Abstract
Like the famous Collyer’s mansion in NY, our genomes have accumulated vast quantities of sequences that have been referred to as ‘junk DNA’, much of which consists of retrotransposons. A recent literature establishes the phenomenology that many retrotransposons become expressed at progressively higher levels during the course of normal aging. This seems to reflect gradual loss of heterochromatin in old age. In addition, retrotransposons appear to be precociously expressed in brains of younger animals that are experiencing neurodegenerative decline. Although it is difficult to distinguish cause from consequence, several recent studies support the contention that retrotransposon expression, and even perhaps transposition, causally contribute to both the normal deterioration seen with age and to the precipitous decline in some neurodegenerative disorders. This may reflect a two hit model in which normal age-dependent loss of heterochromatin synergizes with a disruption to post transcriptional silencing of RTEs caused by genetic and environmental stress.
Introduction
In March of 1947, Homer and Langley Collyer were found dead in their Harlem mansion, surrounded by 120 tons of rubbish. The trash that was later removed from the Collyer brothers’ home included defunct baby carriages, piles of old food, glass chandeliers, inoperable camera equipment, sections of a horse-drawn carriage, rusted bicycles, huge piles of old newspapers, the jaw bone of a horse and a decrepit X-ray machine. In the folklore of New York City, Collyers Mansion Syndrome refers to a psychological obsession with hoarding all manner of items to the point where an apartment is packed from floor to ceiling with junk. A Collyers Mansion is every fire fighters nightmare because the sheer volume of refuse present provides a dangerous source of fuel that can be ignited by any errant spark. Eukaryotic genomes have become a Collyers Mansion of sorts as a byproduct of the evolutionary battle with selfish genetic elements. Indeed our chromosomes are packed floor-to-ceiling with the refuse from ancient viral infections and from the detritus from the replication of mobile elements. Nearly 50% of the human DNA content consists of such accumulated refuse. This is not to say that all of this accumulated sequence is useless. Indeed, evolution has tinkered with this trash and in many cases has turned it to good use. But in this essay I will focus on the potential danger of the repetitive elements in our genomes. I will outline the “retrotransposon (RTE) storm” [7] hypothesis of age-dependent neurodegeneration, and will argue the case that the natural process of aging synergizes with inherited genetic predispositions and with environmental triggers to overwhelm the cellular systems that normally silence RTE expression. This systemic failure of RTE control mechanisms then ignites a storm of toxic RTE expression and perhaps even mobilization, leading to myriad downstream toxic effects, ultimately leading to cell death. I speculate that cell death caused by RTEs may exhibit non-cell autonomous toxic effects, thereby impacting neighboring cells in the brain.
RTE driven genetic heterogeneity: A function for the junk?
Before discussing evidence that unconstrained RTE expression contributes to age-dependent neurobehavioral decline, it is important to acknowledge the hypothesis that regulated RTE expression during developmentally prescribed time windows has the potential to contribute meaningful impact to normal brain function. L1 elements were first established to be capable of replicating and inserting de novo copies within the genomes of hippocampal neurons during development [1]. This startling observation raised the possibility that L1 driven genetic heterogeneity could provide a sort of genomic plasticity to the brain. A series of subsequent publications provided additional evidence that L1s and probably other RTEs are capable of replicating in neurons and glial cells [2–12]. The rate of such de novo events as measured by genomic sequencing experiments remains highly controversial [13–15] and it seems likely that some fraction of reported de novo insertions are artifacts from chimeric sequence reads generated during multiple displacement amplification (MDA) based library preparation [13]. Yet this sort of artifact cannot explain all of the findings. First, the rate of such false positives likely depends on both the analysis pipeline and library preparation method (e.g. not all reports used MDA). But more importantly, there also is overwhelming evidence from a wide array of methods that converge on the conclusion that RTEs are capable of replicating in somatic tissues, including the brain, and even in post-mitotic neurons and glial cells [1,2,4,5,7–10,16–21].
Importantly, this literature includes findings from DNA sequencing using more than one library preparation method, from bulk tissue as well as from single cells, and from several different species and brain region [3–5,10]. In addition, the findings in support of this conclusion include data that is wholly independent of sequencing.
In the case of R2, a LINE like element that inserts specifically in 28S ribosomal RNA genes, a simple but elegant genomic PCR approach was used to document de novo insertions in somatic tissues in Drosophila [21]. In the case of mammalian L1 mobilization, a series of studies used a now classic L1-GFP reporter cassette in which the mRNA encoding a GFP fluorescent protein can only be transcribed after removal of an intron by splicing of the L1 genomic RNA that is then transcribed from the opposite strand after re-insertion {e.g. [1,2,19]}. Thus GFP expression can only take place after the L1 passes through an RNA intermediate, is copied back into a cDNA, and integrates into a new chromosomal location. A second reporter has been developed to reveal de novo transposition of the gypsy LTR retrotransposon/ERV in Drosophila [7]. And as with the L1-GFP reporter, this so-called “gypsy-trap” reporter has been used by several research groups to reveal age-dependent somatic transposition, including brain and other tissues [7,16–18]. Finally, rare L1 mobilizations in neurons also have been validated using somatic cell nuclear transfer to clonally amplify neuronal genomes from adult mice [8,9]. This provided the means to sequence the entire genome of a cloned cell line or mouse strain in which the de novo insertion was incorporated. Together, the above literature provides conclusive evidence that RTEs replicate in normal somatic tissues, including neurons. The rate at which this occurs per neuron, whether the new insertions are essentially random or selective, how it varies across brain regions, cell types, species and individuals still is unresolved, as are the functional consequences.
RTE expression: a hallmark of aging?
Irrespective of whether regulated RTE transposition impacts normal brain development and function, there is accumulating evidence that dysregulated RTE activation contributes to age-dependent neurophysiological decline. Early attempts to characterize RTE expression in aging somatic tissues lead to the discovery that IAP LTR retrotransposons are increasingly expressed in liver from aged mice [22,23]. Although functional consequences were not established, the activity of these potentially mutagenic agents evoked a popular aging hypothesis: that accumulation of DNA damage might contribute to biological aging. More recently, a series of studies have corroborated the idea that expression of some RTEs increases markedly with age in a variety of species and cell types [7,16–18,22,24–30]. This phenomenology has now been documented in yeast, worms, flies, and mammals. And it appears to occur in the germline as well as a variety of somatic tissues including stem cells, fat, liver, and brain. In the case of brain, the new insertions are presumably occurring in non-dividing cells, consistent with ongoing de novo insertions rather than expansion of a lineage of cells that contain a de novo event.
Age dependent expression, and indeed replication, of the Ty1 LTR RTE has been observed in the context of a chronological aging model in S. cerevisiae [24] in which cells are maintained in stationary growth phase. Ty1 elements also are activated in a yeast model of replicative aging in which the number of times a mother cell has divided is progressively increased [29,30]. Similarly, expression of both LINEs and non-autonomous RTEs become aggressively expressed in a mammalian fibroblast culture model of replicative senescence [27] as well as in somatic tissues of aging mice [22,23,28]. Ty3/gypsy family LTR elements also are reportedly active in C. elegans germline in an age dependent manner[25]. Also, both LINE like and LTR retrotransposons become increasingly expressed in adult Drosophila somatic tissues, including adipose/liver [16,17,26], intestinal stem cells[18] and brain [7].
Importantly, in the case of the gypsy ERV/LTR element, age-dependent expression also correlates with de novo replication leading to activation of the gypsy-trap insertion reporter. This reporter makes use of a genomic hotspot for gypsy integrations, which is fused to a Gal80 repressor protein such that de novo integrations disrupt Gal80 expression. This activates Gal4 driven expression of GFP, revealing individual cells in which an RTE has inserted in the Gal80 cassette. Unlike the L1-eGFP reporter, this gypsy-trap reporter offers the advantage that it detects replication of endogenous elements rather than ones that are transgenically supplied. On the other hand, it suffers from the disadvantage that it does not distinguish which gypsy family member may have replicated. Despite this caveat, the gypsy-trap reporter has been used by several groups to detect age dependent increases in de novo transposition events in brain[7], in adipose/liver tissue[16,17], and in intestinal stem cells[18].
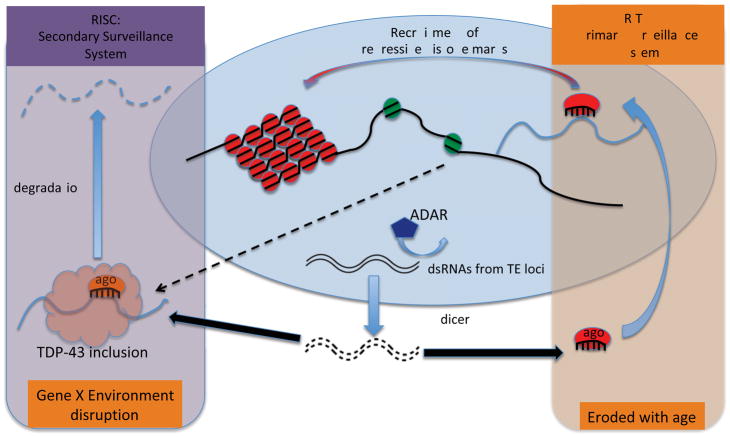
Overall, the picture that is emerging is one in which the progressive de-silencing of RTE transcription is a hallmark of aging that is conserved across species and tissues. This phenomenology of apparent activation of RTEs implies a failure of mechanisms that normally stifle such expression (Fig. 1). Indeed, in both germline and somatic tissues, expression of mobile elements usually is suppressed by multi-layered silencing systems [31–35]. In fission yeast, plants, worms and fly germline, where the silencing systems are better delineated, several general themes have emerged [31–34,36,37]. First, genomic regions that are rich in transposon sequences normally are kept transcriptionally quiescent by packaging into repressive heterochromatin. This primary silencing system likely includes recruitment of repressive histone marks by the RNA induced transcriptional silencing system (RITS). Second, post-transcriptional ‘backup’ silencing systems exist in which transposon RNAs are degraded by the RNA induced silencing complex (RISC). Both RITS and RISC make use of small RNAs with sequence complementarity to mobile elements, loaded onto members of the argonaute protein family (Fig. 1). The source of small RNAs, the mechanisms of their biogenesis, and the identities of argonaute family members vary between species and tissues. Importantly, few details are known about small RNA regulation of RTEs in brain aside from the known expression and requirement of several argonaute proteins[7,38,39] and the presence of various pools of transposon targeted small RNAs. Nevertheless, the observed age-dependent expression of RTEs in brain and in other somatic tissues points to a collapse of the mobile element silencing systems.
Figure 1. Can two hits ignite a fire?
RTE are normally silenced by overlayed systems of repression. First, genomic regions that are rich in RTE sequences are packaged into silent heterochromatin. Heterochromatin formation and maintenance depends on repressive histone modifications, which can be recruited by small RNAs loaded onto an argonaute family member. This loaded RITS complex targets RTE sequences in nascent transcripts. This primary chromatin level silencing appears to be eroded with age [16,27,47,49,53,91,92], perhaps via competition between ADAR and Dicer enzymes for double stranded RNA templates[26]. A secondary system of RTE silencing relies on post transcriptional targeting of RTEs by the RISC complex. This system appears to be stable in the face of normal aging, but is disrupted by TDP-43 proteinopathy[64].
RTE expression: cause or consequence of aging?
The literature is replete with phenomenological changes that correlate with age. As always, the key question is whether a given cellular effect is a cause or consequence of the aging process, and establishing such causality is not a trivial matter. In the case of RTE expression, the jury is not yet in. But three types of observation from the Drosophila system make it clear that contribution of RTE activity to age dependent decline deserves serious consideration. First, the expression of RTEs not only correlates with age, but also is impacted by dietary perturbations that alter the kinetics of aging. Chronic dietary restriction (DR) has a documented lifespan extending impact in a variety of organisms [40–44]. In flies, DR not only extends lifespan, it also significantly delays the onset of age-related RTE expression in adipose/liver tissue [16]. This DR regimen also apparently reduces the rate of de novo RTE insertional mutations, as measured using the gypsy-trap reporter [16]. Thus the kinetics of RTE expression, replication and of aging appear correlated even when the kinetics of aging is altered.
Secondly, the integrity of heterochromatic gene silencing, the primary cellular mechanism responsible for preventing RTE expression, is progressively eroded with age in a variety of organisms including yeast, worms, flies, mice and human cells [45–49]. Here too, genetic manipulations that delay the kinetics of heterochromatin loss result in a concomitant delay in the onset of RTE expression and an extension of lifespan. For example, increasing the efficacy of heterochromatin in old age by expression of Sir2, Dicer-2 or Su(var)3–9 or reducing the gene dosage of ADAR [16] were sufficient extend silencing of transposons in older animals. These same genetic manipulations also extended lifespan.
Although it is not yet clear what causes repressive histone modifications to be lost with age, a hint comes from genetic manipulations of the RNA editing enzyme adenosine deaminase acting on RNA (ADAR), a protein expressed almost exclusively in neurons [26]. In addition to its well-known role in specific editing that changes amino acid coding potential in mRNAs, ADAR (it turns out) also has a promiscuous editing mode in which it attacks long double-stranded RNA templates and copiously edits up to 50% of adenosines. The ADAR gene is able to switch into this promiscuous editing mode by specifically auto-editing its own transcript, leading to production of an ADAR protein isoform that favors promiscuous editing. This regulatory switch provides a brake on chromatin silencing of RTEs (Fig. 1), probably by competing with Dicer-2 for double stranded RNA substrates[26]. This ADAR switch also is associated with elevated levels of expression of heterochromatic sequences, including mobile elements. The levels of ADAR editing thus may provide a critical balance that determines the functional efficacy of RNAi mediated RTE silencing at the transcriptional level (and potentially at the post transcriptional level as well). Importantly, ADAR levels also modulate lifespan[26]. This provides a mechanistic hypothesis to explain the observation that RTEs are progressively de-repressed [16,26–28] and provides further evidence that such de-repression may impact longevity. And as with the case of components of the heterochromatin silencing apparatus, perturbations that alter the balance of ADAR editing simultaneously impact the efficacy of heterochromatin based silencing, the kinetics of mobile element expression during age and lifespan.
Genetic perturbations of argonaute family members that are crucial for RTE silencing also reveal functional impact on aging phenotypes. For example, mutations in argonaute-2 in Drosophila cause precocious expression of several RTEs in brain [7] and other tissues[50–52]. Such mutations also cause rapid age-related memory impairment and shortened lifespan[7]. Similarly, mutations in piwi clade of argonaute proteins lead to increased expression of several RTEs in both brain [38] and adipose/liver [17] of young fruit flies. And as with argonaute-2, mutations in piwi or in the flamenco locus from which many of the piwi loaded small RNAs are derived, cause a dramatic shortening of the animals lifespan[17]. These findings also are consistent with a report that piwi mediated repression of RTEs is required to protect somatic stem cells from age-dependent apoptotic cell death[18]. Thus, although the details of small RNA biogenesis in various somatic tissues and the division of labor between different branches of the argonaute family for somatic RTE suppression are still murky, there is evidence that both siRNAs loaded onto argonaute-2 and piRNAs loaded onto PIWI may each participate in protecting some somatic tissues during aging.
Taken as a whole, the available evidence reveals that RTE expression is progressively increased with age in a variety of species and tissues. In yeast and in Drosophila brain, adipose/liver tissue, and somatic stem cells, the rate of de novo transpositions of Ty1/gypsy family elements also are increased [7,16,18,24]. The effectiveness of heterochromatic silencing of RTEs appears also to fail in older animals, providing a potential mechanistic explanation [16,26,27,53]. Finally, in the face of perturbations to the rate of aging, or to the effectiveness of RTE silencing mechanisms, the correlation between RTE expression and aging are maintained.
Precocious RTE expression: a hallmark of neurodegenerative disorders?
Although age dependent RTE expression and even mobilization has been documented in a variety of somatic cell types and tissues, the potential for impacts in the nervous system is potentially most severe because virtually all of the cells are post-mitotic. This has two major implications. First, with the exception a relatively small percentage of cells in a few brain regions originating via adult neurogenesis [54], the overwhelming majority of neurons cannot be replaced if lost. This is in stark contrast to the situation in most somatic tissues. Second, in non-dividing cells, homologous mechanisms to repair DNA double strand breaks are offline. In post-mitotic neural tissue, such DNA damage thus is more likely handled by non-homologous end joining mediated repair, or by cell death. Hence the DNA damage that replicating RTEs contribute in brain may drive programmed cell death of an irreplaceable pool of cells. The observation that age results in RTE expression and in de novo insertions in brain therefore immediately invoked the possibility of a connection to neurodegeneration [7]. Indeed, the largest known risk factor for onset of neurodegeneration by far, is age. Likewise, while RTE expression appears to be progressively increased in advanced normal aging, there also is accumulating evidence that levels of RTE transcripts are precociously elevated in connection with a suite of neurodegenerative disorders.
For instance, several RTE sequences are elevated in expression in patients with sporadic forms of Creutzfeldt-Jakob, a transmissible prion disease [55] as well as in a hamster scrapie model [56]. Defective dicer-1 and RNA toxicity from expression of non-autonomous elements also has been functionally connected to macular degeneration [57,58]. Expression of the gypsy ERV has been implicated in the toxicity of an rCGG expanded repeat in a Drosophila model of fragile-X tremor ataxia[59]. Tau-related neurodegeneration in fly and mouse models as well as in brain tissue from Alzheimer’s subjects, exhibit a loss of heterochromatin accompanied by expression of heterochromatic transcripts, which include RTEs [60]. Conditional deletion during cortical development of the Uhrf1 gene, which regulates DNA methylation of RTE sequences, also leads to severe postnatal neurodegeneration that correlates with activation of IAP family ERVs [61]. There also is considerable evidence that expression of the HERV-W Env gene is associated with multiple sclerosis, an inflammatory disease leading to CNS demyelination and ultimately to neurodegenerative leasions [see [62] for review].. And finally, a series of recent studies[63–68] establish that activation of ERVs and perhaps other classes of RTEs are associated with disorders that involve protein aggregation pathology of the TAR DNA and RNA binding protein 43 (TDP-43). These disorders include amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). As with normal aging, it is not yet clear whether RTE expression is a cause or a consequence of neurodegenerative toxicity. But in the case of TDP-43 disorders, there is accumulating evidence in support of a causal contribution of RTEs.
A causal role for RTEs in neurodegeneration? Evidence from ALS and FTLD
Protein pathology of TDP-43, an aggregation-prone RNA and DNA binding protein, is a hallmark of a suite of neurodegenerative diseases including ALS and FTLD [see [69] for review]. TDP-43 is a member of the hnRNP family that binds to single stranded RNA and DNA with UG/TG-rich motifs. TDP-43, which was originally identified as a transcriptional repressor that binds to the TAR element of the HIV-1 retrovirus[70], is normally found predominantly in the nucleus in healthy cells. But in cells that are experiencing TDP-43 protein dysfunction, the protein accumulates in dense cytoplasmic inclusions. The functional implications of TDP-43 proteinopathy still are not fully clear, but the disorders are thought to involve some combination of toxicity caused by the cytoplasmic aggregates, generalized interference with normal cytoplasmic functions, and depletion of normal nuclear TDP-43 stores leading to a loss of its normal nuclear function. Such cytoplasmic inclusions of TDP-43 are seen in the vast majority of ALS subjects and in approximately half of FTLD. While rare familial mutations in the TDP-43 protein cause this pathology in some patients, approximately 90% of ALS and a large fraction of FTLD cases are sporadic, meaning that no causal genetic lesion has been identified. Thus in most subjects, TDP-43 protein of apparently normal amino acid sequence forms these abnormal cytoplasmic inclusions in affected neural tissue.
Why does TDP-43 proteinopathy occur in apparently genetically normal individuals with sporadic forms of several neurodegenerative diseases? A hint comes from the observation that TDP-43 contains a low complexity domain in its C-terminal region. This protein motif in fact is a common feature of RNA binding proteins that exhibit aggregation pathology in a variety of neurodegenerative disorders. An emerging literature has established that various forms of cellular stress induce such low complexity domain proteins to undergo concentration dependent phase separation, leading to formation of liquid droplets that over time may drive fibrillization [71–73].
Animal models of TDP-43 related disorders have taken advantage of the concentration dependence of low complexity domain protein aggregation[74]. Using transgenic overexpression to increase protein concentration above endogenous levels, it is possible to reproduce many of the signatures of human disease, including aggregation of TDP-43 protein in cytoplasmic inclusions and downstream neurological defects[69,75,76]. Although such animal models are imperfect representations of what is largely a sporadically occurring disorder, they have enabled the delineation of a myriad of cellular roles for TDP-43 and have provided the means to uncover genetic interactions between TDP-43 and other genes that are implicated in neurodegenerative disorders. TDP-43 pathology in animal models is now understood to cause global alterations in mRNA stability and splicing, de-repression of cryptic splicing, and biogenesis of some microRNAs [69,75–78]. A recent literature indicates that such TDP-43 pathology also may globally disrupt RTE surveillance systems [64], leading to toxic over-expression of ERVs and other RTEs[63–67].
It has been known for some time that ALS subjects and blood relatives often contain elevated levels of reverse transcriptase enzymatic activity in both serum and cerebrospinal fluid [79,80]. The observation that some ALS subjects exhibit elevated levels of HERV-K within affected neural tissue [65,67] suggested the possibility that this reverse transcriptase activity could derive from an endogenous retrovirus rather than from an exogenous source. Although a causative role for HERV-K in ALS has not been established, all major open reading frames encoded by HERV-K appear to be expressed in post mortem cortical tissue from some patients, and expression of HERV-K Env protein is neurotoxic and is sufficient to cause motor neuron degeneration when expressed in mouse [67]. The expression of HERV-K also may be mechanistically linked to TDP-43 by the observation that TDP-43 protein binds to the HERV-K LTR DNA sequences [67]. These findings raise the possibility that HERV-K per se may be functionally relevant to disease progression in some patients. But there also is accumulating evidence that TDP-43 proteinopathy seen in ALS and FTLD may induce widespread derepression of RTEs -- HERV-K may be the tip of the iceberg
Two types of observation suggest the idea that TDP-43 pathology may initiate a full-scale collapse of the RTE surveillance systems. First are phenomenological observations from deep sequencing datasets [63,64,66] and second are observations from functional studies in model organisms [64,68]. A number of groups used RNAseq to characterize transcriptional profiles in post mortem tissue from ALS subjects [81] or mouse [82,83] or fly[64] TDP-43 neurodegeneration models. Several publications also reported the profile of RNA targets that co-purified with the TDP-43 protein from mouse, rat or human brain tissues[82,84,85]. In each case, these studies identified many hundreds of direct binding targets of TDP-43 and also described many transcripts whose levels were altered either in patient tissue or in animal models. Because of the challenges of mapping repetitive sequences from short read technology, the early studies did not describe effects on RTEs [81–85]. Subsequent re-analysis of these repetitive reads [63], however, uncovered a strong connection between TDP-43 pathology and RTEs. In each of the RNA binding datasets from mouse, rat and human tissue, analysis of the multi-mapping sequences revealed that TDP-43 binds broadly to transcripts derived from RTEs. This included not only LTR retrotransposons/ERVs (such as HERV-K), but also many LINE and SINE elements. Second, in tissue from FTLD patients, the association between TDP-43 and RTE derived sequences appeared to be almost completely lost. This was in contrast to the mRNA targets of TDP-43, which were largely unchanged. Finally, levels of RTE sequences were found to be broadly increased in each of two mouse TDP-43 models [63]. This finding, that TDP-43 pathology is correlated with broad upregulation of RTEs has since been confirmed in a fly model [64] and has been observed in post mortem cortical tissue from ALS/FTLD subjects [66]. Taken together, the above findings raise the possibility that TDP-43 protein dysfunction somehow circumvents RTE silencing, leading to their broad up-regulation. Evidence from a Drosophila model provides mechanistic support for this hypothesis and also demonstrates for the first time that an endogenous RTE causally mediates toxic impacts of TDP-43 expression [64].
Overexpression of TDP-43 in Drosophila neurons or glial cells reproduces many pathological hallmarks seen in human subjects and other animal models, likely through dominantly inducing formation of cytoplasmic inclusions[75,76,86,87]. Such expression also causes progressive neurodegenerative effects, cell death, neurobehavioral decline and shortened lifespan. RNA sequencing from flies that express pathological levels of human TDP-43 reveals that such expression also causes a dramatic increase in transcripts from of a broad panel of RTEs, including LINE-like elements, LTR/ERV elements, and non-autonomous elements as well [64]. The broad expression of RTEs in response to pathological levels of TDP-43 expression results from a collapse of the argonaute-2/siRNA mediated gene silencing [64]. The disruptive effects on siRNA mediated silencing was visualized using a reporter system in which an RNAi transgene was used to target either GFP or an eye pigmentation gene.
The toxic expression of TDP-43, either in glial or neuronal cells, was shown to interfere with reporter silencing in an age-dependent manner. The mechanism by which TDP-43 pathology impairs siRNA silencing is not clear, but may involve disruption of small RNA biogenesis pathways [64,68,77,88] (Fig. 1).
Unfettered expression of RTEs can be highly detrimental for a variety of reasons. First, expression of high levels of RTE proteins, RNAs or extrachromosomal cDNA copies can have toxic cellular effects including activation of inflammatory response pathways. Such expression also provides the potential for functional replication of the RTEs, which can lead to insertional mutagenesis and activation of the DNA damage response leading to programmed cell death. In the case of TDP-43 pathology in Drosophila glial cells, expression of the gypsy ERV/LTR element has been demonstrated to contribute to the underlying toxicity from the TDP-43 protein because knocking down the expression of gypsy was sufficient to significantly ameliorate the effects of TDP-43 on lifespan of the animals as well as on cell death [64]. Moreover, the effects of TDP-43 in this context appear to be caused at least in part by DNA damage mediated programmed cell death because knockout of loki, the fly ortholog of Chk2, results in a near complete suppression of the detrimental effects of TDP-43 on both neuronal and glial cell death and on lifespan [64]. This is consistent with a model in which the toxicity of TDP-43 aggregation pathology is mediated at least in part by its disruptive impact on siRNA mediated silencing, leading to a storm of RTE expression.
These findings from the Drosophila model establish a mechanistic connection between TDP-43 proteinopathy and disruption to the RTE silencing systems. Early reports from post mortem brain tissue provides some support for the idea that TDP-43 related disorders exhibit RTE expression [63,65–67], although larger sample sizes are needed as are mechanistic investigations and functional validation to test whether the findings from Drosophila are predictive of the impact in human patients.
Thoughts and Perspectives: A two hit hypothesis
Age is the predominant risk factor for all neurodegenerative disorders. But neurodegeneration is not an inevitable outcome of healthy aging. Importantly the subset of individuals who succumb to neurodegenerative disorders also are of normal health until the point of diagnosis. The onset of neurodegenerative disease appears as an inflection point in a lengthy pre-symptomatic stage, after which the rate of decline increases dramatically. The kinetics of normal aging and of decline after diagnosis of neurodegenerative disorders suggests a two hit model (Figs 1 and 2) in which RTEs might play a contributory role. In this model, normal aging progressively erodes the primary RTE silencing systems, which act at the level of chromatin to block transcription. This leads to some expression of RTEs, an increased background of de novo transposition events, and possibly accumulation of toxic products from RTE expression, which may drive inflammation. On this background of age-dependent heterochromatin decline, any genetic or environmental perturbation that accelerates the effects of age on the primary system [60] or undermines the integrity of the secondary post-transcriptional silencing systems[64], may synergize to unleash toxic RTE expression. In plants, there in fact is evidence for such a synergy between primary and secondary silencing systems. Double mutant combinations in Arabidopsis thaliana of ddm1 and rdr6, which disrupt both transcriptional and post-transcriptional silencing respectively, provide a synergistic effect on transposon expression [89] and on phenotypic consequences [90].
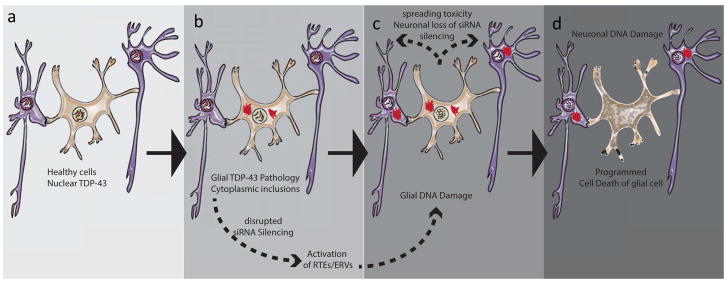
Figure 2. Can the fire spread between glia and neurons?
In healthy glia and neurons, TDP-43 is mostly nuclear in localization (a). For reasons that are not understood, pathological TDP-43 proteinopathy can be initiated, perhaps stochastically in response to cellular stressors (b). When this occurs, TDP-43 is cleared from the nucleus and aggregates in cytoplasmic inclusions that may interfere with argonaute-2 mediated post transcriptional silencing. This leads to a storm of awakened RTEs and ERVs, resulting in DNA damage mediated programmed cell death (c). Non-cell autonomous toxicity of glial cells has been observed in co-culture (see [93] for review). While the mechanisms of such toxicity is not fully explored, the fundamental homology between ERVs and exogenous retroviruses raises the specter of a self amplifying toxic mechanism.
As outlined above, normal aging erodes the efficacy of heterochromatin based gene silencing [47,91,92]. Recent work from Drosophila demonstrates that TDP-43 proteinopathy disrupts RNAi mediated silencing through argonaute-2 [64]. This likely perturbs the secondary RISC pathway, although there may also be cross-talk with transcriptional silencing. Any such disruption to both primary and secondary RTE silencing pathways could explain the observed synergy between age on the one hand and genetic, environmental or stochastic events on the other. When both primary and secondary surveillance systems fail, a given cell crosses a threshold after which expression of RTEs may overwhelm any residual cellular defenses. The ensuing storm of awakened RTE expression then contributes to the detrimental impact of TDP-43 proteinopathy, perhaps via toxicity of accumulated RTE RNAs and extrachromosomal cDNA copies and/or DNA damage, leading programmed cell death within that cell. But TDP-43 dependent neurodegeneration is also known to exhibit a fundamental non-cell autonomous toxicity [93] in which glial cells become toxic to nearby neurons (Fig. 2). Given the evolutionary relationship between exogenous retroviruses and ERVs such as gypsy and HERV-K, it is tempting to speculate that such elements may also contribute to these non-cell autonomous effects. Indeed there is some evidence that both gypsy and HERV-K [94–97] retain the ability to move genetic material between cells.
Acknowledgments
I would like to thank Rob Reenan, Steve Helfand, Yannick Jacobs, Richard Keegan, Yung Heng Chang, Wanhe Li for helpful comments on the manuscript. I also would like to thank the following sources for generous funding support: DART Neuroscience LLC, Ride For Life, NIA R01-RF1AG057338, NINDS R01DC013071.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES CITED
- 1.Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 2.Muotri AR, Zhao C, Marchetto MCN, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heutink P, Carninci P, Jeddeloh JA, Faulkner-Nature GJ. Somatic retrotransposition alters the genetic landscape of the human brain. nature.com. 2011 doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sánchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. This study is one of the clearest examples of single-cell sequencing to detect de novo transpositions of L1 elements in human neurons. The authors used a retrotransposon capture-sequencing approach on individual glia and neurons from hippocampus and cortex to identify de novo L1 insertions. A subset of these events were validated by PCR-- in some cases for both junctions. Rates of de novo events were estimated at about a dozen per neuron. The rate of detected events still is controversial because it has varied between species, cell type, library preparation method, and analysis pipeline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Hazen JL, Faust GG, Rodriguez AR, Ferguson WC, Shumilina S, Clark RA, Boland MJ, Martin G, Chubukov P, Tsunemoto RK, et al. The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning. Neuron. 2016;89:1223–1236. doi: 10.1016/j.neuron.2016.02.004. This study used nuclear transfer to clonally amplify and sequence the genomes of individual MT neurons from mouse olfactory bulb. Remarkably, individual neurons were found to have approximately 100 unique mutations. Most of these neurons also had rare de novo RTE insertions. This study provides an independent sequencing based approach to confirm findings from single cell detection methods. Although the rate of such de novo events has varied widely depending on cell type, species, and method of detection, the conclusion that L1s are capable of replicating in neurons has typically been validated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan AR, Boland MJ, Leibowitz ML, Shumilina S, Pehrson SM, Baldwin KK, Hall IM. Genome Sequencing of Mouse Induced Pluripotent Stem Cells Reveals Retroelement Stability and Infrequent DNA Rearrangement during Reprogramming. Cell Stem Cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treiber CD, Waddell S. Resolving the prevalence of somatic transposition in Drosophila. Elife. 2017:6. doi: 10.7554/eLife.28297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evrony GD, Lee E, Park PJ, Walsh CA. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016:5. doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faulkner GJ, Garcia-Perez JL. L1 Mosaicism in Mammals: Extent, Effects, and Evolution. Trends Genet. 2017;33:802–816. doi: 10.1016/j.tig.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 16**.Wood JG, Jones BC, Jiang N, Chang C, Hosier S, Wickremesinghe P, Garcia M, Hartnett DA, Burhenn L, Neretti N, et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc Natl Acad Sci U S A. 2016;113:11277–11282. doi: 10.1073/pnas.1604621113. This study discovered via RNAseq that many sequences from heterochromatin become expressed with age in both heads and fat tissue from Drosophila. This age-dependent expression of heterochromatin included many RTEs. This expression yields functional de novo transposition events in fat, as detected using the gypsy-trap reporter. A dietary restriction regime that extends lifespan in these animals delayed the onset of RTE expression. Moreover, genetic and pharmacological manipulations that prolonged heterochromatic silencing of RTEs or that disrupted such silencing could extend or shorten lifespan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Jones BC, Wood JG, Chang C, Tam AD, Franklin MJ, Siegel ER, Helfand SL. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan. Nat Commun. 2016;7:13856. doi: 10.1038/ncomms13856. This study reports evidence that the piRNA pathway is functional in fat body of adult fruit flies. Genetic manipulations of Piwi lead to increased TE expression and increased rate of gypsy mobilization in young animals. Such animals also exhibit phenotypes consistent with functionally compromised fat body tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Sousa-Victor P, Ayyaz A, Hayashi R, Qi Y, Madden DT, Lunyak VV, Jasper H. Piwi Is Required to Limit Exhaustion of Aging Somatic Stem Cells. Cell Rep. 2017;20:2527–2537. doi: 10.1016/j.celrep.2017.08.059. This study provides evidence for induction of a functional Piwi system in stem cells under control of the JAK/STAT signaling pathway during stem cell proliferation. Piwi function in the intestinal stem cell niche was shown to be required to suppress active gypsy replication to prevent age-related programmed cell death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macia A, Widmann TJ, Heras SR, Ayllon V, Sanchez L, Benkaddour-Boumzaouad M, Muñoz-Lopez M, Rubio A, Amador-Cubero S, Blanco-Jimenez E, et al. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 2017;27:335–348. doi: 10.1101/gr.206805.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea G-O, Muñoz-Lopez M, Jesuadian JS, Kempen M-JHC, Carreira PE, Jeddeloh JA, et al. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 2017;27:1395–1405. doi: 10.1101/gr.219022.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eickbush MT, Eickbush TH. Retrotransposition of R2 elements in somatic nuclei during the early development of Drosophila. Mob DNA. 2011;2:11. doi: 10.1186/1759-8753-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupressoir A, Puech A, Heidmann T. IAP retrotransposons in the mouse liver as reporters of ageing. Biochim Biophys Acta. 1995;1264:397–402. doi: 10.1016/0167-4781(95)00181-6. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell PH, Burhans WC, Curcio MJ. Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci U S A. 2011;108:20376–20381. doi: 10.1073/pnas.1100271108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis S, Sheth U, Feldman JL, English KA, Priess JR. C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 2012;8:e1002591. doi: 10.1371/journal.ppat.1002591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savva YA, Jepson JEC, Chang Y-J, Whitaker R, Jones BC, St Laurent G, Tackett MR, Kapranov P, Jiang N, Du G, et al. RNA editing regulates transposon-mediated heterochromatic gene silencing. Nat Commun. 2013;4:2745. doi: 10.1038/ncomms3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging. 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson MN, Scannapieco AE, Au PH, Dorsey S, Royer CA, Maxwell PH. Preferential retrotransposition in aging yeast mother cells is correlated with increased genome instability. DNA Repair. 2015;34:18–27. doi: 10.1016/j.dnarep.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol J-H, Chen C-C, Li W, Tyler JK. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582:2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 34.Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Fultz D, Choudury SG, Slotkin RK. Silencing of active transposable elements in plants. Curr Opin Plant Biol. 2015;27:67–76. doi: 10.1016/j.pbi.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 36.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 37.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 38.Perrat PN, DasGupta S, Wang J, Theurkauf W, Weng Z, Rosbash M, Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo L, Wang Z, Tan Y, Chen X, Luo X. piRNAs and Their Functions in the Brain. Int J Hum Genet. 2016;16:53–60. doi: 10.1080/09723757.2016.11886278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapahi P, Kaeberlein M, Hansen M. Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res Rev. 2017;39:3–14. doi: 10.1016/j.arr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 43.Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa S, Lagisz M, Hector KL, Spencer HG. Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell. 2012;11:401–409. doi: 10.1111/j.1474-9726.2012.00798.x. [DOI] [PubMed] [Google Scholar]
- 45.Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Front Genet. 2013;4:274. doi: 10.3389/fgene.2013.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorbunova V, Boeke JD, Helfand SL, Sedivy JM. Sleeping dogs of the genome. Science. 2014;346:1187–1188. doi: 10.1126/science.aaa3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012;22:42–49. doi: 10.1016/j.tcb.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 51.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung W-J, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cecco M, Criscione SW, … Peckham EJ. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging cell. Wiley Online Library. 2013 doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kempermann G, Song H, Gage FH. Neurogenesis in the Adult Hippocampus. Cold Spring Harb Perspect Biol. 2015;7:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong B-H, Lee Y-J, Carp RI, Kim Y-S. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt–Jakob disease. J Clin Virol. 2010;47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 56.Lathe R, Harris A. Differential display detects host nucleic acid motifs altered in scrapie-infected brain. J Mol Biol. 2009;392:813–822. doi: 10.1016/j.jmb.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 57.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan H, Qurashi A, Poidevin M, Nelson DL, Li H, Jin P. Retrotransposon activation contributes to fragile X premutation rCGG-mediated neurodegeneration. Hum Mol Genet. 2012;21:57–65. doi: 10.1093/hmg/ddr437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61*.Ramesh V, Bayam E, Cernilogar FM, Bonapace IM, Schulze M, Riemenschneider MJ, Schotta G, Götz M. Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev. 2016;30:2199–2212. doi: 10.1101/gad.284992.116. This study examined the impact on mouse neural stem cells of genetic disruptions to the ubiquitin-like PHD ring finger-1 (UHRF1) gene. The authors found that conditional deletion of UHRF1 during cortical development had little or no impact on cell proliferation, but lead to global reductions in DNA demethylation. This was accompanied by activation of the IAP family of LTR retrotransposons. This was followed by postnatal neurodegenerative effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morandi E, Tarlinton RE, Tanasescu R, Gran B. Human endogenous retroviruses and multiple sclerosis: Causation, association, or after-effect? Mult Scler. 2017;23:1050–1055. doi: 10.1177/1352458517704711. [DOI] [PubMed] [Google Scholar]
- 63.Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One. 2012;7:e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Krug L, Chatterjee N, Borges-Monroy R, Hearn S, Liao W-W, Morrill K, Prazak L, Rozhkov N, Theodorou D, Hammell M, et al. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 2017;13:e1006635. doi: 10.1371/journal.pgen.1006635. This study used a Drosophila model of TDP-43 related neurodegeneration to investigate the impact of RTEs. RNAseq revealed a broad increase in expression of LINE-like, LTR-retrotransposons and ERVs and non-autonomous elements. This was caused by a morbid loss of argonaute-2 mediated gene silencing. Functional manipulations of the gypsy element demonstrate a causal impact on neurodegenerative phenotypes, which involved activation of DNA damage mediated programmed cell death pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69:141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prudencio M, Gonzales PK, Cook CN, Gendron TF, Daughrity LM, Song Y, Ebbert MTW, van Blitterswijk M, Zhang Y-J, Jansen-West K, et al. Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum Mol Genet. 2017;26:3421–3431. doi: 10.1093/hmg/ddx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Lee M-H, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7:307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stein LD, Blumenthal T, … Petrucelli L. TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. The EMBO. emboj.embopress.org. 2014 doi: 10.15252/embj.201488740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin Y, Protter DSW, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casci I, Pandey UB. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res. 2015;1607:47–74. doi: 10.1016/j.brainres.2014.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gendron TF, Petrucelli L. Rodent models of TDP-43 proteinopathy: investigating the mechanisms of TDP-43-mediated neurodegeneration. J Mol Neurosci. 2011;45:486–499. doi: 10.1007/s12031-011-9610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349:650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacGowan DJL, Scelsa SN, Imperato TE, Liu K-N, Baron P, Polsky B. A controlled study of reverse transcriptase in serum and CSF of HIV-negative patients with ALS. Neurology. 2007;68:1944–1946. doi: 10.1212/01.wnl.0000263188.77797.99. [DOI] [PubMed] [Google Scholar]
- 80.Steele AJ, Al-Chalabi A, Ferrante K, Cudkowicz ME, Brown RH, Jr, Garson JA. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology. 2005;64:454–458. doi: 10.1212/01.WNL.0000150899.76130.71. [DOI] [PubMed] [Google Scholar]
- 81.Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling S-C, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shan X, Chiang P-M, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci U S A. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vanden Broeck L, Callaerts P, Dermaut B. TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol Med. 2014;20:66–71. doi: 10.1016/j.molmed.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Ash PEA, Zhang Y-J, Roberts CM, Saldi T, Hutter H, Buratti E, Petrucelli L, Link CD. Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum Mol Genet. 2010;19:3206–3218. doi: 10.1093/hmg/ddq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emde A, Eitan C, Liou L-L, Libby RT, Rivkin N, Magen I, Reichenstein I, Oppenheim H, Eilam R, Silvestroni A, et al. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J. 2015;34:2633–2651. doi: 10.15252/embj.201490493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature. 2014;508:411–415. doi: 10.1038/nature13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics—A consequence of chromatin damage? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585:2041–2048. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamanaka K, Komine O. The multi-dimensional roles of astrocytes in ALS. Neurosci Res. 2017 doi: 10.1016/j.neures.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 94.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim A, Terzian C, Santamaria P, Pélisson A, Purd’homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]