Abstract
Drug-induced cardiovascular complications are the most common adverse drug events and account for the withdrawal or severe restrictions on use of multitudinous post-marketed drugs. In this study, we developed new in silico models for systematic identification of drug-induced cardiovascular complications in drug discovery and post-marketing surveillance. Specifically, we collected drug-induced cardiovascular complications covering five most common types of cardiovascular outcomes (hypertension, heart block, arrhythmia, cardiac failure, and myocardial infarction) from four publicly available data resources: Comparative Toxicogenomics Database, SIDER, Offsides, and MetaADEDB. Using these databases, we developed a combined classifier framework through integration of five machine-learning algorithms: logistic regression, random forest, k-nearest neighbors, support vector machine, and neural network. The totality of models included 180 single classifiers with area under receiver operating characteristic curves (AUC) ranging from 0.647 to 0.809 on 5-fold cross validations. To develop the combined classifiers, we then utilized a neural network algorithm to integrate the best four single classifiers for each cardiovascular outcome. The combined classifiers had higher performance with an AUC range from 0.784 to 0.842 compared to single classifiers. Furthermore, we validated our predicted cardiovascular complications for 63 anticancer agents using experimental data from clinical studies, human pluripotent stem cell-derived cardiomyocyte assays, and literature. The success rate of our combined classifiers reached 87%. In conclusion, this study presents powerful in silico tools for systematic risk assessment of drug-induced cardiovascular complications. This tool is relevant not only in early stages of drug discovery, but throughout the life of a drug including clinical trials and post-marketing surveillance.
Graphical Abstract
INTRODUCTION
The systematic evaluation of drug cardiovascular (CV) safety profiles is essential for drug development and patient care. Cardiotoxicity is one of the most common severe and life-threatening adverse effects of drug treatments and thus is a major concern in drug discovery and post-marketing surveillance.1 Acute and chronic cardiotoxicity induced by drug treatments has a relatively high incidence rate and is characterized by severe negative symptoms including high blood pressure, heart failure, and death.2 According 1to a study of all safety-related withdrawals of prescription drugs from worldwide markets from 1960 to 1999, heart toxicity is one of the most common reasons for drug withdrawal.3 Numerous otherwise effective drugs, including terfenadine, astemizole, cisapride, vardenafil, and ziprasidone, have been withdrawn from the market owing to CV complications.3 Compounding the problem, cardiotoxicity has been reported for many anticancer drugs including chemotherapies, targeted therapies, and immunotherapies.4–7 These reports likely represent the tip of the iceberg, given the explosion of molecular targeted therapies with few systematic evaluations of cardiotoxicity risk. One of the 10 recommendations for 2016 Cancer Moonshot initiative is to “Accelerate the development of guidelines for monitoring and management of patient symptoms to minimize side effects of therapy.”8 This statement emphasizes the driving imperative to accelerate drug development by systematically identifying drug-induced CV complications.
In the past several decades, tests including radio ligand binding assays, electrophysiology measurements, rubidium-flux assays, and fluorescence-based assays have been used to assess the propensity of compound cardiotoxicity.9 Such experimental methods are not suitable for evaluation of a large number of compounds in early stage drug discovery due to high expense, and poor throughput. Moreover, animal models are limited by significant functional disparities between animal and human cardiomyocytes.10 Recent advances of in silico approaches and tools have promise for systematic evaluation of drug-induced CV complications in both drug discovery and post-marketing surveillance.11–16 For example, a recent study has integrated chemical, biological, and phenotypic properties of drugs to develop predictive and reasonably accurate machine-learning models for evaluation of adverse drug reaction.14 In 2010, Frid and co-workers developed in silico predictive models for prediction of cardiac adverse effects with good sensitivity.15 Building on this, Hitesh and co-workers built classifiers for assessment of drug cardiotoxicity with accuracies ranging from 0.675 to 0.95 by leave-one-out cross validation.16 Reported studies thus far are largely limited by use of only a single machine-learning algorithm with low or moderate accuracy. In order to advance the field of drug development, it is vital to develop robust and effective in silico models with high accuracy for evaluation of drug-induced cardiotoxicity.
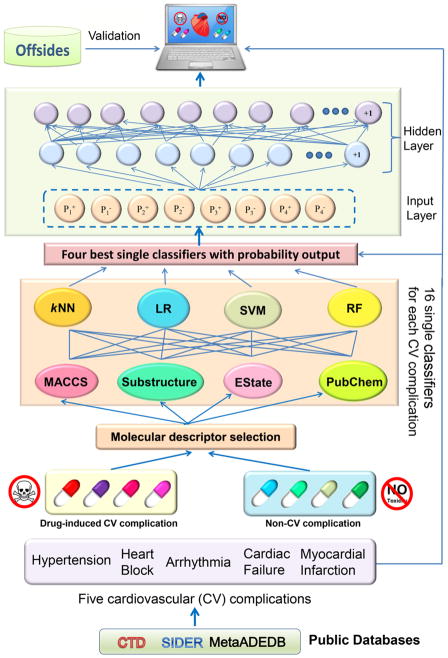
In this study, we proposed a combined classifier framework for prediction of five common CV complications associated with drug treatments (Figure 1). In total, we built 180 single classifiers through integration of molecular fingerprint (FP) and physical descriptors of drugs with four machine-learning algorithms: logistic regression, random forest, k-nearest neighbors, and support vector machine. We then utilized a neural network to combine the four best single classifiers for each CV complication. We showed that the combined classifiers outperformed the single classifiers. Using our combined classifier, we computationally identified multiple CV complications induced by various anticancer agents. We validated the predicted CV complications for various anticancer agents with experimental data. Altogether, the combined classifiers presented here offer a useful computational framework for systematic evaluation of drug-induced CV complications in drug discovery and post-marketing surveillance.
Figure 1. Diagram illustrating a combined classifier framework for prediction of drug-induced cardiovascular (CV) complications.
Five types of drug-induced CV complications are collected from three public databases (CTD, SIDER and MetaADEDB). The single classifiers are built on the basis of molecular fingerprints and the selected physical descriptors using four machine-learning algorithms (logistic regression, random forest, k-nearest neighbors, and support vector machine). The four best single highest performance classifiers were picked for building the combined classifiers using a neural network algorithm. The performance of all models was evaluated by both 5-fold cross-validation and the external validation sets collected from Offsides database 20. kNN: k-nearest neighbors; SVM: support vector machine; RF: random forest; LR: logistic regression.
MATERIALS AND METHODS
Data Preparation
We searched over 20 types of CV events defined by Medical Subject Headings (MeSH) and Unified Medical Language System (UMLS) vocabularies.17 All drug-induced CV complications were collected from four databases: Comparative Toxicogenomics Database (CTD),18 SIDER,19 Offsides,20 and MetaADEDB.21
The protocol of data collection is implemented in five steps: (1) All CV complication terms were further screened based on the publication of “Common Terminology Criteria for Adverse Events” (CTCAE, version 4.03, 2010) released by the U.S. Department of Health & Human Services; (2) Each item was annotated with the most commonly used MeSH and UMLS; (3) The four databases were searched using the MeSH unique ID or UMLS ID to obtain drug-induced CV complication information and then the InChI Keys of according drugs were calculated via Open Babel GUI;22 (4) Drugs annotated in DrugBank23 with unique ID were pinpointed by matching the InChI Keys; and (5) The items with well-annotated clinical report data were used and all the duplicated drugs in the same class of CV complications were excluded. To maintain a sufficient number of drugs with well-annotated CV complication information, we finally obtained five common drug-induced CV complications for building models: hypertension (MeSH ID: D006973), heart block (MeSH ID: D006327), arrhythmia (MeSH ID: D001145), cardiac failure (MeSH ID: D006333), and myocardial infarction (MeSH ID: D009203). The drug information of the five CV complications is provided in Supporting Information, Table S1. Detailed statistical results are listed in Table 1.
Table 1.
Statistical description of five drug-induced cardiovascular (CV) complications collected from four databases.
| CV complication | Number of unique drugs
|
||||
|---|---|---|---|---|---|
| CTD | SIDER | Offsides | MetaADEDB | Total | |
| Hypertension | 289 | 266 | 145 | 481 | 726 |
| Arrhythmia | 473 | 66 | 71 | 660 | 796 |
| Heart block | 159 | 106 | 201 | 237 | 473 |
| Cardiac failure | 135 | 164 | 270 | 267 | 585 |
| Myocardial infarction | 139 | 58 | 89 | 257 | 408 |
Chemical structure representation
In this study, two-dimensional (2D) descriptors of drugs were generated by MOE 2010 software24 to represent molecular descriptors (MD) and structural information. All drug structures were processed in MOE 2010 software by protonating strong bases, deprotonating strong acids, removing inorganic counter ions, adding hydrogen atoms, generating stereo isomers, and validating single 3D conformers by molecular washing and energy minimizing using. Descriptors calculated by MOE consist of 186 2D descriptors, including physical property descriptors, atom count and bond count descriptors, adjacency and distance matrix descriptors, subdivided surface area descriptors, Kier and Hall connectivity and Kappa shape indices descriptors, pharmacophore feature descriptors, and partial charge descriptors. Moreover, four sets of molecular fingerprints were also generated by PaDEL-Descriptor,25 including MACCS, EState, Pubchem, and Substructure fingerprint (SubFP). The more details of the descriptors can be found in recent studies.25,26
Molecular descriptor selection
Pearson correlation coefficient analysis is a common measure for eliminating irrelevant and redundant features (descriptors). In this study, all descriptors were selected based on two criteria: (1) highly relevant label of CV complications (CV complication [1] vs. non-CV complications [−1]); (2) lack of inter-correlation or self-correlation to avoid the over-fitting issue.27,28 Specifically, descriptors that have absolute Pearson correlation coefficient less than 0.1 (|PCC|<0.1) with the label “CV complications” were eliminated to reduce irrelevant descriptors. In addition, for any two descriptors having a pairwise correlation coefficient higher than 0.9 (|PCC|>0.9), the one with the lower correlation coefficient with the CV complication label was excluded. In total, 36, 36, 26, 32 and 37 molecular descriptors (Supporting Information, Table S2) were ultimately selected to build single classifiers for hypertension, heart block, arrhythmia, cardiac failure, and myocardial infarction, respectively.
Description of four single classifiers
Four machine-learning methods, including k-nearest neighbors (kNN), logistic regression (LR), random forest (RF), and support vector machine (SVM), were employed to build single classifiers. We then utilized a neural network (NN) algorithm to construct the combined classifiers based on the single classifiers with the best performance. All tools are freely available in Orange Canvas (version 2.7.6).29
k-nearest neighbors (kNN)
kNN is a non-parametric algorithm to classify objects based on the closest training samples in the feature space.30 It determines the category of a sample based on the categories of its k nearest neighboring samples. In this study, the k value was set to 5, and the distance d between samples x and y was measured by Euclidean distance that is calculated using equation (1) where n is the number of descriptors.
| (1) |
Logistic Regression (LR)
LR is a classification algorithm developed by statistician David Cox in 1958.31 LR calculates the probabilities using a logistic function to measure the relationship between a multitude of independent variables and categorical dependent variables. It maps the result of a linear regression function to a value ranging from 0 to 1 by the sigmoid function, and this value can be modeled as a probability. The realization of LR can be summarized as equation (2) and (3), where a and b are the coefficients determined by LR, n is the number of independent variable x.
| (2) |
| (3) |
Random Forest (RF)
RF is an ensemble algorithm for classification developed by Leo Breiman and Adele Cutler.32 It creates a large number of decision trees by bootstrapping training samples and randomly selecting subsets of original independent variables and predicts the category of new samples via integrating the outputs of the individual trees.33 RF is appealing because it is relatively fast at training models and can be used directly for high-dimensional problems.34 In this study, the number of decision trees in forest is set to 10, the minimal number of instances in a leaf is set to 5, and the considered number of attributes at each split is equal to the square root of the number of attributes in the input dataset.
Support Vector Machine (SVM)
SVM was first developed by Vapnik.35 The core idea of this algorithm is to map data into a higher dimensional space on the basis of the frame of Vapnik-Chervonenkis (VC) theory. SVM defines a decision boundary which is expressed as separating hyperplane based on a linear combination of functions parameterized by support vectors. It seeks the support vectors by maximizing the margin between the instances of different classes. Each molecule is expressed in terms of an eigenvector t, and the chosen patterns t1, t2, … tn are the components of t. The classification label y was introduced in SVM training. The ith molecule in the data set is defined as Mi = (ti, yi), where yi = 1 for the “CV complication” class and yi = −1 for the “non-CV complication” class.36 SVM gives a decision function (classifier) using equation (4)
| (4) |
where αi is the coefficient to be trained, and K is a kernel function. Parameter αi is trained via maximizing the Lagrangian expression using equations (5) and (6).
| (5) |
| (6) |
In this study the commonly used kernel, Gaussian radial basis function (RBF), was utilized. In order to obtain the optimal performance model, the auto searching program “grid” in the LibSVM 3.2 package37 was employed to determine the kernel parameter γ and penalty parameter C by utilizing a grid strategy based on 5-fold cross validation.
The SVM algorithm in this study is provided by an SVM learner in Orange Canvas 2.7, which can provide posterior predictive probability for each prediction. Probabilities are created by directly training an SVM and then training the parameters of an additional sigmoid function to map the SVM outputs into probabilities.38,39 Given training examples xi ∈ Rn, i = 1, … , l, labeled by yi ∈ {+1, −1}, the conversion algorithm for the probability P r(y = 1|x) is described as equation (7)
| (7) |
where fi is an estimate of decision function f(xi) , and parameters A and B are determined by minimizing the negative log likelihood of the training data, which is a cross-entropy error function (with N+ of the yi’s positive, and N− negative):
| (8) |
The detailed description about probability generation in SVM approach are provided in original works.38,39
Description of Combined Classifiers
Combined classifiers can improve prediction accuracy to some extent via comprehensively synergizing the complementary information provided by other methods. This approach is particularly suitable for the cases where single classifiers may not achieve satisfactory predictive accuracy. A combined classifier optimizes the performance of single classifiers by enhancing prediction reliability.36,40,41 In this study, four best single classifiers were combined using the NN algorithm (Figure 1).
Neural Network (NN)
NN is an information processing paradigm that is inspired by biological neural networks.42 A collection of connected units called artificial neurons is the basis of NN. Each connection between neurons can transmit a unidirectional signal with various activating strength hinges on the weight of neurons. The combined incoming signals that exceed a given threshold will promote the propagation of a signal to the downstream neurons and activate them. The output of neuron i is given in Equations (9) and (10).
| (9) |
| (10) |
where wij the connect weighting value from neuron j to neuron i, θ is the threshold, and f is the activation function.
In Orange Canvas 2.7, the NN is implemented by a multilayer perceptron with a single hidden layer. It is performed by minimizing an L2-regularized cost function with SciPy’s implementation of Limited-memory BFGS (L-BFGS), which is often used for parameter estimation in machine-learning.43,44 In this study, the number of hidden layer neurons is set to 11, the regularization factor is set to 3.0, the max iteration is set to 200, and data was normalized prior to learning. Normalization was done by subtracting each column by the mean and dividing by the standard deviation.
Experimental design
Drugs with at least one of five types of CV complications (hypertension, arrhythmia, heart block, cardiac failure and myocardial infarction) were curated from CTD, SIDER, MetaADEDB, and Offsides. The same number of corresponding decoys were then randomly selected from remaining drugs in DrugBank. The drugs collected from CTD, SIDER and MetaADEDB were combined with their decoys as the training sets, while the drugs from Offsides and the corresponding decoys were used as the validation sets after eliminating the ones existing in the training sets. The drugs with CV complications were labeled as “+1” and the decoys (or negative) were labeled as “−1”. Eventually, the training sets of hypertension, arrhythmia, heart block, cardiac failure and myocardial infarction contained 1,162, 1,450, 544, 630 and 638 drugs, and the validation sets included 290, 142, 402, 540 and 178 drugs, respectively (Table 2).
Table 2.
Number of drugs in the training sets and the external validation sets.
| CV complication | Training set
|
External validation set
|
||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | Positive | Negative | Total | |
| Hypertension | 581 | 581 | 1,162 | 145 | 145 | 290 |
| Arrhythmia | 725 | 725 | 1,450 | 71 | 71 | 142 |
| Heart block | 272 | 272 | 544 | 201 | 201 | 402 |
| Cardiac failure | 315 | 315 | 630 | 270 | 270 | 540 |
| Myocardial infarction | 319 | 319 | 638 | 89 | 89 | 178 |
For each CV complication, single classifiers were firstly built using four algorithms (RF, kNN, SVM and LR) and the selected molecular descriptors. Subsequently, four types of fingerprints were introduced to enhance the predictive accuracy. For each algorithm and specific CV complication, the best single classifiers were constructed using the selected molecular descriptors combined with molecular fingerprints. Then the training sets were predicted with the four best single classifiers to obtain positive and negative output probabilities ( and i = 1,2,3,4). After that, the eight output probabilities were chosen as new descriptors to develop the NN classifiers, generating probabilities ( and ) for each drug as final predictions.
Model validation
All classification models were assessed by true positives (TP, drugs with known CV complication were predicted as cardiotoxic drugs), true negatives (TN, drugs with non-CV complication were predicted as non-cardiotoxic drugs), false positives (FP, drugs with non-CV complication were predicted as cardiotoxic drugs), and false negatives (FN, drugs with known CV complication were predicted as non-cardiotoxic drugs). In addition, four metrics (sensitivity [SE], specificity [SP], overall predictive accuracy [Q], and precision [P]) were calculated using equations (11–13) for further evaluation of model performance.
| (11) |
| (12) |
| (13) |
| (14) |
Moreover, receiver operating characteristic (ROC) curves were used to evaluate the performance of the classifiers.45 An ROC curve exhibits the behavior of a model by measuring the relationship between true positive rate and false positive rate. The area under ROC curve (AUC) was computed for each of the classifiers. A perfect classifier yields an AUC of 1, whereas a random model has an expected AUC of 0.5.
RESULTS
GPCRs are the most common target family in training sets
To explore target distribution of drugs in the training sets, we extracted the drug-target interactions for drugs in the training sets from DrugBank23 and examined the drug target family distribution according to the IUPHAR/BPS Guide to PHARMACOLOGY in 2018.46 Here, targets are divided into five categories: kinases, G-protein-coupled receptors (GPCRs), nuclear receptors, ion channels, or others (Figure S1). We found that GPCRs were most highly represented among the four types of target families across five types of CV complications, following by ion channels, kinases, and nuclear receptors.
Molecular descriptors together with molecular fingerprint were the best single classifiers
We built 36 single classifiers for each CV complication: a. 4 classifiers built by 4 machine-learning algorithms (kNN, LR, RF and SVM) with the selected molecular descriptors, b. 16 classifiers built by combining 4 machine-learning algorithms with 4 different types of fingerprints (EState, MACCS, Pubchem and SubFP), and c. 16 classifiers built by 4 machine-learning algorithms with 4 types of integrated features by the selected molecular descriptors and fingerprints. In total, we built 180 single classifiers across 5 types of drug-induced CV complications. The performance of each classifier was assessed by both 5-fold cross validation and external validation. The performances of the 180 single classifiers are provided in Supporting Information, Table S3.
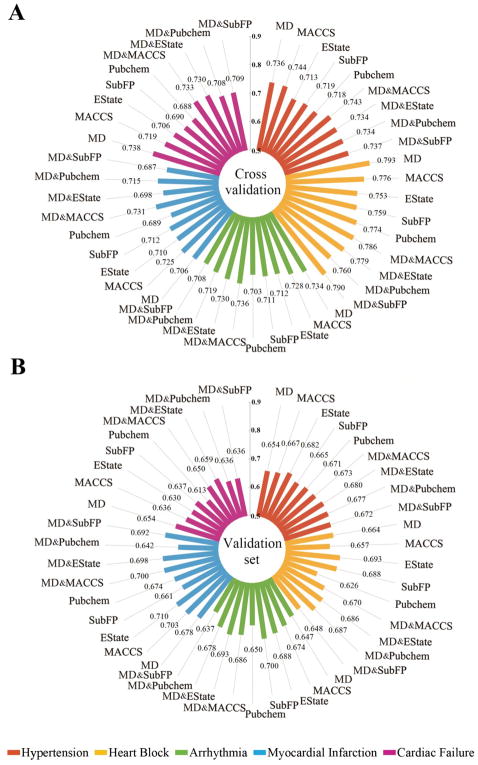
To make a more intuitive comparison between the single classifiers, the average AUC scores of the single classifiers developed using the selected molecular descriptors (MD) only, molecular fingerprint (FP) only, and combination of MD and FP (MD & FP) respectively are shown in Figure 2. The average AUC scores ranged from 0.687 to 0.793 on the 5-fold cross validations and ranged from 0.613 to 0.710 on the external validation sets. The MD&FP classifiers showed better performance on both 5-fold cross validations and the external validation sets compared to classifiers built by the MD or FP alone (Supporting Information, Figure S2). Hence, single classifiers built on MD & FP together were selected as the best single classifiers for further studies. Table 3 illustrates the performance of all single classifiers based on MD & FP across four machine-learning algorithms.
Figure 2.
Comparison of the average area under the receiver operating characteristic curves (AUC) scores across the single classifiers built using three types of descriptors: (i) molecular descriptors only, (ii) molecular fingerprint only, and (iii) molecular descriptors combined with molecular fingerprints, for 5-fold cross validations (A) and the external validation sets (B). Note: MD: molecular descriptors; SubFP: Substructure fingerprint.
Table 3.
Detailed performance of single classifiers built using the selected molecular descriptors (MD) combined with four types of molecular fingerprints across four machine learning algorithms.
| Cardiovascular Complications | Descriptors | 5-fold cross validation
|
External validation set
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| kNN | RF | LR | SVM | kNN | RF | LR | SVM | ||
| Hypertension | MD&MACCS | 0.759 | 0.727 | 0.723 | 0.763 | 0.717 | 0.682 | 0.647 | 0.644 |
| MD&Estate | 0.702 | 0.752 | 0.732 | 0.749 | 0.669 | 0.672 | 0.678 | 0.701 | |
| MD&Pubchem | 0.717 | 0.765 | 0.689 | 0.765 | 0.667 | 0.720 | 0.609 | 0.711 | |
| MD&SubFP | 0.736 | 0.754 | 0.714 | 0.743 | 0.706 | 0.698 | 0.610 | 0.675 | |
|
| |||||||||
| Arrhythmia | MD&MACCS | 0.746 | 0.763 | 0.682 | 0.756 | 0.727 | 0.685 | 0.650 | 0.681 |
| MD&Estate | 0.702 | 0.750 | 0.721 | 0.747 | 0.683 | 0.692 | 0.702 | 0.694 | |
| MD&Pubchem | 0.718 | 0.753 | 0.663 | 0.743 | 0.633 | 0.743 | 0.602 | 0.733 | |
| MD&SubFP | 0.725 | 0.739 | 0.685 | 0.681 | 0.680 | 0.658 | 0.657 | 0.553 | |
|
| |||||||||
| Heart block | MD&MACCS | 0.809 | 0.799 | 0.734 | 0.804 | 0.691 | 0.701 | 0.622 | 0.666 |
| MD&Estate | 0.787 | 0.802 | 0.752 | 0.774 | 0.682 | 0.700 | 0.695 | 0.667 | |
| MD&Pubchem | 0.769 | 0.801 | 0.737 | 0.797 | 0.667 | 0.690 | 0.686 | 0.704 | |
| MD&SubFP | 0.800 | 0.799 | 0.755 | 0.808 | 0.624 | 0.657 | 0.670 | 0.642 | |
|
| |||||||||
| Myocardial infarction | MD&MACCS | 0.765 | 0.746 | 0.647 | 0.766 | 0.718 | 0.716 | 0.667 | 0.700 |
| MD&Estate | 0.708 | 0.714 | 0.712 | 0.731 | 0.664 | 0.702 | 0.709 | 0.718 | |
| MD&Pubchem | 0.732 | 0.720 | 0.654 | 0.754 | 0.645 | 0.680 | 0.599 | 0.645 | |
| MD&SubFP | 0.706 | 0.723 | 0.672 | 0.717 | 0.733 | 0.698 | 0.664 | 0.672 | |
|
| |||||||||
| Cardiac failure | MD&MACCS | 0.757 | 0.741 | 0.667 | 0.767 | 0.641 | 0.667 | 0.632 | 0.659 |
| MD&Estate | 0.728 | 0.737 | 0.709 | 0.744 | 0.643 | 0.663 | 0.685 | 0.646 | |
| MD&Pubchem | 0.708 | 0.720 | 0.665 | 0.739 | 0.615 | 0.664 | 0.648 | 0.615 | |
| MD&SubFP | 0.741 | 0.727 | 0.665 | 0.720 | 0.609 | 0.660 | 0.652 | 0.624 | |
Note: The value in table represents the area under the receiver operating characteristic curve. MD: molecular descriptors; SubFP: Substructure fingerprint; kNN: k-nearest neighbors; SVM: support vector machine; RF: random forest; LR: logistic regression.
Combined classifiers outperform single classifiers
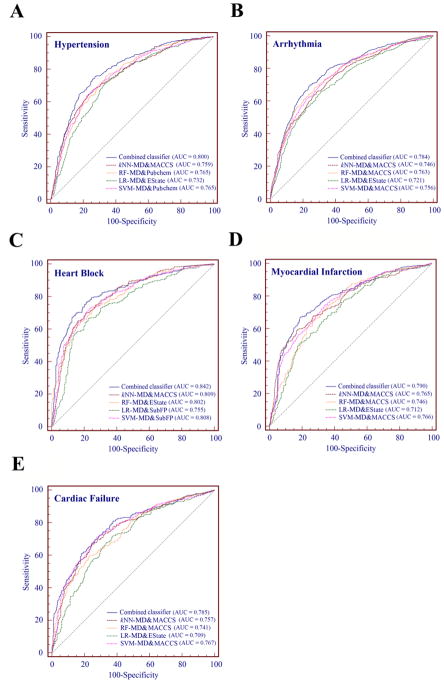
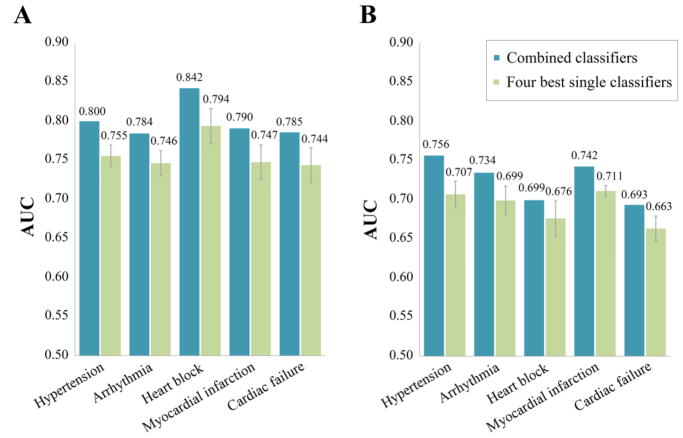
For each type of CV complications, we selected the best single classifiers built by MD & FP and generated by the 4 machine-learning algorithms in order to construct the combined classifiers. From the comparison of AUC values as shown in Figure 3, the combined classifiers outperformed single classifiers in cross validation. For example, the AUC of the combined classifier (AUC = 0.842) is higher than all four single classifiers (kNN AUC = 0.809, LR AUC = 0.755, RF AUC = 0.802, SVM AUC = 0.808) for prediction of drug-induced heart block. Building on this observation, Figure 4 further shows that the combined classifiers outperform the corresponding four best single classifiers for all 5 types of CV complications on both cross validations and external validation sets. Table 4 provides detailed performance of the five combined classifiers. Overall, most of the combined classifiers achieved a satisfactory performance in both cross validation and external validation sets. For example, the AUC values of the combined classifiers for prediction of drug-induced hypertension are 0.800 in the 5-fold cross validation and 0.756 in the external validation set. High cross validation AUC values are also achieved by the combined classifiers on other CV complications including heart block (AUC = 0.842), arrhythmia (AUC = 0.784), myocardial infarction (AUC = 0.790), and cardiac failure (AUC = 0.785). Taken together, the combined classifiers offer potential tools for computational risk assessment of drug-induced CV complications with high accuracy compared to the single classifiers. We hence examined the predicted drug-induced CV complications for anticancer agents via combined classifiers.
Figure 3.
Receiver operating characteristic (ROC) curves of combined classifiers and the four best MD & FP classifiers built by combining the selected molecular descriptors and molecular fingerprints across five types of drug-induced cardiovascular complications on 5-fold cross validation. Note: AUC: the area under the receiver operating characteristic curves; MD: molecular descriptors; SubFP: Substructure fingerprint; kNN: k-nearest neighbors; SVM: support vector machine; RF: random forest; LR: logistic regression.
Figure 4.
Comparison of area under the receiver operating characteristic curves (AUC) of the combined classifiers with the average AUC for four best single classifiers and on 5-fold cross validation (A) and external validation sets (B).
Table 4.
Performance of the five combined classifiers across five types of drug-induced cardiovascular complications.
| Combined classifier | 5-fold cross validation
|
External validation set
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SE | SP | Q | P | AUC | SE | SP | Q | P | AUC | ||
|
|
|
||||||||||
| Hypertension | 0.750 | 0.728 | 0.739 | 0.734 | 0.800 | 0.669 | 0.710 | 0.690 | 0.698 | 0.756 | |
| Arrhythmia | 0.712 | 0.723 | 0.717 | 0.720 | 0.784 | 0.648 | 0.761 | 0.704 | 0.730 | 0.734 | |
| Heart block | 0.721 | 0.813 | 0.767 | 0.794 | 0.842 | 0.507 | 0.746 | 0.627 | 0.667 | 0.699 | |
| Myocardial infarction | 0.690 | 0.765 | 0.727 | 0.746 | 0.790 | 0.596 | 0.708 | 0.652 | 0.671 | 0.742 | |
| Cardiac failure | 0.686 | 0.737 | 0.711 | 0.722 | 0.785 | 0.537 | 0.707 | 0.622 | 0.647 | 0.693 | |
Note: SE: sensitivity; SP: specificity; Q: overall predictive accuracy; P: precision; AUC: the area under receiver operating characteristic curves.
Clinical studies and human pluripotent stem cell-derived cardiomyocyte assays validate combined classifiers
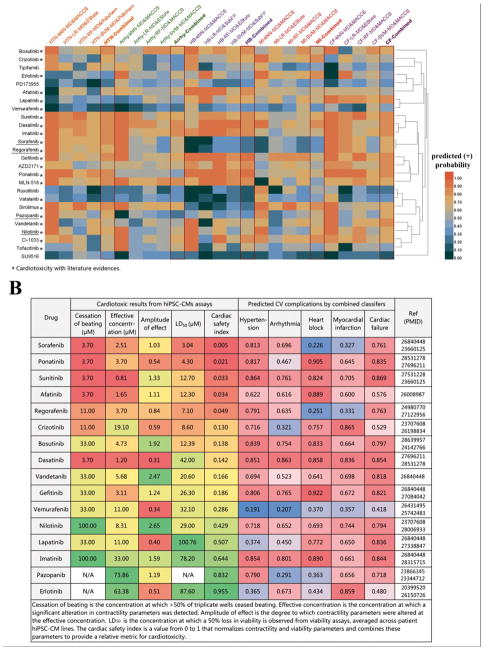
Despite advances in cancer treatments, the frequency of CV complications induced by anticancer agents (i.e., chemotherapy and targeted therapy) has been substantially increasing.47 We applied the four best single classifiers as well as the combined classifiers to predict 5 types of CV complications for 63 anticancer small molecular agents, including 26 targeted therapeutic agents (kinase inhibitors in Figure 5A) and 37 chemotherapeutic agents (non-kinase inhibitors in Supporting Information, Figure S3). According to the known drug-induced CV complications labeled by Drugs@FDA database48, we found a higher success rate of 87% (108/124) for the combined classifiers compared to the four best single classifiers (79%=392/496, Supporting Information, Table S4). For instance, pazopanib-induced cardiac failure and myocardial infarction are severe adverse reactions listed in the Drugs@FDA database and have been reported by several double-blind placebo-controlled trials in patients49,50 and a variety of clinical reports.51–54 Our combined classifiers successfully identified pazopanib-induced cardiac failure and myocardial infarction, while only one of the best single classifiers offered true prediction in this case. An additional example includes Arsenic trioxide (As2O3), which is approved for treating acute promyelocytic leukemia. Here itwas predicted to have potential cardiac failure by the combined classifiers, while only half of the best single classifiers generated the toxicity consistent prediction. Arsenic trioxide was reported to lead to dysfunction of myocardium and reduction of contractility,55 suggesting 100% accuracy of the combined classifiers compared to 50% accuracy of the best single classifiers.
Figure 5.
Validation of the combined classifiers using reported experimental data from human pluripotent stem cell-derived cardiomyocyte assays and literature data. (A) Predicted cardiovascular complications for molecularly targeted cancer therapeutic agents (kinase inhibitors) by four best single classifiers and the combined classifiers respectively. Drugs existing in the training sets are underlined. (B) Comparison of cardiotoxic profiles from human pluripotent stem cell-derived cardiomyocyte assays (cardiac safety indexes described in Supporting Method S1) and the predicted probabilities from the combined classifiers for 16 kinase inhibitors. A lower cardiac safety index represents higher risk of cardiotoxicity. A predicted probability of more than 0.5 denotes an identified probable cardiovascular complication. A higher probability (e.g., 1.0) shows increased likelihood of cardiotoxicity. Red color indicates higher relative likelihood of drug-induced CV complications, while green or blue color indicates a lower relative likelihood of drug-induced CV complications. Note: HTN: hypertension; HB: heart block; Arrhy: arrhythmia; MI: myocardial infarction; CF: cardiac failure; MD: molecular descriptors; SubFP: Substructure fingerprint; kNN: k-nearest neighbors; SVM: support vector machine; RF: random forest; LR: logistic regression.
Human pluripotent stem cell-derived cardiomyocytes (PSC-CMs) are an effective way to assess drug cardiotoxicity in vitro.56 We computationally evaluated CV complications using the combined classifiers for 16 tyrosine kinase inhibitors (TKIs) with known cardiotoxic profiles identified by PSC-CMs assays56 and literature evidence. We calculated cardiac safety indexes (CSI) to provide a relative metric for cardiotoxicity by normalizing 4 contractility and viability parameters (cessation of beating, effective concentration, amplitude of effect, and median lethal dose) as described previously6 (See the details in Supporting Method S1). Then we compared CSI values with the corresponding true probabilities of having CV complications predicted by the combined classifiers. The combined classifiers successfully predicted the reported CV complications for all reported TKIs except vemurafenib (Figure 5B). Altogether, the combined classifiers show high accuracy for identification of drug-induced CV complications.
Combined classifiers predict oncology molecularly targeted therapeutic agent-induced cardiovascular complications
We next used our model to predict several novel CV complications in cancer molecularly targeted therapeutic agents. Figure 5A shows the predicted CV complications for 26 FDA approved kinase inhibitors covering multiple biological pathways, including epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), cyclin-dependent kinases (CDKs), BRAF V600E kinases, mechanistic target of rapamycin (mTOR), mitogen-activated protein kinase (MAPK), janus kinase (JAK), Abelson murine leukemia viral oncogene homolog (Abl), and breakpoint cluster region-Abelson leukemia virus (Bcr-Abl). Erlotinib was predicted to have a high probability of inducing arrhythmia and myocardial infarction by the combined classifiers. In support of our model, recent studies reported that erlotinib induced QTc interval in patients57 and CV damage in rat model.58 In addition, an increasing number of clinical case reports of acute myocardial infarction following treatment by erlotinib in cancer patients also were reported.59–61 Gefitinib, a multi-targeted tyrosine kinase inhibitor, was approved for treating lung cancer. Our combined classifiers predicted that gefitinib induced all five CV complications (hypertension, heart block, arrhythmia, cardiac failure and myocardial infarction), consistent with recent clinical and preclinical studies.62,63 Sunitinib, a FDA-approved tyrosine kinase inhibitor for treatment of renal cell carcinoma and imatinib-resistant gastrointestinal stromal tumor in 2006, was predicted to induce all five CV complications by the combined classifiers. Among them, myocardial infarction and heart block have not yet been listed on its FDA label. Interestingly, a multicenter and randomized phase 3 trial reported that 1.33% (5/375) of sunitinib-treated patients had myocardial infarction, the most common reported CV complication (NCT00098657).64 A multi-parameter in vitro toxicity screening approach based on a human cardiac cell model also reported that sunitinib significantly altered the cardiac beat pattern and selectively blocked the human Ether-a-go-go Related Gene (hERG) channel,65 consistent with our predicted heart block by the combined classifiers. Tandutinib (MLN-518), a novel and selective inhibitor of PDGFR, has no reported cardiotoxic profiles in Drugs@FDA database.48 In stark contrast, tandutinib was predicted to have a high likelihood of cardiotoxicity via the combined classifiers. An in vitro experiment reported that tandutinib potentially caused a progressive increase in rats’ ventricular myocyte damage,66 confirming our prediction.
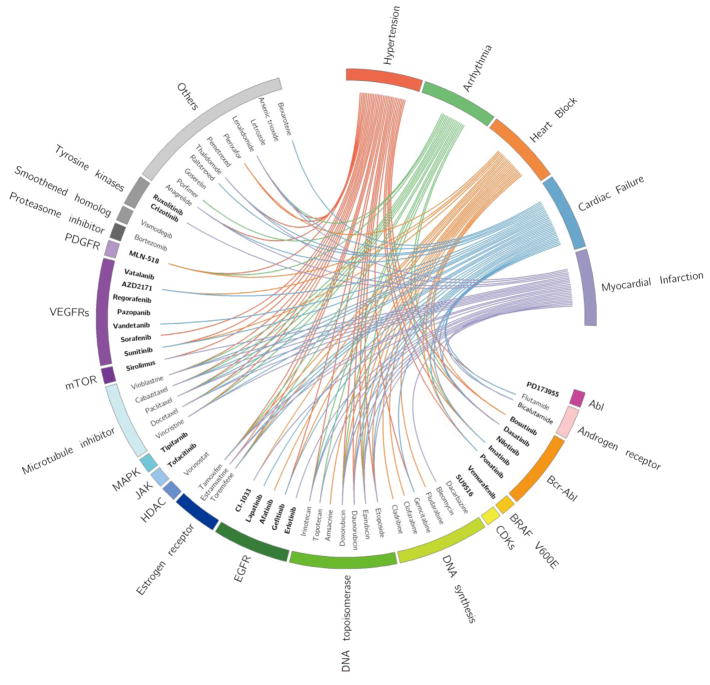
Integration of five machine-learning algorithms uncover cardiovascular complications induced across a diversity of anticancer drugs
It is plausible to hypothesize that drugs in the same class may have similar pharmacological characteristics, of which adverse reactions can be inferred.67,68 Figure 6 presents the relationships among anticancer agents, their target families, and the predicted CV complications. Interestingly, anticancer agents covering Bcr-Abl, DNA topoisomerase, and microtubule inhibitors are clustered together (Figure 6 and Figure S4). Bcr-Abl inhibitors are the first-line treatment for chronic myelogenous leukemia. Recently, cardiovascular safety has been an emerging challenge in patients treated with second-generation Bcr-Abl inhibitors.70 For example, dasatinib reportedly may induce potentially fatal pulmonary hypertension,70 and ponatinib and nilotinib may induce CV disease.71,72 Topoisomerases are ubiquitous enzymes involving in regulating the over- or underwinding of DNA strands.73 Anthracyclines, typical topoisomerase II inhibitors, have demonstrated cardiotoxicity.73 Microtubule inhibitors are anti-mitotic agents and known anticancer agents by inhibiting tubulin polymerization.74 The CV complications induced by several known drugs in this category have been successfully identified by our combined classifiers. For example, paclitaxel and docetaxel, as members of taxanes, lead to dysfunctional microtubules and release massive histamine, resulting in arrhythmias, myocardial ischemia, and conduction disturbances.75 In addition, cabazitaxel, as the fourth taxane, has the potential to induce cardiac-related deaths, including ventricular fibrillation, sudden cardiac death, and cardiac arrest.76,77 Put together, the combined classifiers successfully identified multiple CV complications across multiple pathways targeted by various anticancer agents. Hence, our combined classifiers offer powerful tools for identifying potential cardiotoxicity across drug families
Figure 6.
Circos plot representing the predicted associations between 63 anticancer drugs and the five types of cardiovascular complications. The predicted associations with positively predicted probabilities higher than 0.8 from the combined classifiers are connected lines. Drugs are grouped based on their target families using annotation from DrugBank database.23 Kinase inhibitors are highlighted in bold font. Circos plot was drawn using Circos (v0.69).69
DISCUSSION
In this study, we developed combined classifiers for the systematic identification of 5 types of drug-induced CV complications. We demonstrated that the combined classifiers outperformed the single classifiers in both cross validation and the external validation sets. Moreover, the newly predicted CV complications by the combined classifiers were validated by clinical study experimental data, cardiomyocyte assays and literature. Building on previous studies,15,16 this study adds to the field by: (1) collecting comprehensive CV complications for over 1,000 FDA-approved drugs by integrating clinically reported data from 4 databases, which are reliable and sufficient; (2) improving on the efficiency and accuracy of single classifier algorithms by leveraging a combined classifier infrastructure; (3) demonstrating that our combined classifiers outperformed several traditional ensemble approaches: maximum, minimum, and majority vote (Supporting Information, Method S2 and Table S5); (4) Utilizing our combined classifiers on 63 anticancer agents with high validated accuracy (especially for the top-10 predicted agents, Supporting Information, Table S6).
Several shortcomings should be recognized in the presented current study. First, high quality negative samples is quietly crucial for the accuracy of the machine-learning models. In this work, the decoy sets were randomly extracted from the rest drugs of DrugBank database23 without the known cardiotoxicity, which may bring in potential noise and data bias. Second, data quality of the validation sets may affect the model performance evaluation.78 In this study, the external validation sets were derived from Offsides, which contains computationally inferred adverse drug events from FDA's Adverse Event Reporting System (FAERS).20 This may explain the lower AUC range (0.693 to 0.756) of the combined classifiers on the external validation sets compared to high AUC range in the 5-fold cross validations (0.784 to 0.842). Third, current models were built based on integration of molecular descriptors and fingerprints of drugs. Recent studies have shown that integration of biological descriptors from drug-target networks may further improve model performance.79 In addition, an ideal drug target would be expressed only in disease tissue and sparsely anywhere else.80 In the future, we plan to integrate more relevant biological descriptors and tissue-specific expression profiles of drug targets to further improve the model performance. Furthermore, replacement of the currently used NN algorithm by deep learning algorithms81,82 could further improve accuracy. Finally, the predicted CV complications by the combined classifiers should be further validated by experimental assays or pharmacoepidemiologic analyses from the real-world data (e.g., electronic medical records or health insurance claims databases)83 in the future.
CONCLUSIONS
In this study, four different classification algorithms were applied to develop 180 single classifiers for evaluation of 5 types of drug-induced cardiovascular complications. The best four single classifiers of each cardiovascular complications were used to together construct the combined classifiers with a neural network algorithm. The combined classifiers outperformed the single classifiers not only in 5-fold cross validation but also external validation. Lastly, the combined classifiers were employed to pinpoint anticancer agents with cardiovascular complications. We report novel drug-induced cardiovascular complications which have been discovered and further validated by reported experimental data from clinical studies, in vitro assays, and literature. In summary, the combined classifiers presented here offer powerful in silico tools for systematic evaluation of drug-induced cardiovascular complications throughout the life cycle of a drug.
Supplementary Material
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K99HL138272 to F.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and US Food and Drug Administration. The authors thank Dr. Rebecca Kusko of Immuneering Corporation for editing the English of this manuscript.
Footnotes
Disclaimer: The views expressed in this manuscript do not necessarily represent those of the U.S. Food and Drug Administration.
Competing interests
All authors do not have any conflicts of interest.
The Supporting Information is available free of charge on the ACS Publications website. Detailed description for cardiac safety index is provided in Supporting Method S1, and detailed description of three ensemble approaches (majority vote, maximum and minimum) is provided in Supporting Method S2. Drug information for the five cardiovascular complications used for model building and validation (Table S1), lists of selected molecular descriptors used in this study (Table S2), detailed comparison of performance for single classifiers (Table S3), detailed prediction results of the five cardiovascular complications by the four best single classifiers and combined classifiers, known cardiotoxic profiles derived from Drugs@FDA database, and relevant literature evidence of 63 anticancer agents (Table S4), detailed performance of different ensemble approaches on 5-fold cross validation (Table S5), and predicted list of drug-induced CV complications for the top-10 anticancer agents with highest probability by the combined classifiers (Table S6). Distribution of drugs according to the classification of drug-target pairs in the five training sets covering five types of drug-induced cardiovascular complications (Figure S1), comparison among the area under the receiver operating characteristic curves (AUC) scores of all single classifiers on 5-fold cross validation and the external validation respectively (Figure S2), predicted cardiovascular complications for cancer chemotherapeutic agents (non-kinase inhibitors) by four best single classifiers and combined classifiers respectively (Figure S3), and circos plot representing the predicted associations between 63 anticancer drugs and the five types of cardiovascular complications of which predicted associations with positively predicted probabilities higher than 0.5 from the combined classifiers are exhibited (Figure S4).
References
- 1.Matthews EJ, Frid AA. Prediction of Drug-related Cardiac Adverse Effects in Humans--A: Creation of a Database of Effects and Identification of Factors Affecting their Occurrence. Regul Toxicol Pharmacol. 2010;56:247–275. doi: 10.1016/j.yrtph.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Sandhu H, Maddock H. Molecular Basis of Cancer-therapy-induced Cardiotoxicity: Introducing microRNA Biomarkers for Early Assessment of Subclinical Myocardial Injury. Clin Sci. 2014;126:377–400. doi: 10.1042/CS20120620. [DOI] [PubMed] [Google Scholar]
- 3.Man F, Thornton A, Mybeck K, Wu HH, Hornbuckle K, Muniz E. Evaluation of the Characteristics of Safety Withdrawal of Prescription Drugs from Worldwide Pharmaceutical Markets 1960 to 1999. Ther Innov Regul Sci. 2001;35:293–317. [Google Scholar]
- 4.Moslehi JJ. Cardiovascular Toxic Effects of Targeted Cancer Therapies. N Engl J Med. 2016;375:1457–1467. doi: 10.1056/NEJMra1100265. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DB, Balko JM, Compton ML, Chalkias S, Gorham J, Xu Y, Hicks M, Puzanov I, Alexander MR, Bloomer TL. Fulminant Myocarditis with Combination Immune Checkpoint Blockade. N Engl J Med. 2016;375:1749–1755. doi: 10.1056/NEJMoa1609214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma A, Burridge PW, Mckeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmström A. High-throughput Screening of Tyrosine Kinase Inhibitor Cardiotoxicity with Human Induced Pluripotent Stem Cells. Sci Transl Med. 2017;9:eaaf2584. doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng F, Loscalzo J. Autoimmune Cardiotoxicity of Cancer Immunotherapy. Trends Immunol. 2017;38:77–78. doi: 10.1016/j.it.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Ledford Heidi. Cancer Experts Unveil Wishlist for US Government ‘Moonshot’. Nature. 2016;537:288–289. doi: 10.1038/nature.2016.20535. [DOI] [PubMed] [Google Scholar]
- 9.Polak S, Wişniowska B, Brandys J. Collation, Assessment and Analysis of Literature in Vitro Data on hERG Receptor Blocking Potency for Subsequent Modeling of Drugs' Cardiotoxic Properties. J Appl Toxicol. 2009;29:183–206. doi: 10.1002/jat.1395. [DOI] [PubMed] [Google Scholar]
- 10.Milaninejad N, Janssen PM. Small and Large Animal Models in Cardiac Contraction Research: Advantages and Disadvantages. Pharmacol Ther. 2014;141:235–249. doi: 10.1016/j.pharmthera.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorberbaum T, Sampson KJ, Woosley RL, Kass RS, Tatonetti NP. An Integrative Data Science Pipeline to Identify Novel Drug Interactions that Prolong the QT Interval. Drug Saf. 2016;39:433–441. doi: 10.1007/s40264-016-0393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorberbaum T, Sampson KJ, Chang JB, Iyer V, Woosley RL, Kass RS, Tatonetti NP. Coupling Data Mining and Laboratory Experiments to Discover Drug Interactions Causing QT Prolongation. J J Am Coll Cardiol. 2016;68:1756–1764. doi: 10.1016/j.jacc.2016.07.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins TA, Bergenholm L, Abdulla T, Yates J, Evans N, Chappell MJ, Mettetal JT. Modeling and Simulation Approaches for Cardiovascular Function and Their Role in Safety Assessment. CPT: Pharmacometrics Syst Pharmacol. 2015;4:e00018. doi: 10.1002/psp4.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M, Wu Y, Chen Y, Sun J, Zhao Z, Chen XW, Matheny ME, Xu H. Large-scale Prediction of Adverse Drug Reactions Using Chemical, Biological, and Phenotypic Properties of Drugs. J Am Med Inform Assoc. 2012;19:28–35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frid AA, Matthews EJ. Prediction of Drug-related Cardiac Adverse Effects in Humans--B: Use of QSAR Programs for Early Detection of Drug-induced Cardiac Toxicities. Regul Toxicol Pharmacol. 2010;56:276–289. doi: 10.1016/j.yrtph.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Mistry HB, Davies MR, Di VG. A New Classifier-based Strategy for in-silico Ion-channel Cardiac Drug Safety Assessment. Front Pharmacol. 2015;6:59. doi: 10.3389/fphar.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodenreider O. The Unified Medical Language System (UMLS): Integrating Biomedical Terminology. Nucleic Acids Res. 2004;32:267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A Side Effect Resource to Capture Phenotypic Effects of Drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-Driven Prediction of Drug Effects and Interactions. Sci Transl Med. 2012;4:125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F, Li W, Wang X, Zhou Y, Wu Z, Shen J, Tang Y. Adverse Drug Events: Database Construction and in Silico Prediction. J Chem Inf Model. 2013;53:744–752. doi: 10.1021/ci4000079. [DOI] [PubMed] [Google Scholar]
- 22.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An Open Chemical Toolbox. J Cheminform. 2011;3:1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V. DrugBank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res. 2014;42:1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilar S, Cozza G, Moro S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr Top Med Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 25.Yap CW. PaDEL-descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J Comput Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 26.Klekota J, Roth FP. Chemical Substructures that Enrich for Biological Activity. Bioinformatics. 2008;24:2518–2525. doi: 10.1093/bioinformatics/btn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R. QSAR modeling: where have you been? Where are you going to? J Med Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng F, Li W, Liu G, Tang Y. In silico ADMET prediction: recent advances current challenges and future trends. Curr Top Med Chem. 2013;13:1273–1289. doi: 10.2174/15680266113139990033. [DOI] [PubMed] [Google Scholar]
- 29.Demšar J, Curk T, Erjavec A, Goru Č, Hočevar T, Milutinovič M, Možina M, Polajnar M, Toplak M, Starič A. Orange: Data Mining Toolbox in Python. J Mach Learn Res. 2013;14:2349–2353. [Google Scholar]
- 30.Larose DT. Discovering Knowledge in Data: An Introduction to Data Mining. Wiley-Interscience; 2004. [Google Scholar]
- 31.Cox DR. The Regression Analysis of Binary Sequences. J R Stat Soc. 1958;20:215–242. [Google Scholar]
- 32.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 33.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer. 2009;167:192. [Google Scholar]
- 34.Cutler A, Cutler DR, Stevens JR. Random Forests. Mach Learn. 2004;45:157–176. [Google Scholar]
- 35.Byvatov E, Schneider G. Support Vector Machine Applications in Bioinformatics. Appl Bioinformatics. 2003;2:67–77. [PubMed] [Google Scholar]
- 36.Cheng F, Yu Y, Shen J, Yang L, Li W, Liu G, Lee PW, Tang Y. Classification of Cytochrome P450 Inhibitors and Noninhibitors Using Combined Classifiers. J Chem Inf Model. 2011;51:996–1011. doi: 10.1021/ci200028n. [DOI] [PubMed] [Google Scholar]
- 37.Chang CC, Lin CJ. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. 2007;2:389–396. [Google Scholar]
- 38.PLATT JC. Probabilistic Outputs for Support Vector Machines and Comparisons to Regularized Likelihood Methods. Advances in Large Margin Classifiers. 2000;10:61–74. [Google Scholar]
- 39.Lin HT, Lin CJ, Weng RC. A note on Platt’s probabilistic outputs for support vector machines. Mach Learn. 2007;68:267–276. [Google Scholar]
- 40.Fang J, Pang XC, Yan R, Lian W, Li C, Wang Q, Liu AL, Du G. Discovery of Neuroprotective Compounds by Machine Learning Approaches. RSC Adv. 2016;6:9857–9871. [Google Scholar]
- 41.Fang J, Yang R, Gao L, Yang S, Pang X, Li C, He Y, Liu AL, Du GH. Consensus Models for CDK5 Inhibitors in Silico and their Application to Inhibitor Discovery. Mol Divers. 2015;19:149–162. doi: 10.1007/s11030-014-9561-3. [DOI] [PubMed] [Google Scholar]
- 42.Gurney K. An Introduction to Neural Networks. J Cognitive Neurosci. 1997;8:383. doi: 10.1162/jocn.1996.8.4.383a. [DOI] [PubMed] [Google Scholar]
- 43.Malouf R. A Comparison of Algorithms for Maximum Entropy Parameter Estimation. Proc Conf Natural Language Learning. 2002;20:1–7. [Google Scholar]
- 44.Ghahramani Z. Proceedings of the 24th international conference on Machine learning. International Conference on Machine Learning; 2007. [Google Scholar]
- 45.Fawcett T. An Introduction to ROC Analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 46.Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S, Gray A, Bruce L, Alexander S, Anderton S. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and Expansion to Encompass the New Guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2017;46:D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bansal N, Amdani S, Lipshultz ER, Lipshultz SE. Chemotherapy-induced Cardiotoxicity in Children. Expert Opin Drug Metab Toxicol. 2017;13:817–832. doi: 10.1080/17425255.2017.1351547. [DOI] [PubMed] [Google Scholar]
- 48.US Food and Drug Administration; 2017. [accessed 10 Feb 2017]. Drugs@FDA database. http://www.fda.gov/drugsatfda. [Google Scholar]
- 49.Listed N. Pazopanib and Soft-tissue Sarcomas. Too toxic Prescrire Int. 2013;22:145. [PubMed] [Google Scholar]
- 50.Listed N. Pazopanib. Kidney Cancer: Many Risks, but is there a Benefit for Patients? Prescrire Int. 2011;20:64–66. [PubMed] [Google Scholar]
- 51.Vlmn S, Ime D, van Erp NP, Verwiel J, Sej K, Wta VDG. Fatal Heart Failure in a Young Adult Female Sarcoma Patient Treated with Pazopanib. Acta Oncol. 2017;56:1233–1234. doi: 10.1080/0284186X.2017.1296582. [DOI] [PubMed] [Google Scholar]
- 52.Pandey M, Gandhi S, George S. Heart Failure: A Paraneoplastic Manifestation of Renal Cell Carcinoma - Reversed with Pazopanib. Clin Genitourin Cancer. 2017;15:835–837. doi: 10.1016/j.clgc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Marcke CV, Ledoux B, Petit B, Seront E. Rapid and Fatal Acute Heart Failure Induced by Pazopanib. BMJ Case Rep. 2015;2015:bcr2015211522. doi: 10.1136/bcr-2015-211522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdallah AO, Vallurupalli S, Kunthur A. Pazopanib- and Bevacizumab-induced Reversible Heart Failure in a Patient with Metastatic Renal Cell Carcinoma: A case report. J Oncol Pharm Pract. 2015;22:561–565. doi: 10.1177/1078155215585189. [DOI] [PubMed] [Google Scholar]
- 55.King YA, Chiu YJ, Chen HP, Kuo DH, Lu CC, Yang JS. Endoplasmic Reticulum Stress Contributes to Arsenic Trioxide - Induced Intrinsic Apoptosis in Human Umbilical and Bone Marrow Mesenchymal Stem Cells. Environ Toxicol. 2016;31:314–328. doi: 10.1002/tox.22046. [DOI] [PubMed] [Google Scholar]
- 56.Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC. Human Induced Pluripotent Stem Cell-derived Cardiomyocytes as an in Vitro Model for Coxsackievirus B3-induced Myocarditis and Antiviral Drug Screening Platform. Circ Res. 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kloth JSL, Pagani A, Verboom MC, Malovini A, Napolitano C, Kruit WHJ, Sleijfer S, Steeghs N, Zambelli A, Mathijssen RHJ. Incidence and Relevance of QTc-interval Prolongation Caused by Tyrosine Kinase Inhibitors. Br J Cancer. 2015;112:1011–1016. doi: 10.1038/bjc.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kus T, Aktas G, Sevinc A, Kalender ME, Camci C. Could Erlotinib Treatment Lead to Acute Cardiovascular Events in Patients with Lung Adenocarcinoma after Chemotherapy Failure? Onco Targets Ther. 2015;8:1341–1343. doi: 10.2147/OTT.S84480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding S, Long F, Jiang S. Acute Myocardial Infarction Following Erlotinib Treatment for NSCLC: A Case Report. Oncol Lett. 2016;11:4240–4244. doi: 10.3892/ol.2016.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinquié F, De CG, Urban T, Hureaux J. Maintenance Treatment by Erlotinib and Toxic Cardiomyopathy: A Case Report. Oncology. 2016;90:176–177. doi: 10.1159/000444186. [DOI] [PubMed] [Google Scholar]
- 61.Kumar I, Ali K, Usmansaeed M, Saeed MU. Follow-up of Erlotinib Related Uveitis. BMJ Case Rep. 2012:bcr1220115418. doi: 10.1136/bcr.12.2011.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korashy HM, Attafi IM, Ansari MA, Assiri MA, Belali OM, Ahmad SF, Al-Alallah IA, Anazi FEA, Alhaider AA. Molecular Mechanisms of Cardiotoxicity of Gefitinib in Vivo and in Vitro Rat Cardiomyocyte: Role of Apoptosis and Oxidative Stress. Toxicol Lett. 2016;252:50–61. doi: 10.1016/j.toxlet.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 63.Jacob F, Yonis AY, Cuello F, Luther P, Schulze T, Eder A, Streichert T, Mannhardt I, Hirt MN, Schaaf S. Analysis of Tyrosine Kinase Inhibitor-Mediated Decline in Contractile Force in Rat Engineered Heart Tissue. Plos One. 2016;11:e0145937. doi: 10.1371/journal.pone.0145937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irani J. Sunitinib Versus Interferon-alpha in Metastatic Renal-cell Carcinoma. Prog Urol. 2007;17:996. doi: 10.1016/s1166-7087(07)92405-7. [DOI] [PubMed] [Google Scholar]
- 65.Doherty KR, Wappel RL, Talbert DR, Trusk PB, Moran DM, Kramer JW, Brown AM, Shell SA, Bacus S. Multi-parameter in Vitro Toxicity Testing of Crizotinib, Sunitinib, Erlotinib, and Nilotinib in Human Cardiomyocytes. Toxicol Appl Pharmacol. 2013;272:245–255. doi: 10.1016/j.taap.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 66.Hasinoff BB, Patel D. The Lack of Target Specificity of Small Molecule Anticancer Kinase Inhibitors is Correlated with their Ability to Damage Myocytesin Vitro. Toxicol Appl Pharmacol. 2010;249:132–139. doi: 10.1016/j.taap.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 67.Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Zhou W, Huang J, Tang Y. Prediction of Drug-target Interactions and Drug Repositioning via Network-based Inference. PLoS Comput Biol. 2012;8:e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng F, Li W, Wu Z, Wang X, Zhang C, Li J, Liu G, Tang Y. Prediction of Polypharmacological Profiles of Drugs by The Integration of Chemical, Side effect, and Therapeutic Space. J Chem Inf Model. 2013;53:753–762. doi: 10.1021/ci400010x. [DOI] [PubMed] [Google Scholar]
- 69.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tajiri K, Aonuma K, Sekine I. Cardiovascular Toxic Effects of Targeted Cancer Therapy. Jpn J Clin Oncol. 2017;47:779–785. doi: 10.1093/jjco/hyx071. [DOI] [PubMed] [Google Scholar]
- 71.Steegmann JL, Baccarani M, Breccia M, Casado LF, García-Gutiérrez V, Hochhaus A, Kim DW, Kim TD, Khoury HJ, Le CP. European LeukemiaNet Recommendations for the Management and Avoidance of Adverse Events of Treatment in Chronic Myeloid Leukaemia. Leukemia. 2016;30:1648–1671. doi: 10.1038/leu.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valent P, Hadzijusufovic E, Schernthaner GH, Wolf D, Rea D, Le CP. Vascular Safety Issues in CML Patients Treated with BCR/ABL1 Kinase Inhibitors. Blood. 2015;125:901–906. doi: 10.1182/blood-2014-09-594432. [DOI] [PubMed] [Google Scholar]
- 73.Mordente A, Meucci E, Martorana GE, Tavian D, Silvestrini A. Topoisomerases and Anthracyclines: Recent Advances and Perspectives in Anticancer Therapy and Prevention of Cardiotoxicity. Curr Med Chem. 2017;24:1607–1626. doi: 10.2174/0929867323666161214120355. [DOI] [PubMed] [Google Scholar]
- 74.Sk UH, Dixit D, Sen E. Comparative Study of Microtubule Inhibitors--Estramustine and Natural Podophyllotoxin Conjugated PAMAM Dendrimer on Glioma Cell Proliferation. Eur J Med Chem. 2013;68:47–57. doi: 10.1016/j.ejmech.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 75.Mihalcea DJ, Florescu M, Vinereanu D. Mechanisms and Genetic Susceptibility of Chemotherapy-Induced Cardiotoxicity in Patients With Breast Cancer. Am J Ther. 2017;24:e3–11. doi: 10.1097/MJT.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 76.de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, Gravis G, Bodrogi I, Mackenzie MJ, Shen L. Prednisone plus Cabazitaxel or Mitoxantrone for Metastatic Castration-resistant Prostate Cancer Progressing after Docetaxel Treatment: a Randomised Open-label Trial. Lancet. 2010;376:1147–1154. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 77.de Bono JS, Sartor O. Cabazitaxel for Castration-resistant Prostate Cancer Authors' Reply. Lancet. 2011;377:122–123. doi: 10.1016/S0140-6736(11)60012-3. [DOI] [PubMed] [Google Scholar]
- 78.Fang J, Wu Z, Cai C, Wang Q, Tang Y, Cheng F. Quantitative and Systems Pharmacology. 1. In Silico Prediction of Drug-Target Interaction of Natural Products to Enable of new Targeted Cancer Therapy. J Chem Inf Model. 2017;57:2657–2671. doi: 10.1021/acs.jcim.7b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng F, Zhao Z. Machine Learning-based Prediction of Drug drug Interactions by Integrating Drug Phenotypic, Therapeutic, Chemical, and Genomic Properties. J Am Med Inform Assoc. 2014;21:278–286. doi: 10.1136/amiajnl-2013-002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gayvert KM, Madhukar NS, Elemento O. A Data-Driven Approach to Predicting Successes and Failures of Clinical Trials. Cell Chem Biol. 2016;23:1294–1301. doi: 10.1016/j.chembiol.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lusci A, Pollastri G, Baldi P. Deep Architectures and Deep Learning in Chemoinformatics: The Prediction of Aqueous Solubility for Drug-like Molecules. J Chem Inf Model. 2013;53:1563–1575. doi: 10.1021/ci400187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L. Deep Learning for Drug-Induced Liver Injury. J Chem Inf Model. 2015;55:2085–2093. doi: 10.1021/acs.jcim.5b00238. [DOI] [PubMed] [Google Scholar]
- 83.Fralick M, Kesselheim AS, Avorn J, Schneeweiss S. Use of Health Care Databases to Support Supplemental Indications of Approved Medications. JAMA Intern Med. 2017;178:55–63. doi: 10.1001/jamainternmed.2017.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.