Abstract
Interdisciplinary research focused on biological membranes has revealed them as signaling and trafficking platforms for processes fundamental to life. Biomembranes harbor receptors, ion channels, lipid domains, lipid signals, and scaffolding complexes, which function to maintain cellular growth, metabolism, and homeostasis. Moreover, abnormalities in lipid metabolism attributed to genetic changes among other causes are often associated with diseases such as cancer, arthritis and diabetes. Thus, there is a need to comprehensively understand molecular events occurring within and on membranes as a means of grasping disease etiology and identifying viable targets for drug development. A rapidly expanding field in the last decade has centered on understanding membrane recruitment of peripheral proteins. This class of proteins reversibly interacts with specific lipids in a spatial and temporal fashion in crucial biological processes. Typically, recruitment of peripheral proteins to the different cellular sites is mediated by one or more modular lipid-binding domains through specific lipid recognition. Structural, computational, and experimental studies of these lipid-binding domains have demonstrated how they specifically recognize their cognate lipids and achieve subcellular localization. However, the mechanisms by which these modular domains and their host proteins are recruited to and interact with various cell membranes often vary drastically due to differences in lipid affinity, specificity, penetration as well as protein-protein and intramolecular interactions. As there is still a paucity of predictive data for peripheral protein function, these enzymes are often rigorously studied to characterize their lipid-dependent properties. This review summarizes recent progress in our understanding of how peripheral proteins are recruited to biomembranes and highlights avenues to exploit in drug development targeted at cellular membranes and/or lipid-binding proteins.
Keywords: drug targets, electrostatics, lipid binding, lipid metabolism, membrane penetration, peripheral proteins, phosphoinositides
1. INTRODUCTION
Biomembranes are a site for budding [1], tubulation [2–4], fission [5, 6], and fusion [5, 6], all essential for membrane trafficking and cell growth and differentiation. Interdisciplinary research during the last decade including: biophysics, bioinformatics, chemistry and more recently lipidomics [7–9] have increased the publicity of biomembranes. These fields of work have unveiled the expanding number of enzymes responsible for lipid metabolism and trafficking, which generate distinct lipid compositions of membrane organelles in a spatial and temporal manner. This allows peripheral proteins to transiently translocate to discrete sites in the cell to elicit their function. These reversible binding events are crucial to biological processes and can often be attributed to lipid-binding domains [10–13]. Although there are common mechanisms of lipid-dependent recruitment by these modular domains, the presence of specific lipid targets at distinct biological membranes [14, 15] coupled with variations in electrostatic and hydrophobic residues in and around the lipid-binding site leads to unique binding modes. These subtle differences in binding behaviors are a niche to be taken advantage of in drug design. To this end we review the general themes of lipid-binding domain behaviors at biomembranes and highlight differences within and among the classes as a strategy for designing agonist and/or antagonists of their cellular activity.
2. CELLULAR MEMBRANES
Peripheral proteins that target glycerophospholipids represent the majority of lipid binding discussed in this review. Glycerophospholipids are major structural components of eukaryotic membranes harboring a hydrophobic backbone of diacylglycerol (DAG). These include bulk lipids such as the zwitterionic phosphatdiylcholine (PC) and phosphatdiylethanolamine (PE) and the anionic lipid phosphatidylserine (PS) [16, 17]. The cylindrically shaped PC and conically shaped PE comprise ~ 60% of total lipid while PS makes up ~25% of the inner leaflet of the PM [18]. Thus, PS provides the inner leaflet of the PM with an anionic reservoir to target PS-binding proteins and also has been found in abundant concentrations on the cytoplasmic leaflet of endomembranes [19] where it can target proteins to those sites as well. Phosphatidylinositol is about 4% of cellular membrane phospholipid, and is the building block for the phosphorylated derivatives, the phosphoinositides (PIs). All together the PIs total ~ 1% [20, 21] of cellular lipids where some PIs levels are fairly stable (PI(3)P and PI(4,5)P2) and others are more dynamically regulated (PI(3,4)P2 and PI(3,4,5)P3) [21, 22]. Although recent evidence suggests there are constitutive pools of PI(3,4)P2 and PI(3,4,5)P3 in the PM [23] responsible for targeting their effectors. Nonetheless, these lipids play instrumental roles in recruiting peripheral proteins to different membranes due to differences in their spatial and temporal metabolism. The presence of one specific lipid (such as PI(3)P or PS) in a membrane organelle can be crucial to targeting the effector proteins where there is a complex array of electrostatic (both specific and nonspecific), hydrophobic, and H-bonding between the peripheral protein and the membrane [24]. Table 1 highlights the consensus cellular location of some of these lipids and includes proteins that recognize each target. Sphingolipids are a second class of structural lipids comprised of the hydrophobic backbone of ceramide [25] while sterols such as cholesterol make up non-polar components of cell membranes [26].
Table 1.
Cellular Location of Lipids and their Cognate Ligand
| Lipid | Primary Cellular Location | Lipid-Binding Domain | Drug Target Sites |
|---|---|---|---|
|
| |||
| PS | PM and endomembranes (cytosolic leaflet) | C2 | Ca2+-binding site |
| PS-binding site | |||
| interfacial penetration | |||
|
| |||
| PI(3)P | endosomes | FYVE, PX, PH (PEPP1) | PI(3)P coordination |
| nonspecific electrostatics | |||
| interfacial penetration | |||
|
| |||
| PI(4)P | Golgi | PX (Bem1p), PH (FAPP1), ENTH (EpsinR) | PI(4)P coordination |
| interfacial penetration | |||
|
| |||
| PI(3,4)P2 | PM | PH (Tapp1), PX (p47phox), ENTH (HIP1) | PI(3,4)P2 coordination |
| nonspecific electrostatics | |||
| interfacial penetration | |||
|
| |||
| PI(3,5)P2 | late endosomes | PH (centaurin β2), ENTH (Ent3p, Ent5p) | PI(3,5)P2 coordination |
| nonspecific electrostatics | |||
| interfacial penetration | |||
|
| |||
| PI(4,5)P2 | PM | PH, PX, ENTH, ANTH, C2 | PI(4,5)P2 coordination |
| nonspecific electrostatics | |||
| interfacial/hydrocarbon penetration | |||
|
| |||
| PI(3,4,5)P3 | PM | PH, PX | PI(3,4,5)P3 coordination |
| nonspecific electrostatics | |||
| interfacial penetration | |||
|
| |||
| C1P | Golgi | C2 (cPLA2α) | C1P site |
| Ca2+-binding site | |||
| Hydrocarbon penetration | |||
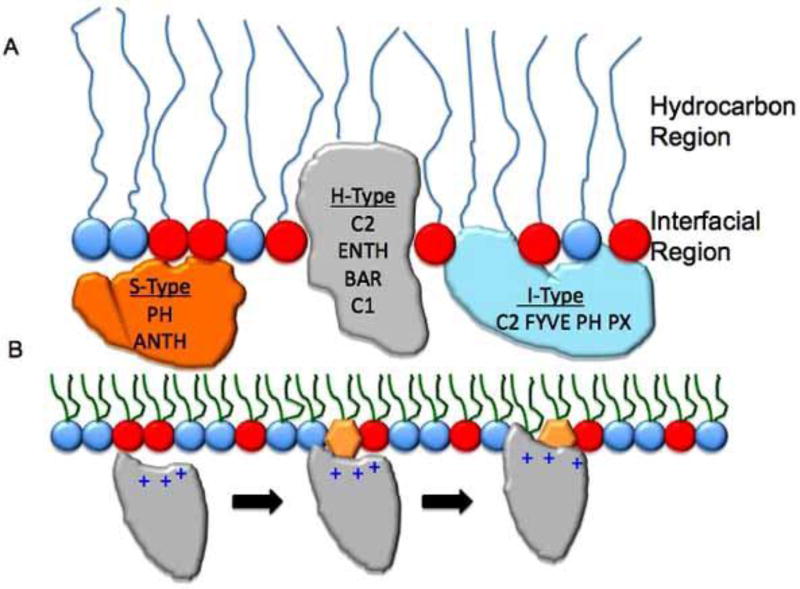
Our understanding of the structure and function of cell membranes has grown considerably since the fluid mosaic model was presented by Singer and Nicolson in 1972 [27]. The lipid bilayer has a highly polarized structure that consists of a central hydrocarbon core and two flanking interfacial regions [28]. The combined hydrocarbon region is ~ 30 Å and can vary in acyl chain length and saturation influencing imbedded peripheral proteins and transmembrane (TM) proteins [29]. The combined interfacial regions have comparable width and are heterogeneous in terms of chemical composition and polarity due to a complex mixture of water, the polar portion of the acyl chains, lipid backbone phosphate groups, and lipid headgroups. The complex natures of the lipid bilayer as well as peripheral protein structure are critical factors that govern the kinetics and energetics of membrane interactions. For instance, the polarity profile in the interfacial region allows proteins with different characteristics to experience a range of membrane interactions such as ion-ion, ion-dipole and van deer Waals forces [30]. Structural, experimental and computational data on lipid-binding proteins has lead to their arbitrary classification based upon their membrane location [10]. The S-type (surface) proteins are localized at the membrane surface (i.e., outside of the level of the backbone phosphate group) and interact pre- dominantly with the polar headgroups. For these proteins the predominant interaction between protein and lipid are ion- ion and ion-dipole interactions. I-type (interfacial) proteins interact with membranes through ion-dipole and H-bonding to penetrate into the level of the phosphate (i.e., interfacial region). H-type (hydrocarbon) proteins penetrate into the hydrocarbon core region of the lipid bilayer using aliphatic residues, which is predominated by van der Waals Forces and H-bonding. It is important to note both I- and H-type peripheral proteins can interact with both the polar head- groups and the interfacial region of the bilayer and often use long-range electrostatics or an electrostatic switch mechanism to drive their localization [10, 24, 31]. See Fig. (1A) for a depiction of proteins imbedded in a bilayer. This classification is a simple view of how peripheral proteins achieve localization to different regions of membranes, however, it can be useful as a first step in designing small molecule mimetics of lipid-binding proteins or lipid-binding inhibitors. These types of reagents would be useful to study and understand lipid binding and signaling or may serve as general inhibitors of binding for peripheral proteins localized to different regions of the membrane. In fact, this simplification has been employed to detect PS containing membranes using functional mimics of Annexin V [32, 33] and to target peptides to the interfacial region through ionic H-bonding [34].
Fig. 1. Lipid binding mechanisms of lipid-binding domains.
(A) A schematic of a lipid monolayer is shown to depict the relative position of lipid-binding domains in membranes. S-type proteins shown in orange do not significantly penetrate membranes and interact primarily through electrostatics with the anionic membranes. H-type proteins shown in gray are able to penetrate into the hydrocarbon region of the bilayer due to exposed aliphatic and aromatic residues. These proteins also can interact with the interfacial region through electrostatics and H-bonding. I-type proteins partially penetrate the membrane at the interfacial phosphate region. These types of proteins generally harbor aromatic and sometimes hydrophobic residues to achieve this binding mode. Taking all three modes into consideration including the types of chemical interactions that occur at each site can serve as a building block for small molecule discovery in the field of lipid binding. (B) A general scheme of lipid binding by a peripheral protein is depicted. In this instance a PI binding domain associates weakly with anionic membranes through nonspecific electrostatic interactions. Upon binding the membrane its dimensionality is reduced as it searches for its cognate PI. The PI coordination to the binding site is responsible for inducing an electrostatic switch (reduction in the desolvation penalty) followed by insertion of aliphatic residues adjacent to the PI binding site. The combination of PI-binding and hydrophobic insertion elongates the membrane residence time of the protein and in many cases is a mode of peripheral protein activation.
Stepping beyond the above classification, biomembrane structure is highly dynamic with both the lateral and transmembrane (TM) distributions of membrane components being heterogeneous. The number of different molecular compounds in biomembranes may number several thousand but essentially polar lipids and proteins are the two primary groups of components. That being said, the three primary intermolecular association partners are: protein-protein, protein-lipid, and lipid-lipid. Thus, a tactful approach to understanding and targeting these interactions can lead to a generation of molecules to promote or antagonize these interactions. The rapidly advancing fields of proteomics [35] and lipidomics [7–9] should help discover and hypothesize means of targeting interactions in and on membranes in a therapeutic fashion. With this in mind, the below sections should serve as summary of general mechanisms of lipid-binding as well as describe subtle differences in mechanisms that can be exploited in disrupting or inducing the lipid-binding.
3. LIPID BINDING PROTEINS
At least 11 lipid-binding domains have been identified to date, which participate in specific and nonspecific interactions with membranes that vary in properties such as affinity, membrane curvature recognition, and induction of membrane curvature changes [11]. These lipid-binding modules, often referred to as membrane-targeting domains, includes C1 [36, 37], C2 [38], PH [39], FYVE [40], PX [40, 41], ENTH [2, 42, 43], ANTH [43], BAR [2–4], FERM [44], PDZ [45], and tubby domains [46]. In addition, some peripheral proteins do not harbor lipid-binding domains but instead utilize a part of their molecular surface (e.g., secretory phospholipase A2s) while others have covalently attached lipid anchors (e.g., Src and Ras proteins) that embed in the lipid bilayer providing a driving force for localization. Table 1 summarizes some of the common properties of these lipid-binding domains including potential targeting regions for small molecules as discussed later.
All intracellular membranes contain a varying degree of anionic lipids and a majority of peripheral proteins contain cationic surfaces, at least locally. Thus, spatial and temporal signals are crucial to regulating targeting of peripheral proteins to the right membrane at the right time. Spatial regulation can be mediated by degree of membrane curvature and inherent differences in bulk lipid compositions among membrane organelles. Temporal regulation can occur through metabolism of PIs and DAG on various membranes and the second messenger Ca2+, which is able to induce the lipid binding of many proteins. Moreover, many proteins are regulated in both a spatial and temporal fashion. For instance, proper targeting may require a certain degree of membrane curvature harboring a temporally regulated lipid. Thus, lipid specificity, affinity, membrane penetration behavior, as well as curvature sensing abilities have important functional consequences with regards to cellular localization and biological activity. There are a number of cases where abrogation of these properties is attributed to disease [47, 48] and therapeutic intervention may be invaluable at the level of protein-lipid interactions.
3.1. Electrostatics and General Binding
In protein-protein interactions the initial formation of nonspecific collisional complexes, driven by diffusion and electrostatic forces, is followed by the formation of tightly bound complexes, which are stabilized by specific interactions [49, 50]. For the ligand binding of human growth hormone receptor, attractive electrostatic forces were shown to enhance the second-order rate constant for association (ka), whereas specific interactions that stabilize tightly bound complexes primarily lowered the dissociation rate constant (kd) [49]. Likewise, electrostatic components have been shown to be essential to protein lipid interactions [51, 52] including nonspecific electrostatic interactions that are crucial to proteins such as MARCKS, K-Ras, and Src [53–55]. This was evident as reducing the electrostatics through mutations was able to abrogate the cellular activity of Src [56]. For most peripheral proteins both specific and nonspecific electrostatic interactions increase the ka of lipid-binding as observed for the promotion of the protein-protein complexes and many peripheral proteins achieve the stable lipid-membrane complex through a combination of electrostatic, both specific and nonspecific, as well as hydrophobic interactions [10, 24]. In consonance with the protein-protein interactions, specific lipid binding and membrane penetration stabilize the complex by lowering the kd [10, 38].
A general schematic of lipid-protein interactions can be described taking into account the aforementioned synopsis of membranes and computational and experimental analysis that has demonstrated proteins bind to anionic membranes in a two-step mechanism. The initial formation of nonspecific collisional complexes, driven by diffusion and electrostatic forces, is followed by the formation of tightly bound complexes, which are stabilized by specific interactions and/or membrane penetration (Fig. (1B)). The long-range electrostatic interactions will increase the chance of protein/membrane association as well as influencing the protein orientation at or near the membrane surface. Once the protein nears the membrane interface, two short-range interactions occur: first, proteins and membranes begin to lose favorable interactions with the polar solvent (cytoplasm for instance). This is a repulsive force called desolvation [24]. Second, penetration of hydrophobic and aromatic residues into the membrane interface according to the interfacial hydrophobicity scale generated by Wimley and White [57] serve as promoters of membrane docking. Essentially, the initial membrane adsorption of peripheral proteins facilitates specific interactions with lipids by reducing the dimensionality of the space through which the protein interacts with its lipid ligand [58, 59]. A clear example of this has been shown for a number of PI-binding domains including: FYVE, PX, and ENTH domains. These domains have low affinity for membranes containing anionic lipids devoid of their cognate PI. However, upon appearance of the target ligand, the nonspecific electrostatic association is essential to forming the weakly bound complex that can undergo a two dimensional search for the PI. However, without PI bound these domains are unable to sufficiently penetrate the membrane due to a high penalty of desolvation [10]. The PI acts as an electrostatic switch when docking to the stereospecific site by reducing the desolvation penalty and inducing the membrane penetration.
The electrostatic switch induced by PIs has been preceded by other well-characterized mechanisms of change in electrostatic potential. For instance, protein phosphorylation can serve as an electrostatic switch. A well-known example is the dissociation of the MARCKS protein from the PM following phosphorylation of three serine residues [63]. This reduces its positive charge through an electrostatic switch mechanism [64]. Similarly, a calcium/electrostatic switch was proposed for the Ca2+ binding C2 domains [65] interactions with acidic phospholipids. Subsequently, computational and experimental investigations expanded the number of C2 domains following this mechanism [66]. These domains have a large negative potential (Ca2+-coordinating Asp residues) surrounding the membrane binding loops providing a large desolvation penalty in the Ca2+-free state. This precludes the insertion of aliphatic residues into the membrane until Ca2+ docks to the C2 domain increasing its positive charge and decreasing the desolvation penalty of insertion. The unique composition of each peripheral protein and/or lipid-binding domain as well as the target membrane provide diverse abilities for protein’s to bind membranes with different specificities, affinities, orientations and more. The different properties of peripheral proteins and membranes will be exploited in the following section as a means of understanding how different drug targets dock on lipid membranes and how understanding these properties can be used as a tool to design small molecules to interfere with or promote the lipid-dependent activation of peripheral proteins.
4. EXAMPLES OF LIPID-BINDING DOMAINS AS DRUG TARGETS
4.1. C1 Domain
The C1 domain (~50 amino acids) or DAG/phorbol ester binding domain, originally identified in protein kinase C (PKC) isozymes [67, 68] are present in multiple signaling families including kinases (PKC, protein kinase D (PKDs), diacylglycerol kinases (DGKs), c-Raf, Kinase Suppressor of Ras (KSR)) and non-kinases (Chimaerins, RasGRPs, RacGAPs, Vav, and Munc). The X-ray crystal structure of the PKC8 C1B domain-phorbol 13-acetate complex revealed how the C1 domain achieves the ligand selectivity with a polar binding pocket for DAG/phorbol ester where main chain peptide groups in the pocket form hydrogen bonds with polar moieties of the phorbol ester [69]. These domains are well conserved containing six Cys- and two His-residues that tightly bind two zinc ions for structural stability. Originally, it was thought DAG bound in the same mode as phorbol ester, however, minor variations in sequence homology are responsible for dramatic affects in affinity for DAG and phorbol ester including abrogation of binding to one ligand and nM affinity for the other [70]. The structural basis of differential DAG and phorbol ester affinities of these C1 domains is not fully understood partially due to lack of structural information on DAG coordination, which may stem from the difficulty associated with purifying these small hydrophobic domains. In addition to binding to DAG/phorbol ester, the C1B domains of PKCδ and PKCε have been also reported to interact with ceramide and arachidonic acid [71, 72], but their binding sites have not been located. Also, the PKC C1 domains have been shown to bind alcohols with high stereospecificity [73] via a non-DAG-binding site [74]. Each of these three sites may serve as a viable site for drug development targeted at C1 domain containing proteins. In fact, a number of compounds have been developed for the DAG site so we refer you to the article by Peter Blumberg and colleagues who do an outstanding job of reviewing these molecules in this issue.
C1 domains play a crucial role in targeting PKC and other molecules from the cytosol to membranes (translocation) in a DAG or phorbol ester dependent fashion [75]. Studies on PKC have shown C1-lipid interactions activated by DAG and also the binding of phorbol ester and related ligands to PKC to be dependent on the presence of negatively charged phospholipids (phosphatidylserine) [76]. Based on this observation it is believed that ligand binding in C1 domains is highly co-operative with respect to phosphatidylserine in typical C1 domain containing proteins. NMR, biochemical and mutagenesis studies suggest a two step membrane-binding mechanism where positively charge residues (non-specific electrostatic interactions) near the DAG binding pocket position the C1 domain near the anionic phospholipids and in the second step, the hydrophobic residues in rim of ligand binding pocket facilitates the membrane penetration. Unlike the high penalty of desolvation associated with insertion of hydrophobic residues for many PI binding domains and the electrostatic switch induced by Ca2+ for many C2 domains, the C1 domain has a low penalty of desolvation and can readily insert in the membrane in a DAG-independent fashion (See Fig. (2A)) [77, 78]. Since hydrophobic residues surrounding the DAG binding pocket are exposed, isolated C1 domains typically have a high tendency to aggregate in solution. Thus, the C1 domain in the full-length protein is expected to be buried in the inactive form of the enzyme and becomes accessible to DAG or phorbol esters only after an inter-domain conformational change [78–80]; in the case of PKCα, C1 domains are exposed upon Ca2+-dependent, C2 domain-mediated membrane binding of the proteins [78, 81].
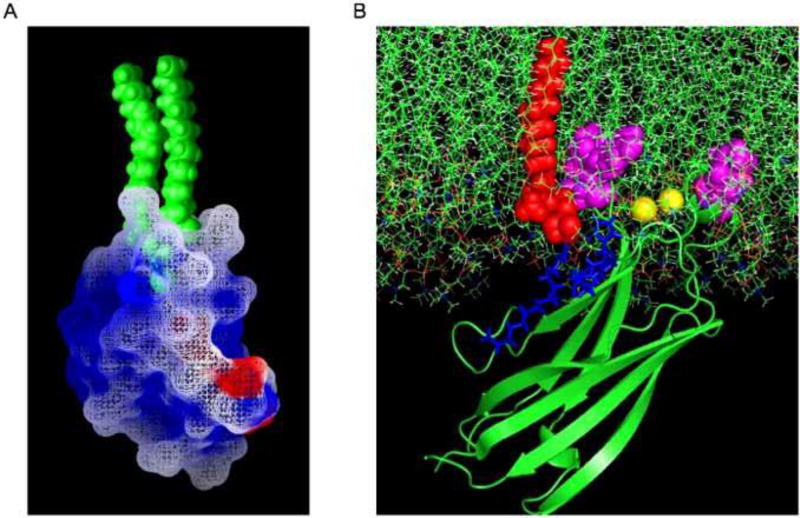
Fig. 2. Lipid binding modes of C1 and C2 domains.
(A) A C1 domain is shown coordinated to DAG. Positively charged residues are shown in blue, negatively charged residues in red, and all other residues in gray. Note the tip of C1 domains harbor hydrophobic and aromatic residues just above the DAG binding pocket that penetrate into the membrane. Nonspecific electrostatic interactions are achieved through the ring of electrostatic residues that line the domain just below the penetrating region. It is surmised these residues are located at the membrane surface and interfacial region upon DAG coordination and may participate in elongating the membrane residence time along with DAG binding. A single Asp shown in red is important for intramolecular interactions within some PKC isoforms. This interaction keeps the C1 domain in a closed state prior to the translocation signal. We would like to thank Diana Murray for help in preparing this figure. (B) The C2 domain of cPLA2α is shown docked to a membrane of PC and C1P. PC molecules are shown in green and C1P in red. This C2 domain penetrates effectively into PC bilayers upon Ca2+-coordination as hydrophobic and aromatic residues shown in magenta are responsible for the deep penetration. C1P binds to an electrostatic cluster shown in blue with high specificity and increases the membrane residence time of the domain on the membrane surface. The C1P interaction is also able to act as an electrostatic switch in a similar manner to Ca2+ promoting the membrane docking of the domain. Lipid binding can be disrupted by inhibiting Ca2+ binding or hydrophobic penetration. Since the C2 domain can translocate to internal membranes in the absence of C1P, disruption of the C1P site may have special therapeutic potential in inflammatory states where C1P is produced at high levels or under conditions where C1P is overproduced and Ca2+ levels are low. We would like to thank Hui Lu and Nitin Bhardwaj for help in preparing this figure.
The surface plasmon resonance (SPR) measurements indicate that DAG binding increases the vesicle affinity (Ka) of the PKC C1 domains by more than two orders of magnitude mainly by reducing the kd [70, 80]. Membrane penetration studies of PKC C1 domains indicated that membrane penetration of the C1 domain is necessary for DAG binding [77, 78], since the glycerol moiety of DAG is expected to be located deep within the interfacial region. The depth of membrane penetration by the C1 domain has not been quantitatively measured yet; however, from the NMR spectra of the PKCψ C1B domain interacting with lipid micelles [82] it is estimated to be at least 10 Å below the level of the lipid phosphate. With this in mind, there are a number of avenues to targeting C1 domains besides the ligand-binding pocket. First, we can think of the C1 domain as participating in the following interactions: ion-ion, ion-dipole, van der Waals Forces, and H-bonding with specific ligand. Molecules targeted toward disrupting the electrostatic association, hydrophobic penetration or even the release of the C1 domain from the full-length protein molecular tether may serve as viable options in disrupting C1 lipid binding. C1 domains may need to be studied on a case-by-case basis to assess ligand selectivity, depth of membrane penetration, orientation at the membrane, intramolecular interactions, alcohol binding, ceramide binding, or arachidonic acid binding. These variations in C1 properties may serve as avenues to selectively target C1 domains in solution or in membranes and may help achieve better selectivity for compounds targeting the DAG binding pocket. As a perspective, Table 2 outlines a number of C1 domains including their function and role they play in disease.
Table 2.
Summary of C1 Domain Containing Proteins, their Cellular Function and Relative Diseases
| Family/Protein | # of C1 Domains | Protein Function | Cellular Roles/ Functions | Relative Diseases |
|---|---|---|---|---|
|
| ||||
| PKCα, βI, βII, γ, δ, ε, η,θ | 2 | Protein kinase | Proliferation, differentiation, migration, neuronal signaling, impaired humoral response and cellular B cell response, ion channel conductance, smooth muscle contraction, transmitter/hormone exocytosis and protein secretion | cancer, cardiomyopathy, cardiac and lung disease, cerebral ischemic and reperfusion injury, delay in wound healing, diabetes, kidney disease, |
| PKCζ | 1 | |||
| PKCι/λ | 1 | |||
|
| ||||
| PKD 1, 2, 3 | 2 | Protein kinase | Cell proliferation, Protein transport from trans-Golgi network to cell surface, cancer cell invasion of tissues | polycystic kidney disease, prostate cancer |
|
| ||||
| DGKα, β, γ, δ, ε, η, ζ, ι | 2 | Lipid kinase | Involved in IL-2 production in T-lymphocytes, involved in the cellular signal transduction, phosphatidic acid turnover | cancer, retinal degeneration, epilepsy, hypertension, autoimmune disease |
| DGKθ | 3 | |||
|
| ||||
| RasGRP-1, -2, -3, -4 | 1 | Ras-GEF | Transformation in fibroblasts and activation of ERK pathway, thymocyte differentiation and TCR signaling, Control of proliferation and transformation in fibroblasts, Control of proliferation, adhesion and transformation in myeloblasts, Neuronal differentiation of PC12 cells | Leukemia |
|
| ||||
| KSR-1, -2 | 1 | Scaffold | Positive regulator of Ras signaling | inflammatory bowel disease, pancreatic cancer |
4.2. C2 Domain
The C2 domain (~130 residues) like the C1 domain was first identified as one of two conserved regulatory domains in PKC [67, 68]. Identification of the C2 domain in other proteins, such as synaptotagmins and group IVA cytosolic phospholipase A2 (cPLA2α), which also bind membranes in a Ca2+-dependent manner, led to the postulation that the C2 domain is involved in Ca2+-dependent membrane binding. A large number of proteins containing the C2 domain have been identified since, and most of them are involved in signal transduction or membrane trafficking. The C2 domain represents the second most abundant lipid-binding domain behind the PH domain with at least 200 examples identified in the Pfam database. While most C2 domain proteins are peripheral and bind reversibly to membranes some C2 parent proteins are actually TM proteins involved in membrane trafficking. However, these C2 domains can also bind reversibly in a Ca2+-dependent fashion [83]. Recently, a number of TM proteins with multiple C2 domains have been identified but the roles of these C2 domains in the protein’s functions are still unknown [83].
The lipid affinity as well as the Ca2+ affinity of the Ca2+-binding C2 domains can vary greatly and some C2 domains are even involved in protein-protein interactions or may play a structural role. Ca2+ can play three different roles in membrane binding of the C2 domain: an electrostatic switch, a Ca2+-bridge, or inducing of intra- or interdomain conformational changes, which in turn trigger lipid binding. On the basis of the diverse Ca2+ affinity of C2 domains, peripheral proteins may be activated in a temporal manner depending upon the extent of the Ca2+ oscillations and their intensities. In other words, does their membrane association and dissociation distinctly follow the cellular Ca2+ oscillations under physiological conditions? While it’s been shown enzymes such as PKC follow the Ca2+ spikes [84], the disparate targeting of C2 domains under the same cellular conditions are yet to be rigorously measured. A significant number of C2 domains with little to no Ca2+ affinity have also been identified. Some of these Ca2+-independent C2 domains have been reported to bind the membrane or other proteins but most of them have not yet been characterized. The general mechanism of how a number of C2 domains associate with membranes in a Ca2+-dependent fashion is well established [85, 86]. However, physiological functions and Ca2+- and membrane-binding properties of a large portion of C2 domains still remain unknown. Structural studies have shown that C2 domains have a common fold of conserved eight-stranded antiparallel β-sandwich connected by surface loops [87, 88]. The disparity in C2 domain targeting arises in the surface loops, which are variable in amino acid sequence and conformation and most often involved in lipid binding. Also of functional consequences is a cationic patch in the concave face of the β-sandwich termed the cationic β-groove, which varies in size and electrostatics among C2 domain [38]. In support of this observation, cationic β–grooves have been shown to bind PIs including PI(4,5)P2 as well as ceramide-1-phosphate (C1P) [89, 90]. Thus, many C2 domains are able to coordinate multiple lipids in both a Ca2+-dependent or independent fashion. The preliminary data available on the multiple lipid recognition mode of C2 domains suggests C2 domains may be multiply regulated by different lipids and Ca2+ signals and in some cases may require coincidence detection (e.g., interaction with multiple lipid targets) to achieve high affinity and cellular localization.
Most lipid binding domains achieve lipid specificity through a specific lipid-binding site formed within a pocket (e.g., C1, PH, PX, and FYVE domains) or with juxtaposed surface cationic residues (e.g., ANTH domain). C2 domains are unique among lipid-binding domains in that they have neither a well-defined lipid-binding pocket nor a conserved cationic patch. The aforementioned sequence variation in the surface loops as well as the cationic β-groove leads to highly variable and relatively low lipid selectivity for some C2 domains and high affinity and specificity for others. Moreover, C2 domains can show different lipid selectivity as a function of Ca2+ concentration because the relative contribution of two lipid binding sites can vary as the β-groove ligand may lower the Ca2+ requirement for lipid docking acting as an electrostatic switch in a similar mode to Ca2+ [Bhardwaj, N., Vora, M., Chalfant, C.E., Lu, H. and Stahelin, R.V., unpublished data]. A majority of Ca2+-dependent C2 domains harbor cationic residues in the Ca2+-binding loops and bind anionic lipids significantly better than zwitterionic ones. Some of these C2 domains bind anionic phospholipids through non-specific electrostatic interactions while others can stereospecifically recognize a lipid headgroup such as PS. For PS coordination, the C2 domains of PKCα [86, 91] and phospholipase C31 (PLC31) [92] harbor residues in the Ca2+-binding loops that can specifically recognize the serine headgroup of PS. C2 domains with aromatic and aliphatic residues in the Ca2+-binding loops are able to associate strongly with zwitterionic lipids such as phosphatidylcholine (PC) where they achieve deep hydrocarbon penetration following Ca2+ coordination [86, 93]. The cationic β-groove was first shown to interact with inositolpolyphosphates for the C2B domains of synaptotagmin II and IV [94, 95]. Thereafter, the C2B domains of synaptotagmin I [96] and II [97] were shown to bind bis- and tris-phosphoinositides in a Ca2+-independent fashion. These first examples of cationic β-grooves do not have high lipid specificity but play a key role in the vesicle fusion activity of host proteins. The role of the cationic β-grooves have been best characterized for PKCα [90] and cPLA2α [89]. PKCα has been reported to bind PIs including PtdIns(4,5)P2 with highest affinity [90]. This binding is attributed to four lysine residues in the β-groove, which at first glance don’t seem to be enough to select PtdIns(4,5)P2 over PI(3,4)P2, PI(3,4,5)P3 or other PIs. In fact, we performed experimental and computational investigation of the PKCα-C2 under various conditions and found that PS and Ca2+ are prerequisite for the PI binding, which augments the PS binding by increasing the membrane residence time [Manna, D., Bhardwaj, N., Vora, M., Stahelin, R.V., Lu, H. and Cho, W., J. Biol. Chem. 2008]. cPLA2α displays clear selectivity in its β-groove coordination of the sphingolipid C1P [89]. C1P is not only able to augment PC binding by increasing membrane residence time but it can increase affinity for membranes at lower Ca2+ levels as C1P acts as an electrostatic compensator for Ca2+ (See Fig. (2B)) [Bhardwaj, N., Vora, M., Chalfant, C.E., Lu, H. and Stahelin, R.V., submitted]. These examples of β-groove binding by C2 domains should not only serve to understand the role of all β-grooves in C2 domains but may also be a site of therapeutic intervention when the β-groove is required for coincidence detection or cellular activity. The disparate roles of this site in C2 domains may also serve as a better target than the more conserved Ca2+-binding sites and surface loop regions. Table 3 serves as insight into a number of C2 domain containing proteins with known cellular functions and importance in disease.
Table 3.
Summary of some C2 Domain Containing Proteins with Known Function in Disease
| Family/Protein | # of C2 Domains |
Protein Function | Cellular Roles/Functions | Relative Diseases |
|---|---|---|---|---|
| cPLA2-α, β, δ | 1 | Phospholipase | cell signaling processes regulating the inflammatory response | Alzheimer’s disease, epilepsy, multiple sclerosis, ischemia, inflammatory disease, cancer and neural trauma, psoriasis |
| 5-, 8R-, and 15-Lipoxygenase | 1 | Dioxygenation of fatty acids | metabolism of prostaglandins and leukotrienes, cancer cell proliferation and survival | cancer, asthma |
| Otoferlin | 1 | Membrane fusion protein | Exocytosis | deafness |
| Perforin | 1 | pore-forming protein | Role in CD8 T-cell and NK cell function, immune response | neurological disease |
| Phosphatidylinositol 3-kinase | 1 | Protein & Lipid Kinase | involved in different signaling pathways and controlling of key functions of the cell, growth and survival, aging, and malignant transformation. | cancer, Alzheimer’s disease |
| PLC-β-I, β-II, β-III, β-IV, γ́ -I, γ-II, δ-I, δ-II, δ -III, δ-IV | 1 | Phospholipase | signal transduction processes: proliferation, differentiation, apoptosis, cytoskeleton remodeling, vesicular trafficking, ion channel conductance, endocrine function, and neuro-transmission. | cancer, apoptosis, hormonal diseases, Alzheimer’s disease, creutzfeldt-jakob disease |
| PKCα, βI, βII, γ, δ, ε, η, θ | 1 | Protein kinase | Proliferation, differentiation, migration, neuronal signaling, impaired humoral response and cellular B cell response, ion channel conductance, transmitter receptor function, smooth muscle contraction, transmitter/hormone exocytosis and protein secretion | apoptotic resistance, cancer, cardiomyopathy, cardiac and lung disease, cerebral ischemic and reperfusion injury, delay in wound healing, diabetes, kidney disease, neurode-generative disease, learning and memory |
| PTEN | 1 | Phosphatase | cell migration and adhesion, apoptosis | brain, breast, and prostate cancer, Cowden syndrome, Bannayan-Riley-Ruvalcaba syndrome, Proteus syndrome, and Proteus-like syndrome |
| Nedd4 | 1 | Ubiquitin Ligases | endocytosis, degradation of membrane proteins, control of cell growth and viral budding. | Hypertension, cancer, and defects in the immune system |
| Synaptotagmin I, II, III, IV, V, VI, VII, VIII, IX, X | 2 | Membrane Fusion | Synaptic vesicle docking, fusion, and exocytosis | Juvenile Parkinson disease and Alzheimer’s disease |
| Tollip | 1 | toll interacting protein | signaling | Huntington disease |
4.3. PH Domain
The PH domain is composed of ~ 100 amino acids and is one of the most common domains in the human proteome and the most abundance lipid-binding domain with >225 examples identified [39]. While it is the most abundant lipid-binding domain in sheer numbers only about 10% of PH domains studied bind PIs with high to moderate affinity. The other 90% bind with low affinity, bind in conjunction with other ligands, such as G-proteins, or their function is simply unknown. Lemmon and Ferguson have proposed that some of these PH domains, such as dynamins, increase their phosphoinositide affinity through oligomerization in host proteins [98, 99]. PH domains can be classified as either S-or I-type proteins as they lack a critical number of hydrophobic residues to achieve deep membrane penetration. Unlike other H- and I-type phosphoinositide-binding domains, such as FYVE, PX and ENTH domains, PH domains do not typically harbor as many aliphatic or aromatic residues around the PI-binding site. The membrane binding of PH domains then is initially driven by non-specific electrostatic interactions, which is followed by specific PI binding [100] to increase the membrane residence time. A few recent reports have demonstrated some PH domains anchor to the membrane through aliphatic residues around the PI-binding site [101–104]. Moreover, a subset of PH domains may be targeted to the membrane under acidic conditions due to the protonation of a key His residue in the binding crevice [104]. Taken together, these data underscore the distinct mechanisms targeting PH domains to membranes.
Of the seven PIs in cells, the PH domain binds specifically to PI(3,4,5)P3, PI(4,5)P2, or PI(3,4)P2 [39]. Reports of binding PI(3)P [105] and PI(4)P [106] are well documented for PH domains but the origin and reliability of this specificity is still controversial. For most of the highly specific PH domains, the vicinal positioning of two phosphate groups is thought to be crucial in the affinity of the PH domain for their cognate ligand as the PH domain does not bind to PIs lacking vicinal phosphates. Affinity of the PH domain for its specific PI is mediated mainly through interactions with conserved basic residues in the conserved β-loops of the domain. The majority of PH domains have a conserved basic motif (K-Xn-(K/R)-X-R) in which the basic lysines and arginines play an important role forming hydrogen bonds with the head group of the PI. Other basic residues, located within the domain or closely surrounding it vary from protein to protein, provide a stronger binding affinity and create a unique binding pocket. Some PH domains are mediated by the PI interaction and the binding of a G-protein to a second site in the PH domain [107]. The twofold binding of these ligands is necessary for a strong interaction, as without the G-protein, the PH domain interacts weakly with membrane. Finally, two distinct members of the PH domain family (TIAM1 and ARHGAP9) [108] were recently discovered as they bind membranes through a site on the opposite side of the β1–β2 loop suggesting there are still novel PH domains to be discovered within the genome. A number of PH domain containing proteins are important drug targets. For instance, mutations in the BTK PH domains that abrogate P(3,4,5)P3 binding cause a disease characterized by inability to produce mature B lymphocytes [47] while mutations in the PH domain of PKB that induce constitutive membrane docking cause cancer [48]. Molecules geared toward promoting or inhibiting PH domain activity could provide therapeutic value in these diseases. Of course, careful consideration to the reversibility (on/off) of the binding must be taken into consideration where understanding lipid specificity, affinity, membrane penetration and coincidence detection (PH-protein and PH-lipid) are of utmost importance.
5. SUMMARY AND CONCLUSIONS
Lipid-protein interactions play important roles in the regulation of many cellular processes, including cell signaling and membrane trafficking [10]. When these interactions are abolished or promoted in a constitutive manner a number of life-threatening diseases can occur. Therapeutics aimed at the protein-lipid interface could serve as an invaluable tool for achieving high selectivity among targets achieving efficacy in abolishing the lipid-dependent activation of the drug target. However, designing small molecule inhibitors of lipid-binding proteins is a challenging task, in part because the driving forces of protein-lipid association are a composite of many chemical interactions. Electrostatic forces, cation-n interactions, van der Waals forces, and hydrogen bonding all play a role in maintaining proper membrane function and in the association of proteins with biological membranes. We still have much to learn about the infusion of these forces into the processes of molecular design that will ultimately produce the compounds necessary to control biological events at the cell membrane. However, progress in our understanding of the membrane binding mechanisms of peripheral proteins has been substantial over the past decade thanks to rapid progress in bioinformatics, computational biology, in vitro biophysical studies, structural biology, and microscopic cell imaging. Furthermore, the advent of proteomics [35] and lipidomics [7–9] has greatly increased our awareness of the large number of molecular targets in a cell membrane that have potential therapeutic value and should only increase our knowledge of targeting events in and on membranes. Thus, the future research on lipid-protein interactions will entail only more interdisciplinary comprehensive studies of both computational predictions and validations of these forecasts. The advent of better high throughput small molecule discovery and computational design should hasten delivery of the first generation of therapeutics. Recently, a proof of concept in silico structure-based virtual ligand screening assay identified compounds that could disrupt the lipid binding of a C2 domain [109]. The simplified view of the lipid-protein interactions within biological membranes presented here provides the basis of small molecule discovery and design to target different interactions at the three main regions of the biological membrane. Combining these concepts with ligand screening should provide the basis of selectivity among and within the lipid-binding domains.
Acknowledgments
We would like to thank the following members of the Stahelin lab for helpful discussions: Serene Samyesudhas, Mohsin Vora, Sean Cullen, Keaton Jones, Elizabeth Seilie, Bradley Wisler, David Nemer, Lauren Buck, and Sophia Cortez. We would also like to thank our collaborators and friends for critical discussions: Wonhwa Cho, Diana Murray, Tatiana Kutateladze, Charles Chalfant, Hui Lu, and Bradley Smith. Research was partially supported by the American Heart Association (#0735350N), American Cancer Society (#IRG-84-002-22), and two Indiana University School of Medicine Biomedical Research Grants (to R.V.S). C.G.S. acknowledges postdoctoral support from the Walther Center for Cancer Research.
ABBREVIATIONS
- C1P
Ceramide-1-phosphate
- cPLA2
Cytosolic phospholipase A2
- DAG
sn-1,2-Diacylglycerol
- PA
Phosphatidic acid
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PI
Phosphoinositide
- PI(3)P
Phosphatidylinositol-3-phosphate
- PI(4)P
Phosphatidylinositol-4-phosphate
- PI(3,4)P2
Phosphatidylinositol-3,4-bisphosphate
- PI(3,4,5)P3
Phosphatidylinositol-3,4,5-trisphosphate
- PI(4,5)P2
Phosphatidylinositol-4,5-bisphosphate
- PKC
Protein kinase C
- PLCo1
Phospholipase Co1
- PM
Plasma membrane
- PS
Phosphatdiylserine
- PTEN
Phosphatase and tensin homolog
- SPR
Surface plasmon resonance
References
- 1.Kaksonen M. J. Cell Biol. 2008;180:1059–1060. doi: 10.1083/jcb.200802174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh T, De Camilli P. Biochim. Biophys. Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Dawson JC, Legg JA, Machesky LM. Trends Cell Biol. 2006;10:493–498. doi: 10.1016/j.tcb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Futterer K, Machesky LM. Cell. 2007;129:655–657. doi: 10.1016/j.cell.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Vassilieva EV, Nusrat A. Methods Mol. Biol. 2008;440:3–14. doi: 10.1007/978-1-59745-178-9_1. [DOI] [PubMed] [Google Scholar]
- 6.Brose N. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00758.x. in press. [DOI] [PubMed] [Google Scholar]
- 7.van Meer G. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meer G, Leeflang BR, Liebisch G, Schmitz G, Goni FM. Methods Enzymol. 2007;432:213–232. doi: 10.1016/S0076-6879(07)32009-0. [DOI] [PubMed] [Google Scholar]
- 9.Torkhovskaya TI, Khalilov EM, Korotaeva AA. Bull. Exp. Biol. Med. 2007;144:408–412. doi: 10.1007/s10517-007-0342-1. [DOI] [PubMed] [Google Scholar]
- 10.Cho W, Stahelin RV. Annu. Rev. Biophys. Biomed. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 11.Lemmon MA. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 12.Teruel MN, Meyer T. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 13.Hurley JH. Biochim. Biophys Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Meer G. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth MG. Physiol. Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 16.Buckland AG, Wilton DC. Biochim. Biophys. Acta. 2000;1483:199–216. doi: 10.1016/s1388-1981(99)00188-2. [DOI] [PubMed] [Google Scholar]
- 17.Feig M, Brooks CL., III Curr. Opin. Struct. Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Zachowski A. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Science. 2007;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon MA. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 21.Rusten TE, Stenmark H. Nat. Methods. 2006;3:251–258. doi: 10.1038/nmeth867. [DOI] [PubMed] [Google Scholar]
- 22.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 23.Weo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulgrew-Nesbitt A, Diraviyam K, Wang J, Singh S, Murray P, Li Z, Rogers L, Mirkovic N, Murray D. Biochim. Biophys. Acta. 2006;1761:812–826. doi: 10.1016/j.bbalip.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Hannun YA, Obeid LM. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 26.Ikonen E. Nat. Rev. Mol. Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 27.Singer SJ, Nicolson GL. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 28.Wiener MC, White SH. Biophys. J. 1992;61:434–437. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Proc. Natl. Acad. Sci. USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanshaw RG, Stahelin RV, Smith BD. Chemistry. 2008;14:1690–1697. doi: 10.1002/chem.200701589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahelin RV, Cho W. Biochemistry. 2001;40:4672–4680. doi: 10.1021/bi0020325. [DOI] [PubMed] [Google Scholar]
- 32.Hanshaw RG, Smith BD. Bioorg. Med. Chem. 2005;13:5035–5042. doi: 10.1016/j.bmc.2005.04.071. [DOI] [PubMed] [Google Scholar]
- 33.Hanshaw RG, Lakshmi C, Lambert TN, Smith BD. ChemBioChem. 2005;6:2214–2220. doi: 10.1002/cbic.200500149. [DOI] [PubMed] [Google Scholar]
- 34.Kooijman EE, Tielman DP, Testerink T, Munnik DTS, Rijkers KNJ, Burger B, de Krujiff B. J. Biol. Chem. 2007;282:11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]
- 35.Suter B, Kittanakom S, Stagljar I. Biotechniques. 2008;44:681–691. doi: 10.2144/000112799. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Fernandez JC, Corbalan-Garcia S. Chem. Phys. Lipids. 2007;148:1–25. doi: 10.1016/j.chemphyslip.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Colon-Gonzalez F, Kazanietz MG. Biochim. Biophys. Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Cho W, Stahelin RV. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Lemmon MA. Biochem. Soc. Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutateladze TG. Prog. Lipid Res. 2007;46:315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seet LF, Hong W. Biochim. Biophys. Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Int. J. Biochem. Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Legendre-Guillemin V, Wasiak S, Hussain NK, Anger A, McPherson SP. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 44.Diakowski W, Grzybek M, Sikorski AF. Folia Histochemi Cytobiol. 2006;44:231–248. [PubMed] [Google Scholar]
- 45.Wu H, Feng W, Chen J, Ling-Nga C, Huang S, Zhang M. Mol. Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 46.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 47.Lindvall JM, Blomberg KE, Valiaho J, Vargas L, Heinonen JE, Berglof A, Mohamed AJ, Nore BF, Vihinen M, Smith CI. Immunol. Rev. 2005;203:200–215. doi: 10.1111/j.0105-2896.2005.00225.x. [DOI] [PubMed] [Google Scholar]
- 48.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 49.Cunningham BC, Wells JA. J. Mol. Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 50.Northrup SH, Erickson HP. Proc. Natl. Acad. Sci. USA. 1992;89:3338–3342. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurley JH, Misra S. Annu. Rev. Biophys. Biomol. Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaughlin S, Murray D. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 53.Abruzova A, Murray D, McLaughlin S. Biochim. Biophys. Acta. 1998;1376:369–379. doi: 10.1016/s0304-4157(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 54.Buser CA, Sigal CT, Resh MD, McLaughlin S. Biochemistry. 1994;33:13093–13101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- 55.Hancock JF, Paterson H, Marshall CJ. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 56.Sigal CT, Zhou W, Buser CA, McLaughlin S, Resh MD. Proc. Natl. Acad. Sci. USA. 1994;91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wimley WC, White SH. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 58.McCloskey MA, Poo MM. J. Cell Biol. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kholodenko BN, Hoek JB, Westerhoff HV. Trends Cell Biol. 2000;10:173–178. doi: 10.1016/s0962-8924(00)01741-4. [DOI] [PubMed] [Google Scholar]
- 60.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 61.Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. J. Biol. Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 62.Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W. J. Biol. Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 63.Seykora JT, Myat MM, Allen LA, Ravetch JV, Aderem A. J. Biol. Chem. 1996;271:18797–18802. doi: 10.1074/jbc.271.31.18797. [DOI] [PubMed] [Google Scholar]
- 64.Kim J, Blackshear JD, Johnson S, McLaughlin S. Biophys. J. 1994;67:227–237. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rizo J, Sudhof TC. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 66.Murray D, Honig B. Mol. Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 67.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Proc. Natl. Acad. Sci. USA. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osada S, Mizuno K, Saido T, Akita Y, Suzuki K, Kuroki T, Ohno S. J. Biol. Chem. 1990;265:22434–22440. [PubMed] [Google Scholar]
- 69.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 70.Ananthanarayanan B, Stahelin RV, Digman MA, Cho W. J. Biol. Chem. 2003;278:46886–46894. doi: 10.1074/jbc.M307853200. [DOI] [PubMed] [Google Scholar]
- 71.Schultz A, Ling M, Larsson C. J. Biol. Chem. 2004;279:31750–31760. doi: 10.1074/jbc.M313017200. [DOI] [PubMed] [Google Scholar]
- 72.Kashiwagi K, Shirai Y, Kuriyama M, Sakai N, Saito N. J. Biol. Chem. 2002;277:18037–18045. doi: 10.1074/jbc.M111761200. [DOI] [PubMed] [Google Scholar]
- 73.Slater SJ, Cook AC, Seiz JL, Malinowski SA, Stagliano BA, Stubbs CD. Biochemistry. 2003;42:12105–12114. doi: 10.1021/bi034860e. [DOI] [PubMed] [Google Scholar]
- 74.Das J, Addona GH, Sandberg WS, Husain SS, Stehle T, Miller KW. J. Biol. Chem. 2004;279:37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- 75.Griner EM, Kazanietz MG. Nat. Rev. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 76.Dries DR, Newton AC. J. Biol. Chem. 2008;283:7885–7893. doi: 10.1074/jbc.M709943200. [DOI] [PubMed] [Google Scholar]
- 77.Medkova M, Cho W. Biochemistry. 1998;37:4892–4900. doi: 10.1021/bi972495j. [DOI] [PubMed] [Google Scholar]
- 78.Medkova M, Cho W. J. Biol. Chem. 1999;274:19852–19861. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- 79.Canagarajah B, Leskow FC, Ho JY, Mischak H, Saidi LF, Kazanietz MG, Hurley JH. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 80.Stahelin RV, Digman MA, Medkova M, Ananthanarayanan B, Rafter JD, Melowic HR, Cho W. J. Biol. Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- 81.Bittova L, Stahelin RV, Cho W. J. Biol. Chem. 2001;276:4218–4226. doi: 10.1074/jbc.M008491200. [DOI] [PubMed] [Google Scholar]
- 82.Xu RX, Pawelczyk T, Xia TH, Brown SC. Biochemistry. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]
- 83.Davis DB, Doherty KR, Delmonte AJ, McNally EM. J. Biol. Chem. 2002;277:22883–22888. doi: 10.1074/jbc.M201858200. [DOI] [PubMed] [Google Scholar]
- 84.Oancea E, Meyer T. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 85.Nalefski EA, Wisner MA, Chen JZ, Sprang SR, Fukuda M, Mikoshiba K, Falke JJ. Biochemistry. 2001;40:3089–3100. doi: 10.1021/bi001968a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stahelin RV, Rafter JD, Das S, Cho W. J. Biol. Chem. 2003;278:12452–12460. doi: 10.1074/jbc.M212864200. [DOI] [PubMed] [Google Scholar]
- 87.Shao X, Davletov BA, Sutton RB, Sudhof TC, Rizo J. Science. 1996;273:248–251. doi: 10.1126/science.273.5272.248. [DOI] [PubMed] [Google Scholar]
- 88.Shao X, Fernandez I, Sudhof TC, Rizo J. Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 89.Subramanian P, Stahelin RV, Szulc Z, Bielawska A, Cho W, Chalfant CE. J. Biol. Chem. 2005;280:17601–176077. doi: 10.1074/jbc.M414173200. [DOI] [PubMed] [Google Scholar]
- 90.Corbalan-Garcia S, Garcia-Garcia J, Rodriguez-Alfaro JA, Gomez-Fernandez JC. J. Biol. Chem. 2003;278:4972–4980. doi: 10.1074/jbc.M209385200. [DOI] [PubMed] [Google Scholar]
- 91.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ananthanarayanan B, Das S, Rhee SG, Murray D, Cho W. J. Biol. Chem. 2002;277:3568–3575. doi: 10.1074/jbc.M109705200. [DOI] [PubMed] [Google Scholar]
- 93.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, Falke JJ, Cafiso DS. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 94.Fukuda M, Aruga J, Niinobe M, Aimoto S, Mikoshiba K. J Biol. Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- 95.Fukuda M, Kojima T, Aruga J, Niinobe M, Mikoshiba K. J. Biol. Chem. 1995;270:26523–26527. doi: 10.1074/jbc.270.44.26523. [DOI] [PubMed] [Google Scholar]
- 96.Schiavo G, Gu QM, Prestwich GD, Sollner TH, Rothman JE. Proc. Natl. Acad. Sci. USA. 1996;93:13327–32. doi: 10.1073/pnas.93.23.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mehrotra B, Myszka DG, Prestwich GD. Biochemistry. 2000;39:9679–86. doi: 10.1021/bi000487o. [DOI] [PubMed] [Google Scholar]
- 98.Lemmon MA, Ferguson KM. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 99.Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA. J. Biol. Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 100.Singh SM, Murray D. Protein Sci. 2003;12:1934–1953. doi: 10.1110/ps.0358803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Flesch FM, Yu JW, Lemmon MA, Burger KN. Biochem. J. 2005;389:434–441. doi: 10.1042/BJ20041721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tuzi S, Uekama N, Okada M, Yamaguchi S, Saito H, Yagisawa H. J. Biol. Chem. 2003;278:28019–28025. doi: 10.1074/jbc.M300101200. [DOI] [PubMed] [Google Scholar]
- 103.Manna D, Albanese A, Park WS, Cho W. J. Biol. Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 104.He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. J. Lipid Res. 2008 doi: 10.1194/jlr.M800150-JLR200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roy A, Levine TP. J. Biol. Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 107.Pitcher JA, Touhara K, Payne ES, Lefkowitz RJ. J. Biol. Chem. 1995;270:11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]
- 108.Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- 109.Segers K, Sperandio O, Sack M, Fischer R, Miteva MA, Rosing J, Nicolaes GA, Villoutreix BO. Proc. Natl. Acad. Sci. USA. 2007;104:12697–12702. doi: 10.1073/pnas.0701051104. [DOI] [PMC free article] [PubMed] [Google Scholar]