Abstract
The Pseudomonas aeruginosa Chp chemosensory system regulates twitching motility, intracellular adenosine 3′′ 5′-cyclic monophosphate (cAMP) levels and is postulated to be involved in directional twitching towards phosphatidylethanolamine (PE). Because PilJ is the only methyl-accepting chemotaxis protein (MCP) identified in the Chp system, we determined the role of PilJ in mediating signal transduction for the distinct outputs of this system. Mutants that lack the periplasmic domain of PilJ (pilJΔ74-273) showed lower levels of cAMP but retained directional twitching towards PE. While initial studies revealed reduced twitching motility by PilJΔ74-273, this was due to decreased cAMP levels. Our data illustrate the importance of the periplasmic domain of PilJ in regulating cAMP. This is the first time a defined domain within PilJ has been identified as having a distinct role in signal transduction.
Keywords: Pseudomonas aeruginosa, twitching motility, cAMP, signal transduction, chemotaxis, PilJ
A distinct region within PilJ is responsible for regulating the levels of a second messenger involved in Pseudomonas virulence.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen that exhibits swimming and twitching motility. The Chp chemosensory system controls type IV pili (T4P) mediated twitching motility and regulates intracellular levels of adenosine 3′, 5′-cyclic monophosphate (cAMP) by altering CyaB activity (Darzins 1995; Kato et al.1999; Whitchurch et al.2004; Fulcher et al.2010). CyaB is one of two intracellular adenylate cyclases that is responsible for synthesis of the majority of cAMP (Fulcher et al.2010). cAMP is an allosteric activator of Vfr, thereby regulating T4P as well as a number of other virulence factors (West, Sample and Runyen-Janecky 1994; Beatson et al.2002; Bertrand, West and Engel 2010). An additional Chp-based signaling mechanism involved in regulation of PilY1 expression and c-di-GMP levels has recently been suggested based on interactions between PilJ and FimS (Luo et al.2015).
Although the Chp chemosensory system has homology to the Escherichia coli chemotaxis system, signal transduction is more complex. Encoded within the chp gene cluster are two CheY-like proteins (PilG and PilH), two CheW coupling proteins (PilI and ChpC), a methyltransferase protein (PilK), a methylesterase protein (ChpB), a complex CheA/Y hybrid protein (ChpA), and one methyl-accepting chemotaxis protein (MCP) PilJ (Whitchurch et al.2004). PilJ is predicted to have two transmembrane domains and a periplasmic domain of 269–273 amino acids (Darzins 1994; Stover et al.2000). The periplasmic domain of PilJ has recently been proposed as the site of interaction with the major pilin (PilA) leading to modulation of CyaB activity (Persat et al.2015). This proposal follows the classical signal transduction model reported in E. coli, where ligands bind to the periplasmic domain of MCPs, ultimately resulting in altered kinase activity (Mowbray and Sandgren 1998; Hazelbauer, Falke and Parkinson 2008; Parkinson 2010).
While the P. aeruginosa genome contains 26 annotated MCPs, PilJ is the only MCP known to function within the Chp chemosensory system. Deletion of pilJ results in loss of surface piliation and twitching motility, and reduction in cAMP levels (Darzins 1994; Whitchurch et al.2004; DeLange et al.2007; Fulcher et al.2010; Luo et al.2015). Piliation and a basal level of motility can be restored through exogenous cAMP (Fulcher et al.2010). The CheY-like proteins, PilG and PilH are involved in regulation of both twitching motility and CyaB activity. Deletion of pilG results in reduced cAMP and surface piliation, and loss of twitching motility, while ΔpilH had increased cAMP and surface piliation, and decreased twitching (Bertrand, West and Engel 2010; Fulcher et al.2010). PilG and PilH function upstream of ATPases PilB and PilT/U, respectively, and likely regulate extension and retraction of T4P (Bertrand, West and Engel 2010). PilG and PilH also modulate CyaB activity, affecting intracellular levels of cAMP and T4P biosynthesis (Wolfgang et al.2003; Fulcher et al.2010). Exactly how PilG and PilH mediate these functions remains unclear.
P. aeruginosa shows directed twitching towards dioleoyl phosphatidylethanolamine (18:1 PE) (Kearns, Robinson and Shimkets 2001). While little is known about PE directed twitching motility, the response is mediated through T4P and requires the extracellular phospholipase PlcB (Barker et al.2004; Miller et al.2008). This preferential migration is specific to unsaturated LCFA (C16-20) and dilauroyl PE (12:0) (Kearns, Robinson and Shimkets 2001; Miller et al.2008). There have been limited investigations into the Chp chemosensory system role in directional twitching; the pilJ mutant is null for twitching and correspondingly cannot exhibit directional twitching, and the pilH mutant is non-viable under directional twitching assay conditions (Kearns, Robinson and Shimkets 2001; DeLange et al.2007).
We determined if intracellular cAMP levels, twitching motility and directional twitching to PE were modulated by classical signal transduction through PilJ. Deletion of the PilJ periplasmic domain reduced intracellular cAMP levels and resulted in a slight cAMP-dependent reduction in twitching, but had no effect on directional twitching. We also found that PilJ is not required for directional twitching, provided sufficient cAMP is present to partially restore surface piliation and twitching motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions
All P. aeruginosa mutants listed (Table S1, Supporting Information) were derived from PAO1. P. aeruginosa and E. coli were grown at 37°C in LB, unless otherwise stated. E. coli S17-1 was used for conjugation of plasmids (listed in Table S1, Supporting Information) into P. aeruginosa. Tetracycline and gentamicin were used at 10 μg ml−1 (E. coli) or 50 μg ml−1 (P. aeruginosa).
Construction of P. aeruginosa mutants and expression strains
In-frame deletions of pilA, cyaB, pilJ, pilT and the putative periplasmic domain of PilJ (pilJΔ74-273) were constructed using splicing by overlap extension PCR, using primers listed in Table S2 (Supporting Information). Colony PCR of PAO1 was used for the initial amplification of each PCR product. Fusion constructs were sequenced to ensure the final deletion alleles were in-frame and no other mutations were introduced. The deletion alleles were cloned into pEX18Tc, transferred into E. coli S17-1, then introduced into P. aeruginosa by conjugation. Merodiploids were selected on 75 μg ml−1 tetracycline and 5 μg ml−1 chloramphenicol. Resolution of the merodiploids was achieved through 10% sucrose counter selection. Following screening on tetracycline and sucrose, the deletions were confirmed by PCR.
Expression plasmids were generated by amplifying cyaB or pilJ-His using the primers listed (Table S2, Supporting Information). The resulting PCR products were cloned into pJN105 and sequenced. To generate cyaBR412H, codons were altered based on the QuikChange mutagenesis kit (Stratagene). The codon change was confirmed through sequencing.
Motility assays
To assay twitching motility, nine individual colonies were stab inoculated into LB agar (1% agar), and incubated at 37°C for 40 h.
Directional twitching assays were performed as described previously (Barker et al.2004; Miller et al.2008). The phospholipid or stearic acid gradient was formed by spotting PE (18:1) or stearic acid (10 mg ml−1 in chloroform) onto the agar and incubating at 30°C for 24 h. All strains were grown in LB until early stationary phase and concentrated to 9 × 109 cells ml−1 in MOPS buffer (pH 7.6). The concentrated cells were spotted 5 mm from the edge of the phospholipids, and incubated at 37°C for 16 h. For each assay, three independent colonies were assayed in triplicate. Images of colonies were obtained using an Olympus SZX16 stereomicroscope and used to determine the length of the leading and lagging zones.
Surface pilus preparation
T4P were sheared from bacterial cell surface by vortexing as previously described (Fulcher et al.2010). Cells were grown on LB agar, scraped from the plates, resuspended in 0.15M NaCl + 0.2% formaldehyde (final OD600 = 23), and vortexed to shear the pili off the bacterial surface. Bacterial cells and debris were removed by centrifugation. The pili were precipitated through overnight incubation at 4°C in 100 mM MgCl2. Pili were resuspended in SDS-PAGE loading dye, resolved on a 15% polyacrylamide gel and stained with a Coomassie brilliant blue G-250—perchloric acid solution (Faguy et al.1996).
β-galactosidase assay
To determine intracellular levels of cAMP, P. aeruginosa strains containing the cAMP reporter construct lacP1-lacZ were grown on LB agar (Fulcher et al.2010). Surface grown cells were resuspended in LB, and β-galactosidase assays were performed as previously described (Wolfgang et al.2003).
Immunoblotting
Whole cell lysates were prepared from mid-log phase (OD600 ∼0.5) cultures, and loading was normalized based on OD600. Protein samples were separated via 15% SDS-PAGE and transferred to PVDF. Membranes were probed with anti-PilA (gift from L. Burrows, McMaster University; 1:10 000) followed by peroxidase conjugated goat anti-rabbit (1:100 000). Immunoblots were developed with SuperSignal West chemiluminescent kit (Thermo Scientific, Waltham, MA) and a Fotodyne Luminary system (Fotodyne, Hartland, WI).
Statistical analysis
Data were analyzed using ANOVA followed by TukeyHSD, or a Student's t-test as indicated in the figure legend. Analysis was done using R (3.2.3, R Foundation for Statistical Computing, Vienna, Austria).
RESULTS AND DISCUSSION
The involvement of the Chp chemosensory system in regulating twitching motility and intracellular cAMP, along with the recent proposal that PilJ—PilA interactions allow T4P to function as a mechanosensor, led us to investigate the role of the periplasmic domain of PilJ in signal transduction. PilJ is the sole MCP encoded within the Chp gene cluster, and deletion of pilJ results in a non-twitching phenotype and reduced levels of intracellular cAMP (Darzins 1994; Whitchurch et al.2004; DeLange et al.2007; Fulcher et al.2010). In the well-studied MCPs of E. coli, the periplasmic domain is the site of ligand binding and is required for classical signal transduction (Parkinson 2010; Pham and Parkinson 2011). We generated an in-frame deletion of the putative periplasmic domain (G74-Q273) and determined the phenotypes of the resulting strain (pilJΔ74-273) with regards to intracellular cAMP levels, twitching motility and directional twitching towards PE.
The putative periplasmic domain of PilJ regulates cAMP levels
As previously reported (Fulcher et al.2010), deletion of full-length pilJ resulted in a loss of twitching motility and reduced levels of intracellular cAMP (∼20% of wild type) (Fig. 1A and D). This reduction in cAMP was similar to that seen in ΔcyaB (P = 0.926). The intermediate twitching motility levels of ΔcyaB contrast with ΔpilJ, which was null for twitching. This difference highlights the importance of PilJ in controlling twitching motility separate from cAMP levels, as previously reported (Fulcher et al.2010). Analysis of surface piliation showed that a residual level of pilus biogenesis occurs even with the low levels of cAMP present in cyaB mutant (Fig. 1B). Deletion of the major type IV pilin (pilA) resulted in a complete loss of twitching motility, and a ∼50% reduction in cAMP levels relative to wild type. Although earlier studies reported that pilA deletion resulted in cAMP levels similar to those found in ΔpilJ (Persat et al.2015), our data show significant differences in cAMP levels between ΔpilJ and ΔpilA (P < 0.001, Fig. 1D), suggesting PilJ plays a larger role in regulating cAMP than PilA.
Figure 1.
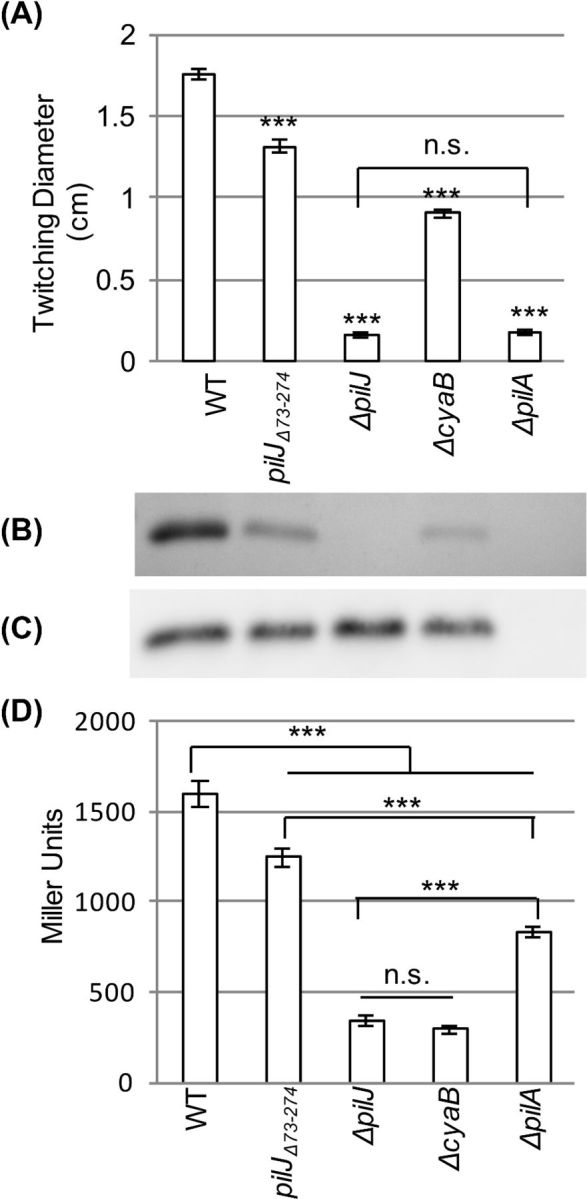
Deletion of the putative periplasmic of PilJ (pilJΔ74-273) reduces twitching motility and intracellular cAMP levels. (A) Diameter of twitching motility zones for the indicated strains. Nine colonies were assayed to determine the average and standard error of the mean. Asterisks (***) indicate values significantly different from wild type (P < 0.001) as determined by ANOVA followed by TukeyHSD. Non-significant differences are indicated by n.s. (B) SDS-PAGE showing the surface levels of PilA obtained from the indicated strains. (C) Whole cell levels of PilA as determined by western blotting for the indicated strains. (D) Intracellular cAMP levels of the indicated strains expressed as Miller Units. Three colonies were assayed in triplicate to determine the average and the standard error of the mean. Asterisks (***) indicate significantly different values (P < 0.001) as determined by ANOVA followed by TukeyHSD. Non-significant differences are indicated by n.s.
To determine the role of the periplasmic domain of PilJ, we tested a mutant that lacks this domain. In contrast to pilJ deletion mutants, surface grown cells of pilJΔ74-273 retained surface pilin levels, twitching motility and intracellular cAMP albeit at significantly reduced amounts relative to wild type (Fig. 1A, B and D). The reduction of surface piliation was not due to unavailability of the major pilin subunit as whole cells retained wild-type levels of PilA (Fig. 1C). The reduction in cAMP suggests that the PilJ periplasmic domain is involved in signal transduction affecting CyaB activity. Surface contact is critical for signal transduction through the periplasmic domain as liquid grown pilJΔ74-273 had wild-type levels of cAMP (data not shown). The mechanosensor model suggests that PilA interacts with PilJ periplasmic domain to trigger signal transduction (Persat et al.2015). However, deletion of the PilJ periplasmic domain did not reduce cAMP to the same levels as the pilA or pilJ deletions, suggesting that additional regions of PilJ are important in regulation of cAMP levels. It is not clear if the periplasmic domain is needed for ligand binding or if its deletion causes a conformational change in PilJ thereby altering ChpA activation and reducing cAMP levels. It is unlikely that regulation occurs through direct interaction between the PilJ periplasmic domain and CyaB, as ΔpilG has reduced cAMP similar to ΔcyaB indicating signal transduction through the Chp system is required (Fulcher et al.2010).
The reduced twitching motility seen in pilJΔ74-273 was partially complemented through plasmid-based expression of wild-type PilJ (Fig. 2A), which corresponded with a slight but insignificant increase in cAMP (P > 0.05, Fig. 2D). This is in contrast to wild-type P. aeruginosa, where overexpression of PilJ resulted in a decrease in twitching motility and cAMP relative to the parent strain (P < 0.05) (Fig. 2A and D). Expression of pilJ in ΔpilJ restored twitching, cAMP and surface piliation to levels similar to or above wild type (Fig. 2A, B and D).
Figure 2.
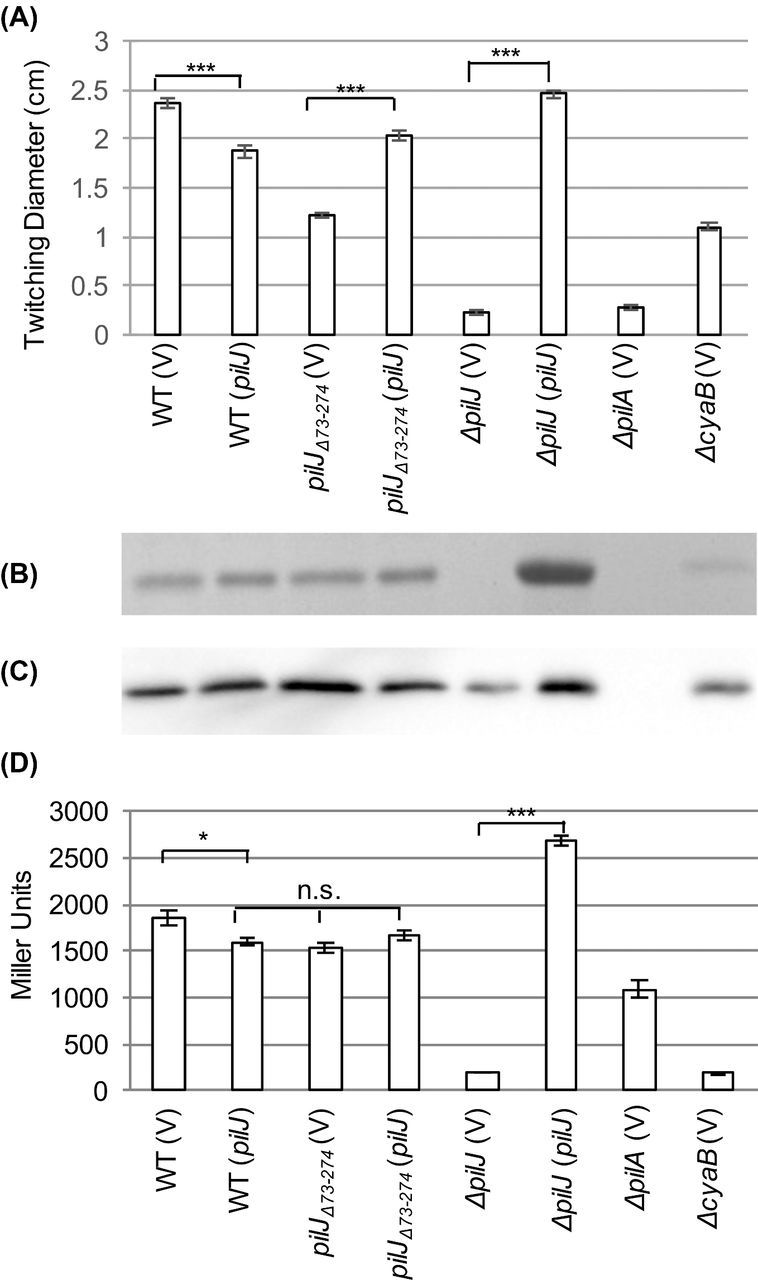
Expression of full-length pilJ partially restores twitching motility and surface piliation in pilJΔ74-273. (A) Zones of twitching motility for the indicated strains. Nine colonies were assayed to determine the average and standard error of the mean. (B) SDS-PAGE showing the surface levels of PilA obtained from the indicated strains. (C) Whole cell levels of PilA as determined by western blotting for the indicated strains. (D) Intracellular cAMP levels of the indicated strains expressed as Miller Units. Three colonies were assayed in triplicate to determine the average and the standard error of the mean. The strains contained either pJN105 (V) or pJN105-pilJ (pilJ) as indicated. In panels A and D, asterisks (***, P < 0.001) indicate values significantly different from the isogenic controls as determined by ANOVA followed by TukeyHSD. Non-significant differences are indicated by n.s.
As chemosensory systems are sensitive to stoichiometry (Li and Hazelbauer 2004), we verified that the impaired signal transduction in pilJΔ74-273 was not due to altered levels of expression or polar effects on the downstream genes. Multiple attempts to quantify levels of epitope-tagged chromosomally encoded PilJ and PilJΔ74-273 were unsuccessful. We therefore overexpressed C-terminal 6 × His-tagged versions of PilJ or PilJΔ74-273 in ΔpilJ background. When protein expression was induced to similar levels using 0.03% arabinose, twitching motility and cAMP levels were reduced in ΔpilJ (pilJΔ74-273-His) relative to the wild type (ΔpilJ (pilJ-His)) (Fig. S1, Supporting Information).
In an attempt to separate the twitching motility and cAMP defects, we expressed a CyaB point mutant to restore intracellular levels of cAMP. CyaBR412H synthesizes cAMP independent of Chp signal transduction and is reported to give cAMP levels most similar to wild type (Topal et al.2012). Expression of cyaBR412H in cyaB and pil mutants resulted in increased cAMP (Fig. 3D), but did not always correspond to large increases in twitching motility (Fig. 3A). The small increase in twitching motility in the absence of PilJ (ΔpilJ (cyaBR412H)) may result from unregulated activity of the extension and retraction ATPases (PilB/T) (Whitchurch et al.1991; Bertrand, West and Engel 2010).
Figure 3.
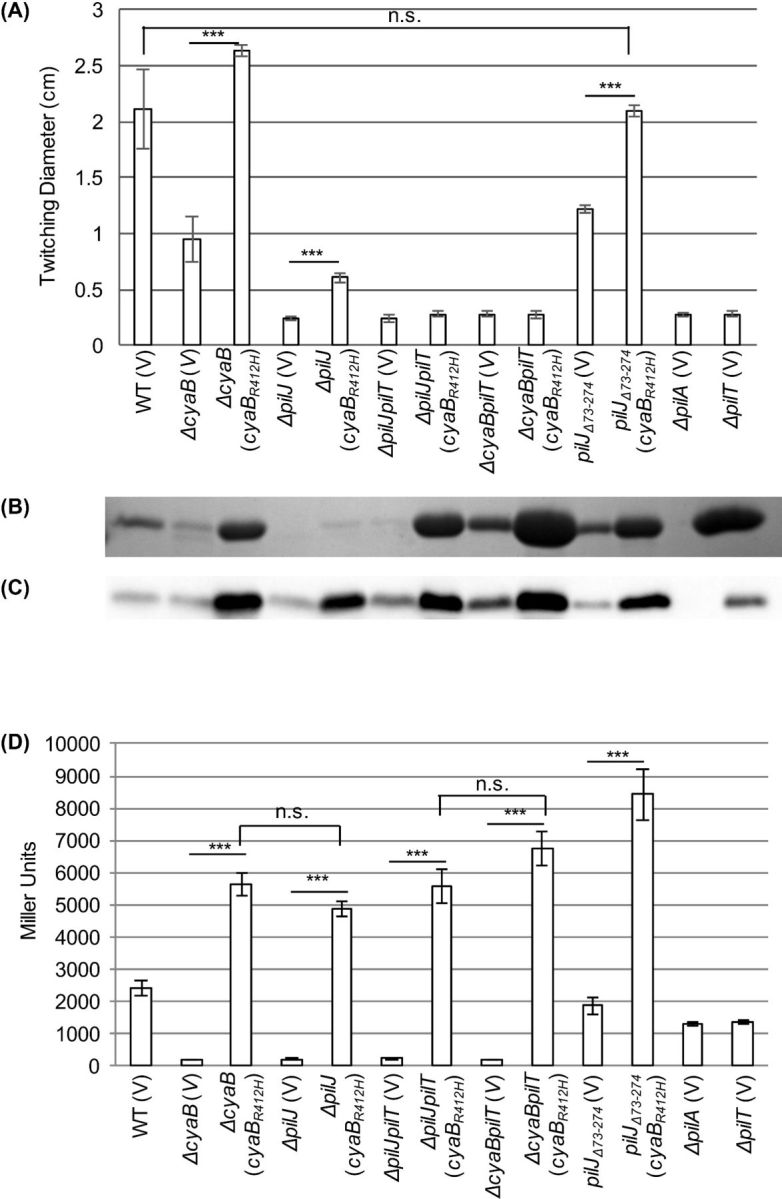
Providing cAMP through a cyaB point mutant restores wild-type levels of twitching motility in pilJΔ74-273. (A) Zones of twitching motility for the indicated strains. Nine colonies were assayed to determine the average and standard error of the mean. (B) SDS-PAGE showing the surface levels of PilA obtained from the indicated strains. (C) Whole cell levels of PilA as determined by western blotting for the indicated strains. (D) Intracellular cAMP levels of the indicated strains expressed as Miller Units. Three colonies were assayed in triplicate to determine the average and the standard error of the mean. The strains contained either pJN105 (V) or pJN105-cyaBR412H (cyaBR412H) as indicated. In panels A and D, asterisks (***, P < 0.001) indicate significantly different values as determined by ANOVA followed by TukeyHSD. Non-significant differences are indicated by n.s.
The inability of increased cAMP to restore wild-type levels of twitching motility in ΔpilJ again highlights the importance of full length PilJ in controlling twitching motility separate from cAMP levels. This separation of cAMP and twitching motility is not seen in pilJΔ74-273, where expression of cyaBR412H increases cAMP and restores wild-type levels of twitching (Fig. 3A and D). This showed that the periplasmic domain is not involved in the regulation of twitching motility, provided sufficient cAMP is present. Additionally, pilJΔ74-273 retains directional twitching towards PE, as evidenced by a leading:lagging ratio >3 (Fig. 4). A positive result for directional twitching is defined as a ratio >2. As a control, these strains were assayed against stearic acid (Kearns, Robinson and Shimkets 2001); both wild type and pilJΔ74-273 twitched uniformly resulting in leading:lagging edge ratios of ∼1 (Fig. 4B). These data indicate that while the periplasmic domain plays an important role in regulating cAMP levels, twitching motility occurs independent of this domain, both in the presence and absence of a proposed chemoattractant. Non-classical signal transduction has previously been reported in E. coli MCPs, where the phenol sensing response is mediated by the transmembrane and HAMP domains (Pham and Parkinson 2011). It is possible that the PilJ transmembrane and/or HAMP domains are involved in the regulation of twitching motility. This is the first time a defined domain within PilJ has been identified as having a distinct role in signal transduction.
Figure 4.
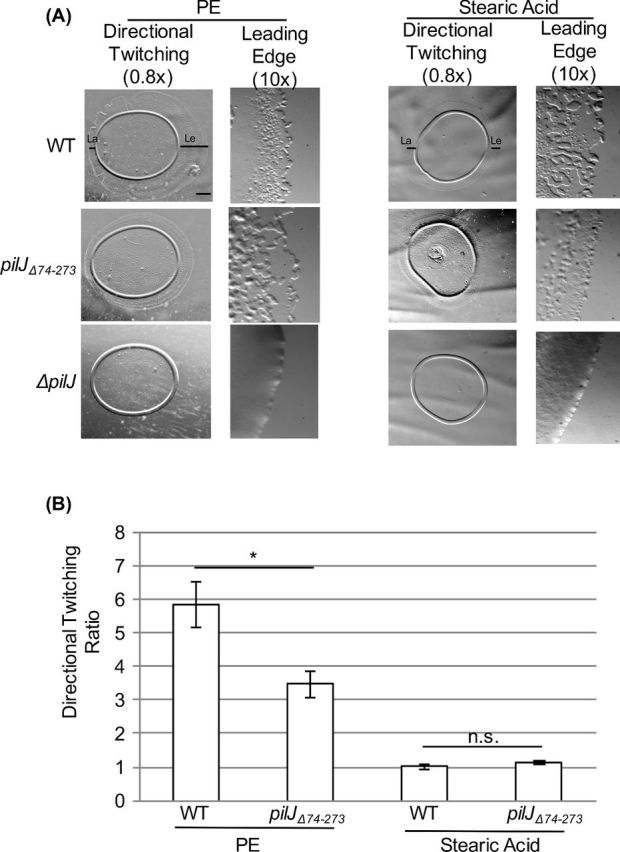
The putative periplasmic domain of PilJ is not required for directional twitching to PE. (A) Representative images of directional twitching results for the indicated strains. The PE or stearic acid was deposited on the plate to the right of where the P. aeruginosa culture was placed. Black bars on the wild-type strain indicates the leading edge (Le) and lagging edge (La) that were measured to determine the directional twitching ratio. Scale bar = 1mm (0.8× magnification) (B) Directional twitching ratios for both wild type and the pilJΔ74-273 strains. Directional twitching ratios were calculated by dividing the length of the leading edge by the length of the lagging edge. Three independent colonies were analyzed in triplicate. A ratio greater than 2 indicates directional twitching. Significantly different values were determined using a student's t-test (paired, *, P < 0.05).
When examining the surface pilin and whole cell pilin levels of these strains, several observations were made (Fig. 3B and C). ΔpilJ (cyaBR412H) has minute amounts of surface piliation. These levels of surface pilin are significantly less than ΔcyaB (cyaBR412H), despite having similar levels of cAMP. This difference is likely due to an extension deficiency as comparable strains lacking the retraction ATPase PilT had unequal amounts of surface piliation (Fig. 3B, compare ΔpilJpilT (cyaBR412H), and ΔcyaBpilT (cyaBR412H)). Additionally, whole cells with increased cAMP had high levels of PilA protein compared to wild type (Fig. 3C), and this increase is not simply due to increased surface piliation (compare ΔpilJ (cyaBR412H) and ΔpilJpilT (V), Fig. 3B and C). PilA expression is thought to be regulated transcriptionally and post-translationally. Transcriptional regulation occurs through a two-component system PilR-PilS, but the signal sensed by PilS remains unidentified (Mikkelsen, Sivaneson and Filloux 2011; Burrows 2012). Additionally, pilA promoter activity is affected by deletion of chpA and pilG (Bertrand, West and Engel 2010). Previous studies did not address if the different promoter activity is due to altered levels of cAMP, or altered levels of PilA in the inner membrane (Bertrand, West and Engel 2010). Our data show a correlation between increased levels of PilA in whole cells and cAMP levels that are significantly higher than wild type. Further studies will be done to determine the relationship behind this pattern.
Excess cAMP enables directional twitching towards PE in the absence of PilJ
Because expression of cyaBR412H partially restores twitching motility in ΔpilJ, this strain was tested for directional twitching towards PE. ΔpilJ (cyaBR412H) demonstrated directional twitching (ratio >2) but with a distinct morphology (Fig. 5A). The rafts extending from the edge of the ΔpilJ (cyaBR412H) colony were thicker than the rafts seen on the other strains assayed. This same morphology was seen on the edges of ΔpilJ (cyaBR412H) colonies showing non-preferential twitching towards stearic acid (Fig. 5A). Therefore, although twitching is restored by providing extra cAMP to ΔpilJ, the appearance of this motility is dramatically different from wild type.
Figure 5.
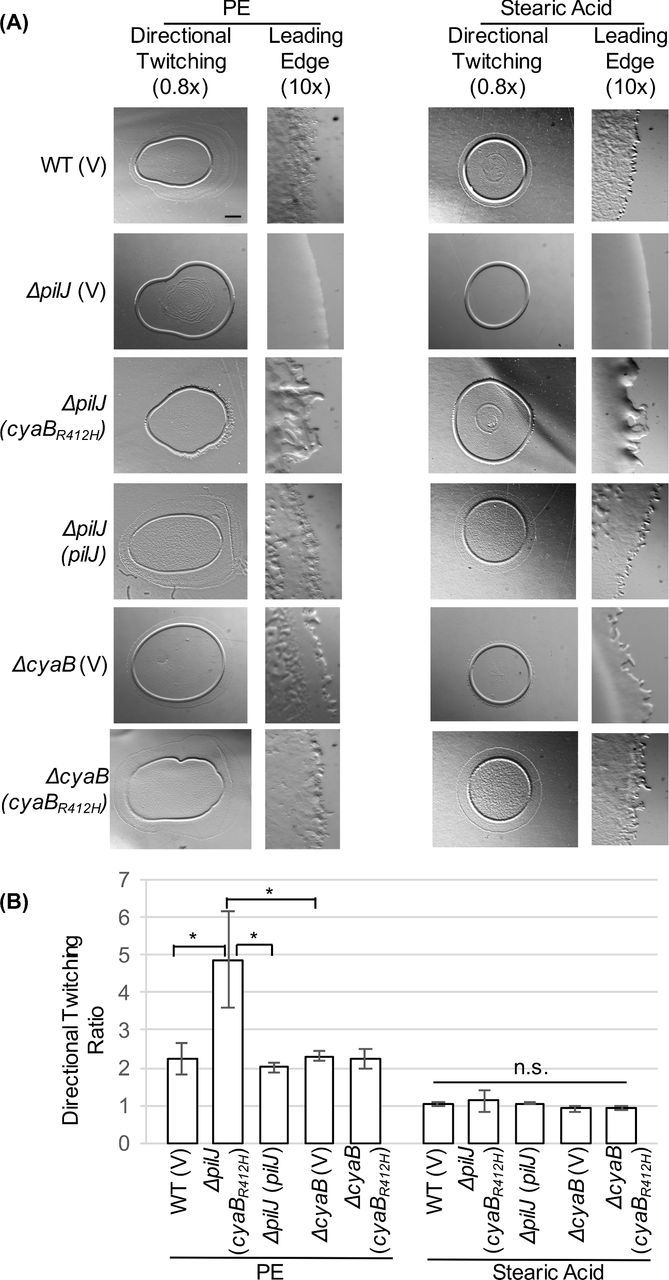
PilJ is not absolutely required for directional twitching to PE provided sufficient cAMP is present. (A) Representative images of directional twitching results. The PE or stearic acid was deposited on the plate to the right of where the P. aeruginosa culture was placed. Scale bar = 1 mm (0.8× magnification) (B) Directional twitching ratios were calculated by dividing the length of the leading edge by the length of the lagging edge. Three independent colonies were analyzed in triplicate. A ratio greater than 2 indicates directional twitching. Significantly different values were determined by ANOVA followed by TukeyHSD (*, P < 0.05).
There are a number of possible explanations for the retention of directional twitching in ΔpilJ (cyaBR412H). Earlier studies focused on the role of PE as a chemoattractant (Barker et al.2004; Miller et al.2008), and it is possible that PE functions as a chemoattractant independent of PilJ. This would suggest that there is another MCP in the Chp system. Previous studies however failed to identify an alternate MCP functioning in directional twitching (Bardy, Vasil and Maddock, unpublished). Although unlikely, it is possible that signal(s) enter the Chp system in an MCP independent manner. ChpA is an atypical histidine kinase with 6 putative histidine phosphotransfer sites, and putative serine/threonine phosphotransfer sites (Whitchurch et al.2004). This large number of phosphotransfer sites may allow for a high degree of regulation or multiple points of signal recognition. Alternatively, it is possible that the directional twitching seen with ΔpilJ (cyaBR412H) is because PE has other properties that result in increased movement. PE may function as a surfactant, similar to rhamnolipids (Glick et al.2010), thereby resulting in increased motility at higher concentrations. This is supported by early studies on directional twitching by P. aeruginosa, wherein uniform concentrations of PE enhanced swarm expansion (Kearns, Robinson and Shimkets 2001). It is also possible that PE triggers signal transduction altering cAMP levels, thereby increasing pilus biogenesis resulting in increased movement. Additional studies are required to further understand this phenotype.
In this study, we have begun to dissect the different domains of PilJ to understand the mechanisms of signal transduction. The periplasmic domain appears to be important for wild-type cAMP levels, and subsequently for twitching motility. This is in agreement with the mechanosensor model, where the periplasmic domain of PilJ was postulated to be important in signal transduction through interaction with PilA in response to surface contact (Persat et al.2015). We propose however that classical signal transduction is not the only mechanism regulating cAMP as pilJΔ74-273 had cAMP levels higher than those detected in ΔpilA indicating that other domains of PilJ are involved.
This is the first reported mutation within the Chp signal transduction system that allows the separation of cAMP levels from twitching motility. A number of recent studies have reported on the roles of second messengers in regulating surface behaviors, including twitching motility, swarming and biofilm formation (Fulcher et al.2010; Luo et al.2015). The ability to separate signal transduction regarding cAMP levels from signal transduction regulating twitching motility will allow us to tease apart the exact roles for each of these outputs in surface sensing and the resultant lifestyle changes.
Supplementary Material
Acknowledgments
We would like to thank D. Saffarini, G. Prasad and anonymous reviewers for helpful comments on the manuscript. We also thank C. Harwood and L. Burrows for the generous gifts of strains and reagents, M. Vasil for advice on the directed twitching motility assay, and H. Owen for assistance with DTA photography.
SUPPLEMENTARY DATA
FUNDING
This work was supported in part by the National Institute of Health ( R00-GM083147) to SLB. GTR was the recipient of an ASM Undergraduate Fellowship and a UWM SURF Undergraduate Research Award.
Conflict of interest. None declared.
REFERENCES
- Barker A, Vasil A, Filloux A, et al. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol Microbiol. 2004;53:1089–98. doi: 10.1111/j.1365-2958.2004.04189.x. [DOI] [PubMed] [Google Scholar]
- Beatson SA, Whitchurch CB, Sargent JL, et al. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3605–13. doi: 10.1128/JB.184.13.3605-3613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JJ, West JT, Engel JN. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J Bacteriol. 2010;192:994–1010. doi: 10.1128/JB.01390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–53. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Darzins A. The Pseudomonas aeruginosa pilK gene encodes a chemotactic methylatransferase (CheR) homologue that is translationally regulated. Mol Microbiol. 1995;15:703–17. doi: 10.1111/j.1365-2958.1995.tb02379.x. [DOI] [PubMed] [Google Scholar]
- DeLange P, Collins T, Pierce G, et al. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol. 2007;55:389–95. doi: 10.1007/s00284-007-9008-5. [DOI] [PubMed] [Google Scholar]
- Faguy DM, Bayley DP, Kostyukova AS, et al. Isolation and characterization of flagella and flagellin proteins from the thermoacidophilic archaea Thermoplasma volcanium and Sulfolobus shibatae. J Bacteriol. 1996;178:902–5. doi: 10.1128/jb.178.3.902-905.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Holliday P, Klem E, et al. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol. 2010;76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick R, Gilmour C, Tremblay J, et al. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol. 2010;192:2973–80. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G, Falke J, Parkinson J. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Nakamura T, Kuroda A, et al. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotech Bioch. 1999;63:155–61. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- Kearns D, Robinson J, Shimkets L. Pseudomonas aeruginosa exhibits directional twitching up phosphatidylethanolamine gradients. J Bacteriol. 2001;183:763–7. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer G. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–94. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhao K, Baker AE, et al. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviours. mBio. 2015;6:e02456–14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13:1666–81. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- Miller R, Tomaras A, Barker A, et al. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J Bacteriol. 2008;190:4038–49. doi: 10.1128/JB.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowbray S, Sandgren M. Chemotaxis receptors: a progress report on structure and function. J Struct Biol. 1998;124:257–75. doi: 10.1006/jsbi.1998.4043. [DOI] [PubMed] [Google Scholar]
- Parkinson J. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–22. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- Persat A, Inclan YF, Engel JN, et al. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. P Natl Acad Sci USA. 2015;112:7563–8. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham HT, Parkinson JS. Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J Bacteriol. 2011;193:6597–604. doi: 10.1128/JB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C, Pham X, Erwin A, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–64. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Topal H, Fulcher N, Bitterman J, et al. Crystal structure and regulation mechanisms of the CyaB adenyl cyclase from the human pathogen Pseudomonas aeruginosa. J Mol Biol. 2012;416:271–86. doi: 10.1016/j.jmb.2011.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Sample A, Runyen-Janecky L. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–42. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Hobbs M, Livingston S, et al. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Leech AJ, Young MD, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2004;52:873–93. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore ME, et al. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–63. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


