Abstract
Bacteria carry a number of genes that cause cell growth arrest or cell lysis upon expression. Notably, defective prophages retain many lysis proteins. Here, we identified a novel lytic gene, ydfD, on the Qin prophage segment of the Escherichia coli genome. YdfD lyses 99.9% of cells within 2 h of its induction. The co-expression of the upstream gene, dicB, encoding a cell division inhibitor, as well as sulA, encoding another cell division inhibitor, abolished YdfD-induced cell lysis. These results imply that YdfD-induced lysis is a cell division-dependent event. We further found that by deleting the hydrophobic 22-residue N-terminal domain, the resulting 42-residue C-terminal domain was still toxic to cause cell lysis. We propose that YdfD, associated with the cytoplasmic membrane, inhibits an essential cellular process(s).
Keywords: YdfD, prophage, lysis, Escherichia coli
Newly characterized putative transmembrane protein YdfD from Escherichia coli can induce cell lysis in a cell division-dependent manner.
INTRODUCTION
Bacterial chromosomes carry multiple prophage segments on their genomes (Casjens 2003). It is estimated that 10%–20% of Escherichia coli genomes are prophage-related sequences. Notably, many of the defective prophage segments retain genes responsible for cell lysis and genes encoding toxic proteins (Srividhya and Krishnaswamy 2007; Yamaguchi, Park and Inouye 2011). Their roles in the host's physiology and evolutionary dynamics remain largely unknown. However, an increasing number of evidences suggest that these toxic proteins encoded by the prophage genes exert a significant impact on the host's fitness (Wang, Kim and Wood 2009; Wang et al. 2010).
At the end of the lytic cycle, phages escape from their hosts by inducing host cell lysis. Bacterial cell lysis is required for the release of viral particles from the host cells. Bacteriophages and active prophages lyse host cells by two distinct mechanisms; the holin–endolysin system and the single protein system (Wang, Smith and Young 2000). The holin–endolysin system is constituted by a protein-forming membrane pores and a cell wall-degrading enzyme. A pore-forming protein allows secretion of cell wall-degrading proteins into the periplasmic space from the cytoplasm, where cell walls will be degraded. In contrast, the single protein system does not require pore-forming proteins (Henrich, Lubitz and Plapp 1982; Coleman, Inouye and Atkins 1983). Examples of the single protein system include lysis protein E from bacteriophage ϕX174, Lysis (L) from MS2 phage and Kil protein of bacteriophage λ (Walderich and Holtje 1989; Conter, Bouche and Dassain 1996; Bernhardt, Roof and Young 2000). Single lysis proteins are small, and typically comprised of fewer than 100 amino acid residues. They are often associated with the membrane through a membrane-spanning domain (Walderich et al. 1988). Lytic proteins themselves do not possess any enzymatic activity (Coleman, Inouye and Atkins 1983).
Lysis proteins lyse the host cells by inducing the host's autolysis system (Lubitz, Halfmann and Plapp 1984; Halfmann and Lubitz 1986; Walderich et al. 1988). The L protein of MS2 expression was shown to induce structural changes of the murein, which triggers autolysis (Walderich et al. 1988). The expression of the L protein was also shown to increase the number of adhesion sites between inner and outer membranes (Walderich and Holtje 1989). However, the mechanisms for the induction of autolysis and the structural changes of cell wall caused by the L protein remain unknown. The lysis protein E from bacteriophage ϕX174 is also a small transmembrane protein (Altman et al. 1985). It inhibits translocase I (MraY), an essential enzyme that catalyzes the formation of the first lipid-linked intermediate in the cell wall biosynthetic pathway (Bernhardt, Roof and Young 2000). Lastly, the Kil protein of λ phage interferes with cell division (Haeusser et al. 2014). The Kil protein blocks the proper assembly of the cell division apparatus through interaction with FtsZ and ZipA. Thus, expression of Kil results in cell filamentation and eventual cell death.
Qin is one of the three lamboid prophages on the E. coli K12 genome. Unlike Rac prophage, another lamboid prophage that remains excision-proficient, Qin is considered to be defective due to a lack of active integrase (Blattner et al. 1997; Liu et al. 2015). The excision of Qin so far has not been reported.
Here, we have identified a novel lytic gene in E. coli, ydfD. The ydfD gene is a part of the Qin defective prophage (Zhou and Rudd 2013). We have shown that its expression causes 99.9% of cells to lyse within 2 h of induction. Interestingly, YdfD-induced cell lysis was abolished by co-expression of its upstream gene, dicB, or another gene for cell division inhibitor, SulA. We, thus, propose that YdfD-induced cell lysis requires functional cell division. We further demonstrated that the cellular target of YdfD is unique, different from targets of other known single-protein lysis systems.
MATERIALS AND METHODS
Strains and growth conditions
Escherichia coli BW25113 (ΔaraBD) (Datsenko and Wanner 2000) and BL21 (DE3) were grown at 37°C in M9 medium supplemented with 0.2% casamino acids, 1 mM thiamine and 0.5% glycerol. For examining the growth pattern, a designated amount of arabinose and/or IPTG was added to the culture when the OD600 reached mid-log phase. Cell morphology was observed using an Olympus BX40 microscope.
Molecular cloning
The full length YdfD was cloned into pBAD24 plasmid and designated as ydfD_pBAD24. The truncated mutant of YdfD, lacking the 22-residue N-terminal domain, was constructed by cloning the segment from residue 23 to residue 66 into pBAD24 (ydfD23–63_pBAD24) or pET21c plasmid (ydfD23–63_pET21c). A fragment containing the coding sequence of both ydfD and dicB was also cloned into pBAD24 (dicBydfD_pBAD24). IPTG inducible expression of dicB or sulA alone using pCN24N was achieved by using the clones obtained from Aska library (Kitagawa et al. 2005).
Transmembrane domain prediction
The transmembrane domain of YdfD was predicted using TMHMM (Sonnhammer, von Heijne and Krogh 1998).
RESULTS AND DISCUSSION
YdfD gene structure and evolutionary conservation
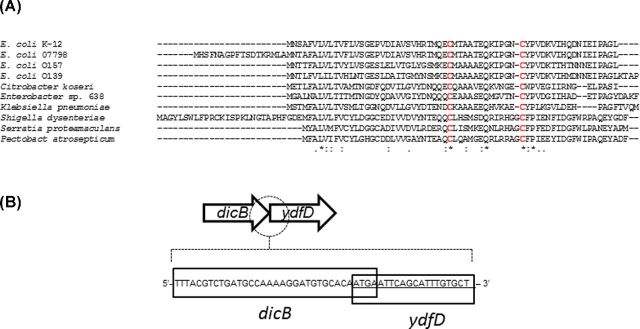
The ydfD gene encodes for a short peptide, consisting of 63 amino acid residues, with two conserved cysteine residues (Fig. 1A). It is commonly found in sequenced genomes of E. coli and related enterobacterial strains and is a part of dicB-ydfDF-insD-intQ operon within the defective Qin prophage segment on the E. coli K-12 genome (Keseler et al. 2013). No homologues were identified in other prophage sequences on the K-12 genome.
Figure 1.
Protein sequence of YdfD and dicB ydfD operon structure. (A) Alignment of YdfD sequences from enterobacteral strains. Sequences from E. coli strains K-12, 07798, O157, O139, Citrobacter koseri ATCC BAA-895, Enterobacter sp. 638, Klebsiella pneumoniae subsp. pneumoniae WGLW2, Shigella dysenteriae Sd197, Serratia proteamaculans 568 and Pectobacterium atrosepticum are shown. (B) Overlapping coding sequence of dicB and ydfD. Sequences corresponding to dicB and ydfD are designated by solid lines.
The ydfD gene is located immediately downstream of a cell division inhibitor, dicB. The coding sequences of dicB and ydfD overlap by 4 bp (Fig. 1B), indicating that the expression of ydfD and dicB are transcriptionally coupled and under the control of transcriptional repressor, DicA (Béjar and Bouché 1985). The function of YdfD has not been investigated to date.
YdfD induces host lysis
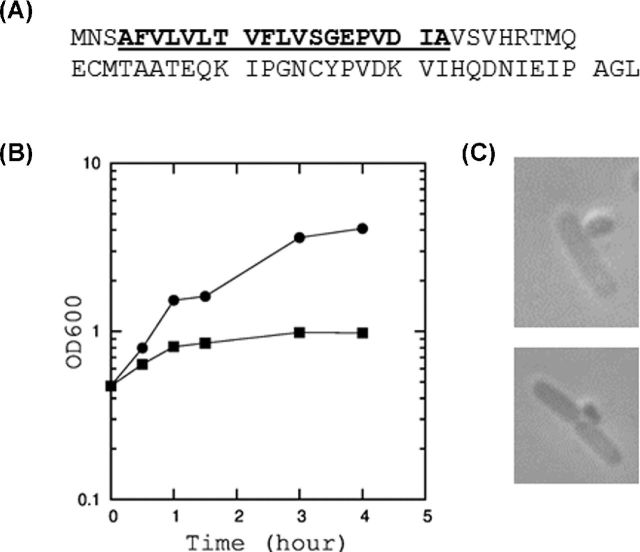
In order to investigate the function of YdfD, we first examined the effects of its expression on E. coli physiology by the ectopic expression of YdfD from the arabinose inducible promoter in pBAD24. As seen in Fig. 2A, the induction of YdfD expression causes a rapid decrease in OD600 within 1 h. The colony-forming unit (CFU) also dropped significantly, killing 99.9% of cells within 2 h of induction (Fig. 2B). CFU continued to decrease down by four orders of magnitude within 4 h of induction. When examined under a phase-contrast microscope, cells became ruptured (Fig. 2C). We observed one or two incisions in the middle or at the posterior of the cells. Leakages of DNA from these incisions were observed using DAPI (4′,6-diamidino-2-phenylindole) staining (Fig. 2D). These results indicate that YdfD expression causes local cell rupture creating one or two large openings in the cell wall through which cytosolic materials were released. YdfD was also toxic to cells when its expression was induced during the growth on solid media (data not shown).
Figure 2.
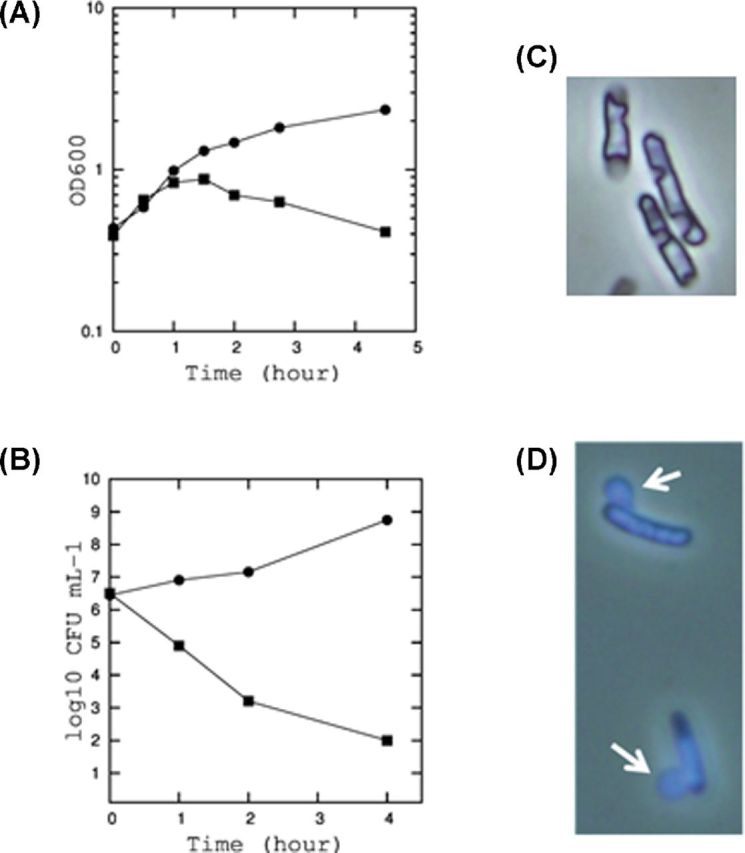
The growth inhibition and cell lysis by YdfD. (A) Growth curves of E. coli BW25113 expressing YdfD from pBAD24. The cells were incubated in M9 medium. When the optical density at 600 nm reached 0.4, 0.2% arabinose was added (square). Arabinose was also added to the cells containing empty pBAD24 vector as a control (circle). (B) The colony-forming unit (CFU) of cells at 0, 1, 2 and 4 h after the induction of YdfD expression (square) and in control (circle). (C) Cell morphology observed by a phase contrast microscope. (D) Cells stained with DAPI.
The C-terminal domain of YdfD is essential for lysis
YdfD is composed of a putative 22-residue N-terminal hydrophobic domain and a 41-residue C-terminal seemingly cytolasmic domain (Fig. 3A). Although the N-terminal domain consists of hydrophobic amino acids, it does not have a feature known for the signal peptide required for protein secretion across the membrane. It seems to be required for the localization of a protein in the inner leaflet of the cytoplasmic membrane. To examine the functional roles of these two domains, we first attempted to clone the C-terminal domain from residue 23 to 63 (YdfD23–63) into expression vectors to express it in a soluble from in the cytoplasmic space. When YdfD23–63 was cloned into pBAD24 and its expression was induced, cell growth was almost completely inhibited after 1 h (Fig. 3B), indicating that this 41-residue cytoplasmic fragment retains the toxicity of YdfD. When cells after 1 h induction of YdfD were examined under a microscope, leakage of the cytoplasm is observed, indicating that the cell wall is locally weakened (Fig. 3C). Truncated ydfD were also cloned into pET21c for a higher level of expression, and even more severe growth retardation was observed (data not shown). Notably, the secretion of the C-terminal domain in the periplasmic space using pCold-V did not show the toxic effect to the cells, indicating that the target(s) of the C-terminal domain of YdfD resides in the cytoplasm to inhibit a cytoplasmic process(s).
Figure 3.
Amino acid sequence of YdfD and the effect of YdfD expression on cell growth and morphology. (A) Amino acid sequence of YdfD. The hydrophobic domain is shown in bold and underlined. (B) Growth curve of E. coli BL21 (DE3) expressing a truncated variant of YdfD, YdfD23–63. 0.2% arabinose was added (square) to induce the expression from ydfD23–63_pBAD24. Arabinose was also added to the cells containing empty pBAD24 vector as a control (circle). (C) Morphology of cells expressing YdfD23–63.
The lower toxicity of YdfD23–63 compared to full length YdfD suggested that the membrane localization of YdfD is important for the full YdfD activity. It remains unknown whether the N-terminal domain merely serves as an anchor to the membrane or possesses additional functions.
YdfD-induced lysis is abolished by the co-expression of upstream gene, dicB
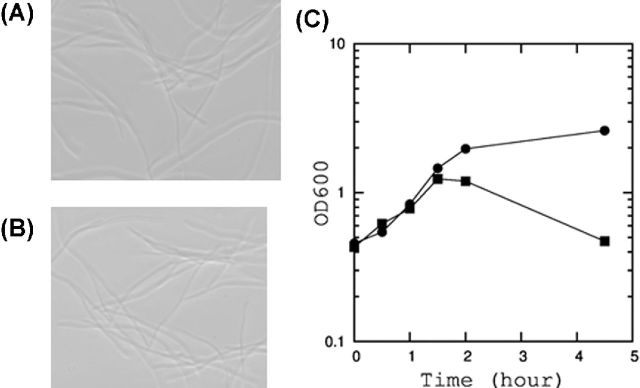
The dicB encodes DicB, an inhibitor of FtsZ polymerization (Cam et al. 1988). FtsZ is an essential protein, forming a ring-like structure at the septum and functioning as a scaffold for the assembly of a cell division complex (Bi and Lutkenhaus 1991). The cell division is inhibited when the polymerization of FtsZ is blocked. Hence, the expression of DicB causes extensive cell elongation (Johnson, Lackner and de Boer 2002), and ultimately cell growth is arrested. DicB overexpression is toxic to cells causing cell elongation, but does not cause immediate cell lysis (Fig. 4A).
Figure 4.
Neutralization of YdfD-induced toxicity by the expression of the upstream gene, dicB. (A) Cell morphology of E. coli BW25113 overexpressing DicB alone from pBAD24. (B) Cell morphology of E. coli BW25113 overexpressing DicB and YdfD from dicBydfD_pBAD24. (C) Growth curve of BW25113 expressing YdfD alone or YdfD and DicB. Cells were grown as described in Fig. 2. YdfD was expressed from ydfD_pBAD24 (circle). YdfD and DicB were co-expressed from dicBydfD_pBAD24 (square) by addition of arabinose at a final concentration of 0.2%.
Since the expression of ydfD and its upstream gene dicB are coupled, we wondered if there is a synergistic or antagonistic relationship between DicB and YdfD. As shown in Fig. 4B and C, when YdfD and DicB are co-expressed, YdfD overexpression no longer caused cell lysis. However, the cells were still elongated, suggesting that DicB still arrested cell division. This result suggested that YdfD-induced lysis cannot occur when cell division is inhibited. The same result was obtained when two proteins were encoded from a single transcript or when they were cloned separately (data not shown).
FtsZ polymerization inhibitor can inhibit YdfD-induced lysis
Multiple inhibitors of FtsZ polymerization have been described (Huisman, D'Ari and Gottesman 1984; Tan, Awano and Inouye 2011; Masuda et al. 2012). We examined if other FtsZ polymerization inhibitors could also abolish YdfD-induced cell lysis. As predicted, SulA did indeed neutralize YdfD-induced cell lysis (Fig. 5), supporting our hypothesis that YdfD-induced lysis is linked to cell division.
Figure 5.
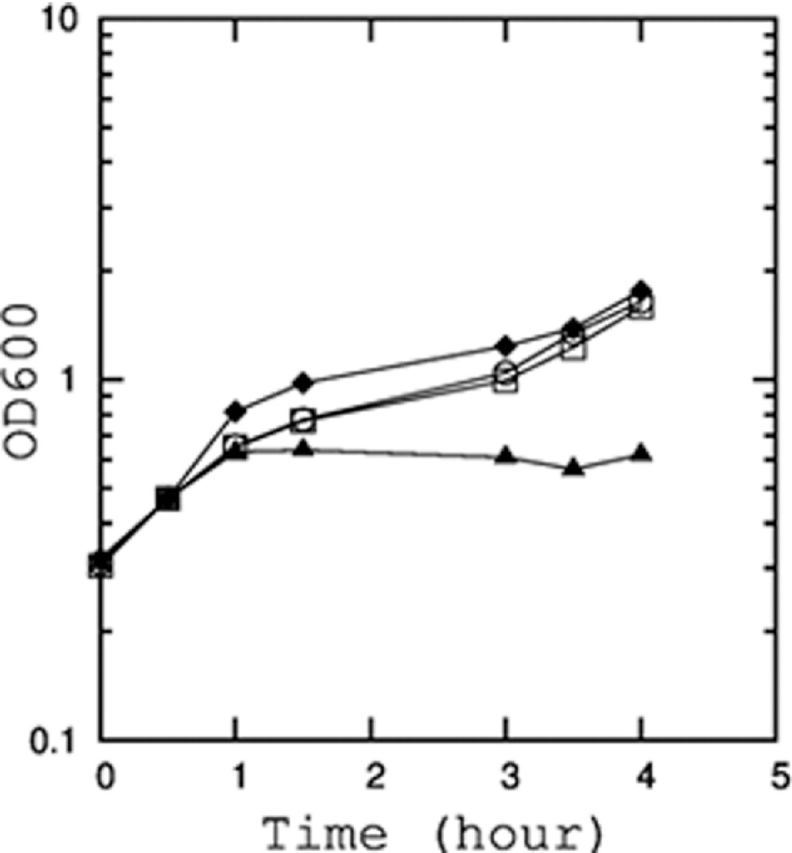
Inhibition of YdfD-induced cell lysis by the co-expression of cell division inhibitor, SulA. Growth curve of E. coli BL21 (DE3) co-transformed with ydfD_pBAD24 and SulA-pCA24N. The cultures were treated with no addition (open square), 1 mM IPTG (open circle), 0.2% arabinose (triangle), or 1 mM IPTG and 0.2% arabinose (diamond).
The cellular target of YdfD is distinct from the known prophage single protein lytic systems
We further examined whether YdfD may inhibit the same cellular target as other known lytic proteins. First, YdfD was compared with the lysis protein E, a membrane-associated lytic protein from bacteriophage ϕX174 (Maratea, Young and Young 1985). Lysis protein E inhibits the MraY-catalyzed step of the peptidoglycan biosynthetic pathway in a KBP-type peptidyl-prolyl cis-trans isomerase (SlyD) dependent manner (Roof and Young 1995). SlyD is proposed to be essential for the stability of lysis protein E (Roof and Young 1995; Bernhardt, Roof and Young 2002), and lysis protein E does not induce lysis in a ΔslyD strain (Maratea, Young and Young 1985). Unlike lysis protein E, YdfD remained toxic in the ΔslyD strain.
Lysis protein L from RNA phage MS2 causes cell lysis at cell membrane adhesion sites (Walderich and Holtje 1989). When cells were grown in media at pH 5, lysis protein L did not induce cell lysis. In contrast, YdfD remained toxic even when cells were grown at pH 5 (data not shown), indicating that YdfD and lysis protein L utilized different mechanisms. Our results, however, do not rule out the possibility that YdfD may also be localized at the membrane-adhesion sites.
The Kil protein of λ phage inhibits cell division, causing cell elongation. Unlike Kil protein, YdfD does not induce cell elongation. Instead, the inhibition of cell division had a negative effect on the function of YdfD. A homologue of Kil protein is also found in another cryptic lambdoid prophage, Rac, on E. coli K-12 genome, located on the equivalent position of dicB in Qin. Although there is no sequence similarity between DicB and Kil, they both inhibit cell division by blocking septum formation. In Rac prophage, a small uncharacterized gene, ydaE, is found immediately downstream of Kil, which is the equivalent genetic location of ydfD. There is also no sequence homology between YdfD and YdaE, however, YdaE also carries conserved cysteine residues. The presence of conserved cysteine residues, small size and transcriptional coupling with cell division inhibitors (DicB and Kil, respectively) suggest that YdfD and YdaE may share a similar function. Further work is required to examine this possibility.
The strong conservation of dicB- ydfD operonic structure, despite diverged gene orders in neighboring regions suggests the strong linkage between the two proteins. The operon consisted of a toxic gene and a gene that neutralize the toxin's function is reminiscent of the toxin–antitoxin (TA) systems. It is possible that these two proteins function as a TA pair, ensuring the maintenance of the Qin prophage.
In summary, we have identified a novel lytic protein on the E. coli chromosome. Its toxicity can be neutralized by DicB as well as by another cell division inhibitor, SulA, indicating that YdfD-induced lysis is a cell division-dependent event. During cell division, many processes, including the degradation of preexisting cell walls and the synthesis of new cell walls, occur. Such processes have to be both spatially and temporally coordinated (Cabeen and Jacobs-Wagner 2005). Uncoupling any of these processes could lead to rapid osmotic lysis. YdfD appears to inhibit or uncouple such processes to induce cell lysis. We also discovered that the C-terminal cytoplasmic domain is important for YdfD-induced lysis. Our data support that YdfD inhibits a unique cellular target, distinguishable from other known phage lytic proteins. Further research is currently being conducted to identify the exact cellular target of YdfD and its mechanisms of action.
Acknowledgments
The authors are grateful to Dr Peter Tupa and Dr Marcia Gillette for critical reading of the manuscript.
FUNDING
The work was supported by the Indiana Academy of Science senior grant (2015-12) and by Grants in Aid from Indiana University Kokomo. HM was supported by NRSA Individual Postdoctoral Fellowship (F32) from National Institute of Health (GM095200). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of interest. None declared.
REFERENCES
- Altman E, Young K, Garrett J, et al. Subcellular localization of lethal lysis proteins of bacteriophages lambda and phiX174. J Virol. 1985;53:1008–11. doi: 10.1128/jvi.53.3.1008-1011.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjar S, Bouché JP. A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol Gen Genet. 1985;201:146–50. doi: 10.1007/BF00425651. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. P Natl Acad Sci USA. 2000;97:4297–302. doi: 10.1073/pnas.97.8.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage phi X174. Mol Microbiol. 2002;45:99–108. doi: 10.1046/j.1365-2958.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–62. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–10. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Cam K, Bejar S, Gil D, et al. Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988;16:6327–38. doi: 10.1093/nar/16.14.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 200349277–300. [DOI] [PubMed] [Google Scholar]
- Coleman J, Inouye M, Atkins J. Bacteriophage MS2 lysis protein does not require coat protein to mediate cell lysis. J Bacteriol. 1983;153:1098–100. doi: 10.1128/jb.153.2.1098-1100.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter A, Bouche JP, Dassain M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage rac. J Bacteriol. 1996;178:5100–4. doi: 10.1128/jb.178.17.5100-5104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichiacoli K-12 using PCR products. P Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Hoashi M, Weaver A, et al. The Kil peptide of bacteriophage lambda blocks Escherichiacoli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLOS Genet. 2014;10:e1004217. doi: 10.1371/journal.pgen.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann G, Lubitz W. Differential induction of Escherichiacoli autolysis by penicillin and the bacteriophage phi X174 gene E product. J Bacteriol. 1986;166:683–5. doi: 10.1128/jb.166.2.683-685.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich B, Lubitz W, Plapp R. Lysis of Escherichiacoli by induction of cloned phi X174 genes. Mol Gen Genet. 1982;185:493–7. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- Huisman O, D'Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. P Natl Acad Sci USA. 1984;81:4490–4. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Lackner LL, de Boer PA. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichiacoli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J Bacteriol. 2002;184:2951–62. doi: 10.1128/JB.184.11.2951-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–12. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–9. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Liu X, Li Y, Guo Y, et al. Physiological function of rac prophage during biofilm formation and regulation of rac excision in Escherichiacoli K-12. Sci Rep. 2015;5:16074. doi: 10.1038/srep16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz W, Halfmann G, Plapp R. Lysis of Escherichiacoli after infection with phiX174 depends on the regulation of the cellular autolytic system. J Gen Microbiol. 1984;130:1079–87. doi: 10.1099/00221287-130-5-1079. [DOI] [PubMed] [Google Scholar]
- Maratea D, Young K, Young R. Deletion and fusion analysis of the phage ϕX174 lysis gene E. Gene. 1985;40:39–46. doi: 10.1016/0378-1119(85)90022-8. [DOI] [PubMed] [Google Scholar]
- Masuda H, Tan Q, Awano N, et al. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol Lett. 2012;328:174–81. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof WD, Young R. Phi X174 lysis requires slyD, a host gene which is related to the FKBP family of peptidyl-prolyl cis-trans isomerases. FEMS Microbiol Rev. 1995;17:213–8. doi: 10.1111/j.1574-6976.1995.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- Srividhya KV, Krishnaswamy S. Subclassification and targeted characterization of prophage-encoded two-component cell lysis cassette. J Bioscience. 2007;32:979–90. doi: 10.1007/s12038-007-0097-x. [DOI] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2011;79:109–18. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walderich B, Holtje JV. Specific localization of the lysis protein of bacteriophage MS2 in membrane adhesion sites of Escherichia coli. J Bacteriol. 1989;171:3331–6. doi: 10.1128/jb.171.6.3331-3336.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walderich B, Ursinus-Wossner A, van Duin J, et al. Induction of the autolytic system of Escherichia coli by specific insertion of bacteriophage MS2 lysis protein into the bacterial cell envelope. J Bacteriol. 1988;170:5027–33. doi: 10.1128/jb.170.11.5027-5033.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, et al. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichiacoli biofilms. ISME J. 2009;3:1164–79. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. Toxin–antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–24. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]