Abstract
Candida albicans is the most common human fungal pathogen causing diseases ranging from mucosal to systemic infections. As a commensal, C. albicans asymptomatically colonizes mucosal surfaces; however, any disruption in the host environment or under conditions of immune dysfunction, C. albicans can proliferate and invade virtually any site in the host. The ability of this highly adaptable fungal species to transition from commensal to pathogen is due to a repertoire of virulence factors. Specifically, the ability to switch morphology and form biofilms are properties central to C. albicans pathogenesis. In fact, the majority of C. albicans infections are associated with biofilm formation on host or abiotic surfaces such as indwelling medical devices, which carry high morbidity and mortality. Significantly, biofilms formed by C. albicans are inherently tolerant to antimicrobial therapy and therefore, the susceptibility of Candida biofilms to the current therapeutic agents remains low. The aim of this review is to provide an overview of C. albicans highlighting some of the diverse biofilm-associated diseases caused by this opportunistic pathogen and the animal models available to study them. Further, the classes of antifungal agents used to combat these resilient infections are discussed along with mechanisms of drug resistance.
Keywords: Candida albicans, biofilms, infections, antifungals, drug resistance
Clinical significance and therapeutic implications of biofilms formed by the fungal pathogen Candida albicans.
INTODUCTION
Candida albicans is the most common fungal human pathogen causing diseases ranging from superficial mucosal to life-threatening systemic infections (Pfaller and Diekema 2007; Ganguly and Mitchell 2011; Calderone 2012). This opportunistic pathogen is part of the commensal human micoflora that asymptomatically colonizes many areas of the human body where its proliferation is controlled by the host immune system (Williams et al. 2013). However, under conditions of immune suppression or any disruption to the host environment, C. albicans can rapidly transition into a pathogen, causing a variety of infections (Finkel and Mitchell 2011; Mathe and Van Dijck 2013). In fact, C. albicans is one of the most often identified agents in nosocomial infections capable of invading virtually any site of the human host from deep tissues and organs to superficial sites (Perlroth, Choi and Spellberg 2007). More significantly, C. albicans is adept at adhering to catheters and various indwelling medical implants, and is currently ranked by the Center for Disease Control as the third most commonly isolated bloodstream pathogen in hospitalized patients with a mortality rate of up to 50% (Wisplinghoff et al. 2004; Tournu and Van Dijck 2012; Mathe and Van Dijck 2013).
As a dimorphic fungus, the ability of C. albicans to transition from commensal to pathogen is primarily the result of its aptitude to morphologically switch between yeast and hyphal forms, a property that is central to its pathogenesis and ability to form biofilms (Jabra-Rizk, Falkler and Meiller 2004; Ramage et al. 2005; Calderone 2012; Chauvel et al. 2012) (Fig. 1A). In fact, the majority of diseases caused by this pathogen are associated with biofilm formation on host or abiotic surfaces (Nett and Andes 2006; Mathe and Van Dijck 2013). Biofilms are structured 3D communities of surface-associated microbial populations embedded in a matrix of extracellular polysaccharides, proposed to provide a structural scaffold and protection for biofilm cells (O'Toole, Kaplan and Kolter 2000; Lewis 2001; Costerton, Montanaro and Arciola 2005; Ghannoum et al. 2015). Therefore, in a biofilm, microbes are afforded a stable environment and can tolerate extremely high concentrations of antimicrobials. The impact of these biofilms on public health is dramatic as cells released from biofilms can migrate into the bloodstream and cause systemic infections with high mortality (Finkel and Mitchell 2011). Importantly, the increase in drug resistance has provided a strong impetus to understand the mechanisms of the enhanced tolerance of biofilm-associated infections to antimicrobial therapy (Tournu and Van Dijck 2012). In this review, we provide an overview of the fungal pathogen C. albicans highlighting some of the diverse biofilm-associated diseases caused by this diverse species and the animal models available to study them. Further, the antifungal classes used to combat these infections are discussed, along with mechanisms of drug resistance.
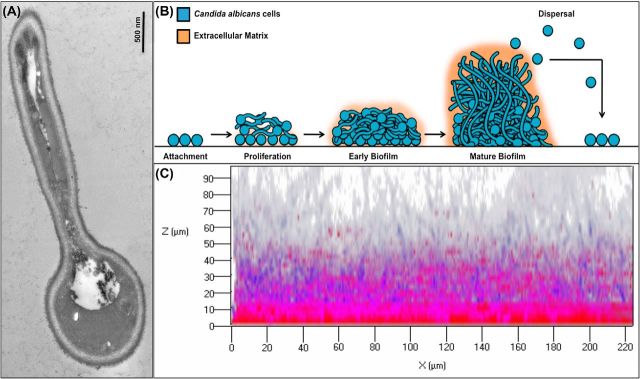
Figure 1.
Candida albicans morphogenesis and biofilm development. (A) Transmission electron micrograph of a germinating yeast cell. Germ tube can be seen as a filament, which continues to elongate forming a hypha. (B) A schematic illustrating the stages of C. albicans biofilm development. Adherence: yeast cells adhere to a substrate forming a yeast basal layer. Initiation: cells propagate and form germ tubes. Maturation: hyphae are formed and extracellular matrix accumulates. Dispersal: cells are released from the biofilm and are dispersed to seed new locations. (C) Confocal scanning laser microscopy of in vitro grown C. albicans biofilm; 48 h biofilm stained with FUN1 (blue) vital fungal stain and the biofilm polysaccharide matrix stain Concanavalin A (ConA) (red). Images were obtained using a Zeiss 710 confocal microscope by LSM 5 Image Browser software at a resolution of 512 × 512 pixels, with an average of 8 images per line. A series of images at ≤1 μm intervals in the z-axis were acquired for the full depth of the biofilm.
Candida albicans biofilm formation and development
The majority of C. albicans infections are associated with its ability to form biofilms (Jabra-Rizk, Falkler and Meiller 2004; Nett and Andes 2006; Finkel and Mitchell 2011; Tournu and Van Dijck 2012; Mathe and Van Dijck 2013) (Fig. 1).In vitro experiments have shown biofilm development to occur in a series of sequential steps over a period of 24–48 h (Ricicova et al. 2010; Mathe and Van Dijck 2013) (Fig. 1B). The initial step consists of the adhesion of single fungal yeast cells to the substrate forming a foundation of a basal yeast cell layer (adherence step) (Nett and Andes 2006; Finkel and Mitchell 2011; Nobile et al. 2012; Tournu and Van Dijck 2012). This is followed by a phase of cell proliferation across the surface and filamentation where cells form elongated projections that continue to grow into the filamentous hyphal forms (initiation step). The production of hyphae is a hallmark of the initiation of biofilm formation followed by the accumulation of an extracellular polysaccharide matrix as the biofilm matures (maturation step) (Fig. 1C). Finally, in the last step, non-adherent yeast cells are released from the biofilm into the surroundings where they can colonize other surfaces (dispersal step). Dispersion of biofilm-associated cells carries great clinical significance as released cells can initiate formation of new biofilms or disseminate into host tissues and therefore, they are associated with candidemia and disseminated invasive disease (Uppuluri et al. 2010; Tournu and Van Dijck 2012).
Interestingly, investigations of quantitative and qualitative properties of cells released from biofilms demonstrated that the majority of dispersed cells are yeast cells suggesting that the transition from yeast to hyphae that occurs during biofilm initiation may be reversed for dispersal. More intriguingly, studies described the dispersed cells to have distinct phenotypes from planktonic cells where they display elevated adherence, filamentation capacity and increased pathogenicity in animal models. Combined, these findings indicate that cells released during the dispersal step are uniquely equipped to seed new biofilms and sites of infection (Uppuluri et al. 2010; Finkel and Mitchell 2011). Of note, genetic analysis indicated that both yeast cells and hyphae are critical for biofilm formation, suggesting that each cell type has unique roles in the process (Nobile and Mitchell 2006). However, biofilms from in vivo catheter infection models appear more complex than those formed in vitro, with yeast and hyphae found to be interspersed. In both in vitro and in vivo model systems however, extracellular matrix material is bound to both yeast and hyphal cells and is typically interspersed throughout the biofilm (Finkel and Mitchell 2011) (Fig. 1B and C). The establishment of the biofilm extracellular matrix represents a unique characteristic of biofilms (Tournu and Van Dijck 2012); C. albicans biofilm matrix is complex, with major polysaccharide constituents being α-mannan, β-1,6 glucan and β-1,3 glucan (Hall and Gow 2013; Mitchell et al. 2015). Although a relatively minor component, β-1,3 glucan is considered the critical matrix polysaccharide as it was linked to biofilm resistance to antifungals by impeding drug diffusion (Taff et al. 2012, 2013). In fact, previous studies have shown elevated β-1,3 glucan levels to be a characteristic of biofilm cells in both the fungal cell wall and as a secreted form.
Another phenomenon that plays a pivotal role in microbial biofilms is quorum sensing or cell–cell communication, where microbial behaviors or responses in a biofilm are governed by cell density mediated by secreted signaling molecules (Mathe and Van Dijck 2013; Demuyser, Jabra-Rizk and Van Dijck 2014). The most well-studied quorum-sensing molecule produced by C. albicans is farnesol, which is secreted extracellularly and accumulates in supernatants of mature biofilms. Significantly, farnesol inhibits hyphal formation and therefore, by promoting yeast cell formation, it may aid in biofilm dispersal (Ramage et al. 2002b; Finkel and Mitchell 2011; Krom et al. 2015). Interestingly, our previous studies demonstrated that at high concentrations, farnesol triggers a classical mammalian-like process of apoptosis in C. albicans (Scheper et al. 2008; Shirtliff et al. 2009). In subsequent studies, we characterized the mechanism to involve oxidative stress and accumulation of intracellular reactive oxygen species (ROS) mediated by farnesol, ultimately resulting in cell death (Zhu et al. 2011). Combined, these findings are intriguing as they paint a sophisticated role for farnesol in fungal biofilms, possibly involving altruistic cell death.
Pathogenesis and clinical significance of C. albicans biofilms
Estimates by the NIH indicate that microbial biofilms account for over 80% of infections in the United States. Similarly, the vast majority of candidal infections are associated with biofilm formation (Ramage et al. 2005). Fungal infectious biofilms can develop on a variety of surfaces including host tissues and implanted biomaterials including vascular catheters. Further, biofilms can potentiate systemic infection, and the presence of a C. albicans biofilm structure has been implicated as a risk factor for increased patient mortality (Mathe and Van Dijck 2013). A number of other properties and virulence factors possessed by C. albicans are known to promote its biofilm-forming ability and contribute to tissue damage and its persistence within the host (Mayer, Wilson and Hube 2013). First and foremost is the property of morphological switching as the distinct morphological states of C. albicans dictate phases of colonization, growth and dissemination; the yeast form has been associated with both initial attachment and dissemination, while the filamentous hyphal form enables C. albicans to invade host tissue and form a mature biofilm (Baillie and Douglas 1999). Hyphal morphogenesis can be triggered by a variety of factors such as temperature, serum, amino acid availability, pH level and CO2 presence. Significantly, hyphal morphogenesis is a key mechanism for C. albicans to evade killing by host phagocytic cells. Phagocytosis induces a switch in morphology from yeast to hyphae, which elongate and eventually puncture the macrophage membrane. This results in lysis and killing of macrophages thereby allowing ingested C. albicans to escape (Berman and Sudbery 2002; Jacobsen et al. 2012; Kong and Jabra-Rizk 2015).
As with other microbial pathogens, the ability of C. albicans to adhere to host surfaces is a prerequisite for both successful colonization and persistence during infection. Within the host, removal of loosely attached C. albicans by the physical flushing action and sloughing off of epithelial cells from mucosal surfaces are important factors in host defense against Candida overgrowth. Therefore, an ability to circumvent these removal mechanisms can be regarded as a virulence attribute. Importantly, adhesion of C. albicans to host tissues is a requisite for infection. Attachment of C. albicans to receptors on host tissues is aided by the expression of a family of cell-wall adhesins known as agglutinin-like sequences (ALS) which have been shown to be differentially regulated in biofilm-grown cells (Hoyer 2001; Williams and Lewia 2011). The ALS family of genes is comprised of eight genes of which ALS3 is the most prominent as it is hyphal specific. Als3 has been implicated in adherence to host tissue as well in the adherence of C. albicans to bacterial species (Phan et al. 2007; Murciano et al. 2012; Peters et al.2012b). Similarly, the hyphal wall protein (Hwp1) is another major adhesin and deletion of either ALS3 or HWP1 results in attenuated virulence (Sundstrom, Balish and Allen 2002; Sundstrom, Cutler and Staab 2002; Naglik et al. 2011). In addition, C. albicans also produces several extracellularly released enzymes such as lipases and esterases as well as hemolysins that are crucial for host tissue invasion and nutrient acquisition. These enzymes facilitate pathogenesis by aiding in the destruction of tissues and tissue barriers and lysing host cells (Naglik, Challacombe and Hube 2003). Most notable are the secreted aspartyl proteinases (Saps), a family of 10 proteinases with a certain degree of functional redundancy between them (Lermann and Morschhäuser 2008). These enzymes can directly induce damage to host cells, facilitate hyphal growth for invasion of tissue, increase adherence following exposure of receptor sites and degrade antibodies and other host defense proteins (Schaller et al. 1999; Williams and Lewia 2011). Phospholipases are another group of enzymes that contribute to the pathogenesis of C. albicans; most notable are the class B phospholipases (PLB) which are secreted and act by disrupting host cell membranes. Accordingly, PLB1 and PLB5 deletions have been implicated in attenuation of systemic Candida infection (Leidich et al. 1998; Theiss et al. 2006).
Biofilm formation on mucosal tissue
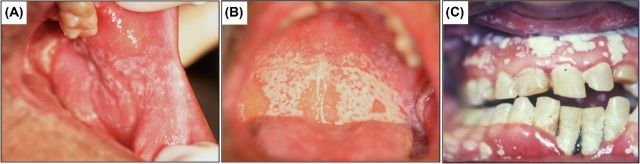
Candida biofilms can form on host mucosal tissue where it resides as part of the commensal microflora with the oral cavity being the primary target (Williams et al. 2013). Oral infections by C. albicans have been recognized throughout recorded history; however, since the 1980s a clear surge of interest in these infections have occurred largely due to the escalation in HIV infection and the AIDS epidemic (Williams and Lewia 2011). In fact, to date, pseudomembranous candidiasis is the most common oral opportunistic infection in HIV+ and other immunocompromised individuals. This condition, commonly known as thrush, presents as creamy white lesions on the palate, buccal mucosa and tongue and may extend into the pharynx (oropharyngeal candidiasis) (Fidel 2006, 2011) (Fig. 2). A diagnostic feature of this infection is that these plaques consisting of desquamated epithelial and immune cells together with yeast and hyphae can be removed by gentle scraping leaving behind an underlying erythematous mucosal surface. Importantly, in addition to HIV+ individuals, candidiasis occurs in 35% of patients with cancer on chemotherapy and radiotherapy, patients with Sjogren's syndrome, the elderly, infants, under conditions of malnutrition and local immune suppression (e.g. steroid inhalers for asthma). In addition, patients with diabetes and other metabolic or hormonal disorders or those on antibiotics are also predisposed to candidiasis (Ramirez-Amador et al. 1997; Redding et al. 1999; Williams and Lewia 2011). Clinical candidiasis is a manifestation of the adherence of yeast cells to mucosal tissue followed by hyphal invasion causing extensive damage, partly mediated by secreted proteolytic enzymes. Significantly, however, in severely immunocompromised patients, mucosal infections can lead to systemic disease, which is associated with high mortality.
Figure 2.
Oral candidiasis. Clinical manifestation of oral candidiasis in a human host characterized by white lesions formed on oral mucosal surfaces: (A) buccal mucosa (B) palate and (C) gingiva.
Similar to the oral cavity, C. albicans is a resident of the normal vaginal microbiota and the leading causative agent of vaginitis. It is estimated that 75% of all women of childbearing age will be afflicted by vulvovaginal candidiasis (VVC) at least once in their lifetime and 5%–8% of these women will suffer from recurring episodes, requiring continued antifungal therapy (Sobel, Muller and Buckley 1984; Peters et al. 2014). Several properties of C. albicans have been proposed to play major roles in causing recurring VVC; notably, strains of C. albicans defective in hyphae formation display significantly reduced vaginitis symptomatology indicating a requirement for hyphae in the pathogenesis of disease (Harriott et al. 2010). Further, estrogen production and microbiota disruption are also considered primary etiologic contributors to this complex disease. However, although Candida vaginitis was believed to result from defects in adaptive immunity, it is now established that an exuberant host innate immune response strongly promotes vaginitis symptomatology. The cognizance that polymorphonuclear leukocyte recruitment into the vagina stimulates acute inflammation highlighted the immunopathological response as a crucial element of vaginitis pathogenesis (Peters et al. 2014). Therefore, studies to identify therapeutic strategies to combat VVC are now also directed toward targeting the host immunopathogenic response.
Biofilm formation on abiotic surfaces
The most serious diseases caused by C. albicans are those associated with biofilm formation on a wide variety of implanted medical devices, which have become an essential component of patient care (Ramage et al. 2005). It is estimated that more than 45 million medical devices are implanted every year in the United States and infection of these devices occurs in up to 60% of patients, with Candida species responsible for up to 20% of these infections (Williams and Lewia 2011). The growing usage and need for implanted medical devices and central venous catheters in managing patient care is an important reason why the incidence of Candida infections has steadily increased. Catheter-associated infections in particular typically emerge from biofilms formed on the surfaces of catheters forming a niche where cells are protected from both the host immune system and antimicrobial therapies. Candida biofilms formed on prosthesis are a major predisposing factor to chronic candidiasis and successful therapy of these foreign-body infections can be a therapeutic challenge, requiring device removal in most instances (Mathe and Van Dijck 2013). Specifically, biofilms on artificial voice box prostheses have been identified as a major cause of their failure. Similarly and in addition to urinary tract infections in catheterized patients, candidal biofilms on hemodialysis and peritoneal dialysis catheters are a common occurrence and are associated with an infection rate of up to 20% in patients undergoing treatment (Raaijmakers et al. 2007; Williams and Lewia 2011; Mathe and Van Dijck 2013). Most significantly, however, biofilms formed on prosthetic heart valves, pacemakers, replacement joints, central nervous system shunts and vascular catheters are the most serious and have been linked with systemic infection. Overall, device-associated Candida infections have mortality rates as high as 30% and the annual cost of antifungal therapies in the United States alone is estimated at $2.6 billion (Williams and Lewia 2011).
Perhaps the most prevalent biofilm-associated candidal infection in healthy individuals is Candida-associated denture stomatitis, which occurs in up to 70% of denture wearers. Denture stomatitis is a chronic disease characterized by localized or generalized inflammation of the denture-bearing mucosa. Candida albicans readily adhere to the polymethylacrylate material of dentures and exploit micro fissures and cracks within the material to facilitate retention. Therefore, infection stems from the adherence of C. albicans to the denture followed by hyphal infiltration of the denture-associated palatal tissue (Williams and Lewia 2011). This is particularly exacerbated in situations where there is an ill-fitting denture and frictional irritation that can damage the normally protective mucosal barrier, allowing infiltration of Candida into the tissue (Williams and Lewia 2011). Further, the continuous seeding of biofilm-associated organisms from the denture is postulated to induce a chronic immune response resulting in inflammation of the oral tissue. Importantly, despite antifungal therapy, this infection is often re-established soon after treatment ceases and therefore tends to be chronic.
Animal models of biofilm-associated Candida infections
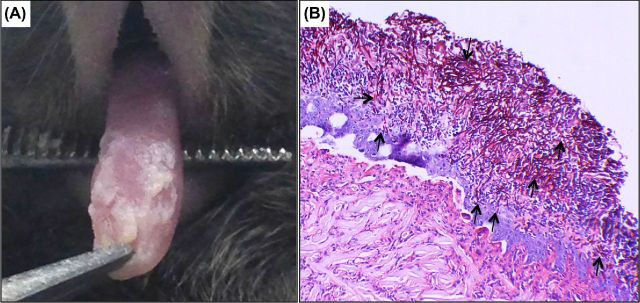
Experimental animal models are crucial to fully understand candidal pathogenesis and to develop new therapeutic approaches particularly in light of the increase in the incidence of fungal infections. The rodent is ideal for studying candidiasis due to the demonstrated similarity to the human disease process and host immune responses. For oral candidiasis, a mouse model is the most commonly used as it is relatively simple and results in a reproducible level of infection and similar histopathology to candidiasis in patients (Solis and Filler 2012) (Fig. 3). Therefore, this model has been widely used for investigating the immune mechanisms against candidiasis, Candida virulence factors and the efficacy of vaccines and antifungal agents. However, due to its small size, the mouse model is not feasible for studying Candida-associated denture stomatitis and therefore, a rat acrylic denture model that recapitulates features of denture stomatitis was recently developed to study this condition in vivo (Nett et al. 2010b; Lee et al. 2011). In this model, a denture device is implanted on the rat palate and following inoculation with C. albicans, biofilm development is monitored over time using microbiological and microscopical analysis. The advantage of this model is that it allows for longitudinal studies and can be used to assess pathogenesis of mutant strains and for designing effective therapeutic strategies. For the study of vaginitis, a well-established estrogen-dependent mouse model is available (Yano and Fidel 2011). Despite the fact that, unlike humans, laboratory rodents do not naturally harbor C. albicans as a commensal, the experimental infection closely parallels the human infection and findings from the animal model are translatable to the human host. Therefore, the mouse model of vaginitis has been an indispensible tool for dissecting disease pathogenesis, the host immunopathological response and drug efficacy.
Figure 3.
Mouse model of oral candidiasis. (A) Four days post sublingual infection with C. albicans, mice develop oral candidiasis characterized by white plaques covering the tongue and other oral surfaces. (B) Histopathology of tongue tissue from mouse with oral candidiasis demonstrating extensive penetration of epithelial tissue by the invasive C. albicans hyphae (arrows).
Fungal infectious biofilms that develop on frequently implanted biomaterials including vascular catheters are the most serious as they can become a potential systemic infection. It has become crucial therefore, to test models of catheter biofilm-associated pathogenesis in vivo. One such model was developed where C. albicans biofilms were formed on catheters implanted in the central venous system of rats for in vivo characterization of biofilm development (Andes et al. 2004). More recently, a similar central venous catheter (CVC) model was developed in mice and used to examine the efficacy of caspofungin to treat CVC-associated candidiasis (Lazzell et al. 2009). In addition, two rabbit models of catheter-associated C. albicans biofilm infection were used to demonstrate the potential of antifungal lock therapy as a therapeutic strategy for catheter infections. Using surgically placed silicone catheters in rabbits, Schinabeck et al. (2004) demonstrated the effectiveness of liposomal amphotericin B against biofilm infections. Similarly, in evaluating systemic and catheter intraluminal lock therapy with amphotericin B deoxycholate and caspofungin using a CVC rabbit model, Schuford et al. (2006) demonstrated the efficacy of caspofungin for the treatment of C. albicans biofilm-associated intravascular catheter infections. The combined findings from these studies are of important clinical relevance as they demonstrated the potential of antifungal lock therapy as a strategy for the successful salvage of catheters infected with C. albicans biofilms.
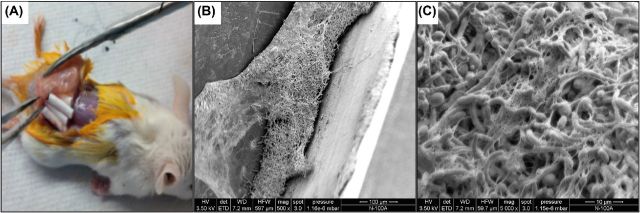
Overall, the benefit of the CVC models is that they take into account the host immune system and provides a means to study novel drug therapies targeting biofilm-associated infections. More recently, a subcutaneous catheter mouse model was developed to study in vivo-grown biofilms (Ricicova et al. 2010). In this model, small CVC catheters inoculated with C. albicans in vitro are implanted under the skin of mice or rats and the biofilm is allowed to form; within 2 days, a mature biofilm structure can be seen developed in the lumen of the explanted catheters (Fig. 4). This catheter model is practical and involves minor surgery to the animals and is readily translatable to applications in the screening and validation of antimicrobial drugs under in vivo conditions.
Figure 4.
Mouse subcutaneous catheter model. (A) Explant of subcutaneous catheters from mouse 48 h post implantation. Candida albicans 48 h biofilm grown in vivo in the lumen of catheters. (B) Scanning electron micrographs demonstrating the thick hyphal matrix formed in the lumen of the recovered implanted catheters. (C) High magnification image of the formed biofilm showing the extracellular matrix material, which appears fibrous.
Classes of antifungals agents
When compared with antibacterial agents, the availability of antifungal agents for the treatment of fungal infections is significantly lower. This is mainly due to the fact that fungi are eukaryotic, and thus identifying drug targets to selectively kill fungal pathogens without toxicity to the host is problematic. Antifungal agents are classified based on their cellular target and mode of action and currently there are only five classes of drugs available for the treatment of fungal infections: azoles, polyenes, echinocandins, allylamines and pyrimidine analogs. (Ghannoum and Rice 1999; Williams and Lewia 2011; Mathe and Van Dijck 2013) (Table 1). Each antifungal compound has advantages and limitations related to its spectrum of activity and mode of action. However, as with microbial biofilms in general, biofilms formed by C. albicans are inherently tolerant to antimicrobial therapy and therefore the susceptibility of Candida biofilms to the current therapeutic agents remains low, with the exception of the echinocandins (Tournu and Van Dijck 2012; Mathe and Van Dijck 2013).
Table 1.
Classes of antifungal agents.
| Antifungal drug classes | Examples | Mechanism of action | Candida albicans means of resistance |
|---|---|---|---|
| Azoles | Fluconazole | Inhibition of lanosterol 14 α-demethylase (ERG11; ergosterol biosynthesis) | Upregulated expression of ERG genes |
| Miconazole | |||
| Ketoconazole | Mutation of ergosterol synthesis enzymes | ||
| Polyenes | Amphotericin B | Binds to ergosterol in fungal cell membranes; transmembrane pore formation, resulting in loss of membrane integrity and ion gradient disruption | Substitution of cell membrane sterols |
| Neutralization of oxidative stress via SODs | |||
| Echinocandins | Caspofungin | Inhibition of β-1,3-glucan synthase | Upregulated expression of glucan biosynthesis genes |
| Micafungin | |||
| Anidulafungin | |||
| Pyrimidine analogs | Flucytosine | Pyrimidine analog that inhibits DNA and RNA synthesis within fungal cells | Mutations in the enzymes that catalyze the pyrimidine analog, such as FUR1 |
| Increased synthesis of nucleotide pyrimidines that competitively inhibit the analogs | |||
| Allylamines | Terbinafine | Inhibition of squalene epoxidase (ERG1; ergosterol biosynthesis) | Mutations in the ERG1 gene |
| Upregulation of drug efflux pumps |
Polyenes include the drugs amphotericin B and nystatin and their mode of action is through direct binding to the sterol ergosterol found within fungal cell membranes (Ghannoum and Rice 1999; Mathe and Van Dijck 2013). Polyene binding to ergosterol induces leakage of electrolytes and cytoplasmic contents via formation of transmembrane channels leading to fungal cell death. However, their use is not widespread due to nephrotoxicity partly due to the similarity of the fungal cell ergosterol to cholesterol, its equivalent sterol in mammalian cell membrane (Ghannoum and Rice 1999; Williams and Lewia 2011; Mathe and Van Dijck 2013). The development of liposomal formulations of amphotericin B reduced the likelihood of toxicity since the usage of a lipid carrier as a delivery system for these drugs results in decreased distribution to the kidneys (Ghannoum and Rice 1999). However, similar to the echinocandins, the prohibitive costs of amphotericin B-liposomal formulation limit their usage for treating fungal infections.
The introduction of the azole antifungals, most notably fluconazole, circumvented many of these toxicity issues, improved bioavailability, spectrum of activity against many fungal pathogens and affordability. However, unlike the polyenes, which are fungicidal, azoles are fungistatic and their mechanism of action involves inhibiting ergosterol biosynthesis via blockage of the enzyme lanosterol 14a-demethylase (Ghannoum and Rice 1999; Mathe and Van Dijck 2013). This inhibition of synthesis and depletion of ergosterol in the fungal cell results in inhibition of fungal growth and impairment of membrane permeability. However, the indiscriminate and over use of fluconazole has led to the emergence of resistant strains, particularly among HIV+ individuals who, given the prevalence of oral candidiasis in this population, were typically administered prolonged prophylactic treatment to prevent infection (Fidel 2006, 2011). As the need to expand antifungal options increases, newer azole drugs such as itraconazole have been used in the treatment of oral candidiasis, while others including voriconazole and posaconazole are alternatives for invasive infections by Candida, albeit rarely used.
Most recently, the echinocandin class of antifungals have emerged as alternatives to the azoles and polyenes (Denning 1997, 2002; Ghannoum and Rice 1999; Walsh 2002). Echinocandins such as caspofungin, micafungin and anidulafungin act through inhibition of the enzyme β-1,3-glucan synthase required for the synthesis of the glucan components in the fungal cell wall, thereby disturbing the integrity of the cell wall. Importantly, as this enzyme is absent from mammalian cells, echinocandins cause little toxicity to the host. Another important advantage of this class of antifungals is that they are fungicidal against Candida and effective against biofilm-associated infections. However, their use is somewhat limited by their large molecular size that dictates the need for intravenous administration and therefore, combined with their high cost, they are used primarily in the treatment of invasive fungal infections (Walsh 2002; Williams and Lewia 2011).
The other two classes of antifungals which are less commonly used are the pyrimidine analogs that incorporate into a growing RNA/DNA strand resulting in arrest of fungal DNA and RNA synthesis and the allylamines that target ergosterol biosynthesis through blocking of the enzyme squalene oxidase (Mathe and Van Dijck 2013). However, for the treatment of biofilms, only the echinocandins and amphotericin B lipid formulations have been shown to be efficacious in vitro and in vivo and therefore, they are the only therapeutic options for the treatment of Candida biofilm infections (Bachmann et al. 2002; Kuhn et al. 2002; Mukherjee and Chandra 2004; Kucharíková et al. 2010; Mathe and Van Dijck 2013; Ghannoum et al. 2015).
Mechanisms of drug resistance
A vast amount of research has been conducted to elucidate the mechanisms underlying increased resistance in biofilm-associated C. albicans cells (White, Marr and Bowden 1998; Baillie and Douglas 1998a,b, 1999, 2000; Cannon et al.2009). Similar to bacterial biofilms, compared to planktonic cells, fungal biofilms exhibit up to 20 000-fold increase in antifungal MICs (minimum inhibitory concentrations) (Hawser 1996; Jabra-Rizk, Falkler and Meiller 2004). Several mechanisms underlying the increased resistance of biofilm-associated cells have been proposed some of which are shared between planktonic and biofilm-associated cells and others are biofilm specific (White, Marr and Bowden 1998; Mathe and Van Dijck 2013). In what follows we highlight the main mechanisms, summarized in Table 2.
Table 2.
Antifungal resistance mechanisms in C. albicans.
| Mechanism | Physiological effect | Citation | |
|---|---|---|---|
| Planktonic | Reduced growth rate | Reduced efficacy of antifungal drugs in inactive/slow-growing cells. | Baillie and Douglas (1998a,b) |
| Both | Modification of drug target | Target substrate has been mutated, disallowing the biding and inhibitory effect of antifungal drugs | Kelly et al. (1997); Martel et al. (2010); Nolte et al. (1997) |
| Upregulation of efflux pumps | Intracellularly accumulated antifungals are transported out of the cell, away from its target substrates | Cannon et al. (2009); Sanglard and Odds (2002); Ramage et al. (2002a); Taff et al. (2013); Mukherjee et al. (2003); Mateus et al. (2004); Lepak et al. (2006); Sanglard et al. (1995) | |
| Cell density | Higher cell numbers require a larger dose of antifungals for efficacy | Perumal et al. (2007) | |
| Biofilms | Persister cells | Metabolic dormancy, cells become highly tolerant of antifungals | LaFleur et al. (2006); Bink et al. (2011) |
| Extracellular polysaccharide matrix | Hinders diffusion of antifungals through biofilm; binds and sequesters fluconazole in the matrix | Al-Fattani and Douglas (2006); Nett et al. (2010a); Mitchell et al. (2015); Nett et al. (2007); Taff et al. (2012) |
Reduced growth rate
It is accepted that slower growing cells are more resistant to antimicrobial therapy as many antimicrobial drugs target active growth and metabolic pathways. However, analysis of planktonic C. albicans cells against biofilms cells at varied growth rates showed conflicting evidence. Whereas planktonic C. albicans cells remained less susceptible to antifungal therapy at reduced growth rates after treatment with amphotericin B, C. albicans biofilm cells exhibited resistance at all growth rates, indicating a more complicated mechanism than just growth rate alone (Baillie and Douglas 1998a,b; Mathe and Van Dijck 2013).
Cell density
It is also commonly accepted that cell density can affect the efficacy of antimicrobial agents. This is especially true in biofilms, where large numbers of cells are concentrated in a small environment. However, although cell density does seem to have an effect on C. albicans resistance to several drugs, it is not likely that this mechanism is biofilm specific since a similar trend was observed in planktonic cells (Perumal, Mekala and Chaffin 2007; Seneviratne et al. 2008).
Altered gene expression
A key feature of biofilms is their ability to dynamically respond to various stressors (Nobile et al. 2012; Mathe and Van Dijck 2013). Exposure of biofilms to antimicrobial agents usually results in the induction of resistance genes. In C. albicans biofilms, this paradigm was studied in the ergosterol and glucan biosynthesis genes following azole and echinocandin drug treatment, respectively. Ergosterol is a major constituent of C. albicans fungal cell membranes and therefore, is considered to be essential to C. albicans growth, which makes it an ideal drug target. Ergosterol biosynthesis in C. albicans is a complex process involving numerous enzymes the most prominent of which are the ERG genes. As the azoles inhibit ergosterol biosynthesis, C. albicans developed several resistance mechanisms to circumvent the deleterious inhibitory effects of fluconazole, most notably is the upregulation of ergosterol biosynthesis genes. In fact, a study by Nailis et al. (2010) examining the expression of ergosterol in treated planktonic C. albicans cells following exposure to fluconazole revealed an upregulation in the ERG9 and ERG11 genes. However, in treated biofilm-associated cells, a significant increase in the expression of ERG1, ERG3, ERG11 and ERG25 was seen indicating a greater capacity for biofilms to respond to antifungal stress possibly explaining why fluconazole treatment is generally ineffective in eradicating C. albicans biofilms (Mathe and Van Dijck 2013).
Modification of drug target substrate
Another means for C. albicans to evade antifungal agents is to alter the drug target substrate, resulting in the inability of the drug to effectively eradicate the pathogen (White, Marr and Bowden 1998). This phenomenon was shown in C. albicans through mutations in the ERG3 and ERG5 genes, as well as point mutations in the ERG11, which resulted in cross-resistance to azoles and amphotericin B (White 1997; White, Marr and Bowden 1998). It is therefore plausible that alterations in gene expression are also responsible for drug resistance in biofilm-associated C. albicans cells. However, although these ERG mutations are more likely involved in the development of resistance in planktonic populations after prolonged treatment with azoles, whether they have implications on pathogenesis and resistance in C. albicans biofilms remains understudied.
Drug efflux pumps
The upregulation of efflux pumps is also correlated with resistance in C. albicans biofilms, primarily with resistance to azole drugs, but does not appear to play a role in resistance to echinocandins. Efflux pumps facilitate the extracellular transport of antifungals, thereby preventing their intracellular accumulation (White 1997; Ramage et al. 2002a). Two major families of efflux pumps have been shown to contribute to drug resistance in C. albicans: the ATP binding cassette transporters encoded by the CDR (Candida drug resistance) genes comprised of CDR1 and CDR2 and the major facilitator superfamily encoded by the MDR genes comprised of MDR1 and FLU1 (Sanglard et al. 1996; White 1997; White, Marr and Bowden 1998; Ramage et al. 2002a; Niimi et al. 2004; Prasad and Panwar 2004). Although these genes can be expressed by resistant planktonic strains, they were shown to be significantly expressed in biofilms. In fact, the expression of CDR and MDR genes appears to be induced upon C. albicans attachment to a substrate surface and therefore, is intrinsic to biofilm formation and not triggered by the presence of azole drugs (Mateus, Crow and Ahearn 2004; Lepak et al. 2006). However, the expression of these efflux pump genes was found to decrease as the biofilm ages, contrary to existing patterns of mechanism of resistance in biofilms where expression of resistance genes increases as the biofilm matures (Mukherjee et al. 2003). These findings strongly support the notion that although present, drug efflux pumps do not play a significant role in biofilm resistance to azole drugs.
Persister cells and stress responses
Although a key feature of biofilms is the ability of the component cells to exhibit differential gene expression, a subset of the biofilm cell population have been described to be phenotypically distinct from the rest (Lewis 2005). These cells termed ‘persister cells’ are not mutants and are often deeply embedded in the biofilm where they become dormant and therefore, unaffected by antimicrobial agents which target metabolically active cells (LaFleur, Kumamoto and Lewis 2006; Lewis 2007; Dawson, Intapa and Jabra-Rizk 2011). This phenomenon is compounded by the ability of these ‘persister cells’ to revert to a metabolically active state and re-establish or repopulate a biofilm following treatment (Lewis 2007). It is currently thought that exposure to stress conditions can trigger the formation of ‘persister cells’ (Dawson, Intapa and Jabra-Rizk 2011).
During colonization of its host, C. albicans is confronted with a wide variety of stresses to which it responds via different conserved signal transduction pathways (Cannon et al. 2007). Amphotericin B treatment was shown to induce oxidative stress in fungal cells, revealing a novel cell death pathway involving ROS generation (LaFleur, Kumamoto and Lewis 2006). However, C. albicans possesses superoxide dismutases (SODs), a family of enzymes important for detoxification of ROS, and the expression of the SODs was shown to be induced upon treatment by amphotericin B or miconazole. Although the generation and survival mechanisms of C. albicans ‘persister cells’ remains largely unknown, the potential role of SODs in this phenomenon has been explored. Indeed, it was found that pharmacological inhibition of SODs and subsequent treatment with amphotericin B or miconazole resulted in significantly greater killing of C. albicans biofilms (Bink et al. 2011). These findings suggest a role for mediating oxidative stress in the generation of ‘persister cells’ and survival of C. albicans biofilms. However, it is clear that the high resistance of biofilm-associated C. albicans cells cannot be attributed to the actions of one mechanism, but is rather a comprehensive mechanism reflecting the complexity of the biofilm lifestyle itself.
Biofilm extracellular polysaccharide matrix
A critical point in the establishment of a fungal biofilm is the production and secretion of an extracellular matrix, which acts as a network that anchors the biofilm, providing structure, stability and protection (Nett et al.2010c; Mitchell et al. 2015). Therefore, production of extracellular matrix is considered one of the key resistance mechanisms in biofilms and recent efforts have been focused on understanding the genetic basis for how matrix production governs drug resistance in biofilm (Zarnowski et al. 2014). In C. albicans, the biofilm matrix is largely composed of polysaccharides such as β-1,3-glucan, β-1,6-glucan, and mannans and to a lesser extent, proteins (Al-Fattani and Douglas 2006; Nett et al. 2007; Nett, Sanchez and Andes 2011; Zarnowski et al. 2014). However, in comparing the exact composition of biofilm matrix material with that produced by planktonic cells, considerate differences were seen in carbohydrate and protein content indicating that there might be some features specific to biofilm matrix material (Hawser 1996; Baillie and Douglas 2000; Al-Fattani and Douglas 2006). It is theorized that the matrix can physically hinder the penetration and diffusion of antimicrobial agents through the biofilm and in C. albicans, the matrix was also shown to bind and sequester fluconazole in the extracellular milieu (Taff et al. 2013). New insights into the role of matrix in biofilm came from studies by Nett et al. (2010a) where the cell walls of biofilm-associated cells were shown to be two times thicker and contained more carbohydrates and β-1,3-glucans than planktonic cells. Further, isolation of matrix material demonstrated the presence of β-1,3-glucans in the biofilm matrix, which was shown to increase over the course of biofilm maturation. More recently, studies by Mitchell et al. (2015) demonstrated that β-1,3-glucans contribute to azole resistance by specific binding. This process was found to be due largely attributed to the FKS1 gene, encoding a β-1,3-glucan synthase, and in part to a series of glucan transferases and exoglucanases which transport glucans into the extracellular space (Nett et al. 2010a). Interestingly, mutations in these genes had no apparent deleterious effects on planktonic C. albicans cells suggesting that this mechanism is specific to C. albicans biofilms (Mitchell et al. 2015). In addition to cell-wall components and similar to what has been described in bacterial biofilms, extracellular DNA was also shown to be a key element in the matrix of C. albicans mature biofilms, important for biofilm structural integrity and maintenance (Martins et al. 2010).
Immune evasion
Unsurprisingly, biofilms are also highly resistant to clearance by the host immune responses, orchestrated by initial detection of the pathogen by pattern recognition receptors (PRR) on leukocytes. In the case of C. albicans, detection of cell-wall components by the host immune system generates a specific cytokine response which signals for the appropriate immune responses to clear the pathogens (Nett and Andes 2006; Netea and Marodi 2010). Although this can prove effective in the clearance of planktonic pathogens, biofilms prove to be far more difficult to handle (Chandra et al. 2007). In C. albicans biofilms it was shown that leukocytes are unable to phagocytose biofilm-associated cells, likely due to interference by the biofilm matrix (Hirschfeld 2014). Intriguingly, C. albicans biofilms appear to trigger a unique form of cell death in neutrophil phagocytes, resulting in rapid expulsion of DNA contents into the extracellular space, named neutrophil extracellular traps or ‘NETosis’. This mechanism is seemingly triggered to further upregulate and augment the host immune response, such as neutrophil degranulation and release of lytic enzymes. However, in the case of C. albicans biofilms, this is of limited efficacy toward eradication and seemingly only serves to propagate inflammation in host tissues (Remijsen et al. 2011; Hirschfeld 2014).
Candida albicans biofilms have also been shown to alter the profile of cytokines secreted by immune cells, thereby manipulating the resultant immune response although, the exact mechanism behind this remains to be elucidated (Chandra et al. 2007; Krysan, Sutterwala and Wellington 2014). However, C. albicans has evolved several immune evasion strategies resulting in its reduced recognition by the host immune system. These include strategies for masking specific cell-wall components to prevent PRR-mediated recognition and secretion of aspartic proteases to inactivate components of the innate immune system (Meiller et al. 2009; Mathe and Van Dijck 2013). Further, the interaction between biofilm-associated C. albicans cells and the immune system was shown to be very distinct from interactions with planktonic C. albicans cells. Specifically, studies by Chandra et al. (2007) showed that peripheral blood mononuclear cells (PBMCs) did not phagocytose biofilm-associated cells. In contrast, the presence of PBMCs during biofilm development resulted in significantly thicker biofilms as a consequence of unknown factors secreted by the immune cells. Additionally, mature biofilms were also shown to not elicit a robust oxidative response, which is one of the main mechanisms by which neutrophils kill pathogens (Mathe and Van Dijck 2013). Taken together, these observations support the hypothesis that biofilm formation might be an adaptation process for survival within the host hostile environment.
Candida albicans–bacterial interactions in polymicrobial biofilms
Biofilms can be polymicrobial in nature where multiple microbes comprise a biofilm and coexist synergistically particularly in the oral cavity where C. albicans adheres to and interacts with oral bacterial species (Jenkinson et al. 2008). In fact, over 20% of Candida bloodstream infections were shown to be polymicrobial (Jabra-Rizk 2011; Peters et al.2012a). Although most known interactions are inhibitory, symbiotic interactions may include augmented adherence or antibiotic resistance (Jenkinson et al. 2008). This poses another hurdle in the treatment and eradication of biofilms, as these polymicrobial biofilms will usually require some form of combination therapy. Although studies into the dynamics of polymicrobial biofilms are in their infancy, ongoing research reveals that such biofilms are even more resistant to antimicrobial therapy, potentially giving rise to novel mechanisms of cross protection (Finkel and Mitchell 2011).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Recent strides in expression profiling and genetic manipulation have driven our understanding of the regulatory pathways and mechanisms that govern biofilm formation and biofilm-based drug resistance. Increasingly, mechanistic studies have provided valuable insights into the regulatory circuitry and networks that control biofilm formation. These studies have revealed new mechanisms and signals that govern C. albicans biofilm formation and associated drug resistance providing therapeutic foresight. Although numerous in vitro biofilm model systems have been crucial for studying biofilm development and cell phenotypes and drug resistance, it is difficult to accurately account for the multitude of host and infection-site variables that are important in humans. As the role and nature of host–pathogen interactions during biofilm formation continue to be unveiled, studies are now focused on developing C. albicans animal biofilm infection models to more accurately reflect the complexity of this host–fungal interaction (Chandra et al. 2001; Kuhn et al. 2002; Nett and Andes 2006; Ricicova et al. 2010).
Importantly, the increase in resistance to existing antifungals has provided a strong impetus to understand the molecular mechanisms of drug resistance with the goal of identifying novel therapeutic targets. Successful treatment of candidiasis can be further hampered where there is an established biofilm and biomaterial infections continue to be an increasingly alarming problem because of their intrinsic recalcitrance to conventional therapy. Therefore, it has become crucial to explore alternative strategies to overcome the limitations of current therapies against resilient biofilm-associated fungal infections. In addition, the ability to adhere, as a unique prerequisite to form a biofilm, is a fast process which makes the prevention of biofilm development difficult with the current tools and strategies (Tournu and Van Dijck 2012). Recent advances elucidating the transcriptional programs that orchestrate biofilm formation by C. albicans have tremendously broadened our knowledge on the complex mechanisms underlying biofilm resistance. Yet, the search for antibiofilm treatments is a multifaceted subject and requires improved understanding of the pathogen, host response to adhesion and biofilm formation and interactions within microbial communities (Tournu and Van Dijck 2012). Recent efforts in the field are directed toward developing new therapeutic approaches such as modification of biomaterials to inhibit adherence, use of catheter lock and combination therapies and exploring natural compounds and immunotherapies. Further, defining the role of quorum sensing in biofilm maturation may lead to the identification of quorum-sensing molecules that are active in vivo which can be harnessed for promoting biofilm disruption. Importantly, understanding the dynamics of formation and key molecular players in mixed fungal-bacterial biofilms is crucial for combating resilient biofilm-associated polymicrobial infections, which are particularly challenging to treat.
Acknowledgments
We would like to thank Dr Valli Meeks and Dr Timothy Meiller for kindly providing us with the clinical images of patients with oral candidiasis attending the HIV PLUS Clinic at the University of Maryland Dental School and the Imaging Core for assistance with electron microscopy.
FUNDING
The work presented was supported by the National Institutes of Health grant ( DE14424); the Scientific Research Flanders (FWO; WO.026.11N); and the Interuniversity Attraction Poles Program, initiated by the Belgian Science Policy Office.
Conflict of interest. None declared.
REFERENCES
- Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- Andes D, Nett J, Pschel P, et al. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–31. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann SP, VandeWalle K, Ramage G, et al. In vitro activity of caspofungin against Candida albicans biofilm. Antimicrob Agents Ch. 2002;46:3591–6. doi: 10.1128/AAC.46.11.3591-3596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas J. Effect of growth rate on resistance of Candidaalbicans biofilms to antifungal agents. Antimicrob Agents Ch. 1998a;42:1900–5. doi: 10.1128/aac.42.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas J. Iron-limited biofilms of Candidaalbicans and their susceptibility to amphotericin B. Antimicrob Agents Ch. 1998b;42:2146–9. doi: 10.1128/aac.42.8.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Douglas J. Candida biofilms and their susceptibility to antifungal agents. Methods Enzymol. 1999;310:644–56. doi: 10.1016/s0076-6879(99)10050-8. [DOI] [PubMed] [Google Scholar]
- Baillie GS, Douglas J. Matrix polymers of Candida biofilms and their possible role in biofilm resistance to antifungal agents. J Antimicrob Chemoth. 2000;46:397–403. doi: 10.1093/jac/46.3.397. [DOI] [PubMed] [Google Scholar]
- Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–30. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- Bink A, Vandenbosch D, Coenye T, et al. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob Agents Ch. 2011;55:4033–7. doi: 10.1128/AAC.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington: ASM Press; 2012. [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, et al. Candida albicans drug resistance another way to cope with stress. Microbiology. 2007;153:3211–7. doi: 10.1099/mic.0.2007/010405-0. [DOI] [PubMed] [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, et al. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Kuhn D, Mukherjee M, et al. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–94. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, McCormick TS, Imamura Y, et al. Interaction of Candidaalbicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun. 2007;75:2612–20. doi: 10.1128/IAI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvel M, Nesseir A, Cabral V, et al. A versatile overexpression strategy in the pathogenic yeast Candida albicans: Identification of regulators of morphogenesis and fitness. PLoS One. 2012;7:e45912. doi: 10.1371/journal.pone.0045912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Montanaro L, Arciola CR. Biofilm in implant infections: its production and regulation. Int J Artif Organs. 2005;28:1062–8. doi: 10.1177/039139880502801103. [DOI] [PubMed] [Google Scholar]
- Dawson CC, Intapa C, Jabra-Rizk MA. ‘Persisters’: survival at the cellular level. PLoS Pathog. 2011;7:e1002121. doi: 10.1371/journal.ppat.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuyser L, Jabra-Rizk MA, Van Dijck P. Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog Dis. 2014;70:219–30. doi: 10.1111/2049-632X.12123. [DOI] [PubMed] [Google Scholar]
- Denning DW. Echinocandins and pneumocandins—a new antifungal class with a novel mode of action. J Antimicrob Chemoth. 1997;40:611–4. doi: 10.1093/jac/40.5.611. [DOI] [PubMed] [Google Scholar]
- Denning DW. Echinocandins: a new class of antifungal. J Antimicrob Chemoth. 2002;49:889–91. doi: 10.1093/jac/dkf045. [DOI] [PubMed] [Google Scholar]
- Fidel PL., Jr Candida-host interactions in HIV disease: relationships in oropharyngeal candidiasis. Adv Dent Res. 2006;19:80–4. doi: 10.1177/154407370601900116. [DOI] [PubMed] [Google Scholar]
- Fidel PL., Jr Candida-host interactions in HIV disease implications for oropharyngealc andidiasis. Adv Dent Res. 2011;23:45–9. doi: 10.1177/0022034511399284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control Of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–18. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14:380–5. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–17. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Roilides E, Katragkou A, et al. The role of echinocandins in Candida biofilm-related vascular catheter infections: in vitro and in vivo model systems. Clin Infect Dis. 2015;61(Suppl 6):S618–21. doi: 10.1093/cid/civ815. [DOI] [PubMed] [Google Scholar]
- Hall RA, Gow N. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol. 2013;90:1147–61. doi: 10.1111/mmi.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott MM, Lilly EA, Rodriguez TE, et al. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–44. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser S. Comparisons of the suceptibilities of planktonic and adherent Candida albicans to antifungal agents: a modified XTT tetrazolium assay using synchronised C. albicans cells. J Med Vet Mycol. 1996;34:149–52. [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–80. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol. 2014;17:26102. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk MA. Pathogenesis of polymicrobial biofilms. Open Mycol J. 2011;5:39–43. [Google Scholar]
- Jabra-Rizk MA, Falkler WA, Jr, Meiller TF. Fungal biofilms and drug resistance. Emerg Infect Dis. 2004;10:14–9. doi: 10.3201/eid1001.030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen ID, Wilson D, Wächtler B, et al. Candida albicans dimorphism as a therapeutic target. Expert Rev Anti-Infe. 2012;10:85–93. doi: 10.1586/eri.11.152. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Barbour ME, Jagger DC, et al. Candida albicans - bacteria interactions in biofilms and disease. Univ Bristol Dent Sch. 2008:1–6. [Google Scholar]
- Kelly SL, Lamb DC, Kelly DE, et al. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 1997;400:80–2. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- Kong E, Jabra-Rizk MA. The great escape: Pathogen versus host. PLoS Pathol. 2015;11:e1004661. doi: 10.1371/journal.ppat.1004661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom BP, Levy N, Meijler MM, et al. Farnesol and Candida albicans: quorum sensing or not quorum sensing? Isr J Chem. 2015;55:1–8. [Google Scholar]
- Krysan DJ, Sutterwala FS, Wellington M. Catching fire: Candida albicans, macrophages, and pyroptosis. PLoS Pathog. 2014;10:e1004139. doi: 10.1371/journal.ppat.1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharíková S, Tournu H, Holtappels M, et al. In vivo efficacy of anidulafungin against mature Candida albicans biofilms in a novel rat model of catheter-associated candidiasis. Antimicrob Agents Ch. 2010;54:4474–5. doi: 10.1128/AAC.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DM, George T, Chandra J, et al. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Ch. 2002;46:1773–80. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MD, Kumamoto C, Lewis L. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Ch. 2006;50:3839–46. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzell AL, Chaturvedi AK, Pierce CG, et al. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J Antimicrob Chemoth. 2009;64:567–70. doi: 10.1093/jac/dkp242. [DOI] [PubMed] [Google Scholar]
- Lee H, Yu A, Johnson CC, et al. Fabrication of a multi-applicable removable intraoral denture system for rodent research. J Oral Rehabil. 2011;38:686–90. doi: 10.1111/j.1365-2842.2011.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidich SD, Ibrahim AS, Fu Y, et al. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J Biol Chem. 1998;273:26078–86. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- Lepak AJ, Nett J, Lincoln L, et al. Time course of microbiologic outcome and gene expression in Candida albicans during and following in vitro and in vivo exposure to fluconazole. Antimicrob Agents Ch. 2006;50:1311–9. doi: 10.1128/AAC.50.4.1311-1319.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermann U, Morschhäuser J. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology. 2008;154(Pt 11):3281–95. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- Lewis K. Riddle of biofilm resistance. Antimicrob Agents Ch. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry. 2005;70:267–74. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Martel CM, Parker JE, Bader O, et al. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5(encoding C22 desaturase) is cross resistant to azoles and amphotericin. Antimicrob Agents Ch. 2010;54:3578–83. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Uppuluri P, Thomas DP, et al. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia. 2010;169:323–31. doi: 10.1007/s11046-009-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus C, Crow SAJ, Ahearn DG. Adherence of Candidaalbicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob Agents Ch. 2004;48:3358–66. doi: 10.1128/AAC.48.9.3358-3366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe L, Van Dijck P. Recent insights into Candida albicans biofilm resistance. Curr Genet. 2013;59:251–64. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenecity mechanisms. Virulence. 2013;4:119–28. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiller TF, Hube B, Schild L, et al. A novel immune evasion strategy of Candida albicans: proteolytic cleavage of a salivary antimicrobial peptide. PLoS One. 2009;4:e5039. doi: 10.1371/journal.pone.0005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KF, Zarnowskia R, Sancheza H, et al. Community participation in biofilm matrix assembly and function. P Nat Acad Sci USA. 2015;112:4092–7. doi: 10.1073/pnas.1421437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J. Candida biofilm resistance. Drug Resist Update. 2004;7:301–9. doi: 10.1016/j.drup.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J, Kuhn DM, et al. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71:4333–40. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano C, Moyes DL, Runglall M, et al. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS One. 2012;7:e33362. doi: 10.1371/journal.pone.0033362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol R. 2003;67:400–28. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Moyes DL, Wächtler B, et al. Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 2011;13:963–76. doi: 10.1016/j.micinf.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nailis H, Kucharikova S, Ricicova M, et al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010;10:114. doi: 10.1186/1471-2180-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MJ, Marodi L. Innate immune mechanisms for recognition and uptake of Candida species. Trends Immunol. 2010;31:346–53. doi: 10.1016/j.it.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Nett JE, Andes DR. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006;9:340–5. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Nett JE, Crawford K, Marchillo K, et al. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Ch. 2010a;54:3505–8. doi: 10.1128/AAC.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Lincoln L, Marchillo K, et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Ch. 2007;51:510–20. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Marchillo K, Spiegel CA, et al. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun. 2010b;78:3650–9. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Sanchez H, Andes DR. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryote Cell. 2011;10:1660–9. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Sanchez H, Cain MT, et al. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis. 2010c;202:171–5. doi: 10.1086/651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi M, Niimi K, Takano Y, et al. Regulated overexpression of CDR1 in Candida albicans confers multidrug resistance. J Antimicrob Chemoth. 2004;54:999–1006. doi: 10.1093/jac/dkh456. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, et al. Recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8:1382–9. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- Nolte FS, Parkinson T, Falconer DJ, et al. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Ch. 1997;41:196–9. doi: 10.1128/aac.41.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–46. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Perumal P, Mekala S, Chaffin L. Role for cell density in antifungal drug resistance in Candidaalbicans biofilms. Antimicrob Agents Ch. 2007;51:2454–63. doi: 10.1128/AAC.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O'May GA, et al. Polymicrobial interactions in biofilms: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012a;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Junko Y, Noverr MC, et al. Candida vaginitis: when opportunism knocks, the host responds. PLoS Pathog. 2014;10:e1003965. doi: 10.1371/journal.ppat.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Ovchinnikova E, Schlecht LM, et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiol. 2012b;158:2975–86. doi: 10.1099/mic.0.062109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–63. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, Myers CL, Fu Y, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Panwar S. Physiological functions of multidrug transporters in yeast. Curr Sci. 2004;86:62–73. [Google Scholar]
- Raaijmakers R, Schroder C, Monnens L, et al. Fungal periotonitis in children on peritoneal dialysis. Pediatr Nephrol. 2007;22:288–93. doi: 10.1007/s00467-006-0289-x. [DOI] [PubMed] [Google Scholar]
- Ramage G, Bachmann S, Patterson TF, et al. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candidaalbicans biofilms. J Antimicrob Chemoth. 2002a;49:973–80. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- Ramage G, Saville S, Thomas DP, et al. Candida biofilms: an update. Eukaryote Cell. 2005;4:633–8. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, Saville S, Wickes P, et al. Inhibition of Candidaalbicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microb. 2002b;68:5459–63. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Amador V, Silverman S, Jr, Mayer P, et al. Candidal colonization and oral candidiasis in patients undergoing oral and pharyngeal radiation therapy. Oral Surg Oral Med O. 1997;84:149–53. doi: 10.1016/s1079-2104(97)90061-5. [DOI] [PubMed] [Google Scholar]
- Redding SW, Zellars RC, Kirkpatrick WR, et al. Epidemiology of oropharyngeal Candida colonization and infection in patients receiving radiation for head and neck cancer. J Clin Microbiol. 1999;37:3896–900. doi: 10.1128/jcm.37.12.3896-3900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q, Kuijpers TW, Wirawan E, et al. Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011;18:581–8. doi: 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricicova M, Kucharíková S, Tournu H, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–19. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Monod M, et al. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Ch. 1996;40:2300–5. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Kuchler K, Ischer F, et al. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Ch. 1995;39:2378–86. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2002;2:73–85. doi: 10.1016/s1473-3099(02)00181-0. [DOI] [PubMed] [Google Scholar]
- Schaller M., Korting HC, Schafer W, et al. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol. 1999;34:169–80. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- Scheper MA, Shirtliff ME, Meiller TF, et al. Farnesol a fungal quorum sensing molecule triggers apoptosis in human oral squamous carcinoma cells. Neoplasia. 2008;10:954–63. doi: 10.1593/neo.08444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinabeck MK, Long LA, Hossain MA, et al. Rabbit model of Candidaalbicans biofilm infection: liposomal amphotericin B antifungal lock therapy. Antimicrob Agents Ch. 2004;48:1727–32. doi: 10.1128/AAC.48.5.1727-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuford JA, Rouse MS, Piper KE, et al. Evaluation of caspofungin and amphotericin B deoxycholate against Candida albicans biofilms in an experimental intravascular catheter infection model. J Infect Dis. 2006;194:710–3. doi: 10.1086/506452. [DOI] [PubMed] [Google Scholar]
- Seneviratne CJ, Jin LJ, Samaranayake YH, et al. Cell density and cell aging as factors modulating antifungal resistance of Candida albicans biofilms. Antimicrob Agents Ch. 2008;52:3259–66. doi: 10.1128/AAC.00541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff ME, Krom BP, Meijering RM, et al. Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Ch. 2009;53:2392–401. doi: 10.1128/AAC.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JD, Muller G, Buckley HR. Critical role of germ tube formation in the pathogenesis of candidal vaginitis. Infect Immun. 1984;44:576–80. doi: 10.1128/iai.44.3.576-580.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012;7:637–42. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom P, Balish E, Allen CM. Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J Infect Dis. 2002;185:521–30. doi: 10.1086/338836. [DOI] [PubMed] [Google Scholar]
- Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun. 2002;70:3281–3. doi: 10.1128/IAI.70.6.3281-3283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff HT, Mitchell KF, Edward JA, et al. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013;8:1325–37. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taff HT, Nett JE, Zarnowski R, et al. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 2012;8:e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiss S, Ishdorj G, Brenot A, et al. Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int J Med Microbiol. 2006;296:405–20. doi: 10.1016/j.ijmm.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournu H, Van Dijck P. Candida biofilms and the host: models and new concepts for eradication. Int J Microbiol. 2012;2012:845352. doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppuluri P, Chaturvedi AK, Srinivasan A, et al. Dispersion an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TJ. Echinocandins—an advance in the primary treatment of invasive candidiasis. N Engl J Med. 2002;347:2070–2. doi: 10.1056/NEJMe020142. [DOI] [PubMed] [Google Scholar]
- White TC. Increased mRNA levels of ERG 16, CDR, and MDR1 correlate with increase in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Ch. 1997;41:1482–7. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Lewia M. Pathogenesis and treatment of oral candidosis. Oral Microbiol. 2011;3 doi: 10.3402/jom.v3i0.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Jordan R, Wei XQ, et al. Interactions of Candidaalbicans with host epithelial surfaces. Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, et al. Nosocomial bloodstream infections in U.S. hospitals: analysis of 24179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Yano J, Fidel P., Jr Protocols for vaginal inoculation and sample collection in the experimental mouse model of Candida vaginitis. J Vis Exp. 2011;58:3382. doi: 10.3791/3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnowski R, Westler WM, Lacmbouh GA, et al. Novel entries in a fungal biofilm matrix encyclopedia. mBio. 2014;5:e01333–14. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Krom BP, Sanglard D, et al. Farnesol-induced apoptosis in Candida albicans is mediated by Cdr1-p extrusion and depletion of intracellular glutathione. PLoS One. 2011;6:2392–401. doi: 10.1371/journal.pone.0028830. [DOI] [PMC free article] [PubMed] [Google Scholar]