Preface
Autophagy is a mechanism by which cellular material is delivered to lysosomes for degradation allowing basal turnover of cell components and providing energy and macromolecular precursors. Autophagy has opposing, context-dependent roles in cancer and interventions to both stimulate and inhibit autophagy have been proposed as cancer therapies. This has caused therapeutic targeting of autophagy in cancer to be sometimes viewed as controversial. Here we suggest a way forward for effective targeting of autophagy by understanding the context-dependent roles of autophagy and capitalizing on modern approaches to clinical trial design.
Introduction
Advancements in the understanding of autophagy and how we can harness this pathway to improve clinical outcomes have come a long way since the introduction of the term by Christian de Duve in 19631 (Fig 1). The importance of autophagy in health and disease was recently highlighted when Yoshinori Ohsumi was awarded the Nobel Prize for Physiology or Medicine for his work elucidating the mechanism of autophagy2. Of particular importance is the role of autophagy in cancer. It is thought that autophagy prevents cancer development. Conversely, once cancer is established, increased autophagic flux often enables tumor cell survival and growth3,4. Thus an important question for cancer therapy is should we try to enhance autophagy or inhibit it? In premalignant lesions much evidence suggests that enhancers of autophagy might prevent cancer development5. Conversely, in advanced cancers, both enhancing autophagy and inhibiting it have been suggested as therapeutic strategies3,6,7.
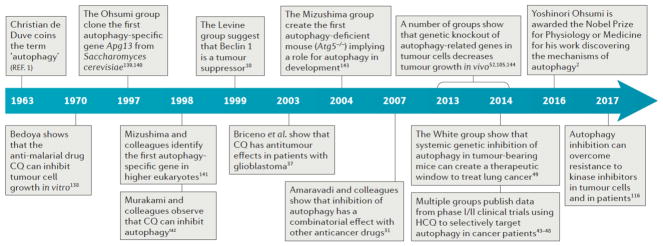
Figure 1. Timeline of the Major Discoveries Leading to the Successful Targeting of Autophagy in Cancer.
De Duve first coined the term ‘autophagy’ during a lysosomal conference in 1963. Since then key discoveries have been made elucidating the mechanisms of the process from yeast to cultured cell lines, into mice, and finally culminating in successful clinical trials and case studies in patient tumors. The timeline concludes with the Nobel Prize awarded to Yoshinori Ohsumi for Physiology or Medicine in 2016, emphasizing the impact of his work as well as that of many others along the way. CQ, chloroquine; HCQ, hydroxychloroquine
1963 - Christian de Duve coins the term “autophagy”1
1970 - Bedoya shows that the anti-malarial drug, CQ can inhibit tumour cell growth in vitro138
1997 - The Ohsumi group clone the first autophagy specific gene, ATG1, from Saccharomyces cerevisiae139,140
1998 - Mizushima and colleagues identify the first autophagy specific gene in higher eukaryotes141
1998 - Murakami and colleagues observe that CQ can inhibit autophagy142
1999 - The Levine group suggest that Beclin 1 is a tumor suppressor gene10
2003 - Briceno et al. show that CQ has anti-tumour affects in patients with glioblastoma37
2004 - The Mizushima group create the first autophagy deficient mouse (Atg5−/−), implicating autophagy in development143
2007 - Amaravadi and colleagues show that inhibition of autophagy has a combinatory effect with other anti-cancer drugs31
2013 - A number of groups show that genetic knock out of autophagy-related genes in tumour cells decreases tumour growth in vivo, e.g. 52,105,144
2014 - The White group show that systemic genetic inhibition of autophagy in tumour-bearing mice can create a therapeutic window to treat lung cancer49
2014 - Multiple groups publish data from Phase I/II clinical trials using HCQ to selectively target autophagy in cancer patients43–48
2016 - Yoshinori Ohsumi is awarded the Nobel Prize for Physiology or Medicine for his work discovering the mechanisms of autophagy2
2017 - Autophagy inhibition can overcome resistance to kinase inhibitors in tumour cells and in patients116
Despite this potential for confusion, clinical interventions to deliberately manipulate autophagy in cancer therapy are already underway7 with the vast majority focused on inhibiting autophagy. Indeed, a search of the ClinicalTrials.gov website in February 2017 using the search term “autophagy and cancer” returned 51 studies focused on inhibiting and evaluating autophagy to improve patient outcomes. As with other areas of cancer biology, such as the potential for the immune system to both promote and inhibit tumor formation and progression, the key to successful autophagy-focused therapeutic intervention comes from understanding the biology of how autophagy effects tumor initiation and progression. Here in this Review we discuss recent studies that clarify and support this concept. By considering past clinical trial results, current clinical trial design, the development of biomarkers of autophagy dependence and response, and the role of autophagy in chemoresistance we will explore how cancer therapy can be maximized by autophagy manipulation. Review of these topics is especially timely now with the continued convergence of a better mechanistic understanding of how autophagy influences therapeutic response both at the tumor cell intrinsic level and within the host, with increasing information from autophagy focused clinical studies. This convergence will allow us to better target autophagy to improve clinical outcomes in oncology patients.
Autophagy
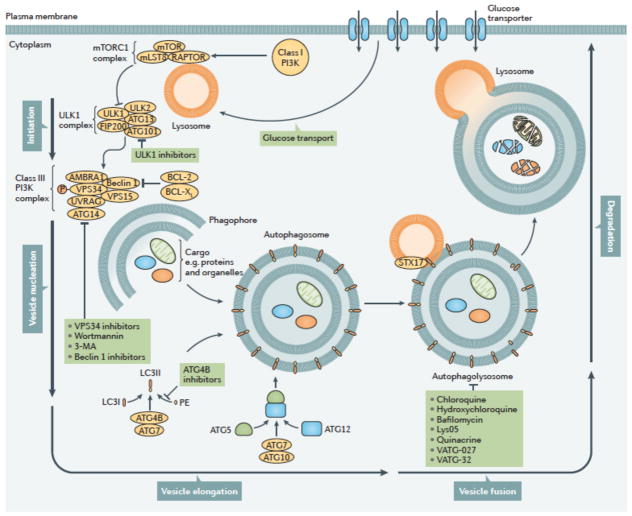
Macroautophagy (referred to hereafter as autophagy) is an evolutionarily ancient and highly conserved catabolic process involving the formation of double membraned vesicles called autophagosomes that engulf cellular proteins and organelles for delivery to the lysosome8,9 (Fig 2). Autophagy is controlled by a highly regulated set of signaling events, occurs at a basal level in all cells, and is induced by diverse signals and cellular stresses7. There may be important differences between stimulus-induced autophagy and basal autophagy but our understanding of such differences is poor. Formation and turnover of the autophagosome involves evolutionarily-conserved genes called autophagy related (ATG) genes9 and is typically divided into distinct stages: initiation, nucleation of the autophagosome, expansion and elongation of the autophagosome membrane, closure and fusion with the lysosome, and concluding with degradation of intravesicular products (Fig 2). Initiation begins with activation of the ULK1 (also known as ATG1) complex (involving ULK1, ULK2, ATG13, FIP200 (also known as RB1CC1) and ATG101), which activates a class III PI3K complex (VPS15, VPS34 (also known as PIK3C3), ATG14, Beclin 1, UV radiation resistance-associated gene protein (UVRAG; also known as p63), and activating molecule in BECN1-regulated autophagy protein 1 (AMBRA1), all of which are scaffolded by a putative tumor suppressor Beclin 110. The ATG5-ATG12 complex conjugates with ATG16 to expand the autophagosome membrane and members of the LC3 and GABARAP families of proteins are conjugated to the lipid phosphatidylethanolamine (PE) and recruited to the membrane. ATG4B, in conjunction with ATG7, conjugates LC3-I and PE to form LC3-II (also known as MAP1LC3B). This lipid-conjugated form of LC3 commonly serves as an autophagosome marker11. Ultimately, the autophagosome fuses with the lysosome, the contents are degraded and macromolecular precursors are recycled or used to fuel metabolic pathways. The adaptor protein sequestosome 1 (also known as p62) that targets specific substrates to autophagosomes and LC3II are degraded along with other cargo proteins and can be used as a measure of autophagic flux11.
Figure 2. Autophagy can be inhibited at multiple stages.
The process of autophagy is divided into five distinct stages: initiation, vesicle nucleation, vesicle elongation, vesicle fusion and cargo degradation. Nonspecific macroautophagy is initiated by upstream activation through either nutrient starvation or growth factors. Under starvation conditions, a drop in glucose transport results in a release of mTOR inhibition of the ULK1 complex, allowing for the progression of autophagy. The ULK1 complex (comprising ULK1, ULK2, FIP200, ATG101 and ATG13) induces vesicle nucleation which is then mediated by a class III PI3K complex consisting of multiple proteins. Beclin 1, a BCL-2 homology (BH)-3 domain only protein, is phosphorylated by ULK1 and acts as an overall scaffold for the PI3K complex, facilitating localization of autophagic proteins to the phagophore. BCL-2 and BCL-XL interact with Beclin 1 at the BH3 domain to decrease the pro-autophagic activity of Beclin 1 by interrupting the Beclin 1–VPS34 complex formation and decreasing the interaction of Beclin 1 with UVRAG. Additional negative regulation of this process occurs with the phosphorylation of VPS34 (also known as PIK3C3), which decreases its interaction with Beclin 1. In contrast, AMBRA binds Beclin 1 and stabilizes the PI3K complex. ATG14 and UVRAG also bind Beclin 1 to promote interactions between Beclin 1 and VPS34 and phagophore formation. VPS15 is required for optimal VPS34 function by enhancing VPS34 interaction with Beclin 1. The growing double membrane undergoes vesicle elongation to eventually form an autophagasome: a process mediated by two ubiquitin-like conjugation systems. The first system involves the conjugation of phosphatidylethanolamine (PE) to cytoplasmic LC3-I to generate the lipidated form, LC3-II which is facilitated by the protease, ATG4B, and the E1-like enzyme, ATG7, whereby LC3-II is incorporated into the growing membrane. The second conjugation system is mediated again by ATG7 as well as the E2-like enzyme, ATG10, resulting in an ATG5-ATG12 conjugate. Subsequently, the SNARE protein, syntaxin 17 (STX17) facilitates autophagosome fusion with the lysosome, resulting in an autophagolysosome. The low pH of the lysosome results in degradation of the autophagosome contents. This process can be targeted pharmacologically upstream by means of direct ULK1, VPS34, or Beclin 1 inhibition. It can also be targeted by wortmannin and 3-methyladenine (3-MA) which act as PI3K inhibitors. Downstream targets include direct ATG4B inhibitors as well as chloroquine or hydroxychloroquine and bafilomycin, which act to prevent autophagosome fusion with the lysosome. PE, phosphatidylethanolamine.
Many of these steps in the autophagy pathway represent potentially druggable targets providing ways to both positively and negatively influence autophagy (Fig 2). Although current efforts in the clinic to inhibit autophagy are focused on inhibiting the lysosome using chloroquine (CQ) or the related hydroxychloroquine (HCQ), inhibitors against other autophagy regulators such as VPS3412–14, ULK115,16 and ATG4B17 have been reported and shown to inhibit tumor cell growth or induce tumor cell death in vitro15–17 and in preclinical mouse models17. Next generation lysosomal inhibitors are also in development including Lys05, a bisaminoquinoline that inhibits autophagy and impairs melanoma and colorectal adenocarcinoma growth as a single agent in preclinical mouse models18. Lys05 is a more potent autophagy inhibitor than HCQ due to a greater deacidification of the lysosome18. Other potent lysosomal inhibitors such as quinacrine and VATG-027 and VATG-032 (novel acridine and 1,2,3,4-tetrahydroacridine derivatives of quinacrine) have also been shown to be effective in patient derived BRAF mutant melanoma cell lines19. Conversely, induction of autophagy is feasible using existing drugs (e.g. BH3 mimetics20 and mTOR inhibitors21) but also nutraceuticals such as trehalose22 and caloric restriction mimetics23 or exercise24.
Other, less studied forms of autophagy include microautophagy and chaperone-mediated autophagy (CMA). Non-selective microautophagy is mediated by direct engulfment of cytoplasm and its components by tubular membrane invaginations into lysosomes. Selective microautophagy involves direct targeting of specific organelles into lysosomes such as peroxisomes (micropexophagy), nonessential components of the nucleus (piecemeal microautophagy of the nucleus), and mitochondria (micromitophagy). While microautophagy has been associated with the development of neurodegenerative disorders such as Alzheimer disease and Huntington’s disease as well as lysosomal glycogen storage diseases such as Pompe disease, it has not been implicated in cancer25. CMA is a form of selective autophagy in which cytosolic proteins with motifs related to the pentapeptide KFERQ are recognized by Heat Shock cognate 70 kDa Protein (HSC70; also known as HSPA8) forming a chaperone complex26,27 that translocates into the lysosome via the lysosomal-associated membrane protein 2A, LAMP2A. CMA has been implicated in cancer28 and drugs targeting the lysosome could affect all types of autophagy.
Substrates that are degraded by autophagy may differ depending on the autophagic stimulus. One example is the role of autophagy in iron homeostasis29. Degradation of ferritin by autophagy is initiated when cells sense that they are deficient in iron and is mediated by nuclear receptor co-activator 4 (NCOA4), allowing release of iron into the cell. Selective autophagy of specific substrates can also occur due to oncogenic stress. For example, degradation of the nuclear lamina occurs in human primary fibroblast cells transformed with oncogenic HRASV12 and genotoxic insults, but not during starvation stress30. It is often implicitly assumed that a measured increase in autophagy must have the same consequence irrespective of the stimulus. These studies suggest that this assumption is wrong, and there may be a high degree of selection with regards to the cargo being degraded, depending on the autophagy stimulus. This could perhaps explain the context-dependent consequences of autophagy on cellular processes and better understanding of such mechanisms in cancer could provide a way to more selectively target autophagy for therapeutic purposes.
Cancer Clinical Trials
Extensive pre-clinical evidence exists to support the idea of inhibiting autophagy to improve clinical outcomes in cancer patients. Animal tumor models driven by specific oncogenes have been shown to cause tumors that regress upon subsequent genetic or pharmacological inhibition of autophagy (see below for further discussion). Similarly, following an initial finding in 2007 by Amaravadi and colleagues31 (Fig 1), a large number of in vitro studies, genetically engineered mouse models (GEMMs) and patient-derived xenograft (PDX) mouse models have demonstrated improved anti-tumor effects when various different types of anti-cancer drug are combined with either genetic or pharmacological autophagy inhibition3,6,32.
CQ and HCQ are currently the only clinically available drugs used to inhibit autophagy. These drugs deacidify the lysosome and block fusion of autophagosomes with lysosomes preventing cargo degradation (Fig 2)33. CQ is also able to sensitize cancer cells to chemotherapeutic agents through autophagy-independent mechanisms34 and has other anti-cancer effects independent of its effect on autophagy35,36. Some of the first clinical evidence of improving outcomes using autophagy inhibition came from a small trial involving 18 patients with glioblastoma. Those treated with CQ in conjunction with radiation and the alkylating agent temozolomide experienced a statistically significant prolonged median survival compared to controls (33 months compared with 11 months)37. Follow up clinical trials, and retrospective data from Briceno et al. supported the findings of the initial study (Table 1)38,39. Additional early studies combining CQ with radiation for brain metastasis also found improved intracranial tumor control40,41.
Table 1.
Published Autophagy Trials in Cancer
| Tumour Type | Autophagy Inhibitor | Clinical trial Phase | Additional treatment | Clinical Response | Grading of Side effects | Biomarker measures | Ref |
|---|---|---|---|---|---|---|---|
| Non-Hodgkin lymphoma | HCQ | I (in Dogs) | Doxorubicin | PFS- 5 months ORR-93.3% |
Grade 1 or 2: Mild lethargy GI upset Grade 3 or 4: None |
Plasma concentrations of HCQ LC3 positive cells by flow cytometry EM of PBMC for AVs |
42 |
| Solid tumors | HCQ | I | Vorinostat | 1 patient (renal cell carcinoma) durable PR 2 patients (colorectal cancer) prolonged SD |
Grade 1 or 2: nausea, diarrhea, fatigue, weight loss, anemia, elevated creatinine Grade 3: fatigue and/or myelosuppression in a minority of patients |
EM of PBMC for AVs IHC for LC3II |
43 |
| Solid tumors and melanoma | HCQ | I | Temsirolimus | 67% SD (solid tumors) 74% SD (melanoma) |
Grade 1 or 2: fatigue, anorexia, nausea, stomatitis, rash, weight loss Grade 3 or 4: Anorexia, fatigue, nausea |
EM of PBMC for AVs | 44 |
| Solid tumors and melanoma | HCQ | I | TMZ | Solid tumor patients: 10% PR 27% SD Metastatic melanoma patients: 14% PR 27% SD |
Grade 2: fatigue, anorexia, nausea, constipation, diarrhea | EM of PBMC for AVs | 45 |
| Solid tumors | HCQ | I | Rapamycin with Metronomic cyproterone and docetaxel | 40% PR 44% SD |
Grade 1 and 2: fatigue, diarrhea, mucositis Grade 3: Fatigue, myelosuppression, diarrhea, nausea, vomiting, cardiotoxicity, hepatic toxicity |
Not evaluated | 145 |
| Sarcoma | HCQ | Case series (10 patients) | Rapamycin | 6 PR 3 SD 1 PD |
Grade 1: rash, nausea, diarrhea, constipation | Evaluation of 18FDG-PET as measure of tumor response after two weeks | 146 |
| Glioblastoma | CQ | III | TMZ and radiation | Median survival 24 months (controls 11 months) | Grade 1: Myelosuppression | Not evaluated | 39 |
| Relapsed Glioblastoma | CQ | Case series (5 patients) | Radiation | 2 month response: 2 PR 1 SD |
None | Not evaluated | 147 |
| Glioblastoma | HCQ | I/II | TMZ and radiation | Median survival 15.6 months | Grade 2 or 3: myelosuppression, nausea, fatigue, constipation, diarrhea Grade 4: myelosuppression, constipation |
Plasma concentrations of HCQ EM of PBMC for mean AVs PBMC LC3-II:Actin ratio |
46 |
| Glioblastoma | CQ | III | TMZ and radiation | Median survival: 33 months (controls 11 months) | Increased seizure frequency | Not evaluated | 37 |
| Brain metastases: Non-small cell lung cancer, small cell lung, breast, ovarian | CQ | Pilot | Radiation | Median OS: 5.7 months PFS of brain metastasis at one year 55% |
Grade 1: radiation dermatitis Grade 2: alopecia |
Not evaluated | 40 |
| Brain metastases: Non-small cell lung cancer, breast cancer | CQ | II | Radiation | ORR 54% (control 55%) PFS of brain metastasis at one year 83.9% (control 55.1%) |
Grade 1 or 2: headache, dizziness, nausea, vomiting, anorexia, myelosuppression Grade 3: nausea, constipation, headache, drowsiness |
Not evaluated | 41 |
| Refractory myeloma | HCQ | I | Bortezomib | 14% very good PR 14% minor response 45% period of SD |
Grade 1 or 2: myelosuppression fatigue, peripheral neuropathy, nausea, vomiting, diarrhea, constipation Grade 3 or 4: nausea, constipation, diarrhea, anorexia, myelosuppression fatigue |
Plasma concentrations of HCQ EM of PBMC and bone marrow plasma cells for mean AVs PBMC LC3-II:Actin ratio |
47 |
| Metastatic PDAC | HCQ | II | None | 2 months PFS 10% Median PFS 46.5 days (1.5 months) OS: 69 days (2.3 months) |
Grade 3 or 4: lymphopenia, elevated alanine aminotransferase | PBMC LC3-II:β-Actin ratio | 48 |
| PDAC | HCQ | I/II | Gemcitabine | 61% with decrease in CA19-9 If >51% increase in LC3-II, improved disease-free survival to 15.03 months vs 6.9 months, OS 34.83 months v 10.83 months |
Grade 3: Myelosuppression, hyponatremia, elevated AST, hypoalbuminemia, hyperbilirubinemia rash hyperglycemia ileus |
CA19-9 as measure of tumor response LC3-II in PBMC |
53 |
| NSCLC | HCQ | I | Erlotinib | 1 PR 4 SD ORR 5% |
Grade 1 or 2: Nausea, fatigue, vomiting, anemia, anorexia Grade 3 or 4: Rash, nausea, nail and skin changes, myelosuppression Grade 5: Pneumonitis |
Plasma concentrations of HCQ | 148 |
AVs: Autophagic vacuoles
CA 19-9: cancer antigen 19-9
CQ: chloroquine
CR: Complete response
EM: Electron microscopy
GI: gastrointestinal tract
HCQ: hydroxychloroquine
IHC: Immunohistochemistry
NSCLC: non-small cell lung cancer
ORR: Overall response Rate
OS: Overall Survival
PBMC: Peripheral blood mononuclear cell
PDAC: pancreatic adenocarcinoma
PD: Progressive disease
FDG-PET: fluorodeoxyglucose-positron emission tomography
PFS: Progression free survival
PR: Partial response
SD: Stable disease
TMZ: temozolomide
Side effects are graded according to the Common Terminology Criteria for Adverse Events using a scale of 0–5 with 0 representing no adverse side effects and 5 representing death as a result of an adverse side effect.
The next major series of clinical trials utilized HCQ and had the additional benefit of attempting to correlate pharmacokinetic (PK)-pharmacodynamic (PD) parameters with autophagy inhiibition42–48. These early phase clinical trials were performed in patients with a wide variety of malignancies and tested multiple drug combinations (Table 1). Notably, these trials provided important lessons on the implementation of autophagy-targeted therapy. A canine lymphoma study of HCQ combined with the chemotherapy doxorubicin modeling a dose escalation phase I human study provided the initial proof of principle that combining HCQ with chemotherapy was safe42. Importantly, it also provided preliminary evidence of the clinical activity of HCQ with an observed objective response rate of 93%42. Additional human studies included a broad range of tumors including advanced solid tumors and melanoma43–45, glioblastoma46, and refractory myeloma47. As predicted, the maximum tolerated dose (MTD) of HCQ varied in relation to the concurrent therapy utilized. A phase I study of vorinostat with HCQ in refractory solid tumors defined the MTD of HCQ to be 600 mg daily when combined with vorinostat at 400 mg daily43. Similar findings related to safety were observed when combining HCQ with concurrent radiation therapy and temozolomide in patients with glioblastoma46. In contrast, combining HCQ with 25 mg daily of the mTOR inhibitor temsirolimus in another solid tumor patient population found the combination to be safe with HCQ used at 600mg twice daily44. Common dose limiting toxicities in these trials included gastrointestinal toxicity and fatigue43–45. Importantly, HCQ-induced neurotoxicity was not observed as might have been predicted from Atg7 gene knockout mouse models wherein mice developed significant neurodegeneration upon complete deficiency of autophagy49. The MTD of HCQ as a single agent has not been measured and 600 mg twice daily of HCQ is the highest dose tested so far when administered in combination with standard chemotherapy agents44. Additional studies of potentially higher HCQ doses or more potent lysosomal autophagy inhibitors such as Lys05, quinacrine, and VATG-032 18,19,50,51 might maximize autophagy inhibition and anti-tumor activity.
Clinical response to autophagy inhibition has varied widely (Table 1). While initial glioblastoma studies utilizing CQ in combination with chemotherapy and radiation found more than a doubling of median survival compared to controls37–39, a phase I/II trial utilizing HCQ in combination with chemotherapy and radiation found no significant improvement in survival of patients with glioblastoma46. Of note, in this particular study with HCQ there was inconsistent inhibition of autophagy between patients and dose-limiting toxicities including myelosuppression that prevented intensification of HCQ therapy, which may explain the different responses in these trials. A phase II trial of HCQ monotherapy in patients with previously treated metastatic pancreatic cancer demonstrated no clinical benefit and inconsistent evidence of autophagy inhibition48. However, this study was performed in patients with advanced disease with limited potential for single agent HCQ to improve end-stage disease outcomes. Pre-clinical data from PDX studies of pancreatic cancer had demonstrated a response to single agent HCQ52. Furthermore, pre-operative treatment with HCQ in combination with gemcitabine resulted in a decrease in the serum tumor marker cancer antigen (CA) 19-9 in 60% of patients with pancreatic adenocarcinoma53. Interestingly, in this same cohort, those patients with a greater than 51% increase in LC3-II puncta labeling in peripheral blood mononuclear cells (PBMC) (suggesting effective autophagy inhibition) experienced both an increase in progression free survival (PFS) (15.03 months compared with 6.9 months) and overall survival (OS) (34.83 months compared with 10.83 months)53.
Biomarkers
A major limitation in all of the clinical studies has been identifying appropriate pharmacodynamic biomarkers evaluating changes in autophagy. Barnard et al.42 showed that increased intra-tumoral HCQ was associated with an expected increase in LC3II puncta formation (a measure of autophagosome turnover) and accumulation of sequestosome 1 compared to treatment naive tumors. This provided evidence that the clinical use of HCQ could inhibit autophagic flux within tumors, and supports the use of LC3II and sequestosome 1 immunohistochemistry as potential biomarkers for future trials. Several human trials have also utilized transmission electron microscopy (TEM) to evaluate the number of double membraned vesicles (presumed to be autophagosomes) in PBMC. However, this was found to be an unreliable method to monitor autophagy inhibition due to a lack of correlation with levels of autophagy inhibition in tumor samples as measured by changes in the lysosomal protease CTSD cathepsin D as well as sequestosome 1 and LC3II by immunohistochemistry43.
There can be up to a 100-fold difference in HCQ uptake in tumors compared to plasma, suggesting that plasma analysis is a poor surrogate for tumor specimen analysis42. Additionally, CQ uptake into tumor tissue is affected by tumor pH, presenting a difficulty in blocking autophagy in more acidic tumors50. Such pH variations could explain some of the differences in accumulation of the drug between tumours. Finally, higher doses of HCQ (1200 mg/daily) may be better at causing an accumulation of autophagic vesicles in both PBMCs and tumor biopsies47, although this cannot always be achieved due to dose limiting toxicities. This highlights the potential benefit of newer autophagy inhibitors. For example, Lys05 (and its parent compound Lys01) more potently accumulate within and deacidify the lysosome, allowing for greater autophagy inhibition at lower doses. These effects can be seen using standard biomarkers including accumulation of LC3II by western blot analysis and the accumulation of autophagosomes as measured by TEM18. Due to the limitations of current autophagy inhibitors, and as we continue to evaluate upcoming new inhibitors, better biomarkers of autophagy manipulation are needed.
Ongoing clinical trials are attempting to define additional biomarkers (Table 2). Functional imaging techniques are being used to correlate intra-tumor hypoxia with autophagy via positron emission tomography (PET)/computed tomography (CT) scans using hypoxia tracers 2-(2-nitro-1H-imidazol-1-yl)-N-(2,2,3,3,3-pentafluoropropyl)-acetamide (EF5) labeled with 18F-fluorine isotope (18F-EF5) and [18F]-HX4 [18F-flortanidazole] (NCT0188145154 and NCT0223338755). Similarly, the relationship between cancer metabolism and autophagy is being evaluated in a clinical trial combining HCQ with chemotherapy in patients with advanced colorectal cancer (NCT0120653056). Other studies plan to correlate the effects of combined proteasome and vorinostat mediated histone deacetylase (HDAC) inhibition on autophagy and serum metabolic profiles (NCT0204298957). HDAC family members have been shown to increase autophagy through several mechanisms including the regulation of gene transcription of essential genes58. Increased activity of autophagy after treatment with HDAC inhibitors has been shown to significantly blunt HDAC anticancer activity58. Induction of autophagy has also been shown to occur in response to proteasome inhibitors and is believed to play a role in resistance59. This is the basis for early phase and ongoing clinical trials inhibiting autophagy in combination with HDAC43 and proteasome47 inhibitors.
Table 2.
Autophagy Biomarker Identification Trials
| Autophagy biomarker | Methods of measurement | Tumor type | Clinical trial ID |
|---|---|---|---|
| Tumor hypoxia | 18F-EF5 PET Tissue LC3II staining Autophagy gene expression |
Clear cell ovarian | NCT0188145154 |
| Tumor hypoxia | 18F-HX4 PET Autophagy gene expression |
Cervical | NCT0223338755 |
| Autophagosomes | Autophagic vesicles in PBMC | Myeloma | NCT01594242149 |
| Metabolic alterations | 18FDG PET Autophagic vesicles in PBMC |
Colorectal | NCT0120653056 |
| Metabolic alterations | Serum metabolic studies | Advanced p53 malignancies | NCT0204298957 |
| Metabolic alterations | MRI including magnetic resonance spectroscopy and diffusion weight imaging | Cervical | NCT01874548150 |
18F-EF5, fluorine 18 (18F)-2-(2-nitro-1H-imidazol-1-yl)-N-(2,2,3,3,3-pentafluoropropyl)-acetamide (EF5); 18 FDG, 18F-fluorodeoxyglucose; HX4, flortanidazole; MRI, magnetic resonance imaging; PBMC, peripheral blood mononuclear cells; PET, positron emission tomography
Pre-clinical studies have also identified the use of the transcriptional regulators belonging to the microphthalmia /transcription factor E (MiT/TFE) family as a potential biomarker of autophagy regulation. Microphthalmia-associated transcription factor (MITF) or TFE3 overexpression was associated with an increase in autophagy and MiT/TFE-dependent autophagy and lysosome gene expression in established pancreatic ductal adenocarcinoma (PDAC) cell lines, primary PDAC tumors, and primary patient-derived PDAC cell lines60. Therefore, evaluating the expression levels of MiT/TFE family members, as well as their associated proteins within tumor samples, has the potential to identify patients with autophagy activation under the control of MiT/TFE proteins. Another interesting study by Follo et al. found that quantification of autophagy initiation by ATG13 puncta was correlated between patient tumor derived ex vivo spheroids and formalin fixed clinical tumor samples, and that differences between ATG13 levels correlated with clinical outcomes in mesothelioma61. This is especially important as current measures of autophagic flux require the use of inhibitors of lysosomal proteases to detect the accumulation of LC3II, which is not possible in formalin-fixed samples11. In contrast, ATG13 is a static marker making it potentially much more of a clinically relevant biomarker of autophagy.
Surrogate markers from peripheral blood could provide another method to assess autophagy inhibition. Autophagy regulates cellular secretion of cytokines and other signaling molecules62. The autophagy-regulated secretome63, e.g. secretion of the cytokine interleukin-6 (IL-6)64, has been suggested as a potential biomarker of autophagic activity. Modern clinical procedures such as endoscopic retrograde cholangiopancreatography (ERCP) allow for sampling of such factors from organ associated ducts65 or the peripheral blood so such an approach is technically feasible. Better understanding of how autophagy regulates secretion, and the molecules secreted, may allow us to incorporate such methods into a biomarker strategy. Together, these studies suggest a potential multi-dimensional biomarker strategy that would incorporate the direct molecular evaluation of autophagy in biopsies, monitoring of autophagy-regulated soluble factors, and functional imaging techniques. While somewhat involved, all these assays are clinically feasible and could be incorporated into clinical trial protocols.
Targeting Autophagy: a good idea?
The collective results of published clinical trials (Table 1) present evidence for the safe use of CQ and HCQ as a cancer therapy. The reported positive clinical outcomes are encouraging for the role of autophagy inhibition in cancer therapy, but care needs to be taken to understand the underlying contexts where autophagy inhibition will be beneficial and where it could potentially be detrimental.
Autophagy is a known survival mechanism conserved from yeast to mammals66. It has also been identified as a survival mechanism across several tumor types67–70. The association between tumor cell survival and autophagy can be explained, in part, by the role of autophagy in protecting cells from undergoing programmed cell death71. This provides a logical rationale for why the inhibition of autophagy could improve the response to other agents and forms the basis for the completed (Table 1) and ongoing (Table 3) clinical trials. However, the effect of autophagy on the ability of tumor cells to undergo apoptosis is not always protective. For example, within the same tumor cell population, autophagy can promote or inhibit apoptosis under different cellular contexts in response to similar death stimuli such as CD95 ligand (CD95L) or tumour necrosis factor-related apoptosis-inducing ligand (TRAIL), which both act as death receptor agonists72. The mechanisms underlying these opposing effects are due to the degradation of different pro- or anti-apoptotic regulators by autophagy72,73. A take home message from this work is that a much better understanding of how autophagy regulates apoptosis sensitivity (i.e. what substrates it degrades) is needed, if the aim is to predict whether a tumor cell is more or less likely to be killed in response to a particular death signal when autophagy is blocked. Moreover, increases in cell death can depend on the stage of the autophagy pathway that is inhibited. For example, prevention of autophagosome maturation can decrease necroptosis while inhibition of autophagosome turnover potentiates necroptosis in the same prostate cancer cells74. These observations exemplify the underlying problem of autophagy manipulation in cancer therapy– autophagy has context dependent and even opposing effects on tumor cell behavior. Such context-dependent effects are poorly understood, emphasizing the importance of a better understanding of the molecular mechanisms that determine how autophagy affects cancer cell behaviors.
Table 3.
Open Trials Targeting Autophagy Dependent Cancers
| Tumor type | Autophagy-dependence marker | Phase | Autophagy Inhibitor | Additional treatment | Clinical Trial ID |
|---|---|---|---|---|---|
| Colorectal | JNK1 | I/II | HCQ | FOLFOX+ Bevacizumab | NCT0120653056 |
| Glioblastoma | EGFRvIII | I/II | CQ | Temozolomide + radiation | NCT02378532114 |
| Pancreatic | Mutant RAS | I/II | HCQ, nab-Paclitaxel | Gemcitabine, nab-Paclitaxel | NCT01506973151 |
| Pancreatic | Mutant RAS | II | HCQ, nab-Paclitaxel | Gemcitabine, nab-Paclitaxel | NCT01978184152 |
| Pancreatic | Mutant RAS | I/II | HCQ | Gemcitabine | NCT01128296153 |
| BRAF Mutant Melanoma | Mutant BRAF | I | HCQ | Vemurafenib | NCT01897116154 |
| BRAF Mutant Melanoma | Mutant BRAF | I/II | HCQ | Dabrafenib, Trametinib | NCT02257424113 |
CQ, chloroquine; EGFRvIII, epidermal growth factor variant III; HCQ, hydroxychloroquine, JNK1, JUN N-terminal kinase 1; nab-paclitaxel, nanoparticle albumin-bound-paclitaxel
Arguments against inhibiting autophagy in cancer therapy
Several studies, especially from Kroemer and colleagues, have suggested that autophagy inhibition is a bad idea in cancer treatment because it would reduce anti-tumor T cell responses75–77. The rationale is that autophagy in dying tumor cells is required for immunogenic cell death leading to efficient recognition by the immune system and activation of an effective anti-tumor immune response78,79. One caveat to these studies is that they focused on highly immunogenic tumor models, including the CT26 colon cancer mouse model80, which may have influenced some of the responses seen. In opposition to this idea, a recent study using less immunogenic B16 mouse melanoma and 4T1 human mammary carcinoma cell mouse models, found equivalent T cell responses between autophagy-competent tumor-bearing mice and tumor-bearing mice wherein autophagy was blocked by either genetic deletion of autophagy genes or pharmacologically through treatment with CQ81. Another study from the Kroemer lab took the idea of autophagy being required for immunogenic cell killing one step further by concluding that enhanced autophagy (using caloric restriction mimetics) could boost anti-tumor immune responses82. This led to the suggestion that not only should autophagy not be inhibited but that interventions aimed at increasing autophagy during cancer therapy should be considered.
Autophagy can stimulate tumor antigen cross- presentation83 providing another potential mechanism by which autophagy inhibition could interfere with a robust anti-tumor immune response. Correlative evidence suggests that these mechanisms may be associated with better outcomes83. Higher LC3II puncta combined with the presence of nuclear high mobility group protein B1 (HMGB1), a non-histone chromatin-binding protein known to stimulate anti-cancer immune responses, in resected breast cancer specimens was associated with improved metastatic free survival and breast cancer specific survival84 and increased immune infiltration of the tumor85. A caveat for these studies is that the available markers (e.g. autophagosome vesicles in PBMCs) are, as noted above, poor measures of the actual level of autophagic flux that is taking place in the tumor tissue. Countering these ideas, other studies report that some anti-tumor immune responses are enhanced by autophagy inhibition86,87. Thus, there are arguments both for and against autophagy inhibition even when just considering the effects on anti-tumor immune responses.
Another potential use of autophagy to influence an immune response has been demonstrated in an ongoing study led by the Second Affiliated Hospital, School of Medicine at Zhejiang University that has proposed the use of the combination of autophagy and proteasome inhibition in ex vivo tumor cells in the development of a tumor vaccine (NCT0305734088). Pre-clinical data has shown that inhibition of autophagy in tumor cells treated with a proteasome inhibitor results in enrichment of short-lived proteins (SLiPs) and misfolded proteins known as defective ribosomal products (DRiPs) in autophagosomes, named DRibble corpuscles89. DRiPs and SLiPs are highly expressed in tumors and have the ability to support an anti-tumor immune response, but are inherently unstable and under normal conditions are degraded by proteasomes. Inhibiting proteasome degradation stabilizes these proteins that are then concentrated in autophagosomes (DRibble corpuscles). Inhibition of autophagy at this stage prevents breakdown of DRiPs and SLiPs that have concentrated in the autophagosome, and allows for fractionation and collection of the DRibble corpuscles to provide the protein needed to create effective DRibble vaccines89. DRibble tumor vaccines developed from these proteins have been shown to induce cross-reactive T-cell responses and tumor antigen cross- protection90. Preliminary analysis of a Phase II trial evaluating the use of DRibble vaccines in patients with non-small cell lung cancer (NSCLC) demonstrated that at 12 weeks, PBMCs from treated patients had multiple induced and increased antibody responses91. Other studies are also attempting to exploit autophagy to improve the efficacy of cancer immunotherapies. For example, a Phase I trial evaluating DNX2401, an oncolytic adenovirus, in glioblastoma patients (NCT0195673492) hypothesizes that autophagy stimulated in response to temozolomide therapy could help viral replication in the tumor cells. This is an example where autophagy inhibition would be counterproductive to the intended purpose of the primary therapy.
In patients, autophagy inhibition is not specifically targeted to tumor cells, thus potential toxicity from global autophagy inhibition represents another reason for pause when considering the value of targeting autophagy. This is exemplified in a study where knockout of an essential autophagy gene (Atg7) in all tissues was achieved in adult mice49. Atg7 deletion led the eventual death of all mice due to severe neuronal toxicity, disruption of glucose homeostasis, and increased susceptibility to infection. However, it is important to remember that while the removal of an essential component of the canonical autophagy pathway in every cell in the body might mimic the effect of a “perfect” autophagy inhibitor, such a strategy is markedly different than that of the clinical application of an autophagy inhibitor, which is unlikely to be as effective at inhibiting autophagy as the complete deletion of an essential autophagy regulator. In support of this idea, chronic use of HCQ for treatment of rheumatological disorders and treatment of some cancer patients with CQ as an autophagy inhibitor for extended time periods without adverse toxicity93 demonstrates that long term treatment with lysosomal autophagy inhibitors is feasible. Most importantly, as long as cancer cells are more dependent on autophagy than normal tissues, even a drug that causes some normal tissue toxicity can have a useful therapeutic window allowing it to be an effective cancer treatment. Indeed, in the inducible Atg7 knockout mouse, the growth of KRAS-driven lung tumors were profoundly inhibited by Atg7 deletion before any signs of neurotoxicity49, indicating that just such a window for autophagy inhibition exists to treat some cancers.
Possible mechanisms and markers of autophagy dependence
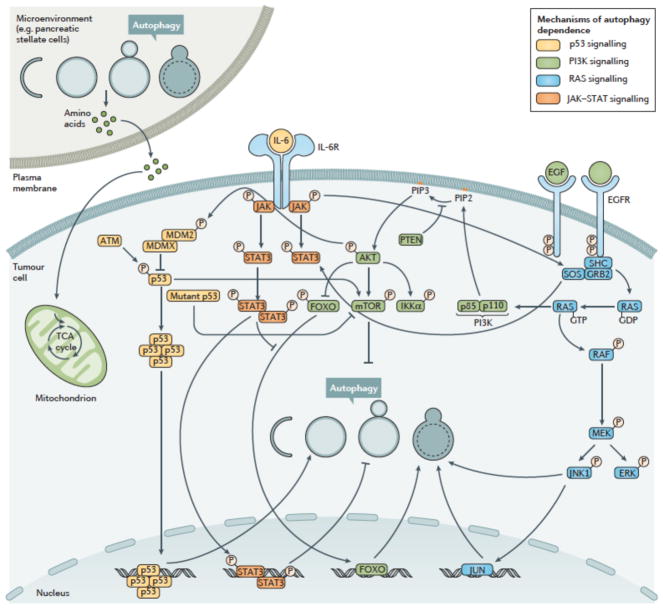
Although autophagy may be functional in many cancer cells and needed to respond to stresses like amino acid deprivation, some cancer cells may be especially dependent on autophagy even in the absence of added stress94. This idea has been called autophagy addiction or autophagy-dependence and is important because in some studies it was recognized that only autophagy-dependent tumors responded to pharmacological autophagy inhibition in vivo94. Moreover, drug synergy between autophagy inhibitors and other anti-cancer drugs can occur in autophagy-dependent tumor cells, while the same drug combination was sometimes even antagonistic in autophagy-independent tumor cells93,94. This implies that if autophagy inhibitors are combined with other drugs in autophagy-independent tumors in the clinic, the effects could be counterproductive. A reliable way to identify autophagy-dependent cancers is now needed to incorporate this concept into clinical decisions. Multiple mechanisms of autophagy addiction are beginning to be uncovered (Fig 3) that may help identify the most autophagy-dependent tumors, and many of these mechanisms are amenable to the development of biomarkers that could potentially be used to select patients whose tumors are most likely to respond to autophagy inhibition therapy.
Figure 3. Molecular Mechanism of Autophagy Dependence.
Pre-clinical and clinical models have indicated that the tumor microenvironment, for example pancreatic stellate cells in the case of pancreatic cancer, p53 status, RAS family status, activation of JAK–STAT and PI3K signaling may all play roles in the determination of autophagy dependence within cancer cells, both in vitro and in patients. These pathways have all been shown to affect autophagy either positively or negatively and many participate in cross-pathway signaling. Signaling through p53 can both promote and inhibit autophagy and may interact with other proteins activated by mutations to enhance autophagy dependence, especially in pancreatic cancer. Activation of EGFR via amplification or mutation leads to the downstream up-regulation of the PI3K-AKT-mTOR pathway as well as activation of STAT3 and the RAS pathway. Although autophagy inhibition can occur through mTOR activation, these downstream effects collectively result in stimulation of autophagy and an increase in autophagy dependence. Mutations or alterations in the RAS family (specifically KRAS) have been shown to promote autophagy, enhancing tumor growth and therapy resistance. Specific mutations in RAF such as BRAFV600E promote autophagy dependence in multiple tumors including central nervous system (CNS) tumors and melanoma. Finally, autophagy regulation of JAK-STAT signaling through IL-6 has been identified as a mechanism of autophagy dependence in breast cancer. All of these pathways are complex and interact on multiple levels. Identification of tumors with these pathways and as of yet to be identified pathways will provide methods of detection of autophagy dependent tumors.
Mutations in the RAS pathway are often associated with high levels of autophagy needed to maintain tumor cell metabolism95–97. For example, pancreatic cancer has very high rates of KRAS mutation and, together with increased activity of transcription factors that promote autophagy60 and pancreatic stellate cells in the tumor microenvironment that use autophagy to fuel tumor cell metabolism98, is thought to cause pancreatic tumors to be especially dependent on autophagy97. Similarly, tumors in mouse models of lung cancer and melanoma driven by the BrafV600E mutation are highly sensitive to Atg7 gene deletion99,100 while autophagy inhibition is sufficient to kill BRAFV600E –expressing, but not wildtype BRAF-expressing brain tumor cell lines93.
These data might lead us to conclude that RAS and BRAF mutant tumors define autophagy-dependency and would be good markers to select patients in whom we should try to inhibit autophagy therapeutically101,102. However, even here there are context-dependent effects that we should keep in mind. Nuclear p53 has been shown to facilitate autophagy while cytoplasmic p53 is associated with inhibition autophagy103,104 indicating that overall the role of p53 in autophagy is complex. Whilst p53 has both autophagy promoting and autophagy inhibiting activities, it is not known if these activities determine whether or not tumor cell growth is increased, or decreased through cell death, upon autophagy inhibition.
In one KRAS mutant mouse pancreatic cancer model, homozygous deletion of Trp53 in the pancreas switched loss of autophagy from being an inhibitor of tumor growth to a promoter105. Based on this study, it was suggested that patients whose tumors had both KRAS and p53 mutations might experience tumor growth following autophagy inhibition106. However, this concern may be unfounded because human pancreatic tumors do not present with homozygous deletion of TP53 occurring simultaneously with activation of KRAS, instead these tumours typically presents as p53 loss of heterozygosity (LOH)52. Subsequent studies performed in mouse models using conditional pancreatic Trp53 LOH that more closely resembles the human disease indicate that p53 status does not affect response to autophagy inhibition in pancreatic cancer52. Huo et al. were able to show that impaired autophagy following monoallelic loss of Becn1 in mice resulted in a reduction of partner and localizer of BRCA2 (Palb2)-associated mammary tumorigenesis (a model of hereditary breast cancer) in the presence of wild type Trp53, but not in a p53 null background107. A similar conclusion was reached using immortalized, HRAS mutant-expressing primary human ovarian surface epithelial cells, skeletal muscle myoblasts and embryonic kidney cells; some displayed growth inhibition when autophagy was blocked, others showed growth promotion108. Moreover, analysis of a large number of human cancer cell lines with KRAS mutations did not find them to be more sensitive to knockdown of ATG genes than tumor cell lines without KRAS mutations35. Taken together these data suggest that while studying RAS and p53 may provide further important insights into the biological mechanism by which autophagy can both promote and inhibit tumor growth, the status of these two genes alone may not identify tumors where autophagy inhibition would be most valuable.
In a panel of breast cancer cell lines selection for or against a library of shRNAs that targeted over 100 autophagy regulators was used to identify those tumor cell lines that could survive and/or proliferate following global genetic interference of the autophagy pathway94. This study revealed that some breast cancer cells grow perfectly well when autophagy is globally inhibited, whilst others are dependent on autophagy for survival. These effects were associated with autophagy regulation of signal transducer and activator of transcription 3 (STAT3) activity and autophagy-dependent secretion of interleukins, especially IL-664. In colon cancer, functional JUN N-terminal kinase 1 (JNK1) was required for hypoxia-induced autophagy109 and ongoing clinical studies are underway to test the use of JNK1 as a marker of autophagy dependence (NCT0120653056). Epidermal growth factor receptor (EGFR) mutated or amplified tumors are another potential target for inhibitors of autophagy. Activation of EGFR leads to the downstream regulation of several pathways that influence autophagy, including PI3K-AKT-mTOR, STAT3, and RAS family signaling and Beclin1-associated signaling pathways110. Specifically, tumors expressing EGFR variant III (EGFRvIII), a common mutation in the extracellular domain of EGFR are shown to require upregulation of metabolism111 and are autophagy dependent112.
Importantly, clinical trials are already utilizing these markers of dependence, or gathering further data for biomarker validation (Table 2). For example, the BRAF, autophagy and MEK inhibition in metastatic melanoma (BAMM) trial (NCT02257424113) is specifically assessing HCQ autophagy inhibition for BRAF V600E or BRAF V600K-expressing metastatic melanoma. An additional trial in glioblastoma will evaluate the role of using EGFRvIII to identify patients who will respond to CQ autophagy inhibition in combination with chemotherapy and radiation (NCT02378532114).
Autophagy in Cancer Escape Mechanisms
There is mounting evidence of the potential role of autophagy in the ability of cancers to develop resistance to chemotherapy. Patients with melanoma whose tumors become resistant to the BRAF inhibitor, vemurafenib, via an ER stress response display higher levels of autophagy115. Moreover, inhibition of autophagy could reverse acquired resistance to vemurafenib that resulted from continued culture of melanoma cell lines in the presence of the drug115. Similarly in the clinical setting, a patient with BRAF mutant brain cancer, who had initially responded to vemurafenib treatment, but then acquired resistance to the drug was successfully treated by a combination of CQ and vemurafenib93. Thus, in this patient, a tumor could be re-sensitized by treatment with the autophagy inhibitor. Importantly though, only the combination therapy of kinase inhibitor with autophagy inhibitor and not autophagy inhibition as a single agent was effective for long-term control of the tumor growth, indicating that the clinical benefit is due to overcoming resistance rather than the acquisition of new sensitivity to autophagy inhibition alone93.
Further laboratory and clinical studies found that genetic and pharmacological autophagy inhibition could overcome multiple molecularly-distinct mechanisms of resistance to BRAF inhibition and was effective in both low and high-grade BRAF mutant brain tumors116. Although only a few patients with clinically-acquired resistance to the BRAF inhibitor have been treated with combinations of CQ and the BRAF inhibitor vemurafenib, it is encouraging that each person obtained clinical benefit suggesting that the autophagy inhibitor is consistently able to overcome resistance to the kinase inhibitor in patients93,116. Additional pre-clinical studies have shown the ability of autophagy inhibition to overcome resistance to tyrosine kinase inhibition in bladder cancer117, thyroid cancer118, NSCLC119,120, and ALK-positive lung cancer121. Because current attempts to circumvent resistance to kinase inhibitors tend to focus on either targeting the same pathway (often the same kinase) in a different way, or targeting a parallel signaling pathway, this strategy of inhibiting an entirely independent process (i.e. autophagy) may represent a fundamentally different way to tackle acquired drug resistance.
Autophagy has also been implicated in resistance to multiple standard chemotherapeutics, often in some of the most difficult to treat tumors. Recent studies have found autophagy induction to be a cause of resistance to the cytotoxic drug paclitaxel in ovarian cancer122. Resistance to the chemotherapy cisplatin has been shown to be due to autophagy induction in ovarian and esophageal cancer123,124, and via hypoxia induced autophagy in lung cancer125. Like in melanoma115, autophagy induction due to an ER stress response results in resistance in primary patient chronic lymphocytic leukemia cells to cyclin dependent kinase (CDK) inhibitors126 and in resistance in glioblastoma cell lines to HDAC inhibitors such as Tubastatin A127. As the link between autophagy and resistance to chemotherapy is strengthened, autophagy will undoubtedly continue to develop as a promising target in cancer therapy128–132.
Autophagy has also been implicated in supporting the survival of dormant tumor cells and, more importantly, may be critical for such tumor cells to start growing again. In pancreatic cancer mouse models where tumor regression was induced by silencing oncogenic KRAS, rare surviving tumor cells that persist after complete inhibition of the oncogenic driver rely, in part, on autophagy133. A recent study using a Drosophila tumor model found that dormant tumors from autophagy-deficient animals reactivate tumor growth when transplanted into autophagy-proficient animals. This suggests that non-tumor cell autonomous autophagy in the surrounding cells of the microenvironment is critical for re-growth of dormant tumors134. If similar effects occur in mammals, this study would suggest that efforts to enhance autophagy after apparently successful treatment of cancer might have the unintended side effect of promoting recurrence from residual dormant tumor cells.
Conclusion
Within the world of oncology, autophagy has competing and context-dependent effects thus a “one size fits all” approach with interventions designed to inhibit or enhance autophagy in cancer therapy will not be successful. Given this situation one might presume the best strategy would be to simply avoid trying to manipulate autophagy at all in cancer therapy. However, altered autophagy is unavoidable. Many of our current treatments (e.g. those that affect the mTOR pathway) themselves affect autophagy. In addition physiological stimuli, especially those that often affect tumors differently compared with normal tissues like nutrient deprivation or hypoxia, will also alter autophagy in the tumor. This means that we need to understand what effect these changes have and try to tailor interventions to the particular situation. Initially at least, such interventions are most likely to revolve around inhibiting autophagy. This means that the key decision is in which patients to offer autophagy inhibition therapy.
Clinical trials utilizing CQ or HCQ as autophagy inhibitors have demonstrated the safety of targeting autophagy for cancer therapy. No devastating neurological toxicities have been observed in patients receiving these agents, suggesting that the neurodegeneration seen in mouse models after complete and irreversible inhibition of autophagy is not necessarily informative of the extent of toxicity that will occur after pharmacological treatment with autophagy inhibitors. The survival benefit associated with combining CQ with the BRAF inhibitor, vemurafenib, in brain tumor93,116 patients provides clinical evidence that autophagy targeted therapy is a feasible clinical strategy in appropriately selected patient populations. To date, the focus of clinical trials has been on the use of lysosomal inhibition with CQ and its derivatives. More potent and autophagy specific inhibitors are in development including better lysosomal inhibitors such as Lys0518 and drugs targeting earlier steps in the autophagy pathway including ULK115,16, VPS3412–14 and ATG4B17 While preliminary data is encouraging, these compounds are still in early pre-clinical studies. Issues with selectivity as well as the need for the use of higher drug concentrations may limit clinical utility and optimization of the lead drugs through chemical modifications of the structures will be needed before moving to clinical trials135.
An important unanswered question that is raised with inhibitors that target early steps in the autophagy pathway is whether it is better to stop the formation of autophagosomes, or to block the degradation of autophagosomes with lysosomal inhibitors. Autophagosomal structures can serve as scaffolds to induce apoptosis136 and necroptosis74,136. Thus, accumulation of autophagosomes might promote such signaling under some circumstances. If this idea is correct, it might be better to block autophagosome degradation with a lysosomal inhibitor rather than inhibit autophagosome formation which might prevent tumor cell killing. And finally, there remains the question of the use of autophagy inducers to prevent oncogenesis. Arguments have been made that increasing autophagy suppresses the development of cancer by limiting genomic mutations, promoting oncogene-induced senescence, and reducing tumor initiating inflammation137. This remains a complex question due to the interaction of autophagy with different genetic backgrounds such as with p53 mutations in pancreatic105 and breast cancer107 where p53 status may influence response to autophagy stimulation, making it either pro- or anti-tumorigenic.
We have begun to combine anti-cancer drugs of many different classes with autophagy inhibitors and inducers, but with little rationale for deciding which combinations to test or a serious attempt to select patients who are most likely to benefit from these therapies. Fortunately, modern clinical trial design often allows collection of samples from tumors and blood before and after treatment. This may aid the development of better biomarkers to serve as pharmacodynamic markers of the efficacy of autophagy inhibitors and better identify which patients we should or should not be treating. If we combine improved clinical studies with detailed molecular and cellular analysis to understand the mechanisms underlying the context-dependent effects of autophagy on cancer it should be possible to develop a more rational basis for deciding when and in which direction we should try to manipulate autophagy during cancer therapy. Since we cannot avoid autophagy being altered in tumors and we know that such alterations will change tumor behavior, ignoring the problem is not a good option; a better answer is to understand the biology and then apply that knowledge in well-designed clinical trials.
Key Points.
Macroautophagy (autophagy) is a highly regulated multi-step process involved in the bulk degradation of cellular proteins and organelles to provide macromolecular precursors that are recycled or used to fuel metabolic pathways.
Autophagy can be targeted for both stimulation and inhibition. Stimulation can be achieved through cellular stress (nutrient deprivation) and mTOR inhibition, while inhibition can be achieved through multiple targets both upstream (ULK1, Beclin 1 and VPS34 inhibitors) as well as downstream at the site of lysosomal fusion with the autophagosome.
Early clinical trials have demonstrated the feasibility and potential benefit of clinically inhibiting autophagy in multiple cancer models including glioblastoma, pancreatic cancer, melanoma, sarcoma, and multiple myeloma.
Ongoing studies are developing novel clinical biomarkers that can be used to monitor autophagy in patients including electron microscopy evaluation of autophagosome number in peripheral blood mononuclear cells and tumor samples, LC3II and ATG13 puncta by immunohistochemistry and novel imaging techniques utilizing positron emission tomography and metabolomics profiles.
The role of autophagy in regulating tumor immune responses is unclear, with arguments both for and against autophagy inhibition. Further research is needed to define the safety and utility of autophagy inhibition while also maximizing tumor immune responses for improved clinical outcomes.
Markers of autophagy dependence have the potential to identify patients who will best respond to autophagy inhibition therapy. Such markers include altered RAS signaling, BRAF mutations, STAT3 activation, autophagy-dependent secretion of interleukins and p53 status.
Autophagy can be an effective cancer escape mechanism and is implicated in the development of resistance is multiple cancers including BRAF mutated central nervous system (CNS) tumors and melanoma, non-small cell lung cancer (NSCLC), bladder cancer, and thyroid cancer. Combination therapy with autophagy inhibition in these cancers has the potential to reduce and reverse resistance to therapy.
Table of Contents Summary.
Autophagy is a process that delivers cytoplasmic components to lysosomes for degradation. This Review discusses clinical interventions to target autophagy in cancer and explains how understanding the context-dependent role of autophagy in cancer should dictate future clinical trial design.
Acknowledgments
Work in our laboratories is supported by: an elope, Inc. St. Baldrick’s Foundation Scholar Award, NIH/NCI (K08CA193982), and the Morgan Adams Foundation (JML). T32 CA190216-1 A1 (CGT) and NIH CA150925, CA190170 and CA197887 (A.T.)
Glossary terms
- Autophagic flux
a measure of the amount of cellular cargo and the rate at which it is degraded through the autophagy pathway.
- Nutraceuticals
a food with medicinal benefit
- Pharmacokinetic (PK)–pharmacodynamic (PD) parameters
the study of the time course of metabolism (PK) and the biochemical and physiological effects (PD) of a drug.
- Maximum tolerated dose (MTD)
the highest dose of a treatment that is effective whilst not causing unacceptable side effects.
- Myelosuppression
a decrease in bone marrow activity resulting in fewer red blood cells, white blood cells, and platelets.
Biographies
Jean M. Mulcahy Levy is an Assistant Professor in the Department of Pediatrics and the Department of Pharmacology at the University of Colorado School of Medicine. She is also a clinical oncologist in the Neuro-Oncology Program in the Center for Cancer and Blood Disorders at Children’s Hospital Colorado. Her laboratory focuses on understanding central nervous system (CNS) tumor biology and the development of new potential therapies for pediatric brain tumors. Specifically, she focuses on the role of autophagy in therapy resistance and autophagy manipulation to improve clinical outcomes in CNS tumors.
Christina G. Towers is a post-doctoral fellow in the Department of Pharmacology at the University of Colorado School of Medicine with Andrew Thorburn. Her research is focused on the role of autophagy in cell death, and the development of novel CRISPR in vitro and in vivo models to better understand the process of autophagy.
Andrew Thorburn is a Professor and Chair of the Pharmacology Department at the University of Colorado School of Medicine. His laboratory focuses on understanding the role of autophagy in cell death, especially as it relates to cancer therapy.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Subject Categories
Biological sciences / Cell biology / Autophagy [URI /631/80/39];
Biological sciences / Cell biology / Cell death [URI /631/80/82]
Biological sciences / Cancer / Cancer therapy [URI /631/67/1059];
Biological sciences / Cancer / Tumour biomarkers [URI /631/67/1857]
References
- 1.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 2.The Nobel Assembly. The Nobel Assembly at Karolinska Instiutet has today decided to award the 2016 Nobel Prize in Physiology or Medicine to Yoshinori Ohsumi. 2016 < https://www.nobelprize.org/nobel_prizes/medicine/laureates/2016/press.html>.
- 3.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, et al. Autophagy in malignant transformation and cancer progression. The EMBO journal. 2015 doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131:130–141. doi: 10.1016/j.pharmthera.2011.03.009. S0163-7258(11)00075-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Towers CG, Thorburn A. Therapeutic Targeting of Autophagy. EBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. A detailed discussion of the protein and membrane interactions required for autophagosome formation. [DOI] [PubMed] [Google Scholar]
- 10.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. Beclin 1 is identified as a putative tumor suppressor gene. [DOI] [PubMed] [Google Scholar]
- 11.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. The definitive consensus of experimental methods appropriate for the study of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bago R, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. The Biochemical journal. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowdle WE, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 14.Ronan B, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 15.Egan DF, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Molecular Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petherick KJ, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. The Journal of biological chemistry. 2015;290:11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akin D, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAfee Q, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodall ML, et al. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014;10:1120–1136. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik SA, et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene. 2011;30:3918–3929. doi: 10.1038/onc.2011.104. [DOI] [PubMed] [Google Scholar]
- 21.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 22.DeBosch BJ, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Science signaling. 2016;9:ra21. doi: 10.1126/scisignal.aac5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino G, Pietrocola F, Madeo F, Kroemer G. Caloric restriction mimetics: natural/physiological pharmacological autophagy inducers. Autophagy. 2014;10:1879–1882. doi: 10.4161/auto.36413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cellular and molecular life sciences : CMLS. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaushik S, et al. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kon M, et al. Chaperone-Mediated Autophagy Is Required for Tumor Growth. Science translational medicine. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou Z, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. Therapy with autophagy inhibition is identified as having combinatory effects with other anti-cancer agents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol. 2014;85:830–838. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YP, et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maycotte P, et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200–212. doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng CH, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci U S A. 2016;113:182–187. doi: 10.1073/pnas.1515617113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maes H, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell. 2014;26:190–206. doi: 10.1016/j.ccr.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurgical focus. 2003;14:e3. doi: 10.3171/foc.2003.14.2.4. Results of the first clinical trial to evaluate the anti-tumour effects of chloroquine showing improved clinical outcomes with autopahgy inhibition in glioblastoma. [DOI] [PubMed] [Google Scholar]
- 38.Briceno E, Calderon A, Sotelo J. Institutional experience with chloroquine as an adjuvant to the therapy for glioblastoma multiforme. Surgical neurology. 2007;67:388–391. doi: 10.1016/j.surneu.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 39.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. 144/5/337 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Eldredge HB, et al. Concurrent Whole Brain Radiotherapy and Short-Course Chloroquine in Patients with Brain Metastases: A Pilot Trial. Journal of radiation oncology. 2013:2. doi: 10.1007/s13566-013-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rojas-Puentes LL, et al. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiation oncology (London, England) 2013;8:209. doi: 10.1186/1748-717x-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnard RA, et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10:1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahalingam D, et al. Combined autophagy and HDAC inhibition: A phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10:1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rangwala R, et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangwala R, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenfeld MR, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogl DT, et al. Combined autophagy and proteasome inhibition: A phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolpin BM, et al. Phase II and Pharmacodynamic Study of Autophagy Inhibition Using Hydroxychloroquine in Patients With Metastatic Pancreatic Adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karsli-Uzunbas G, et al. Autophagy is Required for Glucose Homeostasis and Lung Tumor Maintenance. Cancer discovery. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. An evaluation of the role of genetic knock out of autophagy-related genes and growth of tumour cells in vivo, and the identification of a therapeutic window to inhibit autophagy in lung cancer growth and development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pellegrini P, et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy. 2014;10:562–571. doi: 10.4161/auto.27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, et al. Synthesis of improved lysomotropic autophagy inhibitors. Journal of medicinal chemistry. 2015;58:3025–3035. doi: 10.1021/jm501586m. [DOI] [PubMed] [Google Scholar]
- 52.Yang A, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer discovery. 2014;4:905–913. doi: 10.1158/2159-8290.cd-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boone BA, et al. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01881451?term=NCT01881451&rank=1.
- 55.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02233387?term=NCT02233387&rank=1.
- 56.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01206530?term=NCT01206530&rank=1.
- 57.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02042989?term=NCT02042989&rank=1.
- 58.Fullgrabe J, Heldring N, Hermanson O, Joseph B. Cracking the survival code: autophagy-related histone modifications. Autophagy. 2014;10:556–561. doi: 10.4161/auto.27280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, et al. Next-generation proteasome inhibitor MLN9708 sensitizes breast cancer cells to doxorubicin-induced apoptosis. Scientific reports. 2016;6:26456. doi: 10.1038/srep26456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Follo C, Barbone D, Richards WG, Bueno R, Broaddus VC. Autophagy initiation correlates with the autophagic flux in 3D models of mesothelioma and with patient outcome. Autophagy. 2016;12:1180–1194. doi: 10.1080/15548627.2016.1173799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-Dependent Production of Secreted Factors Facilitates Oncogenic RAS-Driven Invasion. Cancer discovery. 2014;4:466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kraya AA, et al. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy. 2015;11:60–74. doi: 10.4161/15548627.2014.984273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion. Mol Cancer Res. 2015 doi: 10.1158/1541-7786.MCR-14-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varadarajulu S, Bang JY. Role of Endoscopic Ultrasonography and Endoscopic Retrograde Cholangiopancreatography in the Clinical Assessment of Pancreatic Neoplasms. Surg Oncol Clin N Am. 2016;25:255–272. doi: 10.1016/j.soc.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. Basic review of the history and core machinary invovled in the process of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altman JK, et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin Cancer Res. 2014;20:2400–2409. doi: 10.1158/1078-0432.ccr-13-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kun Z, et al. Gastrin enhances autophagy and promotes gastric carcinoma proliferation via inducing AMPKalpha. Oncology research. 2017 doi: 10.3727/096504016x14823648620870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masui A, et al. Autophagy as a Survival Mechanism for Squamous Cell Carcinoma Cells in Endonuclease G-Mediated Apoptosis. PLoS One. 2016;11:e0162786. doi: 10.1371/journal.pone.0162786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan Q, et al. Role of Autophagy as a Survival Mechanism for Hypoxic Cells in Tumors. Neoplasia (New York, NY) 2016;18:347–355. doi: 10.1016/j.neo.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Fitzwalter BE, Thorburn A. Recent insights into cell death and autophagy. FEBS J. 2015;282:4279–4288. doi: 10.1111/febs.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gump JM, et al. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol. 2014;16:47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorburn J, et al. Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep. 2014;7:45–52. doi: 10.1016/j.celrep.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodall ML, et al. The Autophagy Machinery Controls Cell Death Switching between Apoptosis and Necroptosis. Developmental Cell. 2016;37:337–349. doi: 10.1016/j.devcel.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rao S, Yang H, Penninger JM, Kroemer G. Autophagy in non-small cell lung carcinogenesis: A positive regulator of antitumor immunosurveillance. Autophagy. 2014;10:529–531. doi: 10.4161/auto.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–227. doi: 10.1016/j.immuni.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 77.Townsend KN, et al. Autophagy inhibition in cancer therapy: metabolic considerations for antitumor immunity. Immunol Rev. 2012;249:176–194. doi: 10.1111/j.1600-065X.2012.01141.x. [DOI] [PubMed] [Google Scholar]
- 78.Ko A, et al. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ. 2014;21:92–99. doi: 10.1038/cdd.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michaud M, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 80.Lechner MG, et al. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother. 2013;36:477–489. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Starobinets H, et al. Antitumor adaptive immunity remains intact following inhibition of autophagy and antimalarial treatment. J Clin Invest. 2016;126:4417–4429. doi: 10.1172/jci85705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pietrocola F, et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell. 2016;30:147–160. doi: 10.1016/j.ccell.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, et al. The vitamin E analogue alpha-TEA stimulates tumor autophagy and enhances antigen cross-presentation. Cancer Res. 2012;72:3535–3545. doi: 10.1158/0008-5472.CAN-11-31030008-5472.CAN-11-3103. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ladoire S, et al. Combined evaluation of LC3B puncta and HMGB1 expression predicts residual risk of relapse after adjuvant chemotherapy in breast cancer. Autophagy. 2015;11:1878–1890. doi: 10.1080/15548627.2015.1082022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ladoire S, et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy. 2016;12:864–875. doi: 10.1080/15548627.2016.1154244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baginska J, et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc Natl Acad Sci U S A. 2013;110:17450–17455. doi: 10.1073/pnas.1304790110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang X, et al. Inhibiting systemic autophagy during interleukin 2 immunotherapy promotes long-term tumor regression. Cancer Res. 2012;72:2791–2801. doi: 10.1158/0008-5472.can-12-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT03057340?term=NCT03057340&rank=1.
- 89.Page DB, et al. Glimpse into the future: harnessing autophagy to promote anti-tumor immunity with the DRibbles vaccine. J Immunother Cancer. 2016;4:25. doi: 10.1186/s40425-016-0130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu GLY, Cui Z, Morris NP, Weinberg AD, Fox BA, Urba WJ, Wang L, Hu HM. Combinational immunotherapy with allo-DRibble vacciens and anti-OX40 co-stimulation leads to generation of cross-reactive effector T cells and tumor regression. Scientific reports. 2016;6 doi: 10.1038/srep37558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hilton TSR, Boulmay B, Li R, Spieler B, Happel K, Paustian C, Moudgil T, Dubay C, Fisher B, Mederos E, Ochoa A, Urba WJ, Hu HM, Fox B. Preliminary analysis of immune responses in patients enrolled in a Phase II trial of cyclophosphamide wiht allogenic DRibble vaccine alone (DPV-001) or with GM-CSF or imiquimod for adjuvant treatment of stage IIIA or IIIB NSCLC. J Immunother Cancer. 2014;2:249. doi: 10.1186/2051-1426-2-S3-P249. [DOI] [Google Scholar]
- 92.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01956734?term=NCT01956734&rank=1.
- 93.Levy JMM, et al. Autophagy Inhibition Improves Chemosensitivity in BRAFV600E Brain Tumors. Cancer discovery. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maycotte P, et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res. 2014;74:2579–2590. doi: 10.1158/0008-5472.can-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. mbc.E10-06-0500 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sousa CM, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strohecker AM, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer discovery. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie X, Koh JY, Price S, White E, Mehnert JM. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer discovery. 2015;5:410–423. doi: 10.1158/2159-8290.cd-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–1306. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thorburn A, Morgan MJ. Targeting Autophagy in BRAF-Mutant Tumors. Cancer discovery. 2015;5:353–354. doi: 10.1158/2159-8290.CD-15-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang J, Di J, Cao H, Bai J, Zheng J. p53-mediated autophagic regulation: A prospective strategy for cancer therapy. Cancer Lett. 2015;363:101–107. doi: 10.1016/j.canlet.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 105.Rosenfeldt MT, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 106.Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N Engl J Med. 2014;370:1352–1353. doi: 10.1056/NEJMcibr1400189. [DOI] [PubMed] [Google Scholar]
- 107.Huo Y, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer discovery. 2013;3:894–907. doi: 10.1158/2159-8290.cd-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morgan MJ, et al. Regulation of autophagy and chloroquine sensitivity by oncogenic RAS in vitro is context-dependent. Autophagy. 2014;10:1814–1826. doi: 10.4161/auto.32135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vasilevskaya IA, Selvakumaran M, Roberts D, O’Dwyer PJ. JNK1 Inhibition Attenuates Hypoxia-Induced Autophagy and Sensitizes to Chemotherapy. Mol Cancer Res. 2016;14:753–763. doi: 10.1158/1541-7786.mcr-16-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jutten B, Rouschop KM. EGFR signaling and autophagy dependence for growth, survival, and therapy resistance. Cell Cycle. 2014;13:42–51. doi: 10.4161/cc.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo D, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc Natl Acad Sci U S A. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jutten B, et al. EGFR overexpressing cells and tumors are dependent on autophagy for growth and survival. Radiother Oncol. 2013;108:479–483. doi: 10.1016/j.radonc.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 113.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02257424?term=NCT02257424&rank=1.
- 114.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02378532?term=NCT02378532&rank=1.
- 115.Ma X-H, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mulcahy Levy JM, et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. eLife. 2017;6 doi: 10.7554/eLife.19671. The first demonstration of the use of autophagy inhibition to overcome kinase inhibitor resistance in patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kang M, et al. Concurrent Autophagy Inhibition Overcomes the Resistance of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Human Bladder Cancer Cells. International journal of molecular sciences. 2017;18 doi: 10.3390/ijms18020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang W, et al. Targeting autophagy sensitizes BRAF-mutant thyroid cancer to vemurafenib. The Journal of clinical endocrinology and metabolism. 2016:jc20161999. doi: 10.1210/jc.2016-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu JT, et al. Autophagy Inhibition Overcomes the Antagonistic Effect Between Gefitinib and Cisplatin in Epidermal Growth Factor Receptor Mutant Non--Small-Cell Lung Cancer Cells. Clinical lung cancer. 2015;16:e55–66. doi: 10.1016/j.cllc.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 120.Zou Y, et al. The autophagy inhibitor chloroquine overcomes the innate resistance of wild-type EGFR non-small-cell lung cancer cells to erlotinib. J Thorac Oncol. 2013;8:693–702. doi: 10.1097/JTO.0b013e31828c7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ji C, et al. Induction of autophagy contributes to crizotinib resistance in ALK-positive lung cancer. Cancer Biol Ther. 2014;15:570–577. doi: 10.4161/cbt.28162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang SF, et al. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:225–238. doi: 10.1080/15548627.2014.998931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–17173. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yu L, et al. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355:34–45. doi: 10.1016/j.canlet.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 125.Wu HM, Jiang ZF, Ding PS, Shao LJ, Liu RY. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Sci Rep. 2015;5:12291. doi: 10.1038/srep12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mahoney E, et al. ER stress and autophagy: new discoveries in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor flavopiridol. Blood. 2012;120:1262–73. doi: 10.1182/blood-2011-12-400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li ZY, et al. A novel HDAC6 inhibitor Tubastatin A: Controls HDAC6-p97/VCP-mediated ubiquitination-autophagy turnover and reverses Temozolomide-induced ER stress-tolerance in GBM cells. Cancer Lett. 2017;391:89–99. doi: 10.1016/j.canlet.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 128.Aveic S, Tonini GP. Resistance to receptor tyrosine kinase inhibitors in solid tumors: can we improve the cancer fighting strategy by blocking autophagy? Cancer Cell Int. 2016;16:62. doi: 10.1186/s12935-016-0341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen S, et al. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–9. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 130.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–42. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 131.Sannigrahi MK, Singh V, Sharma R, Panda NK, Khullar M. Role of autophagy in head and neck cancer and therapeutic resistance. Oral Dis. 2015;21:283–91. doi: 10.1111/odi.12254. [DOI] [PubMed] [Google Scholar]
- 132.Sui X, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Katheder NS, et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pasquier B. Autophagy inhibitors. Cell Mol Life Sci. 2016;73:985–1001. doi: 10.1007/s00018-015-2104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Basit F, Cristofanon S, Fulda S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013;20:1161–73. doi: 10.1038/cdd.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res (Phila) 2011;4:973–83. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bedoya V. Effect of chloroquine on malignant lymphoreticular and pigmented cells in vitro. Cancer Res. 1970;30:1262–1275. The anti-malarial drug chloroquine is first shown to inhibit tumour cell growth in vitro with a notation of the accumulation of autophagic vacuoles. [PubMed] [Google Scholar]
- 139.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. The Ohsumi group clones the first autophagy specific gene, APG13 (ATG1) [DOI] [PubMed] [Google Scholar]
- 140.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. The Ohsumi group demonstrate direct involvement of protein phosphorylation in regulating autophaygy. [DOI] [PubMed] [Google Scholar]
- 141.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. The first autophagy specific genes in higher eukaryotes are identified. [DOI] [PubMed] [Google Scholar]
- 142.Murakami N, et al. Accumulation of tau in autophagic vacuoles in chloroquine myopathy. Journal of neuropathology and experimental neurology. 1998;57:664–673. doi: 10.1097/00005072-199807000-00003. The first study to observe that chloroquine can inhibit autophagy and the connection between accumulation of cellular proteins can be correlated to inhibition of lysosomal degradation. [DOI] [PubMed] [Google Scholar]
- 143.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. The first autophagy deficient mouse (Atg5−/−) is creating, implicating autophagy in development. [DOI] [PubMed] [Google Scholar]
- 144.Rao S, et al. A dual role for autophagy in a murine model of lung cancer. Nature communications. 2014;5:3056. doi: 10.1038/ncomms4056. Early in vivo model demonstrating the ability of autophagy to repress early oncogeneis, but support late-stage cancer growth. [DOI] [PubMed] [Google Scholar]
- 145.Chi KH, et al. Addition of rapamycin and hydroxychloroquine to metronomic chemotherapy as a second line treatment results in high salvage rates for refractory metastatic solid tumors: a pilot safety and effectiveness analysis in a small patient cohort. Oncotarget. 2015;6:16735–16745. doi: 10.18632/oncotarget.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chi MS, et al. Double autophagy modulators reduce 2-deoxyglucose uptake in sarcoma patients. Oncotarget. 2015;6:29808–29817. doi: 10.18632/oncotarget.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bilger A, et al. FET-PET-based reirradiation and chloroquine in patients with recurrent glioblastoma: first tolerability and feasibility results. Strahlentherapie und Onkologie : Organ der Deutschen Rontgengesellschaft … [et al] 2014;190:957–961. doi: 10.1007/s00066-014-0693-2. [DOI] [PubMed] [Google Scholar]
- 148.Goldberg SB, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01594242?term=NCT01594242&rank=1.
- 150.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01874548?term=NCT01874548&rank=1.
- 151.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01506973?term=NCT01506973&rank=1.
- 152.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01978184?term=NCT01978184&rank=1.
- 153.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01128296?term=NCT01128296&rank=1.
- 154.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01897116?term=NCT01897116&rank=1.