Abstract
Necroptosis has been discovered as a new paradigm of cell death and may play a key role in heart disease and selenium (Se) deficiency. Hence, we detected the specific microRNA (miRNA) in response to Se-deficient heart using microRNAome analysis. For high-throughput sequencing using Se-deficient chicken cardiac tissue, we selected miR-200a-5p and its target gene ring finger protein 11 (RNF11) based on differential expression in cardiac tissue and confirmed the relationship between miR-200a-5p and RNF11 by dual luciferase reporter assay and real-time quantitative PCR (qRT-PCR) in cardiomyocytes. We further explored the function of miR-200a-5p and observed that overexpression of miR-200a-5p spark the receptor interacting serine/threonine kinase 3 (RIP3)-dependent necroptosis in vivo and in vitro. To understand whether miR-200a-5p and RNF11 are involved in the RIP3-dependent necroptosis pathway, we presumed that oxidative stress, inflammation response and the mitogen-activated protein kinase (MAPK) pathway might trigger necroptosis. Interestingly, necroptosis trigger, z-VAD-fmk, failed to induce necroptosis but enhanced cell survival against necrosis in cardiomyocytes with knockdown of miR-200a-5p. Our present study provides a new insight that the modulation of miR-200a-5p and its target gene might block necroptosis in the heart, revealing a novel myocardial necrosis regulation model in heart disease.
Keywords: Selenium, Necroptosis, Cardiomyocytes, miR-200a-5p, RNF11
Graphical abstract
1. Introduction
Selenium (Se) is an essential trace element for the internal environment in the body during homeostasis and pathogenesis [1]. The current issue of Se deficiency in the environment is attracting increasing attention globally, as this deficiency may lead to many diseases, including heart disease, cancer and diabetes. Se deficiency in humans and animals can cause severe myocardial injury, such as Chagas disease in humans [2], Keshan disease [3], mulberry heart of swine [4], chronic heart failure in rats, myocardial hemorrhage and necrosis [5], accompanied by myocardial fiber sarcoplasmic edema and vacuolization of mitochondria in avian species. Research during the last decade of myocardial injury has mainly focused on apoptosis and inflammation. The related literature confirmed that heart disease induced by Se deficiency is mainly due to cardiomyocyte injury caused by oxidative stress and inflammation [6]. Cellular responses to Se deficiency often lead to the induction of inflammation and oxidative overload involving multiple signaling pathways. Jie Y et al. reported that an Se-deficient diet increased the apoptosis of chicken heart in vitro and in vivo, and other researchers have shown that Se deficiency enhances the main pro-apoptotic factors (Bax and p53) in a mouse model [7], [8].
Comprehensive heart disease is intricately connected to myocardial cell death, and the main form of myocardial cell death induced by Se deficiency is necrosis [9]. Necroptosis is a type of non-apoptosis programmed cell death, which is emerging as a new target in inflammation disease and heart disease [10]. Accumulating evidence has revealed that necroptosis is carried out by complex execution pathways and activators, such as TNF-α. Most of our knowledge of necroptosis comes from studies of receptor interacting serine/ threonine kinase 3 (RIP3)-dependent necroptosis, which is mediated by the activation of TNF-α and suppression of Caspase 8 involving receptor interacting serine/threonine kinase 1 (RIP1) and RIP3 complex with the recruitment and phosphorylation of mixed lineage kinase domain-like (MLKL) [11]. MicroRNAs (miRNAs), as non-coding RNAs, can post-translationally modify mRNA by binding to its target sequence to block mRNA translation and protein production, followed by the regulation of different biological reactions, such as cell survival and cell death [12]. Recent studies have shown that miRNA can control necroptosis by modulating related genes [13]. miR-155 can target RIP1 to activate the Akt survival pathway and prevent the occurrence of programmed necrosis of human cardiac progenitor cells after transplantation [14]. In addition, miR-874 plays a key role in cardiomyocyte necrosis and myocardial infarction. miR-874 can mediate necroptosis by targeting Caspase 8 and disable the ability of Caspase 8 to repress the necrotic program [15].
Oxidative stress and cell death occur in the imbalance of reactive oxygen species (ROS) generation, and the high level of ROS production may damage the organelle and DNA and induce necroptosis [16]. Oxidative stress and inflammation have been considered a driving force for RIP3-dependent necroptosis [17]. A related study demonstrated that many ROS produced in mitochondria can activate RIP1 serine residue 161 (S161) autophosphorylation, and this specific process modifies RIP1 to recruit RIP3 to form the necrosome, a key controller of TNF-induced necroptosis [18]. In addition, the importance of excessive inflammation in inducing necroptosis has also been confirmed in vivo in children with inflammatory bowel disease [19]. The ubiquitin-editing enzyme A20 prevents apoptosis induced by the TNF-α pathway as an anti-inflammatory protein. Onizawa et al. reported that A20 restricted necroptosis by limiting the ubiquitination of RIP3 in mouse T cells [20]. As an essential component of the A20 ubiquitin-editing protein, ring finger protein 11 (RNF11) has been implicated in inflammation induced by TNF-α signaling [21]. A specific PPXY motif of RNF11 could bind to the Itch WW domain to facilitate Itch ubiquitination of target proteins, such as RIP1.
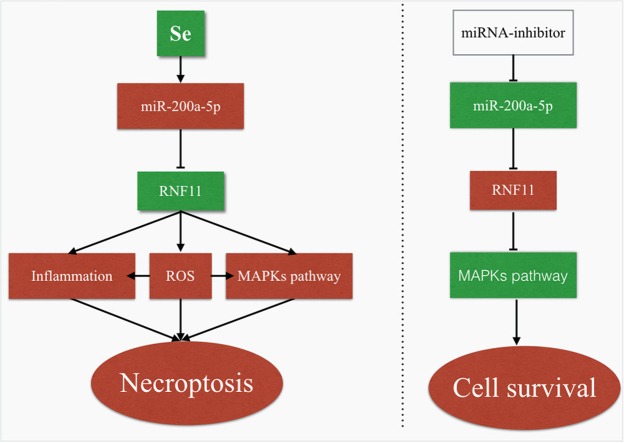
In this setting, the present experiment was designed to investigate whether miRNA can regulate myocardial necrosis induced by Se deficiency. In the proteomics and miRNA genomics detection, miR-200a-5p was identified to be up-regulated under Se-deficient stimulation, and its target gene RNF11 was found by regulating the expression of miR-200a-5p in vitro. We elaborated that the specific Se-deficient-responsive miRNAs likely modulate the target gene RNF11 involved in necroptosis. Thus, through the inhibition of necroptosis and induction, we can suppress necroptosis with knockdown of miR-200a-5p in cardiomyocytes. Our results reveal a novel myocardial necrosis induced by an Se-deficient model composed of miR-200a-5p and RNF11 and new insights for the understanding of heart syndrome treatment.
2. Results
2.1. Se deficiency induces necrotic cell death in the heart
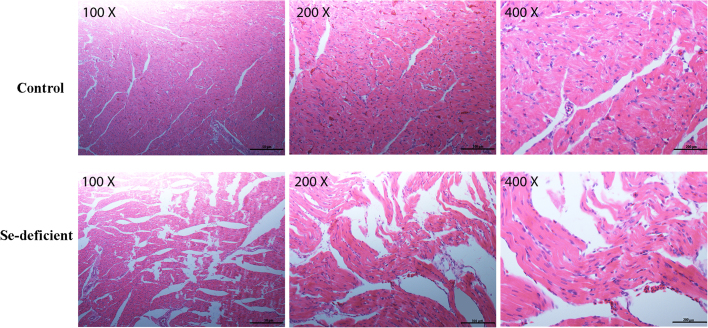
We observed myocardial tissues stained by hematoxylin and eosin (H&E) in the control group and Se-deficient group. The histopathological changes in myocardial tissues are shown in Fig. 1. The myocardial tissues in the control group displayed normal morphologies. However, many typical myocardial necrosis features appeared in the tissues of the Se-deficient group, including myocardial fiber fracture disintegration, myofilament implication and muscle fiber bundles accompanied by inflammatory cell infiltration. All these observations confirmed that Se deficiency induces necrosis in the heart. We reproduced the necrotic model.
Fig. 1.
HE staining for myocardial tissues in the control group and Se-deficient group. Histopathological analysis of the heart in the control group and Se-deficient group.
2.2. miR-200a-5p is involved in Se deficiency-induced cardiac necrosis in the chick through targeting RNF11
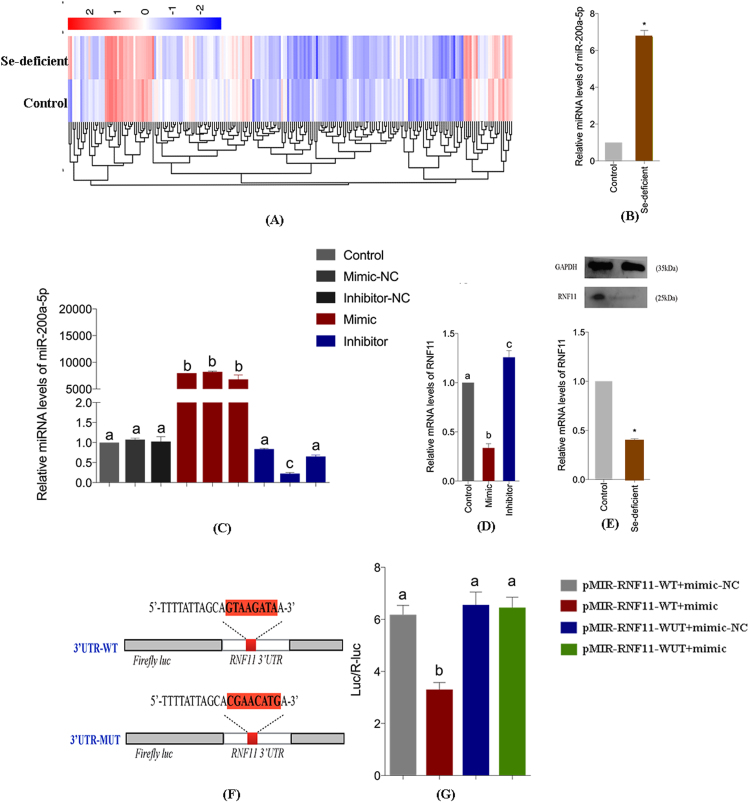
Se deficiency is well known as an important factor inducing necrosis. To determine the effect of Se deficiency on the miRNA expression levels, we performed high-throughput sequencing with Se-deficient chicken cardiac tissue in the miRNA genomics group. Based on miRNA genomics group analysis, we selected the in Se-specific miRNA-miR-200a-5p, which is up-regulated in the Se-deficient cardiac group compared with that in the control group (Fig. 2A, Table. S1). To confirm the genomics group result, we carried out quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to verify the miRNA levels in vivo. Consistent with the predicted results, miR-200a-5p was substantially elevated 3 times in the Se-deficient cardiac compared with the control cardiac (Fig. 2B).
Fig. 2.
Different miRNA and mRNA expression profiles and qRT-PCR detectionin vivoandin vitroand luciferase activity assay. (A) Heat map with intuitive reflection of the features of expression profiles changes in the control group and Se-deficient group. (B) Myocardial tissue miRNA levels of miR-200a-5p in the control group and Se-deficient group. Bars represent the mean ± SD of 30 individuals. Bars represent the mean ± SD of triplicate cell cultures. *p < 0.05 by Student's t-test. (C) To investigate the most appropriate transfection concentration in cardiomyocytes model with transfecting different concentrations of miR-200a-5p-mimic, miR-200a-5p-inhibitor, mimic negative control and inhibitor negative control for 24 h. miRNA levels of miR-200a-5p was determined by RT-PCR. The optimum concentration of miR-200a-5p-mimic and miR-200a-5p-inhibitor used in this paper were always 100 nM and 50 nM, respectively. Bars that do not share the same letters are significantly different (p < 0.05) from each other. The data were expressed as the mean ± SD of triplicate cell cultures. (D) The mRNA levels of RNF11 in the cardiomyocytes transfected with mimic, inhibitor and control cardiomyocytes. Bars that do not share the same letters are significantly different (p < 0.05) from each other. The data were expressed as the mean ± SD of triplicate cell cultures. (E) The mRNA and protein levels of RNF11 in the control myocardial tissue and Se-deficient myocardial tissue. Bars represent the mean ± SD of 30 individuals. Bars represent the mean ± SD of triplicate cell cultures. *p < 0.05 by Student's t-test. (F–G) Correlation between RNF11 and miR-200a-5p in luciferase reporter gene assay results. pMIR-RNF11-WT plasmids was mutated in the miRNA target sites, and designated as pMIR-RNF11-Mut. miR-200a-5p-mimic inhibits RNF11-WT expression but not mutant RNF11-MUT expression. Bars that do not share the same letters are significantly different (p < 0.05) from each other. The data were expressed as the mean ± SD of triplicate cell cultures.
To discovered how miR-200a elicits its effect on cardiac necrosis triggered by Se deficiency, we selected downstream target genes of miR-200a-5p. Using the website-based miRNA target prediction databases, we defined several targets of miR-200a-5p. Some of the predicted targets are implicated in the modulation of inflammation and cell death. We defined target genes through running qRT-PCT in vivo and in vitro (Fig. 2C, Fig. 2D, Fig. 2E). The mRNA expression of RNF11 was reduced by ~ 50% in Se-deficient cardiomyocytes compared with control cardiomyocytes. Additionally, the expression of RNF11 was reduced in response to the increasing level of miR-200a-5p and elevated in response to the decreasing level of miR-200a-5p in cardiomyocytes cultured in vitro. Using the 3’UTR-mediated luciferase activity assay, we found that the miR-200a-5p mimic markedly modulated the luciferase activities driven by RNF11 mRNA 3′UTR plasmids. However, the miR-200a-5p mimic failed to inhibit the luciferase activities driven by the 3′UTR plasmids whose miRNA target sequences were mutated (Fig. 2F, Fig. 2G). These results suggest that RNF11 is a specific downstream target gene of miR-200a-5p and may be involved in cardiac necrosis triggered by Se deficiency.
2.3. miR-200a-5p-mediated modulation of the TNF-α pathway
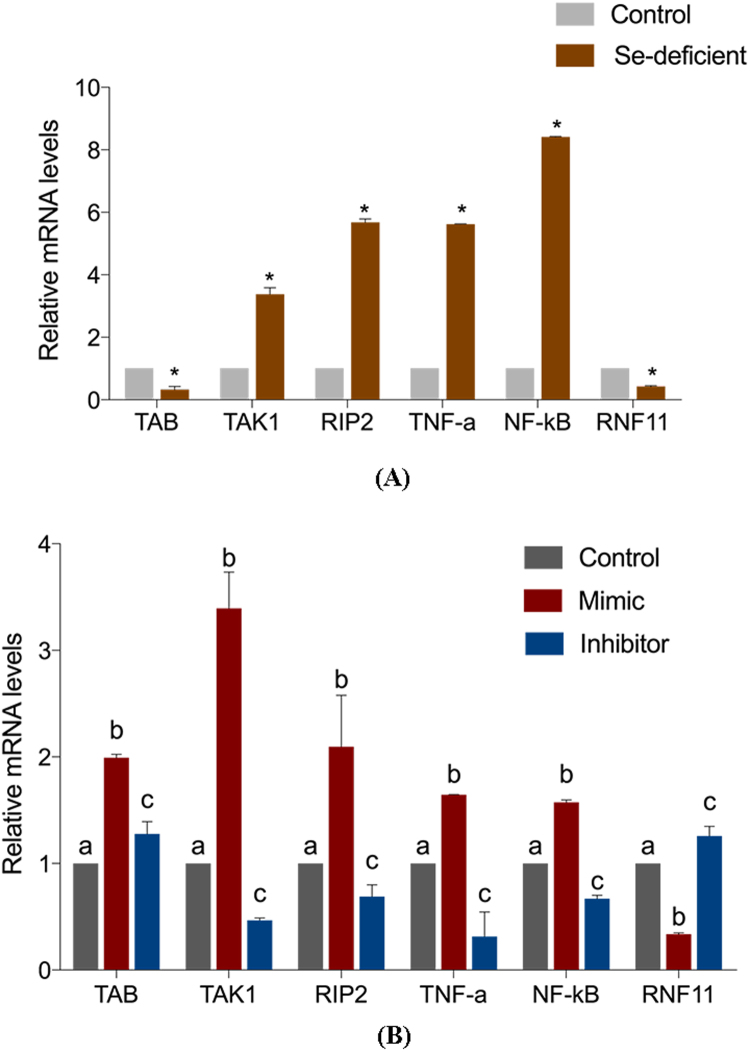
RNF11 is an essential component of the A20 ubiquitin-editing complex and can negatively regulate TNF-α signaling in several cell lines. We have demonstrated that low-grade RNF11 was expressed in Se-deficient heart compared with normal heart in chicken. However, the function of RNF11 whose transcription is inhibited by miR-200a-5p in Se-deficient-triggered cardiac necrosis remains to be determined. Based on the characterization of RNF11, we detected the expression of related genes in the TNF-α pathway by qRT-PCR to explore the role of RNF11 in regulating inflammation and cardiac necrosis. In vitro cultured cardiomyocytes demonstrated reduced abdominal B (TAB) and RNF11 expression, and increased TNF-α, TGF-beta activated kinase 1 (TAK1) and nuclear factor kappa B (NF-κB) expression, resulting in persistent TNF-α signaling in cardiomyocytes over-expressing miR-200a-5p. Opposite results were observed in cardiomyocytes with knockdown of miR-200a-5p. Similar results were validated in an animal model; the levels of TAB and RNF11 were decreased and those of TNF-α, TAK1and NF-κB were increased in the Se-deficient heart compared with the control heart, since the level of miR-200a-5p was elevated over 3 times (Fig. 3).
Fig. 3.
Different mRNA expression of TNF-α-related genes in vivo and in vitro. (A) The mRNA expression of TAB, TAK1, RIP2, TNF-α, NF-κb and RNF11 in the control myocardial tissue and Se-deficient myocardial tissue. Bars represent the mean ± SD of 30 individuals. *p < 0.05 by Student's t-test. (B) The mRNA levels of TAB, TAK1, RIP2, TNF-α, NF-κb and RNF11 in cardiomyocytes transfected with mimic and inhibitor and control cardiomyocytes. Bars represent the mean ± SD of triplicate cell cultures. Bars that do not share the same letters are significantly different (p < 0.05) from each other.
2.4. miR-200a-5p-mediated modulation of RIP3-mediated necroptosis
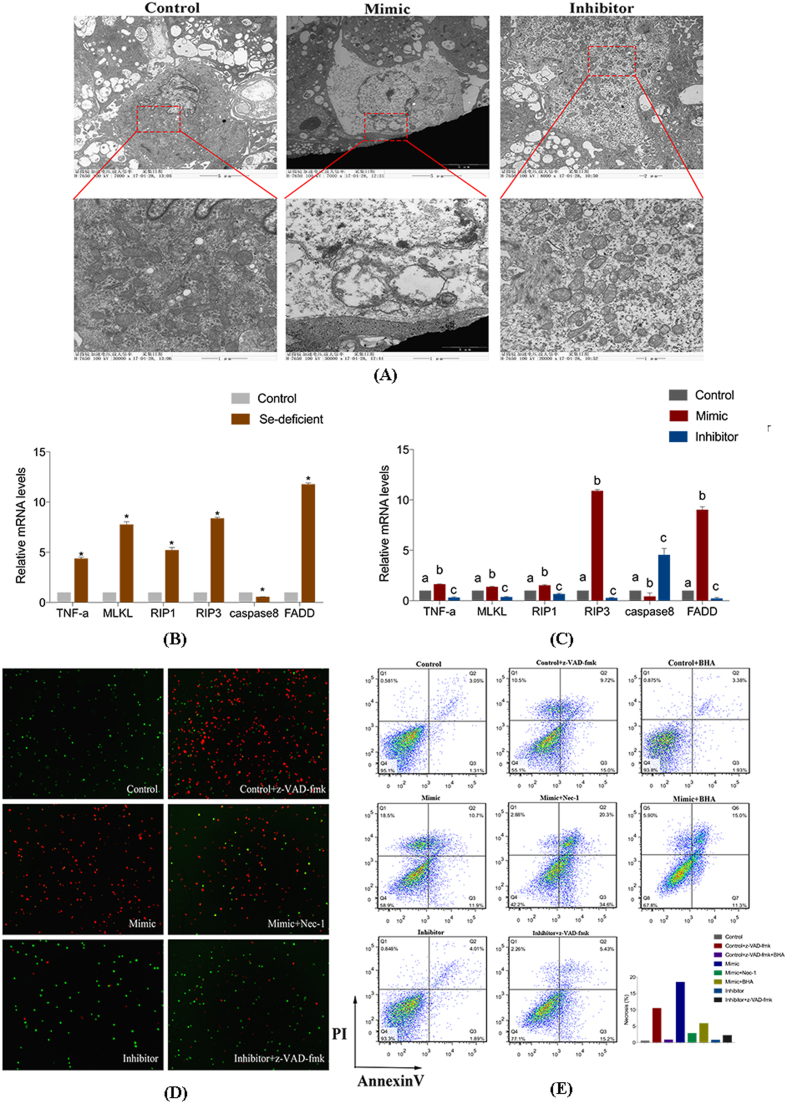
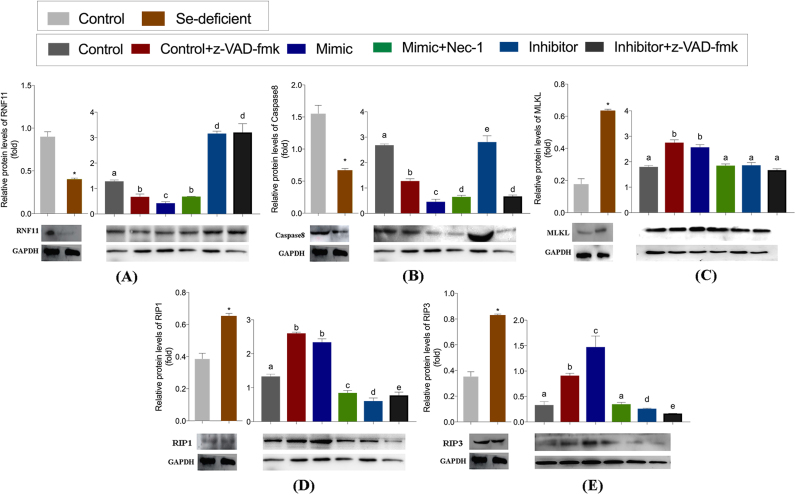
We next determined the type of cardiac necrosis caused by Se deficiency. RIP3 is a key determinant of necroptosis in vivo and in vitro, including cardiomyocytes. We first detected the mRNA levels of RIP1, RIP3, Caspase 8 and MLKL in an animal model. As shown in Fig. 4, Se deficiency increased the release of RIP1, RIP3 and MLKL and inhibited the release of Caspase 8, reflecting necroptosis in the heart. A similar significant effect was observed on the protein expression of RIP1, RIP3, Caspase 8 and MLKL in vivo (Fig. 4B, Fig. 5).
Fig. 4.
miR-200a-mediated modulation of RIP3-mediated necroptosis. (A) The ultrastructural changes of cardiomyocytes transfected with mimic, inhibitor and control cardiomyocytes. (B) The mRNA expression of TNF-α, MLKL, RIP1, RIP3, Caspase8 and FADD in the control myocardial tissue and Se-deficient myocardial tissue. Bars represent the mean ± SD of 30 individuals. *p < 0.05 by Student's t-test. (C) The mRNA levels of TNF-α, MLKL, RIP1, RIP3, Caspase8 and FADD in cardiomyocytes transfected with mimic and inhibitor and control cardiomyocytes. Bars represent the mean ± SD of triplicate cell cultures. Bars that do not share the same letters are significantly different (p < 0.05) from each other. (D) Detection results of staining with the AO/EB of cardiomyocytes. The normal cells are stained green. The early apoptotic cells are stained bright green, and later apoptotic cells are stained orange. The necrotic cells are stained red. (E) Cardiomyocytes were transfected with mimic and inhibitor for 24 h. Cell viabilities and necrosis rate were measured by the fluorescence intensity of Annexin V-FITC and PI to identify healthy (double negative), early apoptotic (Annexin V positive), and necrosis (double-positive) cell populations after treatment of z-VAD-fmk (added TNF-α) with/without BHA or Nec-1.
Fig. 5.
Protein levels of necroptosis-relatedin vivoandin vitro. The protein levels of RNF11, MLKL, RIP1, RIP3 and Caspase8 were detected in vivo and in vitro. Cardiomyocytes were treated with z-VAD-fmk (added TNF-α) with/without Nec-1 after transfected with mimic and inhibitor. Bars represent the mean ± SD. Bars that do not share the same letters are significantly different (p < 0.05) from each other. *p < 0.05 by Student's t-test.
To understand the pathophysiological role of miR-200a-5p and its target gene RNF11 in the heart, we detected whether miR-200a-5p was involved in necroptosis in cardiomyocytes as shown in Fig. 4. AO/EB staining was used to assess necrosis in cardiomyocytes modulated by the level of miR-200a-5p. Knockdown of miR-200a-5p by miR-200a-5p-inhibitor, and overexpress of miR-200a-5p by miR-200a-5p-mimic. Fig. 4D shows that overexpression of miR-200a-5p significantly decreased the number of green cells and increased the number of red cells, and the opposite result was shown in the cardiomyocytes with knockdown of miR-200a-5p and the control cardiomyocytes. Consistent with the results observed by electron microscopy, overpression of the miR-200a-5p cells emerged with membrane rupture and mitochondrial swelling (Fig. 4A). We carried out qRT-PCR to verify the level of RIP1, RIP3, Caspase8 and MLKL to confirm necroptosis. The levels of RIP1, RIP3 and MLKL were elevated and the level of Caspase 8 was reduced by overexpression of miR-200a-5p but not in the cardiomyocytes with knockdown of miR-200a-5p (Fig. 4C). Based on our results, we conclude that miR-200a-5p may have a function role in myocardial necroptosis, since Se-deficient induced cardiac necrosis.
To evaluate the functional relevance of miR-200a-5p and RNF11 in myocardial necroptosis, we used Nec-1, a specific inhibitor of RIP1, to inhibit necroptosis triggered by the miR-200a-mimic. Alternately, z-VAD-fmk was used to prevent necroptosis through the inhibition of miR-200a in vitro. Flow cytometric analysis confirmed these expected observations, exhibiting a 32.3% increase in viability of cells in which miR-200a-5p was inhibited following treatment with z-VAD-fmk, compared with the control cells with z-VAD-fmk treatment (Fig. 4E). No significant difference in the number of surviving cells was observed when comparing cells treated with the miR-200a-5p inhibitor, with or without z-VAD-fmk treatment. Interestingly, cell survival was not significantly improved, but necrosis diminished to a similar extent as control cells, rather apoptosis increased in cardiomyocytes over-expressing miR-200a-5p with necrostatin-1 (Nec-1) treatment. Additionally, the protein levels of RNF11, RIP1, RIP3, Caspase8 and MLKL were examined following stimulation by Nec-1 or z-VAD-fmk in cardiomyocytes (Fig. 5). It was observed that expression of RNF11, Caspase8 and RIP3 did not change significantly following Nec-1 treatment. The only change observed in cells where miR-200a-5p was inhibited with z-VAD-fmk treatment was a decrease in Caspase 8 expression. Therefore, the knockdown of miR-200a-5p may block necroptosis in cardiomyocytes.
2.5. miR-200a-5p-mediated modulation of MAPK activation
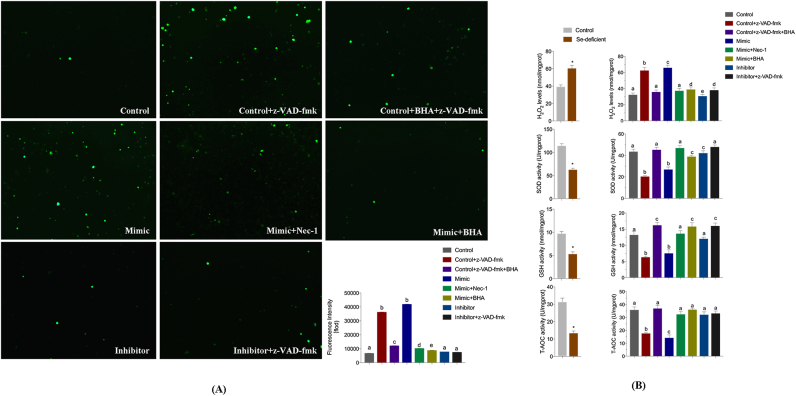
Previously studies have suggested that Se deficiency results in increased oxidative stress in multiple organs associated with high levels of ROS generation, and that mitochondrial ROS are involved in necroptosis in several cell lines. Butylated hydroxyanisole (BHA) is related to various lipophilic compounds that function as antioxidants to scavenge ROS. Here, BHA was used to assay the involvement of ROS in cell death. The inhibitor was used to verify that the regulation of ROS in necroptosis was triggered by the miR-200a-mimic or z-VAD-fmk. Flow cytometric analysis confirmed this observation, as the proportions of necrotic cells decreased by 12.6% in miR-200a-5p-mimic cells with BHA treatment, compared with miR-200a-5p-mimic cells (Fig. 4E).
To assess the relationship between oxidative stress, necroptosis, and miR-200a-5p, the production of ROS, the levels of H2O2, and the activities of GSH, SOD and T-AOC were measured in cardiomyocytes treated with either Nec-1, BHA or z-VAD-fmk. As is presented in Fig. 6, ROS generation and H2O2 was elevated by miR-200a-5p with or without z-VAD-fmk treatment in a dose responsive manner, compared with untreated control cells. In comparison to control cells, GSH, SOD and T-AOC levels decreased when miR-200a-5p was overexpressed in cells and treated with z-VAD-fmk. Interestingly, there was no significant change with BHA treatment and with or without z-VAD-fmk in cardiomyocytes in which expression of miR-200a-5p was knocked down. However, treatment with Nec-1 reduced ROS production in cardiomyocytes overexpressing miR-200a-5p. These results are reflective of the redox states in cardiomyocytes treated with Nec-1, BHA or z-VAD-fmk.
Fig. 6.
miR-200a-5p-mediated modulation of redox states. (A) ROS generation was performed by immunofluorescence using DCFH-DA (green fluorescence, 5 mM) in cells. Cardiomyocytes were treated with z-VAD-fmk (added TNF-α) with/without Nec-1 after transfected with mimic and inhibitor. Cardiomyocytes were visualized using fluorescence microscopy. (B) Oxidative stress markers of H2O2, GSH, SOD and T-AOC contents were measured in vivo and in vitro. Cardiomyocytes were treated with z-VAD-fmk (added TNF-α) with/without BHA or Nec-1 after transfected with mimic and inhibitor. Bars represent the mean ± SD. Bars that do not share the same letters are significantly different (p < 0.05) from each other. *p < 0.05 by Student's t-test.
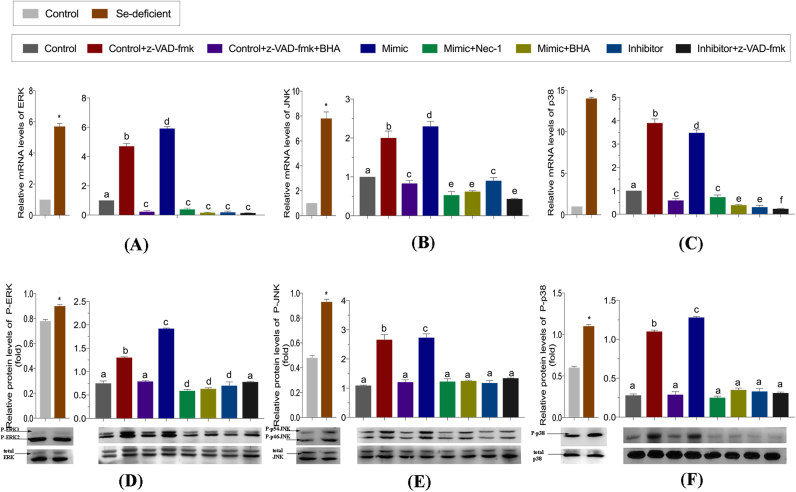
Activation of MAPKs are the key downstream pathways associated with the generation of ROS, and play a significant role in cellular survival. Because miR-200a-5p was demonstrated to induce oxidative stress, we subsequently tested the expression of key kinases in the MAPK pathway. The expression of both the mRNA and phosphorylated proteins of c-jun N-terminal kinase (JNK), p38 and extracellular regulated MAP kinase (ERK) were elevated in miR-200a-5p-mimic treated cells, and was decreased in miR-200a-5p-inhibitor treated cells. This suggests that increasing levels of miR-200a-5p activates the MAPK pathway, and the decreasing of miR-200a-5p inhibits the MAPK pathway (Fig. 7). A reduction in the expression levels of JNK, p38 and ERK were also observed in miR-200a-5p-inhibitor treated cells in the presence of z-VAD-fmk and BHA.
Fig. 7.
miR-200a-5p-mediated modulation of MAPKs activation. (A-C) The mRNA levels of ERK, JNK and p38 were detected in vivo and vitro. Cardiomyocytes were treated with z-VAD-fmk (added TNF-α) with/without BHA or Nec-1 after transfected with mimic and inhibitor. Bars represent the mean ± SD. Bars that do not share the same letters are significantly different (p < 0.05) from each other. *p < 0.05 by Student's t-test. (D-F) The protein levels of phosphorylation ERK, phosphorylation JNK, phosphorylation p38, total ERK, total JNK and total p38 were detected in vivo and in vitro. Cardiomyocytes were treated with z-VAD-fmk (added TNF-α) with/without BHA or Nec-1 after transfected with mimic and inhibitor. Bars represent the mean ± SD. Bars that do not share the same letters are significantly different (p < 0.05) from each other. *p < 0.05 by Student's t-test.
Next, the levels of H2O2 and the activities of GSH, SOD and T-AOC were determined in vivo (Fig. 6B). Activation of MAPKs was also detected in vivo (Fig. 7). ROS generation and H2O2 was significantly increased in the Se-deficient heart tissues (p < 0.05). GSH, SOD and T-AOC content in the Se-deficient group was significantly lower than was observed in the control group (p < 0.05). According to mRNA and protein expression, MAPKs were activated in the Se-deficient group relative to the control group. Taken together, the above results indicate that MAPKs play a critical role in oxidative stress induced by necroptosis in vivo and in vitro.
3. Discussion
Necrosis is the primary form of myocardial injury induced by Se deficiency. However, current research concerning the Se-deficient heart has mainly concentrated on myocardial apoptosis and autophagy instead of necrosis. It is essential to elucidate the pathogenesis and molecular mechanism of myocardial necrosis in heart disease caused by Se deficiency. Our experiments identified the differentially significant miRNA and its target proteins in the Se-deficient animal model. Our present work demonstrated the mechanisms underlying the regulation of miR-200a-5p and its target protein RNF11 on necrosis.
As short chain non-coding RNAs, miRNAs have been shown to play an important role in heart disease in recent years [22]. Pathological over-expression of miR-25 leads to impaired calcium transport and cardiac contractility during severe heart failure, thereby declining cardiac function [23]. Recent works has identified a key gene, FXN, using high-throughput microarray screening in congenital heart disease, that is regulated by miR-145 and that could regulate apoptosis and mitochondrial function and affect the development of congenital heart disease [24]. Ikeda et al. inferred that miRNAs play an important role in different heart diseases by filtering 87 miRNAs that show significant expression levels in heart failure diseases using miRNAome analysis [25]. In this study, we detected a key miRNA in an Se-deficient animal model by miRNAome analysis and confirmed that RNF11 is the downstream target gene of miR-200a-5p in vivo and in vitro. This result may provide a new clue for the study of the miRNA-controlled cellular process in myocardial necrosis.
RNF11 is essential to control inflammatory responses in a negative feedback loop and has strong expression in the heart [26]. Recent studies have established that RNF11 can participate in multiple cellular activities, such as the cell cycle, cell death and cell survival [27]. Dalal et al. confirmed that RNF11 is protective against LPS-induced cell cytotoxicity by negative regulating the NF-κB signaling pathway [28]. RNF11 is the critical component of the ubiquitin-editing protein A20 and limits the duration of the NF-κB pathway and TNF-α pathway [29]. In a paper published in this issue, it was demonstrated that miR-19b expression is inversely related to the RNF11 mRNA levels, indicating a key role for RNF11 in the effect of miR-19b on NF-κB activation [30]. In human LHP-1 monocytes, the depletion of RNF11 prolongs TNF-induced NF-κB and JNK activation and increases RIP1 polyubiquitination [31]. Consistent with our results in vivo and in vitro, overexpression of miR-200a-5p inhibited the expression of RNF11 following the activation of TNF-induced NF-κB and JNK and up regulation of TAK1, TAB, RIP2 and RIP1. RNF11 can bind to the Itch WW domain with its PPXY motif to facilitate Itch ubiquitination of RIP1, which is a key factor in RIPK3-dependent necroptosis [32]. This may be an important link between necroptosis and RNF11 with the involvement of RIP1 in the present study. Moreover, A20, the key building protein of RNF11, can protect cells against necroptosis by restricting the ubiquitination of RIP3 [20]. Taken together, our findings indicate that the depletion of RNF11 might partly induce necroptosis by stimulating inflammation and interacting with RIP1 and A20.
Necroptosis is a necrotic-like cell death that is executed by the ubiquitination of RIP1 and RIP3 during Caspase8 inhibition by recruiting the downstream mediator MLKL [33]. The research confirmed that miRNA can regulate cellular necroptosis by targeting key mediators of the necroptosis pathway, including CLYD, the target gene of miR-19 and miR-181b-1 [34], [35], RIP1, the target gene of miR-155 [14] and Caspase8, the target gene of miR-874 [15]. In addition, It was shown that the regulation of non-necroptosis genes modulates the occurrence of necroptosis. For example, A20 can restrict necroptosis in multiple cell lines [20], and Gpx4 can prevent the occurrence of necroptosis in mouse erythroid precursor cells [36]. These findings indirectly support the results of the present study that Se-deficient diet might cause RIP3-dependent necroptosis in the heart accompanied by enhancement of the level of miR-200a-5p and inhibition of the level of RNF11. In multiple cell types, constitutive and ligand induced necroptosis, such as Caspase8 inhibitor, z-VAD-fmk, enhanced TNF-to induce necroptosis [37]. However, TNF-induced necroptosis can be inhibited by the RIP1 inhibitor, Nec-1, or MLKL inhibitor, NSA [38], [39]. The present study confirmed that Nec-1 can effectively block RIP3-dependent necroptosis induced by miR-200a-5p, and z-VAD-fmk triggers necroptosis with TNF-α treatment in cardiomyocytes. Interestingly, we observed that the depletion of miR-200a-5p prevented myocardial necroptosis induced by z-VAD-fmk/TNF-α treatment, suggesting the inhibition of miR-200a-5p may promote cell survival.
Se-specific induction of miRNA-miR-200a-5p post-transcriptionally modifies RNF11 mRNA, resulting in a modulation of RIP3-dependent necroptosis. The main function of RNF11 is to negatively alter inflammatory pathways, regulating the development of the inflammatory response [26]. In the present study, decreased expression of RNF11 resulting in increased ROS generation, suggesting that RNF11 may be involved in the regulation of ROS and the occurrence of oxidative stress. We previously reported that Se deficiency induced ROS generation increasing and reduced the activity of antioxidant enzymes, resulting in severe oxidative stress in liver, muscle, spleen and vasculature [40], [41], [42]. Excessive production of ROS or an inadequate antioxidant system results in the dysregulation of homeostasis between oxidants and antioxidants in vivo, leading to lipid peroxidation in cellular and mitochondrial membranes and cellular necrosis [43]. ROS include superoxide anion (-O2-), hydroxyl free radical s (-OH) and hydrogen peroxide (H2O2) as well as others. There are 2 categories of antioxidant defense systems in the body, one is the enzyme antioxidant system, the other is the anion-enzyme antioxidant system. The enzyme antioxidant system prevents oxidative stress by the elimination of ROS. For instance, antioxidant enzymes, specifically SOD, capture O2- and convert it into H2O2. This process is believed to be the first line of defense against oxidative stress [44]. GSH is a non-enzymatic antioxidant that participates in the body's redox processes, binding to peroxides and free radicals, to protect tissues against damage caused by free radicals [45]. In this experiment, it was observed that overexpression of miR-200a-5p inhibits transcription of RNF11, resulting in increased ROS production, elevated levels of H2O2, decreased SOD and GSH activity, and decreased activity of T-AOC. These changes were accompanied by inflammation and necroptosis. These changes, however, could be inhibited by treatment with antioxidants or inhibitors of necroptosis. These results indicated that the increased expression of miR-200a-5p induced by Se deficiency likely suppresses RNF11 expression, while the antioxidant enzyme activity and antioxidant capacity is reduced, and ROS production increased. This ultimately resulted in elevated oxidative stress and the induction of myocardial necroptosis.
Oxidative stress caused by the imbalance of cellular antioxidants and reduced generation of ROS is a second messenger of this process. Published data have suggested that TNF-induced ROS generation is dependent on RIP3 [46], and the involvement of ROS is controlled in a positive feedback loop to elevate necrosome formation [47]. High levels of ROS can damage organelles and promote programmed cell death, such as apoptosis and necroptosis [16], [48]. Y. Zhang et al. suggested that mitochondrial ROS resulted in the induction of the positive feedback of necroptosis [18]. This report was consistent with our data, as we observed that the antioxidant system was strengthened and high levels of ROS were produced when RIP3-dependent necroptosis occurred. The MAPK pathway is an important downstream signaling pathway of ROS that is comprised of ERK, p38 and JNK. Related studies have revealed that the MAPK pathways can regulate various biological processes, such as cell growth, cell differentiation, cell death and cell survival, in multiple cell types [49]. A study on colon cancer found that dimethyl fumarate induced necroptosis by depleting cellular, increasing ROS, and activating the MAPK pathway [50]. Metastasis and the tumorigenic ability of cells increased in JNK-deficient progenitor cells [51]. In knockout mouse models, the removal of MAPK p38 resulted in resistance to apoptosis in fibroblasts, and increased cellular survival in cardiomyocytes [52], [53]. In contrast, other researchers have reported that p38 has no effect on cell survival in mouse thymocytes [54]. Together, these findings demonstrate that the MAPK pathway results in different outcomes with respect to cellular death or survival in different cells. In this article, we provided evidence that the knockout of miR-200a-5p induced ROS depletion cooperates with the inhibition of the MAPK pathway and oxidative stress to promote cardiomyocyte survival in response to treatment with triggers of necroptosis. This result sheds light on a possible molecular mechanism for studying the cytoprotective effects of necroptosis via the depletion miR-200a-5p.
In summary, our work identified that miR-200a-5p and its target gene RNF11 could regulate RIP3-dependent necroptosis in cardiomyocytes and animal models. We further revealed that the oxidative stress, the inflammation response and MAPK signaling were involved in miR-200a-5p-induced necroptosis. Accordingly, the regulation of miRNA may provide new insight into the development and treatment of heart disease caused by Se deficiency.
4. Materials and methods
4.1. Treatment of experimental animals
All the procedures used in our experiment were approved by the Institutional Animal Care and Use Committee of the Northeast Agricultural University. Sixty male broilers (1 day old; Weiwei Co. Ltd., Harbin, China) were randomly divided into two groups (30 broilers per group). One group was fed a commercial granulated diet (Control group, 0.2 mg/kg Se in the diet). The other group was fed an Se-deficient granulated diet (Se-deficient group, 0.008 mg/kg Se in the diet). Food and water were provided ad libitum. Broilers were euthanized after 20 days, and myocardial tissues were extracted for inspection. Tissues were frozen immediately with Trizol and stored at − 80 °C until required.
4.2. Cardiomyocyte culture and treatment
Cardiomyocytes were isolated from 12-day-old chicken embryos as described previously by Yablonka and Sato. Briefly, after the dissected hearts were washed, minced, and digested with 0.12% collagenase type II (Invitrogen, Carlsbad, CA, USA), the tissues were triturated by gentle pipetting and filtered to release single cells. Next, cells were subjected to centrifugation and then were suspended in F12 (GIBCO, Grand Island, NY, USA) and were seeded in 6-well plates (Jet, China) coated at a density of 2 × 105cells/cm2 at 37 °C in humidified 95% air and 5% CO2. Necrostatin-1 (Nec-1, Sigma-Aldrich, USA) was dissolved in 10% dimethyl sulfoxide (DMSO, Sigma, USA). Cardiomyocytes were pre-incubated with 200 nM Nec-1 for 24 h or 100 μM Butylated hydroxyanisole (BHA) (Sigma, MO, USA) for 30 min or 30 nM z-VAD-fmk (Calbiochem, San Diego, CA, USA) for 24 h before the addition of TNF-α (R&D, Abingdon, United Kingdom) after transfection for 24 h.
4.3. MicroRNAome analysis
Total RNA was extracted from the myocardial tissues of the control and Se-deficient groups and was processed according to the manufacturer's instructions (Novogene Bioinformatics Technology Co., Ltd.). After quantification and qualification of the total RNA, the RNA was used as input material for the synthesis of microRNA libraries. The sequencing libraries used the NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer's recommendations. Bioinformatics analysis of the differential data from the control and Se-deficient groups were performed using the DESeq R package.
4.4. Sections for electron microscopy
For the tissue ultrastructural examination, the myocardial tissues (size, 1.0 × 1.0 × 1.0 mm) were fixed in 2.5% glutaraldehyde phosphate-buffered saline (v/v, pH 7.2) and post fixed in 1% osmium tetroxide (v/v), and then the tissues were stained with 4.8% uranyl acetate. Next, ultra-thin sections were cut, mounted, washed, impregnated, post-stained and incubated with lead citrate for analysis via microscopy. The microphotographs were acquired using a transmission electron microscope (GEM-1200ES, Japan).
For the cell ultrastructural examination, cardiomyocytes were collected into a tube and centrifuged at 250×g for 10 min, and then the supernatant was discarded. Cells at the bottom of the tube were mixed with 2.5% glutaraldehyde phosphate-buffered saline (v/v, pH 7.2), post fixed in 1% osmium tetroxide (v/v) and dehydrated in a graded series of ethanol concentrations, followed by embedding in Embed812 resin. Next, the ultra-thin sections were cut, mounted, washed, impregnated, post-stained and incubated with lead citrate for analysis by microscopy. The microphotographs were acquired using a transmission electron microscope (GEM-1200ES, Japan).
4.5. Histopathological examination
Myocardial tissues were rapidly fixed in 10% formaldehyde for at least 24 h and were embedded in paraffin for microscopic examination. From the prepared paraffin blocks, sections (5-μm thick) were cut, obtained and stained with hematoxylin and eosin (H&E) for light microscopic observation.
4.6. Cell death assays
Acridine orange/ethidium bromide (AO/EB) was used to detect the live and dead cells (apoptosis and necrosis). Briefly, cells were grown on 6-well plates at a density of 3 × 105 mL−1 and were washed with PBS and stained with AO/EB for 5 min. The cells were then photographed under a fluorescence microscope.
4.7. Transfection of the miR-200a-5p-mimic and miR-200a-5p-inhibitor
Synthetic, chemically modified short RNA oligonucleotides were purchased from Shanghai Gene Pharma Co. Ltd. The miR-200a-5p mimic and mimic negative control (mimic-NC) sequences are 5′-CAUCUUACUAGACAGUGCUGGA-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′, respectively. The miR-200a-5p inhibitor and inhibitor negative control (inhibitor-NC) sequences are 5′-mUmCmCmAmGmCmAmCmUmGmUmCmUmAmGmUmAmAmGmAmUmG-3’ (m represents -Ome) and 5′- CAGUACUUUUGUGUAGUACAA-3′, respectively. Cells were transfected with the miR-200a-5p-mimic, mimic-NC and inhibitor-NC at 100 nM and with the miR-200a-5p-inhibitor at 50 nM. The transfection reactions were performed using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM medium according to the manufacturer's instruction.
4.8. Preparation of the luciferase construct of RNF11 3′UTR and luciferase activity assay
The pMIR-REPORT plasmids for the miR-200a-5p target RNF11 3'UTR were constructed as wild-type (WT) pMIR-RNF11 containing two tandem repeats of miR-200a-5p response elements from RNF11 3'UTR or mutant (MUT) pMIR-RNF11. The sequences of the single-stranded oligo pairs were used to generate the pMIR- RNF11 constructs (WT and MUT). The oligonucleotides were then annealed and inserted into the pMIR-REPORT vector (Thermo Fisher). The empty vector of pMIR-REPORT was used as the negative control. Cells were co-transfected with the plasmid constructs of pMIR-RNF11-WT or pMIR-RNF11-MUT and miR-mimic using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Forty-eight hours after transfection, luciferase activity was performed using the Dual-Luciferase Reporter Assay System (Promega). The activity of Renilla luciferase was normalized to the activity of Firefly luciferase (Renilla LUC/ Firefly LUC).
4.9. Real-time quantitative PCR analysis on mRNA and miRNA levels
For the quantification of miR-200a-5p and mRNA of target genes by RT-PCR, total miRNA and total RNA were isolated from myocardial tissues and cultured cardiomyocytes. Reverse transcription was performed using the miRcute microRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech Co. Ltd., Beijing) according to the manufacturer's instructions (Roche). The primers for the detection of miR-200a-5p and target mRNA by RT-PCR are shown in Table S2. β-actin or U6 was used as an internal reference. qRT-PCR was performed using the Light Cycler®480 System (Roche, Basel, Switzerland) and Fast Universal SYBR Green Master Mix (Roche, Basel, Switzerland). Only one peak for each PCR product was shown in the melting curve analysis. The relative abundance of mRNA was calculated according to the 2-∆∆Ct method, accounting for gene-specific efficiencies and was normalized to the mean expression of the abovementioned index.
4.10. Western blot analysis
Total protein was assessed by SDS-polyacrylamide gel electrophoresis under reducing conditions on 12% gels and then was transferred to nitrocellulose membranes using a tank transfer at 80 mV in Tris-glycine buffer containing 20% methanol for 12 h at 4 °C. Nitrocellulose membranes were blocked for 1 h with 5% bovine serum albumin (BSA) at 37 °C and were incubated for 12 h with the following antibodies: RNF11 (1:800), RIPK1 (1:1500), RIPK3 (1:1500), Caspase8 (1:1000), MLKL (1:1000), JNK (1:2000), ERK (1:1000), p38 (1:1000), PhosphoPlus JNK (1:1000), PhosphoPlus ERK (1:2000) and PhosphoPlus p38 (1:1000) at 4 °C. Next, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody against rabbit IgG (1:1500, Santa Cruz, CA, USA) using the ECL kit (Kangweishiji Biotechnology, Beijing, China). The GAPDH content was analyzed as the loading control with rabbit polyclonal antibody (Sigma, USA).
4.11. Measurement of ROS
The levels of ROS in cardiomyocytes were measured using a detection kit from Beyotime Institute of Biotechnology (Nantong, China) according to the manufacturer's instructions. Fluorescence distribution was detected by fluorospectrophotometer analysis at 488 nm and 525 nm. ROS-associated fluorescent signals were quantified using Image J software (Broken Symmetry Software, Albert, Cardona). The relative intensities were reported in arbitrary units per cell.
4.12. Determination of oxidative stress markers
Cells were grown on 6-well plates at a density of 3 × 105 mL−1 and were collected with jets of saline, centrifuged at 700×g, and the supernatant was collected. Hydrogen peroxide (H2O2), glutathione (GSH), superoxide dismutase (SOD) and total antioxidant capability (T-AOC) contents were measured by the detection kits (Beoytime Bioengineering Institute, Nantong, China), according to the manufacturer protocols. SOD activity was measured at 25 C by the method of auto-oxidation of pyrogallol in 50 mM Tris/HCl, pH 8, with 100 mM pyrogallol (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The total anti-oxidative capacity (T-AOC) in digestive organs was determined by Opara et al.
4.13. Flow cytometry analysis of necrosis
The Annexin V-FITC/PI apoptosis detection kit was used to treat the transfected cardiomyocytes according to the manufacturer's instructions. Live cells were quantitated by flow cytometry of cells negative for DAPI (4, 6-diamidino-2-phenylindole). For apoptosis and necroptosis detection, cardiomyocytes were incubated with 5 μL of Annexin V-FITC and 5 μL of PI in 1 mL of binding buffer for 30 min in the dark. After incubation, apoptotic, necrotic and live cell populations were detected by flow cytometric analysis.
4.14. Statistical analysis
Statistical analyses of all data were performed using GraphPad Prism (version 5.0, Graph Pad Software Inc., San Diego, CA, USA). All the data were analyzed by Student's t-test or one-way ANOVA, had a normal distribution and passed equal variance testing. Quantitative data are presented as the mean ± SD. Samples with different superscript letters represented statistically significant differences (P < 0.05).
Acknowledgments
Shiwen Xu and Xingen Lei conceived and designed the experiments. Tianshu Yang, Changyu Cao, Jie Yang, Tianqi Liu and Ziwei Zhang performed the experiments. Tianshu Yang analyzed the data and wrote the paper. Shiwen Xu assisted in critically revising the manuscript.
Acknowledgments
Disclosure of potential conflicts of interest
The authors declare that there are no conflicts of interest.
Funding
This study was supported by the International (Regional) Cooperation and Exchange Projects of the National Natural Science Foundation of China (31320103920), National Natural Science Foundation of China (31402267) and Degree and Postgraduate Education Teaching Reform Project of Heilongjiang (JGXM_HLJ_201676).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.11.025.
Contributor Information
Xin Gen Lei, Email: xl20@cornell.edu.
Ziwei Zhang, Email: zhangziwei@neau.edu.cn.
Shiwen Xu, Email: shiwenxu@neau.edu.cn.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Eriksson L.C. Selenium homeostasis and thioredoxin reductase induction at long term treatment with selenite in tumor preventive doses. Adv. Funct. Mater. 2006;20(3):377–385. [Google Scholar]
- 2.Yusuf S.W., Rehman Q., Casscells W. Cardiomyopathy in association with selenium deficiency: a case report. J. Parenter. Enter. Nutr. 2002;26(1):63. doi: 10.1177/014860710202600163. [DOI] [PubMed] [Google Scholar]
- 3.Li C. Selenium deficiency and endemic heart failure in China: a case study of biogeochemistry for human health. Ambio. 2013;36(1):90. doi: 10.1579/0044-7447(2007)36[90:sdaehf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Pallarés F.J. Vitamin E and selenium concentrations in livers of pigs diagnosed with mulberry heart disease. J. Vet. Diagn. Investig. 2002;14(5):412. doi: 10.1177/104063870201400509. [DOI] [PubMed] [Google Scholar]
- 5.Lymbury R.S., Marino M.J., Perkins A.V. Effect of dietary selenium on the progression of heart failure in the ageing spontaneously hypertensive rat. Mol. Nutr. Food Res. 2010;54(10):1436. doi: 10.1002/mnfr.201000012. [DOI] [PubMed] [Google Scholar]
- 6.Liu W. Selenoprotein W was correlated with the PRotective Effect Of Selenium On Chicken Myocardial Cells From Oxidative Damage. Biol. Trace Elem. Res. 2016;171(2):419–426. doi: 10.1007/s12011-015-0529-7. [DOI] [PubMed] [Google Scholar]
- 7.Yang J. Interplay between autophagy and apoptosis in selenium deficient cardiomyocytes in chicken. J. Inorg. Biochem. 2017;170:17. doi: 10.1016/j.jinorgbio.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Cui J. Correlation between oxidative stress and L-type calcium channel expression in the ventricular myocardia of selenium-deficient mice. J. Int. Med. Res. 2012;40(5):1677. doi: 10.1177/030006051204000507. [DOI] [PubMed] [Google Scholar]
- 9.Baroldi G. Different types of myocardial necrosis in coronary heart disease: a pathophysiologic review of their functional significance. Am. Heart J. 1975;89(6):742–752. doi: 10.1016/0002-8703(75)90189-1. [DOI] [PubMed] [Google Scholar]
- 10.Andreas Linkermann D.R.G. Necroptosis. N. Engl. J. Med. 2014;370(5):455. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günther C. Caspase-8 regulates TNF-alpha induced epithelial necroptosis and terminal ileitis. Nature. 2011;477(7364):335. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bing C. Roles of microRNA on cancer cell metabolism. J. Transl. Med. 2012;10(1):1–12. doi: 10.1186/1479-5876-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Z S. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6(11):8474. doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J. MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1. J. Cell. Mol. Med. 2011;15(7):1474–1482. doi: 10.1111/j.1582-4934.2010.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4(7):e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berghe T.V. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ. 2010;17(6):922. doi: 10.1038/cdd.2009.184. [DOI] [PubMed] [Google Scholar]
- 17.Shindo R. Critical contribution of oxidative stress to TNFα-induced necroptosis downstream of RIPK1 activation. Biochem. Biophys. Res. Commun. 2013;436(2):212–216. doi: 10.1016/j.bbrc.2013.05.075. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y. RIP1 autophosphorylation is promoted by mitochondrial ROS and is essential for RIP3 recruitment into necrosome. Nat. Commun. 2017;8:14329. doi: 10.1038/ncomms14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierdomenico M. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am. J. Gastroenterol. 2014;109(2):279–287. doi: 10.1038/ajg.2013.403. [DOI] [PubMed] [Google Scholar]
- 20.Onizawa M. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 2015;16(6):618. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pranski E.L. Neuronal RING finger protein 11 (RNF11) regulates canonical NF-κB signaling. J. Neuroinflamm. 2012;9(1):67. doi: 10.1186/1742-2094-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda S., Pu W.T. Expression and function of microRNAs in heart disease. Curr. Drug Targets. 2010;11(8):913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 23.Wahlquist C. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508(7497):531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L. MiRNA-145 regulates the development of congenital heart disease through targeting FXN. Pediatr. Cardiol. 2016;37(4):629–636. doi: 10.1007/s00246-015-1325-z. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda S. Altered microRNA expression in human heart disease. Physiol. Genom. 2007;31(3):367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 26.Kitching R. The RING-H2 protein RNF11 is differentially expressed in breast tumours and interacts with HECT-type E3 ligases. Biochim. Biophys. Acta. 2003;1639(2):104. doi: 10.1016/j.bbadis.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Azmi P., Seth A. RNF11 is a multifunctional modulator of growth factor receptor signalling and transcriptional regulation. Eur. J. Cancer. 2005;41(16):2549–2560. doi: 10.1016/j.ejca.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Dalal N.V. RNF11 modulates microglia activation through NF-κB signalling cascade. Neurosci. Lett. 2012;528(2):174. doi: 10.1016/j.neulet.2012.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shembade N. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. EMBO J. 2009;28(5):513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gantier M.P. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 2012;40(16):8048. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shembade N. Essential role for TAX1BP1 in the termination of TNF-alpha-, IL-1- and LPS-mediated NF-kappa B and JNK signaling. EMBO J. 2007;26(17):3910. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacque E., Ley S.C. RNF11, a new piece in the A20 puzzle. EMBO J. 2009;28(5):455–456. doi: 10.1038/emboj.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau A. RIPK3-mediated necroptosis promotes donor kidney inflammatory injury and reduces allograft survival. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transplant. Surg. 2013;13(11):2805–2818. doi: 10.1111/ajt.12447. [DOI] [PubMed] [Google Scholar]
- 34.Ye H. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012;40(12):5201. doi: 10.1093/nar/gks175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iliopoulos D. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010;39(4):493. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canli Ö. Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood. 2016;127(1):139. doi: 10.1182/blood-2015-06-654194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong K., Sun X., Ke G. Involvement of autophagy in z-VAD-FMK induced photoreceptor necroptosis, a caspase-independent cell death, after experimental retinal detachment. Arvo Meet. Abstr. 2013;54(6) [Google Scholar]
- 38.Liao D. Necrosulfonamide inhibits necroptosis by selectively targeting the mixed lineage kinase domain-like protein. Med. Chem. Commun. 2014;5(3):333–337. [Google Scholar]
- 39.G. Trichonas, et al., Identification of Necroptosis as a Mechanism of Photoreceptor Damage After Retinal Detachment and Nec-1 as a Potential Treatment, 2009.
- 40.Yao L. Roles of oxidative stress and endoplasmic reticulum stress in selenium deficiency-induced apoptosis in chicken liver. Biometals. 2015;28(2):255–265. doi: 10.1007/s10534-014-9819-3. [DOI] [PubMed] [Google Scholar]
- 41.Yu J. The role of nitric oxide and oxidative stress in intestinal damage induced by selenium deficiency in chickens. Biol. Trace Elem. Res. 2015;163(1–2):144. doi: 10.1007/s12011-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 42.Cao C. Dietary selenium increases the antioxidant levels and ATPase activity in the arteries and veins of poultry. Biol. Trace Elem. Res. 2016;172(1):222–227. doi: 10.1007/s12011-015-0584-0. [DOI] [PubMed] [Google Scholar]
- 43.Ferrer M.D. Enzyme antioxidant defences and oxidative damage in red blood cells of variegate porphyria patients. Redox Rep. Commun. Free Radic. Res. 2009;14(2):69. doi: 10.1179/135100009X392502. [DOI] [PubMed] [Google Scholar]
- 44.Spector A. Oxidative stress-induced cataract: mechanism of action. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1995;9(12):1173. [PubMed] [Google Scholar]
- 45.Giblin F.J. Glutathione: a vital lens antioxidant. J. Ocul. Pharmacol. Ther. Off. J. Assoc. Ocul. Pharmacol. Ther. 2000;16(2):121. doi: 10.1089/jop.2000.16.121. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D.W. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325(5938):332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 47.Schenk B., Fulda S. Reactive oxygen species regulate Smac mimetic|[sol]|TNF|[alpha]|-induced necroptotic signaling and cell death. Oncogene. 2015;34(47):5796. doi: 10.1038/onc.2015.35. [DOI] [PubMed] [Google Scholar]
- 48.D'Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8(10):813. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 49.Ballif B.A., Blenis J. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2001;12(8):397. [PubMed] [Google Scholar]
- 50.Xie X. Dimethyl fumarate induces necroptosis in colon cancer cells through GSH depletion/ROS increase/MAPKs activation pathway. Br. J. Pharmacol. 2015;172(15):3929. doi: 10.1111/bph.13184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atala A. Re: JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc. Natl. Acad. Sci. USA. 2013;189(5):1989. doi: 10.1016/j.juro.2013.01.065. [DOI] [PubMed] [Google Scholar]
- 52.Roux P.P., Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. Mmbr. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee I. Role of TLR4/NADPH oxidase/ROS-activated p38 MAPK in VCAM-1 expression induced by lipopolysaccharide in human renal mesangial cells. Cell Commun. Signal. 2012;10(1) doi: 10.1186/1478-811X-10-33. (33-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirois J. The atypical MAPK ERK3 controls positive selection of thymocytes. Immunology. 2014;145(1):161–169. doi: 10.1111/imm.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material