Abstract
BACKGROUND
Nutritional metabolomics is rapidly evolving to integrate nutrition with complex metabolomics data to discover new biomarkers of nutritional exposure and status.
CONTENT
The purpose of this review is to provide a broad overview of the measurement techniques, study designs, and statistical approaches used in nutrition metabolomics, as well as to describe the current knowledge from epidemiologic studies identifying metabolite profiles associated with the intake of individual nutrients, foods, and dietary patterns.
SUMMARY
A wide range of technologies, databases, and computational tools are available to integrate nutritional metabolomics with dietary and phenotypic information. Biomarkers identified with the use of high-throughput metabolomics techniques include amino acids, acylcarnitines, carbohydrates, bile acids, purine and pyrimidine metabolites, and lipid classes. The most extensively studied food groups include fruits, vegetables, meat, fish, bread, whole grain cereals, nuts, wine, coffee, tea, cocoa, and chocolate. We identified 16 studies that evaluated metabolite signatures associated with dietary patterns. Dietary patterns examined included vegetarian and lactovegetarian diets, omnivorous diet, Western dietary patterns, prudent dietary patterns, Nordic diet, and Mediterranean diet. Although many metabolite biomarkers of individual foods and dietary patterns have been identified, those biomarkers may not be sensitive or specific to dietary intakes. Some biomarkers represent short-term intakes rather than long-term dietary habits. Nonetheless, nutritional metabolomics holds promise for the development of a robust and unbiased strategy for measuring diet. Still, this technology is intended to be complementary, rather than a replacement, to traditional well-validated dietary assessment methods such as food frequency questionnaires that can measure usual diet, the most relevant exposure in nutritional epidemiologic studies.
Recent high-throughput technologic developments in molecular biology, namely, genomics, transcriptomics, proteomics, and metabolomics, are leading us toward a new era in epidemiologic research. In the past few years, the scientific community has focused on a more integrated systems epidemiology approach, in which several fields converge to integrate traditional knowledge with novel -omics techniques (1). Nutritional epidemiology has not been the exception, and several studies in this field have incorporated -omics data in the past decade (1). Along these lines, the concept of precision nutrition has recently emerged (2) and refers to the integration of -omics techniques to personalize diets based on individual genetic makeup to achieve better prevention or management of disease. Among all the -omics, metabolomics plays a crucial role in the field of nutrition because it is more time sensitive than other -omics and can reflect the current biological status of an individual (3). The human metabolome can be influenced by several factors, such as age, diseases, drugs, environment, genetic factors, lifestyle, and nutrition (3).
Metabolomics can provide a comprehensive picture of overall dietary intake by measuring the full profile of small molecule metabolites in biological samples such as saliva, blood, and urine. Thus, it could help deepen our knowledge of metabolic pathways relevant to human nutrition (3). Importantly, because nutritional epidemiologic studies frequently rely on self-reported dietary assessment methods that are subject to recall bias and measurement error and because objective biomarkers do not exist for all nutrients and foods (4), metabolomics can be a promising technique to objectively identify dietary biomarkers. Metabolite profiling accounts for intrinsic variability in metabolism by measuring downstream components or metabolic products of foods, and might therefore accurately reflect true exposure as compared with traditional methods that measure individual food intake (5). Thereby, nutritional metabolomics, which refers to the integration of metabolic profiling with nutrition in complex biosystems, can be applied to discover new biomarkers of nutritional exposure and status and can help disentangle the molecular mechanisms by which diet affects health and disease. Diet can have effects on 2 different components of the metabolome: the endogenous metabolome, referring to all metabolites present in a biological sample of the host, and the food metabolome, which includes metabolites that are derived from food consumption and their subsequent metabolism in the human body (6). Food metabolome not only includes metabolites of known micro-and macronutrients but also nonnutrient food compounds with biological roles yet to be elucidated.
To date, several studies have identified metabolomic signatures associated with the intake of specific foods and food groups, including fruits, vegetables, meat, fish, nuts, whole-grain bread, wine, coffee, and cocoa (6). A growing body of evidence has also emerged relating metabolic profiles with overall dietary patterns. Therefore, the purpose of this review is 2-fold: (a) to provide a broad overview of the metabolite measurement techniques, study designs, and statistical approaches used in nutrition metabolomics studies, and (b) to describe the current knowledge from epidemiologic studies identifying metabolite profiles associated with the intake of individual nutrients, foods, and dietary patterns.
Methods and Approaches in Studies of Nutritional Metabolomics
MEASUREMENT OF METABOLITES
Metabolomics refers to the systematic analysis of low molecular weight biochemical compounds in a biological sample. Urine, serum, and plasma samples are the most common biofluids used in nutritional metabolomics studies. One of the major differences between urine and plasma is that urine contains a higher concentration of nonmetabolites and nonnutrient compounds (representing noncontributory information or noise) that are derived from food phytochemicals and other chemicals. However, most of the metabolites in urine are excreted faster than those from plasma and can serve as acute markers of frequently consumed foods. For example, urine excretion of proline betaine is known to peak within a few hours after intake and be almost completely excreted within 24 h (7). Blood, on the other hand, contains a higher concentration of metabolically active compounds, and lipid-soluble metabolites are present only in plasma, not in urine (3).
In general, 2 different metabolomics techniques have been applied: (a) mass spectrometry coupled with gas- or liquid-phase chromatography, and (b) proton (1H) nuclear magnetic resonance (NMR)4 spectroscopy (8). Other platforms, such as inductively coupled plasma mass spectrometry, are also used to detect trace minerals and other electrolytes in biological samples (9). Targeted approaches that focus on a specific subset of predefined metabolites, as well as more agnostic untargeted approaches that analyze many measurable compounds in the sample, including chemical unknowns, have been implemented (8). Although targeted approaches are usually less expensive and follow a hypothesis-driven approach of metabolites of known identity, untargeted metabolomics detect thousands of unknown metabolites that may provide novel information on biological pathways with clinical relevance. However, because of the higher cost of the latter approach, the high density of the data acquired, and the methods required for complex statistical analysis, several epidemiological studies so far have relied solely on targeted metabolite profiling. Human metabolic databases, such as the Human Metabolome Database, can be a useful resource for nutritional metabolomics. The Human Metabolome Database includes more than 6800 fully annotated metabolites, such as metabolic intermediates, hormones, drugs, and food components (10). Despite the existence of many metabolites, the nutrition community is particularly interested in metabolic pathways in which nutrients are involved, including carbohydrate, lipids, amino acids, and energy metabolism pathways, along with mineral, trace elements, and vitamin metabolism pathways (3).
STUDY DESIGNS USED IN NUTRITIONAL METABOLOMICS
Intervention studies
Metabolomics can be used as a key tool in the search for novel biomarkers of dietary intake. Studies of nutritional metabolomics need to account for intersubject metabolic variation and should be able to deal with measurements of subtle metabolic modulations against relatively low doses of bioactive food nutrients or supplements (11). One method is to conduct controlled dietary intervention trials; in acute feeding studies, participants consume the food of interest in a single meal. For short- to medium-term trials, participants typically consume the food of interest in repeated meals over a given period, ranging from a few days up to several months. For this purpose, a crossover study design has traditionally been favored over a parallel design because it effectively deals with intersubject variation as each participant serves as his or her own control. Biofluids can be collected before and after the consumption of the food of interest in acute studies; in short- and medium-term trials, biofluids are usually collected at baseline and at the end of the intervention period (6). Any biomarker identified in acute studies must ideally be validated with longer intervention studies and replicated in different populations. Given the need to detect accurate measurements of dietary exposures, nutritional metabolomics can be a useful tool for identifying objective markers of relatively short-term response to diet interventions, as well as compliance with such dietary interventions. In addition, well-conducted clinical trials are less prone to confounding and measurement errors compared with observational studies.
Observational studies
One of the advantages of nutritional metabolomics studies is the possibility to identify a wide range of dietary biomarkers instead of a single biomarker. Indeed, multimetabolite biomarker panels can offer a better estimation than single biomarkers and increase the accuracy and precision of dietary assessment when combined with a food frequency questionnaire (FFQ). In this context, observational studies with repeated measurements of diet over time, which provide information of usual diet, play an important role. Epidemiologic studies in this field compare low and high consumers of nutrients/foods using FFQ, food records, and other dietary assessment tools, and then characterize objective biomarkers that are reflective of habitual intake or related to the intake of specific nutrients and food groups. These studies can also be used to detect metabolite signatures associated with overall dietary patterns. It is important to note that many of the foods consumed are highly correlated, and there is a risk of identifying biomarkers that are not specific to the particular food of interest (6). For example, vitamin C, several carotenoids, and flavonoids are common to many fruit and vegetables; therefore, they can be used as generic biomarkers of total fruit and vegetable intake but not specific to individual fruits or vegetables (12).
A number of these observational studies have applied cross-sectional designs comparing groups of participants at a single time point (i.e., consumers vs nonconsumers). Another possible approach to identify dietary biomarkers through metabolomics is the study of longitudinal variations in metabolite concentrations and their associations with diet and particular health outcomes or markers in population-based studies and clinical trials (6). With these prospective designs, individuals or populations exposed to different environments, lifestyles, or dietary patterns can be distinguished and metabolic differences can be identified.
Nevertheless, it is worth mentioning that most of the observational studies in nutritional metabolomics have small sample sizes and have not been replicated. One of the reasons that may account for the lack of replication in metabolomics studies is the fact that such analyses have not yet been standardized and homogenized, especially compared with genome-wide, large-scale studies (13). Several differences across studies exist, including storage of biological samples, different platforms for analyses of specimens, quality control and data preparation, and relative vs absolute quantification of metabolites. Therefore, there is an urgent need to evaluate metabolite stability and biological variability in large populations before markers of food intake or dietary patterns can be properly validated (13).
STATISTICAL APPROACHES
Analyses of high-throughput nutritional metabolomics data require the use of advanced bioinformatics and computational tools. Challenges with metabolomics analyses include data preparation and normalization, data reduction into fewer dimensions, and interpretation. In the section below, we briefly provide an overview of the statistical approaches commonly used in nutritional metabolomics studies.
Data preparation
The first step in metabolomics analysis is to prepare the metabolic profile of raw data generated by the analysis of biological samples. Raw data undergo data preprocessing, data alignment, data normalization, and signal correction before application of statistical methods (14). The first step is preprocessing; in mass spectrometry, this step includes peak detection, peak matching, retention time alignment, peak integration, and peak filling (13). Data alignment consists of matching peaks (m/z) and retention times to standards to align the different sample profiles. Some software tools also include gap filling, which checks raw data for any peak that has not been detected in a sample but was found in others. Quality control is also undertaken in this phase with the use of repeatability filters (i.e., filtering out features with coefficient of variation >30% or lower cut-points for targeted biomarkers) (13). In most laboratories, internal standards and pooled study samples are used to standardize data across data sets. In NMR, preprocessing needs to account for peak overlap and peak shift. Binning (or grouping) of metabolite signals based on correlation structure is commonly used to account for peak shift (15). Peak fitting can address peak shifting and overlap, but it is time consuming (16). Batch normalization, scaling, and outlier removal are also important parts of data preparation; normalization is used to account for uncontrolled metabolome-wide effects like dilution. Normalization is aimed to remove any unwanted variation in the spectrometric signal that cannot be controlled for or removed in any other way (17). Annotation of metabolites is usually the last step before applying statistical analysis. Comparing peaks with standards, databases, and commercial software is an important part of the process to avoid “putative annotation” because each feature could correspond to multiple metabolites. Although these methods are generally applied to targeted and untargeted data, identification of individual metabolite from untargeted peaks is challenging. Although online databases of annotated metabolites are growing, many unknown compounds still need to be identified.
Missing values in metabolomics data sets occur widely and can arise from different sources, including technical and biological reasons. Several methods to deal with missing data in metabolomics analysis have been examined (18–21). In mass spectrometry data analysis, a common approach is to remove from the analysis individual metabolites when a large proportion of participants have missing values or to remove a participant with many missing metabolites (21). Other standard methods of missing value imputation include the replacement of missing values with a nonzero value while maintaining data structure (i.e., minimum value, mean, or median value between 0 and the detection limit). Importantly, results and interpretation of the data can vary depending on the imputation method used (18). Other algorithms, including K-nearest neighbors, Bayesian principal component analysis (PCA), local least-squares regression, singular value decomposition, and random forest, can also be used to impute missing data in metabolomics analysis (19, 22). In a study comparing 5 different imputation methods on unsupervised and supervised analyses, random forest provided better classification rates for PCA and partial least-squares discriminant analysis (PLS-DA) (19). However, in a recent study comparing 7 imputation methods, no imputation method was perfect, but the simple substitution methods (half minimum and mean) consistently performed poorly (22).
Statistical methods to derive metabolomics fingerprints of diet
To maximize the recovery of information and to help in the interpretation of high-dimensional data sets, advanced statistical and bioinformatics tools are applied (11). Clustering methods are often used to reduce and divide the data into groups with a high degree of similarity (23). The most popular clustering techniques in nutritional metabolomics include multivariate analysis such as simple unsupervised clustering algorithms, PCA, and supervised techniques like PLS-DA and its variant combining a data-filtering step such as orthogonal signal correction and orthogonal partial least-squares discriminant analysis (O-PLS-DA) (24). These techniques create subgroups of metabolites without a priori hypotheses of metabolic pathways or the association with dietary information. Unsupervised PCA derives a linear transformation that preserves as much of the variance in the original data as possible while maximizing intergroup variation and minimizing within-group variation. PLS-DA, on the other hand, maximizes the covariance between scores in x (predictor) and y (outcome) spaces, and it accounts for both systematic variations in the metabolic profiles and correlations between metabolomics data and the outcome. Partial least-squares analysis is often used for discriminant analysis to classify metabolic profiles according to categories (i.e., dietary patterns) (13).
PCA is usually the starting point for an exploratory analysis, as it allows visualization of biological sets based on the resemblance of samples with respect to their biochemical composition, as well as allowing the extraction of information on factors contributing to the difference among samples (25). PCA creates a reduction summary of the data, which can be analyzed graphically using scores plot and loading plots (11). PCA can facilitate the comparison of many complex data such as biofluid spectra and can provide information on metabolite changes. This technique is particularly efficient for the identification of outliers. Although PCA can help to reduce the dimension of the data set, it does not give an insight of the association between metabolite signatures and dietary components. Other techniques such as correlations and multivariate regression analyses are then applied to test the associations between the factors extracted from PCA with dietary patterns and food groups.
Supervised multivariate analyses are commonly used in nutritional metabolomics. PLS-DA provides a way to filter out metabolic information that is not correlated to the predefined classes, whereas the PLS-DA loadings, similarly to PCA loadings, yield information on which spectral signals are associated with the observed clustering (11). The O-PLS-DA method is similar to PLS-DA, but the interpretation of the models is improved because the structured noise is modeled separately from the variation common to the matrices. O-PLS-DA enables a more straightforward and accurate interpretation of metabolomics data (24). O-PLS-DA models the metabolic profile that is linearly predictive of the dietary component of an individual but also captures external factors not linearly related to the outcome, such as age, sex, and batch effects. These techniques are useful in exploring the relation of any features in the metabolomics profiling data set with an external variable (i.e., intake of a specific food based on a questionnaire or any biological outcome marker).
All these statistical methods require rigorous model validation using cross-validation and permutation testing [i.e., using a proportion of the data as the test set (usually 10%) and building the model in the remaining training set] and ideally external validation in independent studies to avoid false-positive discoveries and to ensure model robustness (13). Moreover, as a comprehensive way of interpreting the data, it is highly recommended to report Bonferroni adjustment and false-positive discovery rate procedures to account for multiple comparison testing, as well as P values and CIs, effect size, and adjustment for covariates (26).
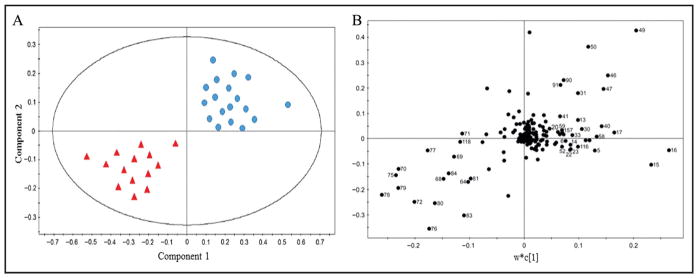
Fig. 1 depicts an example of scores (A) and loading plots (B) obtained from O-PLS-DA. Data in Fig. 1 are hypothetical and used only for the purpose of illustrating scores and loading plots. Fig. 1A could represent O-PLS-DA of 1H NMR of urine data and dietary interventions. Blue dots in the figure represent control diet, and red triangles represent dietary pattern intervention. Component 1 and component 2 are extracted from O-PLS-DA analysis. Loading scatterplot (Fig. 1B) shows the individual compounds.
Fig. 1. Example of scores (A) and loading plots (B) of O-PLS-DA.
Data in this figure are hypothetical and used for the only purpose of illustrating scores and loading plots. (A) could represent O-PLS-DA of 1H NMR of urine data and dietary interventions. Blue dots represent control diet, and red triangles represent dietary pattern intervention. Component 1 and component 2 are extracted from O-PLS-DA analysis. Loading scatterplot (B) shows the individual compounds.
Pathway analyses and other systems biology approaches
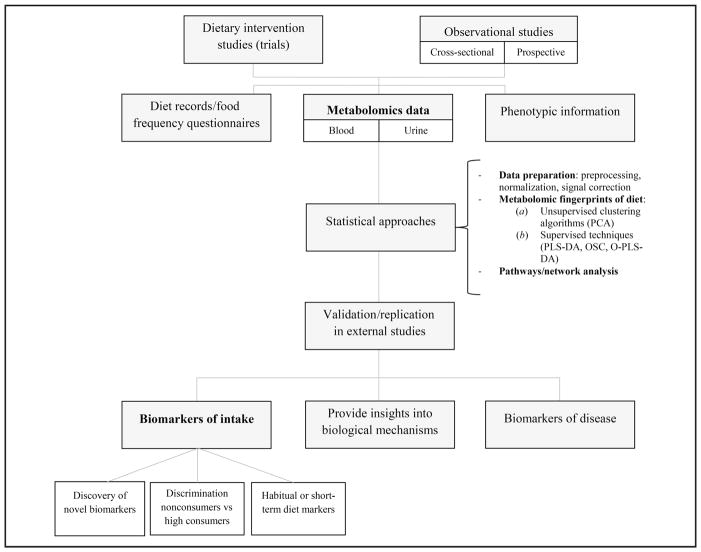
Because of the efforts to analyze complex genomic data and their integration with bioinformatics in the past decades, several metabolic databases, such as KEGG, MetaCyc, and BiGG families, have been developed (27, 28). These databases can help to elucidate underlying metabolic pathways and integrate targeted and untargeted metabolomics data. Semisupervised pathway analysis, such as the web-based tool Metabolic Set Enrichment Analysis (29) (which includes >1000 predefined metabolite sets covering various metabolic pathways, biofluids, and tissue locations), follows the principle of gene enrichment analysis to derive metabolic sets. These techniques, often used for the prediction of health outcomes, can also be applied to nutritional metabolomics to evaluate the associations between these enriched metabolite sets and dietary patterns. Agnostic network models are commonly used to combine significant metabolites identified from the targeted approach with the untargeted method to discover and validate metabolomics signatures. Correlation networks, a systems biology tool that enables visualization of the complex correlation structure between metabolites and clinical parameters (30), can be combined with algorithms for detecting active subnetworks to integrate data into metabolic pathways. These software tools, although not yet widely used in nutrition metabolomics, can help in the understanding and visualization of metabolic networks, as well as the description and prediction of metabolic pathways, and also constitute an important resource for nutrition research (25). Most of the statistical approaches that are currently used for genomics and metabolomics data treatment can serve as a foundation for nutritional studies, for which high-throughput metabolomics data are becoming extensively available. A schematic summary work flow of nutritional metabolomics approaches is presented in Fig. 2.
Fig. 2.
Work flow of nutritional metabolomics approaches.
Metabolomic Fingerprinting of Diet
Metabolomic techniques have been applied to several clinical and population settings to characterize metabolic effects of nutrients, foods, and dietary patterns. Biomarkers identified with the use of high-throughput metabolomics (usually mass spectrometry and NMR) have been measured in urine, plasma, and serum, and fall into different subclasses, including polar metabolites (amino acids, acylcarnitines, carbohydrates, bile acids, metabolites from purine and pyrimidine) and apolar metabolites (lipid classes). Briefly, most studies focused on the identification of dietary biomarkers of specific food and food groups, whereas fewer identified biomarkers of single nutrients (6). The most extensively studied food groups include fruits, vegetables, meat, fish, bread, whole-grain cereals, nuts, wine, coffee, tea, cocoa, and chocolate (6). The number of participants in these studies ranged from 4 to 500 participants, and a majority used a cross-sectional study design to identify specific metabolites of a food or food group differing between consumers and nonconsumers (6).
BIOMARKERS AND METABOLITES OF SINGLE NUTRIENTS
Several biomarkers of single nutrients including fiber and protein intake have been identified. In a randomized intervention study, concentrations of metabolite markers 2,6-dihydroxybenzoic acid and 2-aminophenol sulfate increased after participants followed a high-fiber diet (mean intake 48 g compared with the low-fiber group at 30.2 g) for 5 weeks (31). In another controlled intervention study in which participants were randomized to a high or low glycemic index diet for 6 months, hippuric acid was correlated with dietary fiber intake (32). However, none of the biomarkers identified in the first study were confirmed in the second. The lack of consistency across studies may be explained because of differences in study duration (5 weeks vs 6 months), interventions (high-fiber diet in the first study consisted of oat bran, rye bran, and sugar beet fiber incorporated into test food products, whereas the intervention diets in the second study were designed to achieve a difference of 15 glycemic index points), or the age of participants (37–45 years vs 30–70 years). This highlights the need to validate and replicate biomarkers in different populations to identify consistent biomarkers for the same nutrient.
In another crossover feeding trial of two 28-day diet periods of high and low glycemic load diet, concentrations of kynurenate and trimethylamine-N-oxide (TMAO) were found to be significantly higher after consumption of a low glycemic load diet (33). A cross-sectional analysis of 1003 participants from the Twins UK cohort showed that protein intake was positively associated with several plasma amino acids, including valine, phenylalanine, and tyrosine, and inversely associated with glutamine (34). In a randomized, crossover feeding study including 11 obese nondiabetic participants, plasma concentrations of tryptophan, phenylalanine, and kynurenine were increased after a high-fat meal with whey protein isolate (35).
METABOLOMIC FINGERPRINTS OF SPECIFIC FOOD GROUPS
Several studies have identified biomarkers of total polyphenol intake and polyphenols from specific food groups, including polyphenols from cocoa powder (36–38), red wine and grape juice (39, 40), almond skin (41), nuts (42), and orange juice (43). In the PRE-DIMED study, a randomized, parallel, clinical trial for primary prevention of cardiovascular disease, using an untargeted metabolomics analysis of urine from 32 consumers of cocoa-derived products and 32 controls, the discriminant biomarkers of cocoa consumption were related to the metabolic pathways of theobromine and polyphenols (38). In another study of 57 volunteers at high cardiovascular risk, tartrate was associated with wine polyphenol consumption (40). Hydroxyphenylvaleric, hydroxyphenylpropionic, and hydroxyphenylacetic acids were identified in human urine samples after the consumption of flavan-3-ols from almond skin in an acute feeding study (41). Betonicine, stachydrine, methyl glucopyranoside (i + β), dihydroferulic acid, and galactonate were proposed as new metabolic signatures to distinguish the intake of orange juice with different polyphenol content in the BIONAOS study, which compared a normal-polyphenol orange juice with a high-polyphenol orange juice for 12 weeks in a randomized parallel study (43). Moreover, a recent study evaluating metabolic profiles in urine of 481 subjects from the European Prospective Investigation into Cancer (EPIC) and Nutrition cohort identified >80 polyphenol metabolites associated with the consumption of 6 polyphenol-rich foods (coffee, tea, red wine, citrus fruit, apples and pears, and chocolate products) (44).
As extensively reviewed by Scalbert et al. (6), many biomarkers identified using metabolomics were associated with the consumption of fruits, vegetables, and specific markers of citrus fruit and cruciferous vegetables. As an example, proline and betaine were identified as biomarkers of citrus intake in an acute feeding study of 8 participants and then validated with 499 participants from INTERMAP (7) and another cross-sectional study (45). At the same time, urine concentrations of S-methyl-L-cysteine sulfoxide were found to be related to the intake of cruciferous vegetables in a short-term intervention study that included 20 healthy men who consumed 250 g/day of cruciferous vegetables (broccoli and Brussels sprouts) for 14 days (46). Metabolite biomarkers of tomato sauce consumption have also been characterized (47). Specifically, serum concentrations of creatine, creatinine, leucine, choline, methionine, and acetate were found to be increased after a 4-week intervention with 160 g/day of high-lycopene tomato sauce, and ascorbic acid, lactate, pyruvate, isoleucine, and alanine were increased after the intervention with 160 g/day of normal-lycopene content tomato sauce (47).
Potential markers of nut intake identified in intervention studies ranging from 12 weeks to 6 months (42, 48, 49) include conjugated fatty acids, serotonin metabolites, and microbial-derived phenolic metabolites. In the PREDIMED study, walnut consumption was characterized by the presence of 18 urinary metabolites, including markers of fatty acid metabolism, ellagitannin-derived microbial compounds, and intermediate metabolites of the tryptophan/serotonin pathway (38).
Metabolomics fingerprints of beverage consumption have been extensively examined as well. Coffee has been reported to be positively associated with specific classes of sphingomyelins and negatively associated with long- and medium-chain acylcarnitines (50). In a case–cohort study of type 2 diabetes incidence including 1610 EPIC-Potsdam participants, in which 163 metabolites were targeted, coffee consumption was inversely associated with diacylphosphatidylcholine C32:1 in both men and women and phenylalanine in men, whereas in women, coffee intake was positively associated with acyl-alkyl-phosphatidylcholines C34:3, C40:6, and C42:5 (51). Other confirmed metabolite markers of coffee exposure included methylxanthines and reduced, sulfated, and methylated forms of hydroxycinnamates (52–54), which are coffee-derived metabolites. Recently, among participants of a 3-stage coffee trial in which 47 participants refrained from drinking coffee in the first month, consumed 4 cups of filtered coffee per day in the second month, and 8 cups of coffee per day in the third month, 115 plasma metabolites, evaluated with a nontargeted metabolomic profiling approach, were significantly associated with coffee intake (55). Five metabolic pathways were significantly enriched: (a) xanthine metabolism, which includes caffeine metabolites, (b) benzoate metabolism that reflects polyphenol metabolite products of gut-microbiota metabolism, (c) steroid, which is novel but may reflect sterol content of coffee, (d) fatty acid metabolism (acyl choline), a novel link to coffee; and (e) endocannabinoid, a novel link to coffee (55).
Metabolites linked with carnitine metabolism and sulfation of tyrosine are among the set of metabolites linked to cocoa intake (37, 38). Findings from the Atherosclerosis Risk in Communities study suggested that sugar-rich foods and beverages were inversely associated with 5 metabolites in the 2-hydroxybutyrate-related sub-pathway and 7 unsaturated long-chain fatty acids, and positively associated with 5-γ-glutamyl dipeptides (56). Hippuric acid, previously identified as a marker of fiber intake (32), has also been identified as a marker of tea consumption, especially green and black tea, in several intervention and cross-sectional studies (48, 57, 58). The nonspecificity of the biomarkers to 1 specific food highlights the limitation of using nutritional metabolomics for the characterization of markers of food intake. As another illustration, hydroxytyrosol, which is a minor dopamine metabolite that derives from oleuropein, is a well-established biomarker of olive oil consumption (59). Plasma and urinary hydroxytyrosol levels have been shown to increase in a dose-dependent manner with the phenolic content of food (i.e., olive oil) (60). However, recently, findings from the PREDIMED study have revealed a direct association between red wine consumption and urinary hydroxytyrosol, independent of the amount of olive oil consumed (59). Thus, hydroxytyrosol cannot be considered a specific biomarker, as it cannot differentiate between olive oil and red wine consumption.
In a randomized, crossover, feeding study of 19 post-menopausal women comparing the intake of refined wheat, whole-meal rye, and refined rye breads providing 50 g of carbohydrates, and in which 189 metabolites were targeted, 8 amino acids (leucine, isoleucine, citrulline, ornithine, proline, asparagine, methionine, and lysine) were significantly influenced by intervention (P < 0.01). Branched-chain amino acids (BCAA) were higher after refined wheat bread consumption compared with the other breads at 45 and 60 min (61). Other metabolites, including 3-(3,5-dihydroxyphenyl)-1-propanoic acid sulfate, enterolactone glucuronide, azelaic acid, and 2-aminophenol sulfate, have also been detected as markers of whole-grain rye bread intake (62). Finally, a targeted metabolomics study of >300 lipid metabolites suggested that lysophosphatidylcholine, lyso-platelet-activating factor, and several phospholipid fatty acids were associated with consumption of full-fat dairy products (63). In addition, a recent 24-week energy-restricted intervention study with low [0–1 dairy products/day (<600 mg calcium/day)] or high [4–5 dairy products/day (approximately 1200 mg calcium/day)] dairy intake showed that high dairy consumption increased urinary citrate and creatinine and decreased the urinary excretion of TMAO and hippurate (64).
METABOLOMICS SIGNATURES OF DIETARY PATTERNS
Several studies have evaluated metabolite profiles related to overall dietary patterns. We searched PubMed for studies published in English from database inception to April 2017 using the search terms {[“metabolomics” (MeSH Terms) OR “metabolomics” (All Fields)] OR [metabolite (All Fields)] AND profiling (All Fields)]} AND {[“diet” (MeSH Terms) OR “diet” (All Fields) OR “dietary” (All Fields)] AND [pattern (All Fields)] OR [“diet therapy” (MeSH Terms)] OR [“diet” (All Fields) AND “therapy” (All Fields)] OR “diet therapy” (All Fields) OR [“dietary” (All Fields) AND “intervention” (All Fields)] OR “dietary intervention” (All Fields)} NOT [“review” (Publication Type) OR “review literature as topic” (MeSH Terms) OR “review” (All Fields)] AND “humans” (MeSH Terms). We excluded nonhuman studies, studies in children, reviews, and commentary articles. We further excluded studies focusing on dietary supplementation and single foods because of a recent comprehensive review by Scalbert et al. (6) on metabolomics studies on food groups. After exclusions, the search identified 130 studies, of which 16 (summarized in Table 1) evaluated biomarkers of dietary patterns or dietary interventions using metabolomics profiling.
Table 1.
Studies evaluating dietary patterns and dietary interventions using metabolomics profiles.
| Author, journal, year | Study population | Study design, n of participants and follow-up | Dietary interventions/patterns | Biofluid | Analytical technique | Biomarkers identified |
|---|---|---|---|---|---|---|
| Stella et al., J Proteome Res, 2006 | White, nonsmoking men, of normal body weight (ages 25–74 years) | Randomized, crossover, n = 12, 15- day diets with 7-day washout |
|
Urine | 1H NMR | Taurine, carnitine, acetylcarnitine, 1-methylhistidine, 3-methylhistidine, TMAO were elevated in high-meat period p-Hydroxyphenylacetate was higher in vegetarian diet than meat diets |
| Xu et al., Anal Bioanal Chem, 2010 | Healthy men and women (ages 18–55 years) | Cross-sectional, habitual diet, n = 161 |
|
Urine | 1H NMR | TMAO, dimethylamine, phenylalanine, methylhistidine were characteristic of omnivorous diet Citrate was characteristic of lactovegetarian diet |
| O’Sullivan et al., Am J Clin Nutr, 2011 | Healthy men and women (ages 18–63 years) | Participants from a double-blind, randomized, placebo-controlled dietary intervention on vitamin D and probiotics, n = 160, results for metabolomics from a cross-sectional analysis of dietary records and biofluids | Cluster 1: favorable food highest in vegetables, whole-grain breads, and fish Cluster 2: higher fruit intake Cluster 3: unhealthiest with high intakes of meat products, white bread, butter, and lower fruits and vegetables, and whole-meal bread |
Plasma and urine | 1H NMR | Phenylacetylglutamine marker of vegetables intake, O-acetylcarnitine marker of red meat Glycine, phenylacetylglutamine, acetoacetate higher in cluster 1 TMAO, O-acetylcarnitine and N,N-dimethylglycine higher in cluster 3 |
| Altmaier et al., Eur J Epidemiol, 2011 | Healthy males (ages 55–79 years) from KORA | Cross-sectional, n = 284 | Dietary component 1: high poultry, fish, rice, vegetables, fruit, chocolate, flaked oat, curds, cheese, milk and low meat and sausages Component 4: traditional Bavarian with high meat, poultry, fish, snacks and eggs and low potatoes, pasta, whole-grain bread and flaked oats Component 6: high in whole-grain bread, eggs, milk, low consumption of fish, fresh fruits, and cakes |
Plasma | MS/MS | Component 1 associated with diacyl-phosphatidylcholines with 4 and 5 bounds and acyl-alkyl-phosphatidylcholines with 1 or 2 double bonds, and a decrease in saturation of fatty acids of glycerol-phosphatidylcholines Components 4 and 6 associated with several phospholipids |
| May et al., Br J Nutr, 2013 | Healthy, nonsmoking men and women (ages 21–45 years) | Randomized trial, n = 10, 2- week and cross-sectional, n = 93 |
|
Urine | LS-MS/MS | Proline betaine marker of citrus intake Phytochemical diet characteristic by higher sulforaphane, hippuric acid, genistein, daidzein, equol, glycitein, O-desmethylangolensin, enterolactone, trigonelline |
| Bouchard-Mercier et al., Br J Nutr, 2013 | Healthy participants (ages 18–50 years) | Cross-sectional, n = 210 |
|
Plasma | MS | Western dietary pattern associated with short-chain acylcarnitines, leucine, phenylalanine Prudent pattern composed of medium- to long-chain acylcarnitines |
| Floegel et al., Eur J Clin Nutr, 2013 | Healthy participants (ages 35–65 years) from EPIC-Potsdam | Cross-sectional, n = 2380 |
|
Serum | (FIA)-MS/MS | Acylcarnitines, amino acids, hexoses, diacyl-, acyl- alkyl- lysophosphatidylcholines, and sphingomyelins were related to habitual diet Diet high in red meat and fish, and low in whole-grain bread and tea related to hexoses and phosphatidylcholines Diet high in potatoes, dairy, and cornflakes high in methionine and branched-chain amino acids |
| Menni et al., Metabolomics, 2013 | Healthy women (mean age 59.5 years) from Twins UK | Cross-sectional, n = 1003 | Nutritional scores:
|
Serum | (FIA)-MS/MS | Fruits and vegetables related to glycerophospholipid (phosphatidylcholine-dyacil C38:6 and acylcarnitine C9 associated with hypocaloric dieting) |
| Andersen et al., Anal Bioanal Chem, 2014 | Participants with increased waist circumference and ≥1 metabolic syndrome risk factors (ages 18–65 years) | Randomized, controlled, parallel, trial, n = 181, 6 months |
|
Urine | UPLC-qTOF- MS | TMAO was related to seafood consumption; Nordic diet characterized by TMAO, hydroquinone-glucuronide, hippuric acid, 3,4,5,6-tetrahydrohippurate |
| Lankinen et al., PloS One, 2014 | Participants with impaired glucose metabolism and features of metabolic syndrome (ages 40–70 years) | Randomized, controlled, parallel, trial, n = 106, 12 weeks |
|
Serum | NMR | Healthy diet increased polyunsaturation of plasma fatty acids, increase in n-3 fatty, whereas n-6 and n-7 fatty acids decreased Greater fish intake associated with large HDL lipid components |
| Hanhineva et al., J Nutr, 2015 | Participants with impaired glucose metabolism and features of metabolic syndrome (ages 40–70 years) | Randomized, controlled, parallel, trial, n = 106, 12 weeks |
|
Plasma | LC-qTOF-MS | Glucuronidated alk(en)ylresorcinols elevated in both intervention diets and correlated with whole-grain products Healthy diet increased furan fatty acids, hippuric acid, and various lipid species such as polyunsaturated fatty acids Furan fatty acids related with fish intake |
| Schmidt et al., Am J Clin Nutr, 2015 | Healthy men (ages >20 years), from EPIC-Oxford | Cross-sectional, n = 379 |
|
Plasma | LC-MS/MS | 79% of metabolites differed significantly by diet group. Meat eaters had the highest concentrations of 6 acylcarnitines, 61 glycerophospholipids, and 12 sphingolipids, and vegans had the lowest concentration of these metabolites. Fish eaters and vegetarians had the highest concentrations of amino acids and biogenic amines |
| Vázquez-Fresno R et al., J Proteome Res, 2015 | Nondiabetic participants at high cardiovascular risk from the PREDIMED Study (ages 55–80 years) | Randomized, controlled, parallel trial, n = 98, 1 and 3 years of follow-up |
|
Urine samples at baseline, 1 and 3 years | 1H NMR | Mediterranean diet groups: carbohydrates (3-hydroxybutyrate, citrate, and cisaconitate), creatine, creatinine, amino acids (proline, N-acetylglutamine, glycine, branched-chain amino acids, and derived metabolites), lipids (oleic and suberic acids), and microbial cometabolites (phenylacetylglutamine and p-cresol) Control low-fat diet: hippurate, TMAO, histidine, and derivates (methylhistidines, carnosine, and anserine), and xanthosine |
| Garcia-Perez I et al., Lancet Diabetes Endocrinol, 2015 | Healthy volunteers (ages 21–65 years) | Randomized, controlled, crossover trial, n = 19, 4 inpatient stays (72 h each, separated by at least 5 days) | Four diets differing in compliance to the WHO healthy eating guidelines: Decreased sugar, salt, and total fat consumption, and increased intake of whole grains, fruits, vegetables, and dietary fiber, with diet 1 being the most concordant with guidelines and diet 4 the least concordant |
Urine samples during 3 periods that were then pooled to obtain 24-h urine | 1H NMR | Diet 1: Higher concentration of urinary biomarkers from individual healthy foods, e.g., hippurate (marker of fruit and vegetable consumption), N- acetyl-S-methyl-cysteine-sulfoxide (cruciferous vegetables), dimethylamine, and TMAO (fish), and 1-methylhistidine and 3-methylhistidine (oily fish and chicken) Diet 4: Higher concentration of nine urine metabolites, which had higher amounts of red meat (O-acetylcarnitine, carnitine, and creatine) and sugars (glucose) |
| Bondia-Pons et al., Mol Nutr Food Res, 2015 | Participants with metabolic syndrome features from the RESMENA Study | Randomized, controlled, trial, n = 72, 2 and 6 months | Two energy-restricted diets
|
Plasma | LC-QTOF/MS | Mediterranean diet: plasmalogen phosphatidylcholine (P-18:1/20:3) after the 2-month intervention, and palmitic acid after the 6-month intervention. PC (P-18:1/20:3) significantly increased in the RESMENA group and decreased in the Control group after 2 months |
| Playdon et al., Am J Clin Nutr, 2017 | Healthy male smokers (≥5 cigarettes/day) ages 50–69 years | Nested case-control studies in the ATBC study (randomized, double-blind, placebo-controlled primary prevention trial of cancer) | Four dietary patterns and their components:
|
Serum | UPLC-LC-MS/MS | HEI-2010 was associated with 17 identifiable chemical structure metabolites: 3 amino acids, 2 cofactors or vitamins, 9 lipids, and 3 exogenous xenobiotics aMED was associated with 21 identifiable metabolites: 4 amino acids, 1 carbohydrate, 2 cofactors or vitamins, 11 lipids, and 3 xenobiotics HDI was associated with 11 metabolites: amino acids, 2 were cofactors or vitamins, 4 were lipids, and 2 were xenobiotics BSD associated with 10 metabolites: 2 amino acids, 1 carbohydrate, 3 cofactors or vitamins, and 4 lipids The lysolipid pathway contained the largest number of metabolites associated with diet quality |
aMED, Alternate Mediterranean Diet Score; ATBC, Alpha-Tocopherol, Beta-Carotene Cancer Prevention; BSD, Baltic Sea Diet; FIA, flow injection analysis; HDI, Healthy Index Dietary Indicator; HEI, Healthy Eating Index; 1H NMR, proton nuclear magnetic resonance; KORA, Collaborative Health Research in the Region of Augsburg; LC-QTOF/MS, liquid chromatography coupled to quadrupole time of flight-MS; PREDIMED, Prevención con DIeta MEDiterránea; RESMENA, Metabolic Syndrome Reduction in Navarra.
Most studies included healthy participants and applied mass spectrometry or NMR in blood and urine to extract metabolite profiles. Nine articles included cross-sectional analyses discriminating consumers from non-consumers of specific dietary patterns. Nine studies were intervention studies that evaluated the effect of overall dietary patterns on metabolic profiles. Several dietary patterns have been studied in the context of nutrition metabolomics, including vegetarian and lactovegetarian diets, omnivorous diet, Western dietary patterns, prudent dietary patterns, Nordic diet, and Mediterranean diet.
Using a cross-sectional study design, Bouchard-Mercier et al. investigated the associations between dietary patterns and metabolic profiles (65). In a targeted mass spectrometry analysis of 14 amino acids and 41 acylcarnitines, a PCA-derived Western dietary pattern was associated with a metabolite signature characterized by high levels of BCAA (leucine), aromatic amino acids (phenylalanine), and short-chain acylcarnitines (65). In another cross-sectional analysis of 2380 EPIC-Potsdam participants (127 serum metabolites were analyzed using mass spectrometry), a dietary pattern characterized by high intakes of red meat and fish and low intakes of whole-grain bread and tea was directly related to high plasma levels of hexose and phosphatidylcholines (66). A pattern consisting of high intake of potatoes, dairy products, and cornflakes was associated with higher methionine and BCAA concentrations (66). Another cross-sectional study of women from the Twins UK cohort, characterized by metabolites associated with several nutritional scores, including fruit and vegetable intake, high alcohol intake, low meat intake pattern, hypocaloric dieting, and traditional English diet; phosphatidylcholine-dyacil C38:6 and acylcarnitine C9 were significantly and positively associated with hypocaloric dieting (34).
Another interesting study aimed to investigate the differences in concentrations of 118 circulating metabolites between male meat eaters, fish eaters, vegetarians, and vegans was conducted in the Oxford EPIC cohort. The study found that concentrations of 79% of metabolites differed significantly by diet groups. Concentrations of acylcarnitines, C-0, C-4, and C-5 were highest among meat eaters, followed by fish eaters, vegetarians, and vegans. At the same time, concentrations of acylcarnitines, C-3 and C-16, 61 glycerophospholipids, and 12 sphingolipids were highest among meat eaters and lowest among vegans. In contrast, fish eaters and vegetarians had the highest concentrations of amino acids (such as leucine, valine, lysine, methionine, tryptophan, and tyrosine) and biogenic amines (67).
Recently, Playdon et al. (58) identified metabolomic fingerprints of diet quality [evaluated using different dietary scores (Healthy Eating Index-2010, Alternate Mediterranean Diet Score, WHO Healthy Dietary Indicator, and Baltic Sea Diet)] in healthy male smokers from 5 nested case–control studies of the Alpha-Tocopherol, Beta-Carotene Cancer Prevention study. Healthy Eating Index-2010 was associated with 17 identifiable chemical structure metabolites: 3 amino acids, 2 cofactors or vitamins, 9 lipids, and 3 exogenous xenobiotics. Alternate Mediterranean Diet Score was associated with 21 identifiable metabolites: 4 amino acids, 1 carbohydrate, 2 cofactors or vitamins, 11 lipids, and 3 xenobiotics. WHO Healthy Dietary Indicator was associated with 11 metabolites: 3 amino acids, 2 cofactors or vitamins, 4 lipids, and 2 xenobiotics. Baltic Sea Diet associated with 10 metabolites: 2 amino acids, 1 carbohydrate, 3 cofactors or vitamins, and 4 lipids. The lysolipid pathway contained the largest number of metabolites associated with diet quality (68).
Several randomized intervention trials evaluated the effect of dietary interventions on metabolic profiles; however, most of these studies had small samples sizes and short follow-up periods (Table 1). The effect of a Mediterranean diet intervention on the urinary metabolome was assessed in the PREDIMED trial by comparing a subsample of nondiabetic subjects at 1 and 3 years of follow-up (69). Findings from this study showed that the most relevant metabolic signatures related to a Mediterranean diet intervention were metabolites of carbohydrate pathways (3-hydroxybutyrate, citrate, and cis-aconitate), creatine, creatinine, amino acids (proline, N-acetylglutamine, glycine, BCAA, and derived metabolites), lipids (oleic and suberic acids), and microbial co-metabolites (phenylacetylglutamine and p-cresol) (69).
The RESMENA study was an intervention study that included 72 subjects with metabolic syndrome features who consumed either an energy-restricted Mediterranean diet or an energy-restricted control diet (low fat) for 6 months (70). Metabolomics in plasma samples showed that the Mediterranean diet intervention resulted in significant changes in the metabolic profile at 2 months (mainly phospholipids and lysophospholipids), but differences were attenuated at 6 months (70).
Garcia-Perez et al. published the findings of a controlled, crossover, feeding study of 19 healthy participants who followed 4 different diets for 72 h separated by 5 days (71). The 4 diets differed in compliance to the WHO healthy eating guidelines: decreased sugar, salt, and total fat consumption and increased intake of whole grains, fruits, vegetables, and dietary fiber. Diet 1 was the most concordant with the guidelines and diet 4, the least concordant. The authors noted that 19 urinary metabolites were significantly increased after consumption of diet 1 compared with diet 4. Specifically, diet 1 resulted in higher concentration of urinary biomarkers from individual healthy foods like hippurate (marker of fruit and vegetable consumption), (N-acetyl-)S-methyl-L-cysteine-sulfoxide (cruciferous vegetables), dimethylamine and TMAO (fish), and 1-methylhistidine and 3-methylhistidine (oily fish and chicken). On the contrary, diet 4 was associated with higher concentration of 9 urinary metabolites, related to higher amounts of red meat (O-acetylcarnitine, carnitine, and creatine) and sugars (glucose) (71).
DIETARY BIOMARKERS DERIVED FROM GUT MICROBIOTA METABOLISM OF DIETARY COMPONENTS
Microbial species in human gut can directly deliver compounds from their metabolome, which are absorbed and contribute to human metabolism (such as amino acids, bile acids, short-chain fatty acids, vitamins, and energy substrates) (3). On the other hand, the gut microbiota can change constituents in food and make them available to themselves or the host for additional metabolism (3).
A number of metabolites that are related to diet and can be metabolized by gut microbiota have been identified. For example, microbial enzymes can hydrolyze soy isoflavones and release several metabolites, including aglycons, daidzein, genistein, and glycetin (3). The gut microbiota can also transform polyphenols to phenolic breakdown products, including benzoate and various derivatives of hydroxyphenylacetic and hydropropionic acids (3). In a randomized, parallel, controlled study designed to compare a high-soy diet [104 (24) mg total isoflavones/day] with a low-soy diet [0.54 (0.58) mg total isoflavones/day] in 76 healthy young adults followed for 10 weeks, concentrations of isoflavones and their gut flora metabolites in plasma, urine, and feces were significantly increased in participants who consumed the high-soy diet. Fecal β-glucosidase activity was significantly higher in the subjects who consumed the high-soy diet than in those who consumed the low-soy diet (72).
Some of the most studied gut microbiota-dependent metabolites are those related to TMAO and its precursors choline and carnitine. Foods such as meat and meat products, egg yolks, and high-fat dairy products, which are high in phosphatidylcholine, choline, carnitine, and trimethylamine (TMA), serve as precursors to TMAO (73). TMAO has been found to be a potential biomarker of meat intake, but it has also been reported as a biomarker of fish and seafood intake, and more recently, it has been reported to be related to plant foods like soybeans (71, 74, 75). Although certain microbial metabolites can be putative food biomarkers, there is a complex relation between the food sources, gut microbiota, and the food metabolites derived; thus, these biomarkers should be interpreted with caution.
Application of Metabolomics in the Field of Nutritional Epidemiology
Widely used dietary assessment methods such as diet records and FFQ have been instrumental in their ability to measure diet in large populations and in examining the role of diet in human health (4). Because diet represents a complex set of exposures that are intercorrelated, and because foods are mixtures of known and unknown constituents, assessing dietary intakes in the context of relatively large within-individual variations is an ongoing challenge in nutritional epidemiology (76). The integration of metabolomics into traditional nutritional epidemiology can, to some extent, overcome the limitations of traditional dietary assessment methods and can provide important insights into mechanistic pathways. In Table 2, we briefly summarize and compare traditional dietary assessment methods with biomarkers of dietary intake. Traditional assessment methods are noninvasive and have the advantage of providing useful information on long-term dietary intake (FFQs and multiple-week diet records). FFQs are easy to administer with a low respondent burden; multiple-week records provide open-ended dietary data and do not rely on memory (4). Nevertheless, they are subject to some biases, such as recall errors, health consciousness bias, and errors in nutrient estimation from food composition diets. On the other hand, objective biomarkers, usually measured in serum, plasma, and urine, can objectively assess dietary intakes, represent the true “bioavailable” dose, and can serve as validation markers of other assessment methods. However, they may not be sensitive to intakes, can have low specificity, and are not available for many nutrients and foods. In addition, they are expensive, more invasive, and are subject to laboratory errors (4). Some examples of these biomarkers include urinary nitrogen for protein intake, 24-h urinary potassium and sodium, doubly labeled water for energy intake, plasma vitamin D, serum and plasma folate, essential fatty acids and other vitamins, trace minerals, and metabolite biomarkers.
Table 2.
Comparison of traditional dietary assessment methods and biomarkers.
| Self-reported measures of diet
|
Objective measures of diet
|
|
|---|---|---|
| Validated FFQ | Multiple-week diet records | Biomarkers identified through metabolomics approaches |
| Provides information on usual long-term intake | Detailed dietary data that do not rely on memory | Objective assessment of intake and represents bioavailable dose |
|
| ||
| Estimating and recall errors | Errors from incorrect estimate of portion size and omission of foods | Subject to laboratory errors |
|
| ||
| Easily administered and low respondent burden | Participant burden is high | Can be measured in stored biospecimens |
|
| ||
| Least expensive method and noninvasive | Noninvasive but expensive | Expensive and more invasive |
|
| ||
| Can assess usual dietary intake | Can assess usual dietary intake if measured multiple times over the course of a year | May not be time-integrated or represent usual long-term intake; may not be sensitive or specific to intakes |
|
| ||
| Health consciousness bias | Health consciousness bias | Not applicable |
|
| ||
| Errors in nutrition estimation from food composition tables | Errors in nutrition estimation from food composition tables | Biomarkers are not available for many nutrients and most foods |
|
| ||
| Culture and population specific | Needs literate and motivated participants | Biomarker variations may exist between cultures and populations |
|
| ||
| Association analyses in large epidemiologic studies | Validation of other methods and assess compliance | Can be used to assess associations in cohort or nested case–control studies |
Adapted with permission from Satija et al. (4).
Nutritional metabolomics holds considerable promise for the development of a more robust and unbiased strategy for measuring diet. However, there are important issues to consider. Certain metabolites have a short half-life and may, therefore, not represent usual intake, which is the most relevant exposure in nutritional epidemiology. Of note, a single measurement of metabolites is not sufficient to represent usual intake. For a metabolite to be a valid biomarker of dietary intake, it needs to be sensitive to intake and should be relatively easy to measure in biofluids.
So far, an extensive list of potential biomarkers related to the intake of nutrients, foods, and diets has been revealed by metabolomics. Compared with single biomarkers of food consumption, nutritional metabolomics is contributing to the discovery of biomarker patterns. Metabolomics can also generate biomarker patterns to evaluate the efficacy of nutritional interventions for maintaining and improving health at the individual level. However, it is unlikely that metabolomics biomarkers will replace traditional dietary assessments using self-reported methods because for most foods and nutrients, sensitive and specific biomarkers are not available or have not yet been identified. In addition, metabolomics assays are expensive, rendering them infeasible to assess dietary habits among hundreds and thousands of participants in large cohort studies. Therefore, biomarkers identified from metabolomics and traditional self-reported methods such as validated FFQs should be used in a complementary fashion. In the future, reproducible metabolomics biomarkers may be used to validate self-reported measurements of dietary intake, calibrate estimates of dietary intake, identify novel biomarkers of food consumption, and provide objective biomarkers of adherence to dietary interventions and dietary patterns. However, more efforts are needed to develop, validate, and fine-tune assessment methods that can capture the multidimensional nature of diet.
From the standpoint of public health, the incorporation of nutritional metabolomics into traditional nutritional epidemiology can help to identify subgroups that differ in their response to specific dietary components so that interventions can be tailored to those who will benefit the most, reducing the cost and side effects for those who will not (77). However, before nutritional metabolomics can have a real impact on public health, there is an urgent need to establish reference intervals based on absolute metabolite concentrations in defined human bio-fluids, improve the specificity of metabolite biomarkers of certain foods, and to conduct studies with adequate statistical power with independent replications in diverse cohorts while considering ethnic and regional differences.
Finally, because recent metabolomics efforts have focused on the analysis of known metabolites, current efforts to characterize unknowns may enable more comprehensive investigations and the discovery of novel metabolic pathways. The field of nutritional epidemiology will greatly benefit from the integration of other -omics technologies, such as genomics, proteomics, epigenomics, and metagenomics. Global initiatives are needed to standardized data collection and analytic methods for metabolomics in human nutrition (3) and to create consortia of metabolomics studies including well-assessed dietary data across diverse populations in the world.
Acknowledgments
Research Funding: NIH grants HL60712, HL118264, and DK102896. M. Guasch-Ferré, postdoctoral fellowship granted by the Agency for Administration of University and Research Grants (AGAUR) of the Autonomous Government of Catalonia (2014-BP-A 00017); S.N. Bhupathiraju, Career Development Grant from the National Institutes of Health (K01 DK107804).
Footnotes
Nonstandard abbreviations: NMR, nuclear magnetic resonance; FFQ, food frequency questionnaire; PCA, principal component analysis; PLS-DA, partial least-square discriminant analysis; O-PLS-DA, orthogonal partial least-squares discriminant analysis; TMAO, trimethylamine-N-oxide; EPIC, European Prospective Study into Cancer; BCAA, branched-chain amino acids.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared. Consultant or Advisory Role: None declared. Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: None declared.
References
- 1.Cornelis MC, Hu FB. Systems epidemiology: a new direction in nutrition and metabolic disease research. Curr Nutr Rep. 2013;2:10. doi: 10.1007/s13668-013-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohlmeier M, De Caterina R, Ferguson LR, Görman U, Allayee H, Prasad C, et al. Guide and position of the International Society of Nutrigenetics/Nutrigenomics on personalized nutrition: part 2— ethics, challenges and endeavors of precision nutrition. J Nutrigenet Nutrigenomics. 2016;9:28–46. doi: 10.1159/000446347. [DOI] [PubMed] [Google Scholar]
- 3.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 4.Satija A, Yu E, Willett WC, Hu FB. Understanding nutritional epidemiology and its role in policy. Adv Nutr. 2015;6:5–18. doi: 10.3945/an.114.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guertin KA, Moore SC, Sampson JN, Huang W-Y, Xiao Q, Stolzenberg-Solomon RZ, et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet–disease relations in populations. Am J Clin Nutr. 2014;100:208–17. doi: 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Draper J, et al. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99:1286–308. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 7.Heinzmann SS, Brown IJ, Chan Q, Bictash M, Dumas M-E, Kochhar S, et al. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. Am J Clin Nutr. 2010;92:436–43. doi: 10.3945/ajcn.2010.29672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455:1054–6. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 9.Konz T, Migliavacca E, Dayon L, Bowman G, Oikonomidi A, Popp J, Rezzi S. ICP-MS/MS-based ionomics: a validated methodology to investigate the biological variability of the human ionome. J Proteome Res. 2017;16:2080–90. doi: 10.1021/acs.jproteome.7b00055. [DOI] [PubMed] [Google Scholar]
- 10.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezzi S, Ramadan Z, Fay LB, Kochhar S. Nutritional metabonomics: applications and perspectives. J Proteome Res. 2007;6:513–25. doi: 10.1021/pr060522z. [DOI] [PubMed] [Google Scholar]
- 12.Mennen LI, Sapinho D, Ito H, Bertrais S, Galan P, Hercberg S, Scalbert A. Urinary flavonoids and phenolic acids as biomarkers of intake for polyphenol-rich foods. Br J Nutr. 2006;96:191–8. doi: 10.1079/bjn20061808. [DOI] [PubMed] [Google Scholar]
- 13.Tzoulaki I, Ebbels TMD, Valdes A, Elliott P, Ioannidis JPA. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol. 2014;180:129–39. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M. Bioinformatics tools for mass spectroscopy-based metabolomic data processing and analysis. Curr Bioin-form. 2012;7:96–108. doi: 10.2174/157489312799304431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaise BJ, Navratil V, Domange C, Shintu L, Dumas M-E, Elena-Herrmann B, et al. Two-dimensional statistical re-coupling for the identification of perturbed metabolic networks from NMR spectroscopy. J Proteome Res. 2010;9:4513–20. doi: 10.1021/pr1002615. [DOI] [PubMed] [Google Scholar]
- 16.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 17.De Livera AM, Sysi-Aho M, Jacob L, Gagnon-Bartsch JA, Castillo S, Simpson JA, Speed TP. Statistical methods for handling unwanted variation in metabolomics data. Anal Chem. 2015;87:3606–15. doi: 10.1021/ac502439y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrydziuszko O, Viant M. Missing values in mass spectrometry based metabolomics: an undervalued step in the data processing pipeline. Metabolomics. 2011;8:161–74. [Google Scholar]
- 19.Gromski PS, Xu Y, Kotze HL, Correa E, Ellis DI, Armitage EG, et al. Influence of missing values substitutes on multivariate analysis of metabolomics data. Metabolites. 2014;4:433–52. doi: 10.3390/metabo4020433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Guida R, Engel J, Allwood JW, Weber RJM, Jones MR, Sommer U, et al. Non-targeted UHPLC-MS metabolomic data processing methods: a comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics. 2016;12:93. doi: 10.1007/s11306-016-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah JS, Rai SN, DeFilippis AP, Hill BG, Bhatnagar A, Brock GN. Distribution based nearest neighbor imputation for truncated high dimensional data with applications to pre-clinical and clinical metabolomics studies. BMC Bioinformatics. 2017;18:114. doi: 10.1186/s12859-017-1547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SL, Ruhaak LR, Kelly K, Weiss RH, Kim K. Effects of imputation on correlation: implications for analysis of mass spectrometry data from multiple biological matrices. Brief Bioinform. 2017;18:312–20. doi: 10.1093/bib/bbw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (GPM) Anal Bioanal Chem. 2004;380:419–29. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- 24.Cloarec O, Dumas M-E, Craig A, Barton RH, Trygg J, Hudson J, et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem. 2005;77:1282–9. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 25.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu Rev Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavalarias D, Wallach JD, Li AHT, Ioannidis JPA. Evolution of reporting P values in the biomedical literature, 1990–2015. JAMA. 2016;315:1141–8. doi: 10.1001/jama.2016.1952. [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–3, 119–28, 244–52. [PubMed] [Google Scholar]
- 28.Karp PD, Caspi R. A survey of metabolic databases emphasizing the MetaCyc family. Arch Toxicol. 2011;85:1015–33. doi: 10.1007/s00204-011-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–7. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehlenborg N, O’Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 31.Johansson-Persson A, Barri T, Ulmius M, Onning G, Dragsted LO. LC-QTOF/MS metabolomic profiles in human plasma after a 5-week high dietary fiber intake. Anal Bioanal Chem. 2013;405:4799–809. doi: 10.1007/s00216-013-6874-5. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen LG, Winning H, Savorani F, Ritz C, Engelsen SB, Astrup A, et al. Assessment of dietary exposure related to dietary GI and fibre intake in a nutritional metabolomic study of human urine. Genes Nutr. 2012;7:281–93. doi: 10.1007/s12263-011-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton S, Navarro SL, Buas MF, Schwarz Y, Gu H, Djukovic D, et al. Targeted plasma metabolome response to variations in dietary glycemic load in a randomized, controlled, crossover feeding trial in healthy adults. Food Funct. 2015;6:2949–56. doi: 10.1039/c5fo00287g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menni C, Zhai G, MacGregor A, Prehn C, Römisch-Margl W, Suhre K, et al. Targeted metabolomics profiles are strongly correlated with nutritional patterns in women. Metabolomics. 2013;9:506–14. doi: 10.1007/s11306-012-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanstrup J, Schou SS, Holmer-Jensen J, Hermansen K, Dragsted LO. Whey protein delays gastric emptying and suppresses plasma fatty acids and their metabolites compared to casein, gluten, and fish protein. J Proteome Res. 2014;13:2396–408. doi: 10.1021/pr401214w. [DOI] [PubMed] [Google Scholar]
- 36.Llorach R, Urpi-Sarda M, Jauregui O, Monagas M, Andres-Lacueva C. An LC-MS-based metabolomics approach for exploring urinary metabolome modifications after cocoa consumption. J Proteome Res. 2009;8:5060–8. doi: 10.1021/pr900470a. [DOI] [PubMed] [Google Scholar]
- 37.Llorach R, Urpi-Sarda M, Tulipani S, Garcia-Aloy M, Monagas M, Andres-Lacueva C. Metabolomic finger-print in patients at high risk of cardiovascular disease by cocoa intervention. Mol Nutr Food Res. 2013;57:962–73. doi: 10.1002/mnfr.201200736. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Aloy M, Llorach R, Urpi-Sarda M, Jáuregui O, Corella D, Ruiz-Canela M, et al. A metabolomics-driven approach to predict cocoa product consumption by designing a multimetabolite biomarker model in free-living subjects from the PREDIMED study. Mol Nutr Food Res. 2015;59:212–20. doi: 10.1002/mnfr.201400434. [DOI] [PubMed] [Google Scholar]
- 39.van Dorsten FA, Grün CH, van Velzen EJJ, Jacobs DM, Draijer R, van Duynhoven JPM. The metabolic fate of red wine and grape juice polyphenols in humans assessed by metabolomics. Mol Nutr Food Res. 2009;54:897–908. doi: 10.1002/mnfr.200900212. [DOI] [PubMed] [Google Scholar]
- 40.Vázquez-Fresno R, Llorach R, Perera A, Mandal R, Feliz M, Tinahones FJ, et al. Clinical phenotype clustering in cardiovascular risk patients for the identification of responsive metabotypes after red wine polyphenol intake. J Nutr Biochem. 2016;28:114–20. doi: 10.1016/j.jnutbio.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Llorach R, Garrido I, Monagas M, Urpi-Sarda M, Tulipani S, Bartolome B, Andres-Lacueva C. Metabolomics study of human urinary metabolome modifications after intake of almond (Prunus dulcis (Mill.) D.A. Webb) skin polyphenols. J Proteome Res. 2010;9:5859–67. doi: 10.1021/pr100639v. [DOI] [PubMed] [Google Scholar]
- 42.Tulipani S, Llorach R, Jáuregui O, López-Uriarte P, Garcia-Aloy M, Bullo M, et al. Metabolomics unveils urinary changes in subjects with metabolic syndrome following 12-week nut consumption. J Proteome Res. 2011;10:5047–58. doi: 10.1021/pr200514h. [DOI] [PubMed] [Google Scholar]
- 43.Rangel-Huerta OD, Aguilera CM, Perez-de-la-Cruz A, Vallejo F, Tomas-Barberan F, Gil A, Mesa MD. A serum metabolomics-driven approach predicts orange juice consumption and its impact on oxidative stress and inflammation in subjects from the BIONAOS study. Mol Nutr Food Res. 2017;61:1600120. doi: 10.1002/mnfr.201600120. [DOI] [PubMed] [Google Scholar]
- 44.Edmands WM, Ferrari P, Rothwell JA, Rinaldi S, Slimani N, Barupal DK, et al. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am J Clin Nutr. 2015;102:905–13. doi: 10.3945/ajcn.114.101881. [DOI] [PubMed] [Google Scholar]
- 45.Pujos-Guillot E, Hubert J, Martin J-F, Lyan B, Quintana M, Claude S, et al. Mass spectrometry-based metabolomics for the discovery of biomarkers of fruit and vegetable intake: citrus fruit as a case study. J Proteome Res. 2013;12:1645–59. doi: 10.1021/pr300997c. [DOI] [PubMed] [Google Scholar]
- 46.Edmands WMB, Beckonert OP, Stella C, Campbell A, Lake BG, Lindon JC, et al. Identification of human urinary biomarkers of cruciferous vegetable consumption by metabonomic profiling. J Proteome Res. 2011;10:4513–21. doi: 10.1021/pr200326k. [DOI] [PubMed] [Google Scholar]
- 47.Bondia-Pons I, Cañellas N, Abete I, Rodríguez MÁ, Perez-Cornago A, Navas-Carretero S, et al. Nutri-metabolomics: subtle serum metabolic differences in healthy subjects by NMR-based metabolomics after a short-term nutritional intervention with two tomato sauces. Omi A J Integr Biol. 2013;17:611–8. doi: 10.1089/omi.2013.0027. [DOI] [PubMed] [Google Scholar]
- 48.Andersen M-BS, Kristensen M, Manach C, Pujos-Guillot E, Poulsen SK, Larsen TM, et al. Discovery and validation of urinary exposure markers for different plant foods by untargeted metabolomics. Anal Bioanal Chem. 2014;406:1829–44. doi: 10.1007/s00216-013-7498-5. [DOI] [PubMed] [Google Scholar]
- 49.Mora-Cubillos X, Tulipani S, Garcia-Aloy M, Bulló M, Tinahones FJ, Andres-Lacueva C. Plasma metabolomic biomarkers of mixed nuts exposure inversely correlate with severity of metabolic syndrome. Mol Nutr Food Res. 2015;59:2480–90. doi: 10.1002/mnfr.201500549. [DOI] [PubMed] [Google Scholar]
- 50.Altmaier E, Kastenmüller G, Römisch-Margl W, Thorand B, Weinberger KM, Adamski J, et al. Variation in the human lipidome associated with coffee consumption as revealed by quantitative targeted metabolomics. Mol Nutr Food Res. 2009;53:1357–65. doi: 10.1002/mnfr.200900116. [DOI] [PubMed] [Google Scholar]
- 51.Jacobs S, Kröger J, Floegel A, Boeing H, Drogan D, Pischon T, et al. Evaluation of various biomarkers as potential mediators of the association between coffee consumption and incident type 2 diabetes in the EPIC-Potsdam Study. Am J Clin Nutr. 2014;100:891–900. doi: 10.3945/ajcn.113.080317. [DOI] [PubMed] [Google Scholar]
- 52.Stalmach A, Mullen W, Barron D, Uchida K, Yokota T, Cavin C, et al. Metabolite profiling of hydroxycin-namate derivatives in plasma and urine after the ingestion of coffee by humans: identification of biomarkers of coffee consumption. Drug Metab Dispos. 2009;37:1749–58. doi: 10.1124/dmd.109.028019. [DOI] [PubMed] [Google Scholar]
- 53.Redeuil K, Smarrito-Menozzi C, Guy P, Rezzi S, Dionisi F, Williamson G, et al. Identification of novel circulating coffee metabolites in human plasma by liquid chromatography-mass spectrometry. J Chromatogr A. 2011;1218:4678–88. doi: 10.1016/j.chroma.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 54.Nagy K, Redeuil K, Williamson G, Rezzi S, Dionisi F, Longet K, et al. First identification of dimethoxycin-namic acids in human plasma after coffee intake by liquid chromatography-mass spectrometry. J Chromatogr A. 2011;1218:491–7. doi: 10.1016/j.chroma.2010.11.076. [DOI] [PubMed] [Google Scholar]
- 55.Cornelis M, Erlund I, Herder C, Westerhuis J, Tuomilehto J. Metabolomics of coffee consumption. FASEB J. 2017;31(Suppl):42.1. [Google Scholar]
- 56.Zheng Y, Yu B, Alexander D, Steffen LM, Boerwinkle E. Human metabolome associates with dietary intake habits among African Americans in the Atherosclerosis Risk in Communities study. Am J Epidemiol. 2014;179:1424–33. doi: 10.1093/aje/kwu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Dorsten FA, Daykin CA, Mulder TPJ, Van Duynhoven JPM. Metabonomics approach to determine metabolic differences between green tea and black tea consumption. J Agric Food Chem. 2006;54:6929–38. doi: 10.1021/jf061016x. [DOI] [PubMed] [Google Scholar]
- 58.Law WS, Huang PY, Ong ES, Ong CN, Li SFY, Pasikanti KK, Chan ECY. Metabonomics investigation of human urine after ingestion of green tea with gas chromatography/mass spectrometry, liquid chromatography/mass spectrometry and 1H NMR spectroscopy. Rapid Commun Mass Spectrom. 2008;22:2436–46. doi: 10.1002/rcm.3629. [DOI] [PubMed] [Google Scholar]
- 59.Schröder H, de la Torre R, Estruch R, Corella D, Martínez-González MA, Salas-Salvadó J, et al. Alcohol consumption is associated with high concentrations of urinary hydroxytyrosol. Am J Clin Nutr. 2009;90:1329–35. doi: 10.3945/ajcn.2009.27718. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Jimenez F, Alvarez de Cienfuegos G, Badimon L, Barja G, Battino M, Blanco A, et al. International conference on the healthy effect of virgin olive oil. Eur J Clin Invest. 2005;35:421–4. doi: 10.1111/j.1365-2362.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 61.Moazzami AA, Shrestha A, Morrison DA, Poutanen K, Mykkänen H. Metabolomics reveals differences in post-prandial responses to breads and fasting metabolic characteristics associated with postprandial insulin demand in postmenopausal women. J Nutr. 2014;144:807–14. doi: 10.3945/jn.113.188912. [DOI] [PubMed] [Google Scholar]
- 62.Bondia-Pons I, Barri T, Hanhineva K, Juntunen K, Dragsted LO, Mykkänen H, Poutanen K. UPLC-QTOF/MS metabolic profiling unveils urinary changes in humans after a whole grain rye versus refined wheat bread intervention. Mol Nutr Food Res. 2013;57:412–22. doi: 10.1002/mnfr.201200571. [DOI] [PubMed] [Google Scholar]
- 63.Nestel PJ, Straznicky N, Mellett NA, Wong G, De Souza DP, Tull DL, et al. Specific plasma lipid classes and phospholipid fatty acids indicative of dairy food consumption associate with insulin sensitivity. Am J Clin Nutr. 2014;99:46–53. doi: 10.3945/ajcn.113.071712. [DOI] [PubMed] [Google Scholar]
- 64.Zheng H, Lorenzen J, Astrup A, Larsen L, Yde C, Clausen M, Bertram H. Metabolic effects of a 24-week energy-restricted intervention combined with low or high dairy intake in overweight women: an NMR-based metabolomics investigation. Nutrients. 2016;8:108. doi: 10.3390/nu8030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouchard-Mercier A, Rudkowska I, Lemieux S, Couture P, Vohl M-C. The metabolic signature associated with the Western dietary pattern: a cross-sectional study. Nutr J. 2013;12:158. doi: 10.1186/1475-2891-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Floegel A, von Ruesten A, Drogan D, Schulze MB, Prehn C, Adamski J, et al. Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. Eur J Clin Nutr. 2013;67:1100–8. doi: 10.1038/ejcn.2013.147. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt JA, Rinaldi S, Ferrari P, Carayol M, Achaintre D, Scalbert A, et al. Metabolic profiles of male meat eaters, fish eaters, vegetarians, and vegans from the EPIC-Oxford cohort. Am J Clin Nutr. 2015;102:1518–26. doi: 10.3945/ajcn.115.111989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Playdon MC, Moore SC, Derkach A, Reedy J, Subar AF, Sampson JN, et al. Identifying biomarkers of dietary patterns by using metabolomics. Am J Clin Nutr. 2017;105:450–65. doi: 10.3945/ajcn.116.144501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vázquez-Fresno R, Llorach R, Urpi-Sarda M, Lupianez-Barbero A, Estruch R, Corella D, et al. Metabolomic pattern analysis after Mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J Proteome Res. 2015;14:531–40. doi: 10.1021/pr5007894. [DOI] [PubMed] [Google Scholar]
- 70.Bondia-Pons I, Martinez JA, de la Iglesia R, Lopez-Legarrea P, Poutanen K, Hanhineva K, Zulet M, de los Á. Effects of short- and long-term Mediterranean-based dietary treatment on plasma LC-QTOF/MS metabolic profiling of subjects with metabolic syndrome features: the Metabolic Syndrome Reduction in Navarra (RES-MENA) randomized controlled trial. Mol Nutr Food Res. 2015;59:711–28. doi: 10.1002/mnfr.201400309. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH, Vestergaard H, et al. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol. 2017;5:184–95. doi: 10.1016/S2213-8587(16)30419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiseman H, Casey K, Bowey EA, Duffy R, Davies M, Rowland IR, et al. Influence of 10 wk of soy consumption on plasma concentrations and excretion of isoflavonoids and on gut microflora metabolism in healthy adults. Am J Clin Nutr. 2004;80:692–9. doi: 10.1093/ajcn/80.3.692. [DOI] [PubMed] [Google Scholar]
- 73.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu J, Yang S, Cai S, Dong J, Li X, Chen Z. Identification of biochemical changes in lactovegetarian urine using 1H NMR spectroscopy and pattern recognition. Anal Bioanal Chem. 2010;396:1451–63. doi: 10.1007/s00216-009-3338-z. [DOI] [PubMed] [Google Scholar]
- 75.Lloyd AJ, Fave G, Beckmann M, Lin W, Tailliart K, Xie L, et al. Use of mass spectrometry fingerprinting to identify urinary metabolites after consumption of specific foods. Am J Clin Nutr. 2011;94:981–91. doi: 10.3945/ajcn.111.017921. [DOI] [PubMed] [Google Scholar]
- 76.Bhupathiraju SN, Hu FB. One (small) step towards precision nutrition by use of metabolomics. Lancet Diabetes Endocrinol. 2017;5:154–5. doi: 10.1016/S2213-8587(17)30007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franks PW, Poveda A. Lifestyle and precision diabetes medicine: will genomics help optimise the prediction, prevention and treatment of type 2 diabetes through lifestyle therapy? Diabetologia. 2017;60:784–92. doi: 10.1007/s00125-017-4207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]