Abstract
Macrophages have been identified in the periodontium. Data have phenotypically described these cells, demonstrated changes with progressing periodontal disease, and identified their ability to function in antigen-presentation critical for adaptive immune responses to individual oral bacterium. Recent evidence has emphasized an important role for the plasticity of macrophage phenotypes, not only in the resulting function of these cells in various tissues, but also clear differences in the stimulatory signals that result in M1 (classical activation, inflammatory) and M2 (alternative activation/deactivated, immunomodulatory) cells. This investigation hypothesized that the oral pathogens, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans induce M1-type cells, while oral commensal bacteria primarily elicit macrophage functions consistent with an M2 phenotype. However, we observed that the M1 output from P. gingivalis challenge, showed exaggerated levels of pro-inflammatory cytokines, with a much lower production of chemokines related to T-cell recruitment. This contrasted with A. actinomycetemcomitans infection that increased both the pro-inflammatory cytokines and T-cell chemokines. Thus, it appears that P. gingivalis, as an oral pathogen, may have a unique capacity to alter the programming of the M1 macrophage resulting in a hyperinflammatory environment and minimizing the ability for T-cell immunomodulatory influx into the lesions.
Keywords: macrophage, oral bacteria, pathogens, commensals, cytokines
Plasticity of macrophage populations related to microbial stimulation and ability of commensal bacteria to help regulate maturation towards inflammatory and tissue destructive cells via oral pathogens.
INTRODUCTION
Mucosal tissues are colonized by an extremely dense and diverse microbiota of commensal bacteria, are often the first site of interaction with pathogenic microorganisms (Socransky et al.1998; D'Aiuto et al.2004; Dye et al.2005; Tatakis and Kumar 2005; Pedron and Sansonetti 2008). These sites continuously sample foreign material via various cells types, including macrophages (MΦ), which are innate immune cells at epithelial surfaces that respond rapidly to infection, carrying crucial information about the infection to lymph nodes to trigger an immune response (Nestle et al.1994; Jotwani et al.2001; Makino et al.2001; Kopitar, Ihan Hren and Ihan 2006). Historically, the MΦ were identified to effectively engage microbes using a repertoire of pattern recognition receptors (PRRs) (Hemmi and Akira 2005; Benko et al.2008), which recognize distinct classes of microorganism-associated molecular patterns (MAMPs), including a range of bacterial, viral and fungal pathogens, through engagement of LPS, LTA and nucleic acid (e.g. CpG, DNA, dsRNA) ligands (Wollenberg et al.2002; Blach-Olszewska 2005; Kawai and Akira 2006; Kumar, Kawai and Akira 2009). This enables an avid uptake of these foreign materials by the macrophages (Lauvau and Glaichenhaus 2004).
Macrophages are effective as antigen presenting cells (APCs) and are particularly adept at stimulating T cells for controlling the quality of the T effector cells (Girardi 2006; Gray and Cyster 2012). These cells also play a critical role in innate immunity, responding to microbial challenge and producing elevated levels of various cytokines that contribute to host innate defenses. These cells recognize and respond to microbial structures using the PRRs, which decorate the surface of the mΦ enabling uptake of antigenic components (Allavena et al.2004; Blach-Olszewska 2005; den Haan and Kraal 2012; Zanoni and Granucci 2012; Locati, Mantovani and Sica 2013; Striz et al.2014) and triggering activation and phenotype plasticity after engagement of microbial or viral pathogens as ligands (Kawai and Akira 2006; Schaible, Schaffer and Taylor 2010; Ferrante and Leibovich 2012; Salomao et al.2012). This recognition of microbial components by the APCs triggers the production of selected cytokines, e.g. IL-1ß and TNFα that enhance cellular differentiation and maturation and are also linked to upregulation of a repertoire of cytokines and chemokines, enabling communication with both B and T cells as major effector cells in adaptive immunity (Guiney, Hasegawa and Cole 2003; Gervassi et al.2004; Karlsson et al.2004; Kranzer et al.2004; Hart et al.2005; Hu et al.2006).
Recent evidence has emphasized an important role for the plasticity of MΦ phenotypes, not only in the resulting function of these cells in various tissues, but also clear differences in the stimulatory signals that result in M1 (classical activation, inflammatory) and M2 (alternative activation/deactivated, immunomodulatory) cells (Gratchev et al.2001; Martinez et al.2008; Sica and Mantovani 2012; Mantovani et al.2013; Rodriguez et al.2014). These different MΦ phenotypes perform distinct and crucial functions in innate and adaptive immunity in local tissues (Benoit, Desnues and Mege 2008; Labonte, Tosello-Trampont and Hahn 2014). It is now clear that the innate and adaptive immune response outcomes of antigen recognition depends upon these functions of subpopulations of MΦ (Ivashkiv 2013; Locati, Mantovani and Sica 2013; Mantovani et al.2013; Labonte, Tosello-Trampont and Hahn 2014; Zhou et al.2014). These variations are regulated by the types of microorganisms providing the stimulus and the local host factor microenvironment (Banchereau et al.2000). The resulting signaling pathways activated through these receptors and processes lead to different immune cell response patterns. Through ‘classical activation’, the MΦ expresses an inflammatory function that leads to cytotoxicity, tissue injury and fibrosis (Locati, Mantovani and Sica 2013). This differentiation is related to host derived IFNγ as either an autocrine or paracrine factor and LPS (Labonte, Tosello-Trampont and Hahn 2014). The ‘alternative activation’ process is driven by host factors, IL-4 and IL-13, that can also be autocrine or paracrine derived and is an immunomodulatory cell type that controls the response, aids in tissue repair and cellular regeneration (Mantovani et al.2013). Finally, the ‘deactivated’ macrophage phenotype is triggered by IL-10 and is highly phagocytic contributing to tissue remodeling, and parasite encapsulation (Zhou et al.2014).
Periodontitis is a chronic immunoinflammatory lesion of mucosal surfaces triggered by a polymicrobial challenge derived from subgingival biofilms in local host tissues, that undermines soft tissue integrity and progresses to resorption of alveolar bone (Tatakis and Kumar 2005). The lesion is a result of a complex host response comprising inflammatory cells, cytokines, chemokines and mediators produced by resident gingival cells and inflammatory cells that infiltrate into the infected tissues (Kornman, Page and Tonetti 1997; Kantarci and Van Dyke 2005; Salvi and Lang 2005; Tatakis and Kumar 2005; Trombelli et al.2006). A range of APCs, including MΦ, have been identified in the periodontium, with data providing phenotypic descriptions of these cells, detecting changes in these cell populations with progressing periodontal disease, and demonstrating in vitro that these APCs can function in antigen-presentation critical in controlling the adaptive antibody response patterns in periodontal disease to individual bacteria (Cutler et al.1999; Cohen, Morisset and Emilie 2004; Tanaka et al.2006; Cutler and Teng 2007; Zelkha, Freilich and Amar 2010; Nanbara et al.2012; Papadopoulos et al.2013). However, the regulatory role of these cell types is of particular importance at mucosal surfaces as they are in constant association with external antigenic stimuli. While much emphasis has been expended examining host responses to members of the oral ecology purported to contribute to the pathogenic potential of the biofilm, little information is available examining the characteristics of host responses to commensal bacteria that represent ‘early colonizers’ and how this horde of bacteria compete, co-exist and/or synergize with opportunistic pathogens to initiate this chronic disease process (Paster et al.2001). Numerous biomarkers of innate immunity are observed in gingival tissues, e.g. LBP, CD14, TLRs, irrespective of the health of the tissues, although changes in TLR2/TLR4 appear in diseased gingiva (Ren et al.2005). Combined these results support the likely role of MΦ in diseased tissues, and suggest that the development and function of these APCs, may actually differ between the forms of periodontal disease (Cutler and Jotwani 2004; Gonzalez et al.2014, 2015). The mechanisms of action at mucosal surfaces include ignoring commensal MAMPs, compartmentalized TLR expression and commensal-driven attenuation of proinflammatory signaling (Kelly et al.2004; Ramos, Rumbo and Sirard 2004). In this regard, intestinal epithelial cells have been shown to be ‘tolerant’ of commensal MAMPs through additional mechanisms that regulate MAMP binding and PRR signaling (Sirard, Bayardo and Didierlaurent 2006). However, how commensal bacteria and pathogens are distinguished by these cells in the periodontium is not completely understood, and some new concepts are beginning to emerge. Gaps exist in our knowledge of how the host discriminates among these microorganisms, specifically as related to the interaction of MΦ with the polybacterial challenge that can occur in the subgingival sulci of the oral cavity. Although, while both types of macrophages would be expected to be functioning in the mucosal gingival tissues, there is little information on the distribution in health and disease, and more importantly, how pathogens like Porphyromonas gingivalis versus commensal bacteria might be critical in molecularly programming the tissue macrophages towards differing phenotypes.
This report focuses on our findings of the capacity of the major periodontal pathogen, P. gingivalis, to alter the functional/phenotypic characteristics of macrophages that would help to create a hyperinflammatory environment in the gingival tissues and, as such, affect the characteristics of the microbial ecology (e.g. dysbiosis), as well as undermine normal host response maturation that would enhance resolution of the chronic inflammatory tissue damage.
METHODS
Cell line and culturing conditions
The THP-1 cell line was used to generate M1 macrophages. For M1 polarization, THP-1 cells were cultured in medium (RPMI 1640 + 10% FBS; Gibco). The cells were expanded prior to an experiment for 24 hr, harvested by centrifugation and resuspended to the appropriate concentration to be evaluated in 12-well culture plates. One ml of media containing 1 × 106 cells was added to wells coated with PEI (polyethyleneimine) and allowed to incubate overnight to allow attachment and phenotype change reflecting a macrophage-like cell (Finlin et al.2013). For polarization to an M1 phenotype the cells are treated for 16 hr with 50 ng/ml of Escherichia coli LPS (Sigma) and 1000 U/ml of recombinant IFNγ (R&D Systems, Minneapolis, Minn.). The treated macrophages were harvested in 3 ml of PBS, lysed using 100 μl lysis buffer, and the lysates stored frozen at −80°C until analyzed.
Microorganisms and cell culture
The bacterial strains used in this study were Porphyromonas gingivalis ATCC 33277, Aggregatibacter actinomycetemcomitans JP2, Fusobacterium nucleatum ATCC 25586, Prevotella intermedia ATCC 25611, Streptococcus mutans ATCC 33535, S. gordonii ATCC 10558 and S. sanguis ATCC 10556. All bacteria were grown, harvested, washed sonicates prepared and protein levels determined by BCA assay (Pierce, Rockford, IL, USA) as we have described previously (Huang et al.2011b). Macrophages (105 cells/well) were stimulated in duplicate with sonicates or live bacteria (P. gingvalis, S. gordonii) at various MOI for 16 hrs.
Analysis of mRNA
Total RNA was isolated from THP-1, untreated and treated M1 macrophages using pure Link RNA Mini Kit (Ambion), and reverse transcription reaction was carried out using Transcriptor First Strand cDNA Synthesis Kit (Roche). Conditions were optimized using 2 μg of total RNA and 2.5 μM oligo(dT). Real-time PCR primers and probes were designed using Universal Probe Library for humans (Roche). A primer and probe for GAPDH was designed to use it as an internal control. The real-time PCR experiment was performed in 96-well plates using Light Cycler 480 (Roche) at conditions which were all uniformed for each probe and primers MonoColor Hydrolysis probe-UPL probe 96. QPCR was performed with pre-incubation for 30 s at 95°C, amplification: denaturation 95° 10 sec, annealing 60° for 30 sec and elongation 72°-1 s for 45 cycles. A LightCycler 480 (Roche) software was used to perform the analysis. Each reaction was performed in triplicate and the average number of cycles required to detect DNA (Cp) was plotted and used to calculate the quantitative value of real-time RT-PCR (Gonzalez et al.2013, 2016). Table 1 provides a summary of the primers and conditions for each of the mRNA expression assessments.
Table 1.
Primers for qPCR for gene expression analysis.
| Target gene | Primer | Amplicon Size (bp) |
|---|---|---|
| CCL5 | Forward – TGCCCACATCAAGGAGTATTT | 72 |
| Reverse – TTTCGGGTGACAAAGACGA | ||
| CXCL10 | Forward – GAAAGCAGTTAGCAAGGAAAGGT | 132 |
| Reverse – GACATATACTCCATGTAGGGAAGTGA | ||
| CD86 | Forward – CAGAAGCAGCCAAAATGGAT | 97 |
| Reverse – TCAGGTTGACTGAAGTTAGCAGA | ||
| IL1ß | Forward – TACCTGTCCTGCGTGTTGAA | 76 |
| Reverse – TCTTTGGGTAATTTTTGGGATCT | ||
| IL6 | Forward – GATGAGTACAAAAGTCCTGATCCA | 130 |
| Reverse – CTGCAGCCACTGGTTCTGT | ||
| IL8 | Forward – GAGCACTCCATAAGGCACAAA | 90 |
| Reverse – ATGGTTCCTTCCGGTGGT | ||
| IL12 | Forward – CACTCCCAAAACCTGCTGAG | 91 |
| Reverse – CAATCTCTTCAGAAGTGCAAG | ||
| IL23 | Forward – AGCTTCATGCCTCCCTACTG | 71 |
| Reverse – CTGCTGAGTCTCCCAGTGGT | ||
| TNFα | Forward – CAGCCTCTTCTCCTTCCTGAT | 123 |
| Reverse – GCCAGAGGGCTGATTAGAGA | ||
| GAPDH | Forward – GGTGTGAACCATGAGAAGTATGA | 112 |
| Reverse – GAGTCCTTCCACGATACCAAAG |
Analysis of chemokines/cytokines
Culture supernatants were evaluated for IL-8, TNFα, IL-6, IL-12 heterodimer (p70) or IL-10 by standard sandwich ELISA (eBioscience, San Diego, CA) according to the manufacturer's instructions as we have described previously (Huang et al.2011b). All samples were tested in triplicate.
Statistical analysis
Statistical analyses were performed using a Mann–Whitney U or Kruskal–Wallis analysis of variance on ranks with a post hoc Dunn's test for multiple testing (SigmaStat 3.5, Point Richmond, CA, USA). An alpha value of p < 0.05 was accepted as statistically significant when comparing the mediator levels under test conditions to media derived from untreated cells.
RESULTS
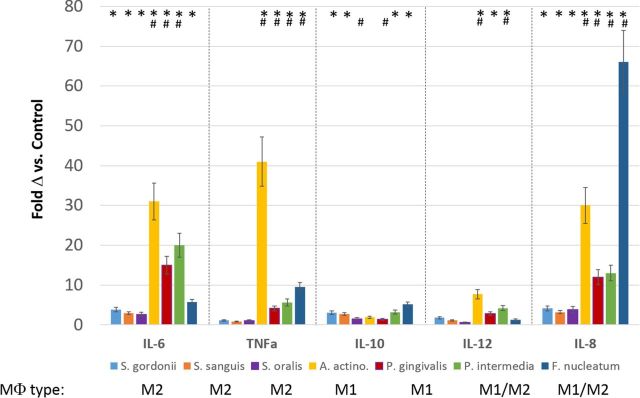
We examined the outcomes of stimulation of the THP-1 myelomonocytic human cell line (M0 type of cell), with various oral bacteria. Figure 1 summarizes the variations in responses related to the types of bacteria [e.g. Gram(+) vs. Gram(−); pathogen vs. commensal]. The data were derived from experiments that challenged THP-1 cells with 50 μg/ml of sonicates of each bacteria. Supernatants were harvested after 24 hr and assayed. While these analytes provide only a rough snapshot of the macrophage responses, the data are consistent with the oral streptococci stimulating primarily an M2 response, and both A. actinomycetemcomitans and P. gingivalis eliciting responses consistent with M1 macrophages. Interestingly, both P. intermedia and F. nucleatum sonicates stimulated a mixed response with markers for both M1 and M2-type cells.
Figure 1.
Cytokine/chemokine profiles from THP-1 macrophages induced by challenge with oral bacterial sonicates. Values denote mean fold difference (Δ) from control unstimulated THP-1 cells and vertical brackets enclose 1 SD from triplicate determinations. * denotes significantly different from control levels at least at p < 0.05. # denotes significantly different from levels with S. gordonii challenge at least at p < 0.05. Patterns of cytokines that were used to identify the MΦ type identified for each bacterial species in the figure, indicated that the Gram-positive commensal oral bacteria primarily elicit an M2 macrophage phenotype, while the oral pathogens (A. actinomycetemcomitans, P. gingivalis) induced an M1 type of macrophage. The oral bacteria (P. intermedia, F. nucleatum) associated with gingival inflammation and development of a pathogenic biofilm elicited a patterns of cytokines/chemokines reflective of both M1 and M2 phenotypes.
We then evaluated in greater detail macrophage responses following challenge with P. gingivalis and S. gordonii, as a prototype commensal microorganism (Fig. 2). The data focused on the phenotype of the M1 type cells following challenge with live bacteria of these two species. The results demonstrated high levels of IL-6, TNFα and IL-1ß following P. gingivalis stimulation, all markers of TLR engagement and NFκB activation of proinflammatory responses. Of interest was the decrease in IL-6 mRNA levels with the highest dose of P. gingivalis. Since this pathogen has the capacity to interact with various cell surface receptors, beyond TLRs, for example Protease Activated Receptors (PARs), and macrophages tend to display enhanced expression of PAR1, PAR2 and PAR3, this outcome could be a composite of the complex P. gingivalis bacterium engaging multiple receptors leading to complementary or competing signaling events in the cells (Chung et al.2004; Holzhausen, Spolidorio and Vergnolle 2005; Holzhausen et al.2006; Hajishengallis and Sahingur 2014). Streptococcus gordonii challenge did stimulate a response of these cytokines, but at much lower levels. In contrast, IL-12 and IL-23 products of the M1 phenotype, that are response markers of interferon-gamma receptor (IFNGR) engagement and activation through interferon regulatory factor (IRF)5 were not increased, and appeared even below the normal M1 cell levels following P. gingivalis challenge. CD86 a more pan-marker of macrophage activation was decreased somewhat by both types of bacteria, with rather consistent observations that higher concentrations of the commensal bacteria appeared to decrease this expression to a greater degree. It is clear that these types of co-stimulatory molecules can be elicited during innate immune responses to activate adaptive immune responses (Kobayashi and Flavell 2004). However, the literature also supports that various bacteria including pathogens and commensals can regulate the expression of CD86 (Shklovskaya et al.2011; Xin et al.2014; Christoffersen et al.2015; Gaikwad and Agrawal-Rajput 2015; Li et al.2015a) with numerous reports of commensal bacteria down-regulating these surface receptors to modify the characteristics of T-cell activation and create a more tolerigenic environment for the commensal bacteria to permanently colonize (Kobayashi and Flavell 2004; Shklovskaya et al.2011; Xin et al.2014; Li et al.2015a) similar to the results with these oral bacteria. Our data are consistent with these previous concepts about regulation by commensals, and as P. gingivalis also appeared to decrease the CD86 levels, it may suggest another mechanisms for virulence and dysbiotic regulation by this primary oral pathogen.
Figure 2.
Message levels of cytokines/chemokines/receptors following challenge of M1 macrophages (i.e. IFNγ+LPS) with different oral bacteria at different MOI (MΦ:bacteria). Values denote mean fold difference (Δ) from control unstimulated THP-1 cells and vertical brackets enclose 1 SD from triplicate determinations. * denotes significantly different from control levels at least at p < 0.05. # denotes significantly different from levels with IFNγ+LPS along at least at p < 0.05.
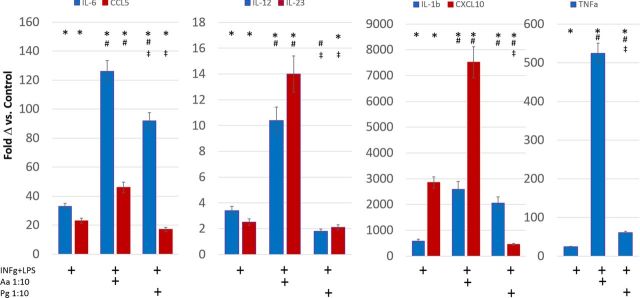
We extended these studies to more specifically address pathogenic features of the P. gingivalis interaction with the macrophages. As shown in Fig. 3, the oral pathogen A. actinomycetemcomitans elicited a pattern of substantially elevated gene expression representing both the pro-inflammatory (e.g. IL-1β, IL-23 etc.) and T-cell engagement activities [e.g. CCL5 (regulated on activation, normal T cell expressed and secreted; RANTES), CXCL10 (interferon-gamma induced protein 10; IP-10)] of M1 macrophages. This contrasted with the P. gingivalis challenge that appeared to elicit a ‘hyperinflammatory’ type of response with decreased levels of chemokines related to T-cell activation and immunoregulation.
Figure 3.
Message levels of cytokines/chemokines from M1 macrophages (i.e. INFγ+LPS) challenged with oral pathogens at MOI (MΦ:bacteria). Values denote mean fold difference (Δ) from control unstimulated THP-1 cells and vertical brackets enclose 1 SD from triplicate determinations. * denotes significantly different from control levels at least at p < 0.05. # denotes significantly different from levels with IFNγ+LPS alone at least at p < 0.05. ‡ denotes significantly different from A. actinomycetemcomitans levels at least at p < 0.05.
DISCUSSION
A summary of the literature demonstrates that MΦ are present in the periodontium, respond to the environment at diseased sites, and likely contribute crucial functions to maintaining or re-establishing homeostasis of these oral tissues. The differential induction of host cell responses by commensals and pathogens, and the ability of the host to differentiate and respond to trigger adaptive immunity remains unclear (Lu, Kurago and Brogden 2006). The characteristics of gingival MΦ interactions with the complex microbial biofilms in the oral cavity, triggering innate and adaptive immune response specificity to members of the biofilms must be presumed to play a role in the maintenance of homeostasis, formation of chronic destructive inflammatory lesions, and the adaptive immune response contribution to correcting the dysregulated inflammatory response of disease.
The results of this study demonstrate that commensal bacteria primarily elicit an M2 phenotype macrophage. This might be predicted based upon the existing literature describing the type of immune cell activities that occur within the periodontium (Lappin et al.1999; Hillmann et al.2001). However, of interest are reports suggesting a limited production of IL-4, albeit these reports focused on changes with periodontitis (Yamazaki et al.1997; Duarte et al.2012). Additionally, elevated levels of IL-13 in gingival tissues with periodontal lesions could contribute to the molecular environment to drive M2 formation (Roberts, McCaffery and Michalek 1997; Johnson and Serio 2007). Additionally, challenge of macrophages that have been driven towards an M1 phenotype with INFγ and LPS, with commensal bacteria appears to provide some modulation of the inflammatory nature of the M1 cells. There have been rather limited reports on the modulation of M1 macrophage maturation/functions related to specific bacterial infections. Recently, Christoffersen et al. (2014) demonstrated that Gram-negative gastrointestinal pathogens tended to elicit M1 macrophages compared with Gram-positive probiotic bacteria identified by targeted gene expression (e.g. iNOS, ARG), as well as cytokine secretion. Escherichia faecalis was found to polarize colon macrophages to the M1 phenotype and contribute to a ‘bystander effect’ mediating DNA damage to neighboring cells (Yang et al.2013). Using a THP-1 monocyte cell line-derived model, Habil et al. (2011) examined the immunomodulatory effects of probiotic bacteria and their secreted proteins on a macrophage subset-specific inflammatory marker profile. The cells were stimulated by enteropathic lipopolysaccharides in the presence or absence of the probiotic bacteria. The probiotics differentially regulated M1 and M2 production of TNFα, whereas M2 IL-6 production was suppressed.
An interesting observation was the identification of the apparent capacity of both F. nucleatum and P. intermedia to induce a profile of functions in the macrophages that exhibited characteristics of both M1 and M2 types. These bacteria are often identified as representative of the Socransky and colleagues ‘orange complex’ (Socransky et al.1998). As such, they are considered to represent the change in microbial ecology that occurs with biofilm accumulation leading to inflammation, clinically described as gingivitis. Additionally, they have been identified by Kolenbrander and co-workers (Kolenbrander, Palmer and Rickard 2006) to display an array of surface biomolecules that enable cognate interactions with putative pathogens, enabling co-aggregation, accretion and emergence of the pathogens at sites that progress to periodontitis. We and others have found that F. nucleatum, while thought of as a less pathogenic member of the subgingival ecology, has the in vitro capability to elicit very high levels of an array of cytokines and chemokines from multiple cell types (Gonzalez, Ebersole and Huang 2011; Huang et al.2011a,b; Peyyala et al.2012). Thus, it may actively contribute to changing the local nutritional milieu through this induced inflammation that allows the opportunistic pathogens to emerge and create a climax microbial community that is periodontopathic (Uzel et al.2011; Teles et al.2012). Also of interest is periodontal microbiological data that indicated a somewhat inverse relationship between levels of P. intermedia and P. gingivalis in subgingival plaque samples from health through periodontitis (Zambon 1996; Haffajee et al.2004; Sakamoto, Umeda and Benno 2005). This was interpreted as P. intermedia providing a resource for enhancing P. gingivalis binding and growth in the transition from gingivitis to periodontitis; however, P. gingivalis could outcompete P. intermedia for a similar ecological niche and thus emerge in the disease ecology. Consequently, the ability of both of these transition or ‘bridging’ microorganisms to activate the macrophage population more broadly, would in theory provide some ability of the host to mount an antimicrobial effort through M1 cells, contributing to local inflammation although inducing chemokines that would help engage the adaptive immune response cells. This combined with the induction of M2-like activities could then enable the local tissue environment to enhance the armamentarium of host immune responses to more effectively manage the noxious bacterial challenge. A caveat to these interpretations is that the commensal bacteria tested were Gram-positive, with the pathogens being Gram-negative reflecting the predominate distribution of these morphotypes of bacteria in periodontal health and disease. We have also developed some additional preliminary information suggesting that selected Gram-negative commensal bacteria also induce a more M2-like response profile (unpublished data). Thus, this polarization may reflect more general characteristics of the oral bacteria, not simply the cell wall/membrane structure of the microorganisms.
Based upon the existing literature we had predicted that P. gingivalis, as an oral pathogen contributing to the chronic inflammatory lesions of periodontitis, would primarily induce an M1 polarized macrophage population. This type of macrophage is primarily associated with inflammatory responses to bacterial infections, and is a primary cell type for combatting these infections (Zhou et al.2014). However, a collateral aspect of the induction of M1 cells, particularly through engagement of TLRs, is signaling through the NF-κB pathway and production of an array of proinflammatory mediators (Martinez et al.2008). While it is clear that inflammation is required as a presage to the development of adaptive immunity, chronic elevated levels of these biomolecules in the local tissues is associated with undermining epithelium integrity, enzymatic degradation of connective tissue matrix and loss of fibroblast function/viability, and activation of osteoclastogenesis leading to alveolar bone resorption (Souza and Lerner 2013; Hajishengallis 2014). Our data demonstrate that P. gingivalis clearly has the ability to trigger this pathway in macrophages and synergizes with host factors, i.e. IFNγ and extrinsic LPS to induce significant elevations in M1-produced inflammatory mediators. The finding that was rather unexpected was the apparent capacity of P. gingivalis to down-regulate/block M1 production of an array of cytokines and chemokines (e.g. IL-12, CCL5, CXCL10) that would help engage immunoregulatory cells and adaptive immunity (Gemmell, Carter and Seymour 2001). Recent results from Foey and Crean (Foey and Crean 2013) examined the impact on MΦ subsets of challenge with heat-killed P. gingivalis or LPS prior to stimulation by bacterial PRRs (e.g. lipoteichoic acid, peptidoglycan). Both P. gingivalis pretreatments suppressed PRR-induced TNFα, IL-6 and IL-10, but not IL-1β expression in both M1 and M2 MΦs. In addition, suppressed NFκB activation in M2 MΦs, but not in pro-inflammatory M1 MΦs, was noted. Thus, the authors suggested that this oral pathogen appeared to selectively tolerize MΦ subsets that could facilitate immunopathology and marginalize immunity. Recent results have supported the importance of macrophages in alveolar bone resorption elicited by P. gingivalis infection of mice, which also emphasized the profile of periodontal infiltrating macrophages to be dominantly an M1 type cell (Lam et al.2014). This same group also demonstrated that P. gingivalis LPS only weakly activated macrophage polarization, while still inducing pro-inflammatory mediators via TLR2 engagement (Holden et al.2014). These findings are generally consistent with our observations regarding the capacity of P. gingivalis to induce a specific response profile in macrophages with some predilection for polarizing towards an M1 phenotype.
In order to assess if our findings were a specific characteristic of P. gingivalis or a more general characteristic of oral pathogenic bacteria, we performed similar assessments examining A. actinomycetemcomitans, as the likely etiologic agent in many cases of aggressive periodontitis (Kononen and Muller 2014). The results showed that A. actinomycetemcomitans appeared to synergize with the M1 cell maturation and polarization process to induce a macrophage that has inflammation regulatory mechanisms intact, as well as the capacity to actively interface with the adaptive immune responses that would be predicted to help reestablish tissue homeostasis.
These results suggested that the P. gingivalis challenge was stimulating M1 cells towards a somewhat different functional phenotype that could have ramifications on the local environment in diseased gingival tissues, while commensal bacteria tend to polarize these cells towards an M2 phenotype. Multiple mechanisms could be contributing to inducing this plasticity in the periodontium, with potential targets including that P. gingivalis has the capacity to regulate the level and function of STAT1 (Signal Transducer and Activator of Transcription-1) (Matsukawa 2007) as a crucial molecule for generation of an array of host responses to external stimuli during inflammation that includes numerous autocrine/paracrine factors. This critical cellular outcome could occur via P. gingivalis effects on multiple molecular controls. ShP-1/2 can negatively regulate the Jak/STAT pathway in the nucleus, as well as by interacting with cytosolic STAT1 and preventing the recruitment of STAT1 to IFNγR, thus specifically inhibiting STAT1 signaling (Christophi et al.2009; Wu et al.2012). PIAS proteins sumoylate various transcription factors to modulate their function with PIAS1 as a transcriptional corepressor of STAT1 (Liu et al.2004, 2013). Suppressors of cytokine signaling (SOCS) negatively regulate cytokine signaling, and form part of a negative feedback loop for inflammation via STATs and NFκB (Baetz, Zimmermann and Dalpke 2007; Delgado-Ortega et al.2013; Carow and Rottenberg 2014). Several phosphatases have also been implicated in negative regulation of cytokine signaling via STAT1 (PTP1B; TCPTP). These enzymes can dephosphorylate JAK and TYK kinases that are crucial for STAT phosphorylation and cellular responses to IFN (Heinonen et al.2009; Ma et al.2011). Finally, an additional mechanism for these varied cellular responses to P. gingivais is through epigenetic actions of histone deacetylases (HDACs) on targeted gene transcription related to inflammatory phenotypes and various diseases, ie. septic shock, rheumatoid arthritis, asthma and colitis (Hawtree, Muthana and Wilson 2013; Royce and Karagiannis 2014; Cantley et al.2015; Li et al.2015a,b; Wendling et al.2015). Histone modifications have also been shown to govern multiple aspects of inflammation and immunity, including impacts on the functions and polarization of macrophages (Halili et al.2010; Sweet et al.2012; Turgeon et al.2013), and P. gingivalis has been shown to alter HDAC levels (Cantley et al.2011; Imai, Ochiai and Okamoto 2009; Imai and Ochiai 2011).
These findings support the importance of understanding the molecular events that are triggered by various oral bacteria resulting in a polarization of macrophages in the gingival tissues that would proscribe an environment exacerbating destructive inflammation or one oriented towards a ‘wound healing’ and resolution of the chronic inflammatory response. Detailed mechanisms of this polarization modulation within the context of a polymicrobial challenge need to be evaluated to better understand how the host reacts in the complex microbial milieu.
FUNDING
This work was supported by USPHS grants P20 RR020145 from the National Center for Research Resources, P20 GM103538 from the National Institute of General Medical Sciences, and UL1TR000117 from the National Center for Advancing Translational Sciences of the NIH, and the Center for Oral Health Research.
Conflict of interest. None declared.
REFERENCES
- Allavena P, Chieppa M, Monti P, et al. From pattern recognition receptor to regulator of homeostasis: the double-faced macrophage mannose receptor. Crit Rev Immunol. 2004;24:179–92. doi: 10.1615/critrevimmunol.v24.i3.20. [DOI] [PubMed] [Google Scholar]
- Baetz A, Zimmermann S, Dalpke AH. Microbial immune evasion employing suppressor of cytokine signaling (SOCS) proteins. Inflamm Allergy Drug Targets. 2007;6:160–7. doi: 10.2174/187152807781696446. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Benko S, Magyarics Z, Szabo A, et al. Dendritic cell subtypes as primary targets of vaccines: the emerging role and cross-talk of pattern recognition receptors. Biol Chem. 2008;389:469–85. doi: 10.1515/bc.2008.054. [DOI] [PubMed] [Google Scholar]
- Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–9. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- Blach-Olszewska Z. Innate immunity: cells, receptors, and signaling pathways. Arch Immunol Ther Ex. 2005;53:245–53. [PubMed] [Google Scholar]
- Cantley MD, Bartold PM, Marino V, et al. Histone deacetylase inhibitors and periodontal bone loss. J Periodontal Res. 2011;46:697–703. doi: 10.1111/j.1600-0765.2011.01392.x. [DOI] [PubMed] [Google Scholar]
- Cantley MD, Fairlie DP, Bartold PM, et al. Inhibiting histone deacetylase 1 suppresses both inflammation and bone loss in arthritis. Rheumatology. 2015;54:1713–23. doi: 10.1093/rheumatology/kev022. [DOI] [PubMed] [Google Scholar]
- Carow B, Rottenberg ME. SOCS3, a major regulator of infection and inflammation. Front Immunol. 2014;5:58. doi: 10.3389/fimmu.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen TE, Hult LT, Kuczkowska K, et al. In vitro comparison of the effects of probiotic, commensal and pathogenic strains on macrophage polarization. Probiotics Antimicrob Proteins. 2014;6:1–10. doi: 10.1007/s12602-013-9152-0. [DOI] [PubMed] [Google Scholar]
- Christoffersen TE, Olsen Hult LT, Solberg H, et al. Effects of the non-commensal Methylococcus capsulatus Bath on mammalian immune cells. Mol Immunol. 2015;66:107–16. doi: 10.1016/j.molimm.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Christophi GP, Panos M, Hudson CA, et al. Macrophages of multiple sclerosis patients display deficient SHP-1 expression and enhanced inflammatory phenotype. Lab Invest. 2009;89:742–59. doi: 10.1038/labinvest.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WO, Hansen SR, Rao D, et al. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:5165–70. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- Cohen N, Morisset J, Emilie D. Induction of tolerance by Porphyromonas gingivalis on APCS: a mechanism implicated in periodontal infection. J Dent Res. 2004;83:429–33. doi: 10.1177/154405910408300515. [DOI] [PubMed] [Google Scholar]
- Cutler CW, Jotwani R. Antigen-presentation and the role of dendritic cells in periodontitis. Periodontol 2000. 2004;35:135–57. doi: 10.1111/j.0906-6713.2004.003560.x. [DOI] [PubMed] [Google Scholar]
- Cutler CW, Jotwani R, Palucka KA, et al. Evidence and a novel hypothesis for the role of dendritic cells and Porphyromonas gingivalis in adult periodontitis. J Periodontal Res. 1999;34:406–12. doi: 10.1111/j.1600-0765.1999.tb02274.x. [DOI] [PubMed] [Google Scholar]
- Cutler CW, Teng YT. Oral mucosal dendritic cells and periodontitis: many sides of the same coin with new twists. Periodontol 2000. 2007;45:35–50. doi: 10.1111/j.1600-0757.2007.00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aiuto F, Parkar M, Andreou G, et al. Periodontitis and systemic inflammation: control of the local infection is associated with a reduction in serum inflammatory markers. J Dent Res. 2004;83:156–60. doi: 10.1177/154405910408300214. [DOI] [PubMed] [Google Scholar]
- Delgado-Ortega M, Marc D, Dupont J, et al. SOCS proteins in infectious diseases of mammals. Vet Immunol Immunop. 2013;151:1–19. doi: 10.1016/j.vetimm.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan JM, Kraal G. Innate immune functions of macrophage subpopulations in the spleen. Journal Innate Immun. 2012;4:437–45. doi: 10.1159/000335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte PM, Miranda TS, Lima JA, et al. Expression of immune-inflammatory markers in sites of chronic periodontitis in patients with type 2 diabetes. J Periodontol. 2012;83:426–34. doi: 10.1902/jop.2011.110324. [DOI] [PubMed] [Google Scholar]
- Dye BA, Choudhary K, Shea S, et al. Serum antibodies to periodontal pathogens and markers of systemic inflammation. J Clin Periodontol. 2005;32:1189–99. doi: 10.1111/j.1600-051X.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- Ferrante CJ, Leibovich SJ. Regulation of macrophage polarization and wound healing. Adv Wound Care. 2012;1:10–6. doi: 10.1089/wound.2011.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin BS, Zhu B, Starnes CP, et al. Regulation of thrombospondin-1 expression in alternatively activated macrophages and adipocytes: role of cellular cross talk and omega-3 fatty acids. J Nutr Biochem. 2013;24:1571–9. doi: 10.1016/j.jnutbio.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foey AD, Crean S. Macrophage subset sensitivity to endotoxin tolerisation by Porphyromonas gingivalis. PLoS One. 2013;8:e67955. doi: 10.1371/journal.pone.0067955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaikwad S, Agrawal-Rajput R. Lipopolysaccharide from Rhodobacter sphaeroides attenuates microglia-mediated inflammation and phagocytosis and directs regulatory T cell response. Int J Inflam. 2015;2015:361326. doi: 10.1155/2015/361326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell E, Carter CL, Seymour GJ. Chemokines in human periodontal disease tissues. Clin Exp Immunol. 2001;125:134–41. doi: 10.1046/j.1365-2249.2001.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervassi A, Alderson MR, Suchland R, et al. Differential regulation of inflammatory cytokine secretion by human dendritic cells upon Chlamydia trachomatis infection. Infect Immun. 2004;72:7231–9. doi: 10.1128/IAI.72.12.7231-7239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol. 2006;126:25–31. doi: 10.1038/sj.jid.5700003. [DOI] [PubMed] [Google Scholar]
- Gonzalez OA, Ebersole JL, Huang CB. The oral commensal, Streptococcus gordonii, synergizes with Tat protein to induce HIV-1 promoter activation in monocytes/macrophages. Cell Immunol. 2011;269:38–45. doi: 10.1016/j.cellimm.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, John Novak M, Kirakodu S, et al. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis. 2013;18:249–59. doi: 10.1007/s10495-013-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Kirakodu S, Novak MJ, et al. Effects of aging in the expression of NOD-like receptors adn inflammasome-related genes in the oral mucosa. Mol Oral Microbiol. 2016;31:18–32. doi: 10.1111/omi.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Novak MJ, Kirakodu S, et al. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol. 2014;41:327–39. doi: 10.1111/jcpe.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Novak MJ, Kirakodu S, et al. Differential gene expression profiles reflecting macrophage polarization in aging and periodontitis gingival tissues. Immunol Invest. 2015;44:643–64. doi: 10.3109/08820139.2015.1070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratchev A, Schledzewski K, Guillot P, et al. Alternatively activated antigen-presenting cells: molecular repertoire, immune regulation, and healing. Skin Pharmacol Appl Skin Physiol. 2001;14:272–9. doi: 10.1159/000056357. [DOI] [PubMed] [Google Scholar]
- Gray EE, Cyster JG. Lymph node macrophages. Journal Innate Immun. 2012;4:424–36. doi: 10.1159/000337007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney DG, Hasegawa P, Cole SP. Helicobacter pylori preferentially induces interleukin 12 (IL-12) rather than IL-6 or IL-10 in human dendritic cells. Infect Immun. 2003;71:4163–6. doi: 10.1128/IAI.71.7.4163-4166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habil N, Al-Murrani W, Beal J, et al. Probiotic bacterial strains differentially modulate macrophage cytokine production in a strain-dependent and cell subset-specific manner. Beneficial microbes. 2011;2:283–93. doi: 10.3920/BM2011.0027. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Bogren A, Hasturk H, et al. Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J Clin Periodontol. 2004;31:996–1002. doi: 10.1111/j.1600-051X.2004.00597.x. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014;29:248–57. doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Sahingur SE. Novel inflammatory pathways in periodontitis. Adv Dent Res. 2014;26:23–9. doi: 10.1177/0022034514526240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halili MA, Andrews MR, Labzin LI, et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–14. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- Hart AL, Al-Hassi HO, Rigby RJ, et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology. 2005;129:50–65. doi: 10.1053/j.gastro.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Hawtree S, Muthana M, Wilson AG. The role of histone deacetylases in rheumatoid arthritis fibroblast-like synoviocytes. Biochem Soc Trans. 2013;41:783–8. doi: 10.1042/BST20130053. [DOI] [PubMed] [Google Scholar]
- Heinonen KM, Bourdeau A, Doody KM, et al. Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-gamma signaling. P Natl Acad Sci USA. 2009;106:9368–72. doi: 10.1073/pnas.0812109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Akira S. TLR signalling and the function of dendritic cells. Chem Immunol Allergy. 2005;86:120–35. doi: 10.1159/000086657. [DOI] [PubMed] [Google Scholar]
- Hillmann G, Krause S, Ozdemir A, et al. Immunohistological and morphometric analysis of inflammatory cells in rapidly progressive periodontitis and adult periodontitis. Clin Oral Investig. 2001;5:227–35. doi: 10.1007/s00784-001-0134-3. [DOI] [PubMed] [Google Scholar]
- Holden JA, Attard TJ, Laughton KM, et al. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect Immun. 2014;82:4190–203. doi: 10.1128/IAI.02325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzhausen M, Spolidorio LC, Ellen RP, et al. Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am J Pathol. 2006;168:1189–99. doi: 10.2353/ajpath.2006.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzhausen M, Spolidorio LC, Vergnolle N. Role of protease-activated receptor-2 in inflammation, and its possible implications as a putative mediator of periodontitis. Mem Inst Oswaldo Cruz. 2005;100(Suppl 1):177–80. doi: 10.1590/s0074-02762005000900030. [DOI] [PubMed] [Google Scholar]
- Hu L, Bray MD, Osorio M, et al. Campylobacter jejuni induces maturation and cytokine production in human dendritic cells. Infect Immun. 2006;74:2697–705. doi: 10.1128/IAI.74.5.2697-2705.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CB, Altimova Y, Strange S, et al. Polybacterial challenge effects on cytokine/chemokine production by macrophages and dendritic cells. Inflamm Res. 2011a;60:119–25. doi: 10.1007/s00011-010-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CB, Alimova YV, Strange S, et al. Polybacterial challenge enhances HIV reactivation in latently infected macrophages and dendritic cells. Immunology. 2011b;132:401–9. doi: 10.1111/j.1365-2567.2010.03375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Ochiai K. Role of histone modification on transcriptional regulation and HIV-1 gene expression: possible mechanisms of periodontal diseases in AIDS progression. J Oral Sci. 2011;53:1–13. doi: 10.2334/josnusd.53.1. [DOI] [PubMed] [Google Scholar]
- Imai K, Ochiai K, Okamoto T. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J Immunol. 2009;182:3688–95. doi: 10.4049/jimmunol.0802906. [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34:216–23. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RB, Serio FG. The contribution of interleukin-13 and -15 to the cytokine network within normal and diseased gingiva. J Periodontol. 2007;78:691–5. doi: 10.1902/jop.2007.060204. [DOI] [PubMed] [Google Scholar]
- Jotwani R, Palucka AK, Al-Quotub M, et al. Mature dendritic cells infiltrate the T cell-rich region of oral mucosa in chronic periodontitis: in situ, in vivo, and in vitro studies. J Immunol. 2001;167:4693–700. doi: 10.4049/jimmunol.167.8.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci A, Van Dyke TE. Resolution of inflammation in periodontitis. J Periodontol. 2005;76:2168–74. doi: 10.1902/jop.2005.76.11-S.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson H, Larsson P, Wold AE, et al. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect Immun. 2004;72:2671–8. doi: 10.1128/IAI.72.5.2671-2678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–7. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Flavell RA. Shielding the double-edged sword: negative regulation of the innate immune system. J Leukoc Biol. 2004;75:428–33. doi: 10.1189/jlb.0703321. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Rickard AH, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- Kononen E, Muller HP. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65:46–78. doi: 10.1111/prd.12016. [DOI] [PubMed] [Google Scholar]
- Kopitar AN, Ihan Hren N, Ihan A. Commensal oral bacteria antigens prime human dendritic cells to induce Th1, Th2 or Treg differentiation. Oral Microbiol Immunol. 2006;21:1–5. doi: 10.1111/j.1399-302X.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- Kornman KS, Page RC, Tonetti MS. The host response to the microbial challenge in periodontitis: assembling the players. Periodontol 2000. 1997;14:33–53. doi: 10.1111/j.1600-0757.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- Kranzer K, Eckhardt A, Aigner M, et al. Induction of maturation and cytokine release of human dendritic cells by Helicobacter pylori. Infect Immun. 2004;72:4416–23. doi: 10.1128/IAI.72.8.4416-4423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Labonte AC, Tosello-Trampont AC, Hahn YS. The role of macrophage polarization in infectious and inflammatory diseases. Mol Cells. 2014;37:275–85. doi: 10.14348/molcells.2014.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam RS, O'Brien-Simpson NM, Lenzo JC, et al. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J Immunol. 2014;193:2349–62. doi: 10.4049/jimmunol.1400853. [DOI] [PubMed] [Google Scholar]
- Lappin DF, Koulouri O, Radvar M, et al. Relative proportions of mononuclear cell types in periodontal lesions analyzed by immunohistochemistry. J Clin Periodontol. 1999;26:183–9. doi: 10.1034/j.1600-051x.1999.260309.x. [DOI] [PubMed] [Google Scholar]
- Lauvau G, Glaichenhaus N. Mini-review: Presentation of pathogen-derived antigens in vivo. Eur J Immunol. 2004;34:913–20. doi: 10.1002/eji.200424944. [DOI] [PubMed] [Google Scholar]
- Li Q, Ma Y, Li L, et al. Flagellin influences the expression of a variety of important cytokines and chemokines without affecting the immune status of umbilical cord mesenchymal stem cells. Mol Med Reports. 2015a;12:6955–61. doi: 10.3892/mmr.2015.4276. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhao T, Liu B, et al. Inhibition of histone deacetylase 6 improves long-term survival in a lethal septic model. Journal Trauma Acute Care Surg. 2015b;78:378–85. doi: 10.1097/TA.0000000000000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Mink S, Wong KA, et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–8. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang YD, Guo L, et al. Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein beta (C/EBPbeta) during adipogenesis. Mol Cell Biol. 2013;33:4606–17. doi: 10.1128/MCB.00723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol. 2013;120:163–84. doi: 10.1016/B978-0-12-417028-5.00006-5. [DOI] [PubMed] [Google Scholar]
- Lu X, Kurago Z, Brogden KA. Effects of polymicrobial communities on host immunity and response. FEMS Microbiol Lett. 2006;265:141–50. doi: 10.1111/j.1574-6968.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Ma YM, Tao RY, Liu Q, et al. PTP1B inhibitor improves both insulin resistance and lipid abnormalities in vivo and in vitro. Mol Cell Biochem. 2011;357:65–72. doi: 10.1007/s11010-011-0876-4. [DOI] [PubMed] [Google Scholar]
- Makino M, Utsunomiya A, Maeda Y, et al. Association of CD40 ligand expression on HTLV-I-infected T cells and maturation of dendritic cells. Scand J Immunol. 2001;54:574–81. doi: 10.1046/j.1365-3083.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Biswas SK, Galdiero MR, et al. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–85. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Sica A, Mantovani A, et al. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- Matsukawa A. STAT proteins in innate immunity during sepsis: lessons from gene knockout mice. Acta Med Okayama. 2007;61:239–45. doi: 10.18926/AMO/32897. [DOI] [PubMed] [Google Scholar]
- Nanbara H, Wara-aswapati N, Nagasawa T, et al. Modulation of Wnt5a expression by periodontopathic bacteria. PLoS One. 2012;7:e34434. doi: 10.1371/journal.pone.0034434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Thompson C, Shimizu Y, et al. Costimulation of superantigen-activated T lymphocytes by autologous dendritic cells is dependent on B7. Cell Immunol. 1994;156:220–9. doi: 10.1006/cimm.1994.1166. [DOI] [PubMed] [Google Scholar]
- Papadopoulos G, Weinberg EO, Massari P, et al. Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J Immunol. 2013;190:1148–57. doi: 10.4049/jimmunol.1202511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedron T, Sansonetti P. Commensals, bacterial pathogens and intestinal inflammation: an intriguing menage a trois. Cell Host Microbe. 2008;3:344–7. doi: 10.1016/j.chom.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Peyyala R, Kirakodu SS, Novak KF, et al. Oral microbial biofilm stimulation of epithelial cell responses. Cytokine. 2012;58:65–72. doi: 10.1016/j.cyto.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HC, Rumbo M, Sirard JC. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12:509–17. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Ren L, Leung WK, Darveau RP, et al. The expression profile of lipopolysaccharide-binding protein, membrane-bound CD14, and toll-like receptors 2 and 4 in chronic periodontitis. J Periodontol. 2005;76:1950–9. doi: 10.1902/jop.2005.76.11.1950. [DOI] [PubMed] [Google Scholar]
- Roberts FA, McCaffery KA, Michalek SM. Profile of cytokine mRNA expression in chronic adult periodontitis. J Dent Res. 1997;76:1833–9. doi: 10.1177/00220345970760120501. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Domingo E, Municio C, et al. Polarization of the innate immune response by prostaglandin E2: a puzzle of receptors and signals. Mol Pharmacol. 2014;85:187–97. doi: 10.1124/mol.113.089573. [DOI] [PubMed] [Google Scholar]
- Royce SG, Karagiannis TC. Histone deacetylases and their inhibitors: new implications for asthma and chronic respiratory conditions. Curr Opin Allergy Clin Immunol. 2014;14:44–8. doi: 10.1097/ACI.0000000000000029. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Umeda M, Benno Y. Molecular analysis of human oral microbiota. J Periodontal Res. 2005;40:277–85. doi: 10.1111/j.1600-0765.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- Salomao R, Brunialti MK, Rapozo MM, et al. Bacterial sensing, cell signaling, and modulation of the immune response during sepsis. Shock. 2012;38:227–42. doi: 10.1097/SHK.0b013e318262c4b0. [DOI] [PubMed] [Google Scholar]
- Salvi GE, Lang NP. Host response modulation in the management of periodontal diseases. J Clin Periodontol. 2005;32(Suppl 6):108–29. doi: 10.1111/j.1600-051X.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- Schaible B, Schaffer K, Taylor CT. Hypoxia, innate immunity and infection in the lung. Resp Physiol Neurobi. 2010;174:235–43. doi: 10.1016/j.resp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Shklovskaya E, O'Sullivan BJ, Ng LG, et al. Langerhans cells are precommitted to immune tolerance induction. P Natl Acad Sci USA. 2011;108:18049–54. doi: 10.1073/pnas.1110076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard JC, Bayardo M, Didierlaurent A. Pathogen-specific TLR signaling in mucosa: mutual contribution of microbial TLR agonists and virulence factors. Eur J Immunol. 2006;36:260–3. doi: 10.1002/eji.200535777. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Souza PP, Lerner UH. The role of cytokines in inflammatory bone loss. Immunol Invest. 2013;42:555–622. doi: 10.3109/08820139.2013.822766. [DOI] [PubMed] [Google Scholar]
- Striz I, Brabcova E, Kolesar L, et al. Cytokine networking of innate immunity cells: a potential target of therapy. Clin Sci. 2014;126:593–612. doi: 10.1042/CS20130497. [DOI] [PubMed] [Google Scholar]
- Sweet MJ, Shakespear MR, Kamal NA, et al. HDAC inhibitors: modulating leukocyte differentiation, survival, proliferation and inflammation. Immunol Cell Biol. 2012;90:14–22. doi: 10.1038/icb.2011.88. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Fakher M, Barbour SE, et al. Influence of proinflammatory cytokines on Actinobacillus actinomycetemcomitans specific IgG responses. J Periodontal Res. 2006;41:1–9. doi: 10.1111/j.1600-0765.2005.00829.x. [DOI] [PubMed] [Google Scholar]
- Tatakis DN, Kumar PS. Etiology and pathogenesis of periodontal diseases. Dent Clin North Am. 2005;49:491–516, v.. doi: 10.1016/j.cden.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Teles FR, Teles RP, Uzel NG, et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2012;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombelli L, Scapoli C, Tatakis DN, et al. Modulation of clinical expression of plaque-induced gingivitis: response in aggressive periodontitis subjects. J Clin Periodontol. 2006;33:79–85. doi: 10.1111/j.1600-051X.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- Turgeon N, Blais M, Gagne JM, et al. HDAC1 and HDAC2 restrain the intestinal inflammatory response by regulating intestinal epithelial cell differentiation. PLoS One. 2013;8:e73785. doi: 10.1371/journal.pone.0073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzel NG, Teles FR, Teles RP, et al. Microbial shifts during dental biofilm re-development in the absence of oral hygiene in periodontal health and disease. J Clin Periodontol. 2011;38:612–20. doi: 10.1111/j.1600-051X.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling D, Abbas W, Godfrin-Valnet M, et al. Dysregulated serum IL-23 and SIRT1 activity in peripheral blood mononuclear cells of patients with rheumatoid arthritis. PLoS One. 2015;10:e0119981. doi: 10.1371/journal.pone.0119981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg A, Mommaas M, Oppel T, et al. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J Invest Dermatol. 2002;118:327–34. doi: 10.1046/j.0022-202x.2001.01665.x. [DOI] [PubMed] [Google Scholar]
- Wu X, Guo W, Wu L, et al. Selective sequestration of STAT1 in the cytoplasm via phosphorylated SHP-2 ameliorates murine experimental colitis. J Immunol. 2012;189:3497–507. doi: 10.4049/jimmunol.1201006. [DOI] [PubMed] [Google Scholar]
- Xin L, Jiang TT, Chaturvedi V, et al. Commensal microbes drive intestinal inflammation by IL-17-producing CD4+ T cells through ICOSL and OX40L costimulation in the absence of B7-1 and B7-2. P Natl Acad Sci USA. 2014;111:10672–7. doi: 10.1073/pnas.1402336111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Nakajima T, Kubota Y, et al. Cytokine messenger RNA expression in chronic inflammatory periodontal disease. Oral Microbiol Immunol. 1997;12:281–7. doi: 10.1111/j.1399-302x.1997.tb00392.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang X, Huycke T, et al. Colon macrophages polarized by commensal bacteria cause colitis and cancer through the bystander effect. Trans Oncol. 2013;6:596–606. doi: 10.1593/tlo.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon JJ. Periodontal diseases: microbial factors. Ann Periodontol. 1996;1:879–925. doi: 10.1902/annals.1996.1.1.879. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Granucci F. Regulation and dysregulation of innate immunity by NFAT signaling downstream of pattern recognition receptors (PRRs) Eur J Immunol. 2012;42:1924–31. doi: 10.1002/eji.201242580. [DOI] [PubMed] [Google Scholar]
- Zelkha SA, Freilich RW, Amar S. Periodontal innate immune mechanisms relevant to atherosclerosis and obesity. Periodontol 2000. 2010;54:207–21. doi: 10.1111/j.1600-0757.2010.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Huang C, Lin Z, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–7. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]