the differing interpretations of ploidy and gene divergence in parasitic root-knot nematodes obtained in two different studies (3–5 vs hypotriploidy, several vs one hybridization event, and 4.7–6% vs 3% inter-homeolog divergence in the coding regions), assemblies of chromosome-length scaffolds utilizing long-read technologies would be required.
Rotifers: Diverse transposon families kept at bay
Early studies of putatively homologous gene pairs in bdelloid rotifers, small freshwater invertebrates best known for their asexual mode of reproduction and a high degree of resistance to desiccation and ionizing radiation, revealed signs of degenerate tetraploidy [28]. Indeed, completion of the genome of the first bdelloid species, Adineta vaga, helped to establish that 40% of the coding sequences are present in quartets formed by homeologous (ohnologous) pairs [29]. In comparison with nematodes, the WGD event in the class Bdelloidea (phylum Rorifera) is much more ancient, since gene quartets (with average 25% nucleotide divergence between ohnologs) can be found in representatives of different bdelloid families, Adinetidae and Philodinidae, which diverged at least 40 Mya and possibly earlier [30]. We also found an unusually low TE content for a metazoan (ca. 3% of the genome), but a very high degree of family diversity (255 families), with most families represented by very few members. The low TE abundance was accompanied by a high degree of duplication and diversification of the principal components of the RNA-mediated genome defense machinery, such as Dicers, Argonaute/Piwi, and RNA-dependent RNA polymerases (RdRP), present in 8, 15, and 20 copies, respectively. Only some of these copies were arranged in quartets, suggesting that in other cases differential loss of homeologs may have occurred. Overall, these findings suggested that TEs in A. vaga may be under tight control to prevent their unchecked proliferation in the genome.
Indeed, our analysis of genome-wide distribution of pi-like RNAs, which in A. vaga are 25–31 nucleotides in length with a strong 5′-uridine bias, showed that almost every active TE family displays high levels of piRNA coverage: 71% of mapped reads corresponded to annotated transposons, with 93% of these reads being in the antisense orientation with regard to TE transcript annotation [31]. This is largely correlated with low levels of transcriptional activity, as determined by RNA-seq, in agreement with the expected role of piRNAs in TE silencing. Unexpectedly, a significant fraction of piRNAs originated from predicted coding regions corresponding to genes of putatively foreign origin, which are abundant in bdelloid genomes and constitute over 8% of the annotated gene set, and in many cases are co-localized with TEs [29,31,32]. The fortified gene silencing machinery may have played an important role in adapting to the combined transposon load that may have arisen from the ancient merger of two genomes, and apparently continues to resist horizontal invasions of TEs and other foreign genetic material.
In the relatively compact genomes such as rotifers and nematodes (Fig. 2b), only the most recently active TE families are present: the decayed copies usually do not accumulate to provide a fossil record, as they do in vertebrates, and TE landscape divergence plots such as in Fig. 3 place most TEs into the lowest-divergence bins (not shown). The high rate of TE removal in A. vaga, similar to that observed in plants, is due to microhomology-mediated deletions arising during DSB repair by non-homologous end-joining, and to ectopic recombination between homologous stretches of DNA, including LTR-LTR recombination in LTR retrotransposons. Similar processes have likely played a role in post-WGD diversification of homeologous pairs, which do not display obvious subgenome asymmetry: the regions between ohnologs have apparently experienced gene losses and/or acquisitions in both subgenomes, and consist of entirely dissimilar sets of genes without obvious expansion/contraction bias in either of the homeologous pairs (somewhat akin to “unbiased fractionation” instead of “subgenome dominance” observed in different plants [33]).
As mentioned above, the high degree of diversity of TE families (with nearly all known TE types and superfamilies present, except for Polintons/Mavericks) is accompanied by very low copy numbers, accounting for the overall low TE content. This applies even to TEs of previously unknown types, which could not initially be recognized as such. Recently, we described a new type of giant retrotransposable elements in bdelloids, called Terminons, which in addition to Athena-like reverse transcriptases [34] contain numerous other ORFs and can reach 40 kb in length [35]. Their 3′-terminal hammerhead ribozyme structures may help to expose short stretches of telomeric repeats for attachment to G-rich overhangs to prime reverse transcription. Telomeric repeats are added by telomerase, which is present in two copies but was missed by gene annotation pipelines, leading another research group to erroneously declare its absence [36]. While the other homeologous pair may have been lost from one of the subgenomes during genome fractionation, it is also possible that one of the ancestral species in the hybrid initially lost its telomerase and had it replaced with telomere-specific retrotransposons, similarly to the well-known case of drosophilid insects [37]. The latter possibility would explain the co-existence of interspersed telomerase-mediated and transposon-mediated addition of DNA observed at telomeres, which may have helped to avoid conflict between different telomerases in a hybrid.
Concluding remarks
Comparative genomics has reached the point where it is no longer constrained by the genome complexity or the degree of ploidy. Further, the methodology for TE annotation and analysis in complex genomes has also improved a lot, although it has not yet reached the point where it can be easily and uniformly applied across the diversity of life forms [38,39], as is largely the case for gene annotation methods. Several polyploid genomes were recently sequenced in the phylum Arhtropoda, including horseshoe crabs, house spider, and bark scorpion [40,41]. However, the reported genome analyses did not include TEs, and therefore we were unable to include those species in this review; besides, the WGD events involving these species are quite ancient (>135 Mya for horseshoe crabs, and >430 Mya in the common ancestor of spiders and scorpions). With several additional rotifer genomes on the way, it should be possible to compare the subgenomes of different families with regard to differential gene loss/acquisition, and to scan their genomes for unfamiliar TE types. An exploration of the ploidy series in Xenopus, which may include octoploids and even dodecaploids [42], may provide a quantitative side to the observations made in diploids vs. tetraploids. There is much to be learned from in-depth epigenetic studies of polyploid genomes, which are still in their infancy, since most attention on this front is being paid to warm-blooded vertebrates unable to form healthy and viable polyploids. Epigenetic changes in hybrids often involve TE derepression which may lead to hybrid breakdown, and further studies of these phenomena may shed light on the underlying basis of genetic incompatibilities leading to speciation [20,43]. Finally, the availability of the newest long-read sequencing and scaffolding technologies is revolutionizing our ability to assemble chromosome-length scaffolds end-to-end, one haplotype at a time. The wealth of information from these efforts will greatly assist in uncovering the full impact of TEs on the genetic and epigenetic aspects of evolution in polyploids and in eukaryotic species in general.
Acknowledgments
We thank W. Reznikoff for critical reading of the manuscript. Work in the laboratory is supported by R01GM111917 from the U.S. National Institutes of Health to I.A.
Glossary
- Polyploidy
having more than the usual diploid set of homologous chromosomes
- Allopolyploidy
a form of polyploidy that results from interspecific hybridization
- Autopolyploidy
a form of polyploidy that results from chromosome redoubling in the same species
- Paleopolyploidy
ancient polyploidy
- Neopolyploidy
recent polyploidy
- Homeologs
pairs of genes originated by speciation and brought back together by allopolyploidization
- Ohnologs
same as homeologs, named after S. Ohno who introduced the concept
- Subgenome
genome of the progenitor species which now constitutes part of the polyploid genome
- Apomixis
asexual reproduction
- Automixis
combination of two haploid gametes from the same meiosis
Footnotes
Conflicts of interest
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.White MJD. Animal Cytology and Evolution. 3. Cambridge, UK: Cambridge University Press; 1977. [Google Scholar]
- 2.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 3.Alix K, Gerard PR, Schwarzacher T, Heslop-Harrison JSP. Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann Bot. 2017;120:183–194. doi: 10.1093/aob/mcx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Jaillon O, Aury JM, Wincker P. “Changing by doubling”, the impact of Whole Genome Duplications in the evolution of eukaryotes. C R Biol. 2009;332:241–253. doi: 10.1016/j.crvi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Young MW. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979;76:6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vicient CM, Casacuberta JM. Impact of transposable elements on polyploid plant genomes. Ann Bot. 2017;120:195–207. doi: 10.1093/aob/mcx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendel JF, Jackson SA, Meyers BC, Wing RA. Evolution of plant genome architecture. Genome Biol. 2016;17:37. doi: 10.1186/s13059-016-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Springer NM, Lisch D, Li Q. Creating order from chaos: Epigenome dynamics in plants with complex genomes. Plant Cell. 2016;28:314–325. doi: 10.1105/tpc.15.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendel JF, Lisch D, Hu G, Mason AS. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Dev. 2018;49:1–7. doi: 10.1016/j.gde.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Arkhipova IR, Rodriguez F. Genetic and epigenetic changes involving (retro)transposons in animal hybrids and polyploids. Cytogenet Genome Res. 2013;140:295–311. doi: 10.1159/000352069. [DOI] [PubMed] [Google Scholar]
- 13.Allendorf FW, Thorgaard GH. In: Tetraploidy and evolution of salmonid fishes. Turner BJ, editor. Plenum Press; 1984. [Google Scholar]
- 14.Freeling M, Woodhouse MR, Subramaniam S, Turco G, Lisch D, Schnable JC. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol. 2012;15:131–139. doi: 10.1016/j.pbi.2012.01.015. [DOI] [PubMed] [Google Scholar]
- **15.Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. Waves of transposon activity in salmon were correlated with initiation of rediploidization and may have assisted in driving the genome back to the diploid state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer JG, Yazawa R, Davidson WS, Koop BF. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics. 2007;8:422–422. doi: 10.1186/1471-2164-8-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noel B, Bento P, Da Silva C, Labadie K, Alberti A, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalopin D, Volff J-N. Analysis of the spotted gar genome suggests absence of causative link between ancestral genome duplication and transposable element diversification in teleost fish. J Exp Zool B Mol Dev Evol. 2017;328:629–637. doi: 10.1002/jez.b.22761. [DOI] [PubMed] [Google Scholar]
- 20.Dion-Côté A-M, Renaut S, Normandeau E, Bernatchez L. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol Biol Evol. 2014;31:1188–1199. doi: 10.1093/molbev/msu069. [DOI] [PubMed] [Google Scholar]
- 21.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- **22.Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. The two subgenomes in the allotetraploid X. laevis are marked with different transposon subsets, permitting full subgenome partition and illuminating the asymmetry of preferential gene loss from the S subgenome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *23.Elurbe DM, Paranjpe SS, Georgiou G, van Kruijsbergen I, Bogdanovic O, Gibeaux R, Heald R, Lister R, Huynen MA, van Heeringen SJ, et al. Regulatory remodeling in the allo-tetraploid frog Xenopus laevis. Genome Biol. 2017;18:198. doi: 10.1186/s13059-017-1335-7. Dissection of the epigenetic component and contribution of transposons to asymmetrical regulatory evolution characterizing the X. laevis subgenomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Blanc-Mathieu R, Perfus-Barbeoch L, Aury JM, Da Rocha M, Gouzy J, Sallet E, Martin-Jimenez C, Bailly-Bechet M, Castagnone-Sereno P, Flot JF, et al. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet. 2017;13:e1006777. doi: 10.1371/journal.pgen.1006777. Differential expression of homeologs and increase in transposon content are observed in the genomes of asexual plant parasitic nematodes with elevated ploidy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci U S A. 2008;105:14802–14807. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *26.Szitenberg A, Salazar-Jaramillo L, Blok VC, Laetsch DR, Joseph S, Williamson VM, Blaxter ML, Lunt DH. Comparative genomics of apomictic root-knot nematodes: hybridization, ploidy, and dynamic genome change. Genome Biol Evol. 2017;9:2844–2861. doi: 10.1093/gbe/evx201. Apomictic and automictic root-knot nematodes shared a common hybrid origin, with apomicts evolving into hypotriploids and the homozygous automict losing most of its homeologous pairs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szitenberg A, Cha S, Opperman CH, Bird DM, Blaxter ML, Lunt DH. Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biol Evol. 2016;8:2964–2978. doi: 10.1093/gbe/evw208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mark Welch DB, Mark Welch JL, Meselson M. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc Natl Acad Sci U S A. 2008;105:5145–5149. doi: 10.1073/pnas.0800972105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A, Henrissat B, Koszul R, Aury JM, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500:453–457. doi: 10.1038/nature12326. The first paleotetraploid invertebrate genome revealed low transposon content and highly diversified RNA-mediated silencing machinery. [DOI] [PubMed] [Google Scholar]
- 30.Hur JH, Van Doninck K, Mandigo ML, Meselson M. Degenerate tetraploidy was established before bdelloid rotifer families diverged. Mol Biol Evol. 2009;26:375–383. doi: 10.1093/molbev/msn260. [DOI] [PubMed] [Google Scholar]
- *31.Rodriguez F, Arkhipova IR. Multitasking of the piRNA silencing machinery: Targeting transposable elements and foreign genes in the bdelloid rotifer Adineta vaga. Genetics. 2016;203:255–268. doi: 10.1534/genetics.116.186734. Transposable elements and foreign genes in the bdelloid rotifer are silenced by the piwi-mediated pathway, which helps to maintain genome integrity and may facilitate foreign gene acquisition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 33.Garsmeur O, Schnable JC, Almeida A, Jourda C, D’Hont A, Freeling M. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 2014;31:448–454. doi: 10.1093/molbev/mst230. [DOI] [PubMed] [Google Scholar]
- 34.Gladyshev E, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Arkhipova IR, Yushenova IA, Rodriguez F. Giant reverse transcriptase-encoding transposable elements at telomeres. Mol Biol Evol. 2017;34:2245–2257. doi: 10.1093/molbev/msx159. Giant telomere-associated retrotransposable elements have been a peculiar feature of bdelloid rotifer genomes since the ancient polyploidization event. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai AG, Pouchkina-Stantcheva N, Di Donfrancesco A, Kildisiute G, Sahu S, Aboobaker AA. The protein subunit of telomerase displays patterns of dynamic evolution and conservation across different metazoan taxa. BMC Evol Biol. 2017;17:107. doi: 10.1186/s12862-017-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci U S A. 2011;108:20317–20324. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier AS, Hua-Van A, Hubley R, Kapusta A, et al. A call for benchmarking transposable element annotation methods. Mob DNA. 2015;6:13. doi: 10.1186/s13100-015-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arkhipova IR. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob DNA. 2017;8:19. doi: 10.1186/s13100-017-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, Chan TF, Kwan HS, Holland PW, Chu KH, et al. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity (Edinb) 2016;116:190–199. doi: 10.1038/hdy.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, Akiyama-Oda Y, Esposito L, Bechsgaard J, Bilde T, et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017;15:62. doi: 10.1186/s12915-017-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobel HR, Du Pasquier L. Genetics of polyploid Xenopus. Trends Genet. 1986;2:310–315. [Google Scholar]
- 43.Kelleher ES, Barbash DA. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 2013;30:1816–1829. doi: 10.1093/molbev/mst081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015. 2015 < http://www.repeatmasker.org>.