Abstract
We have previously shown that aberrant promoter methylation of ZNF331 is a potential biomarker for colorectal cancer detection with high sensitivity (71%) and specificity (98%). This finding was recently confirmed by others, and it was additionally suggested that promoter methylation of ZNF331 was an independent prognostic biomarker for colorectal cancer (n = 146). In the current study, our initial colorectal cancer sample series was extended to include a total of 423 cancer tissue samples. Aberrant promoter methylation was found in 71% of the samples, thus repeatedly suggesting the biomarker potential of ZNF331 for detection of colorectal cancer. Furthermore, multivariate Cox’s analysis indicated a trend towards inferior overall survival for colorectal cancer patients with aberrant methylation of ZNF331.
Keywords: Colorectal cancer, Diagnosis, DNA methylation, Prognosis, ZNF331
Introduction
In cancer, increased promoter DNA methylation is a frequent event commonly occurring early in tumor development. Methylated DNA sequences may serve as tumor biomarkers in liquid biopsies for detecting cancer and for predicting patient prognosis [1].
In 2011, we filed a patent application covering methylation of ZNF331 (Zinc finger protein 331) as a biomarker for gastrointestinal cancers [2]. ZNF331 was shown by Yu et al. to be inactivated by promoter methylation in gastric cancer, providing the cancer cells with increased growth potential and invasiveness [3]. We also found a high methylation frequency in patients with gastric cancer (80%) and to a lesser extent in patients with pancreatic cancer (40%) and cholangiocarcinomas (26%) [4]. Most importantly, we reported high sensitivity (71%) and specificity (98%) for ZNF331 methylation in colorectal cancer early 2015, strengthening the potential of ZNF331 as a biomarker for colorectal cancer detection [4]. Interestingly, these findings were recently confirmed, further supporting the biomarker potential of ZNF331 in colorectal cancer [5]. The same study also suggested aberrant promoter methylation of ZNF331 as an independent prognostic marker for colorectal cancer, analyzing 146 samples [5]. In the present study, we analyzed the effect of ZNF331 methylation on overall survival, including altogether 423 colorectal tissue samples.
Results and discussion
Methylation of the ZNF331 promoter was found in 71% (301/423) of the patients with colorectal cancer and was associated with localization in the right colon, microsatellite instability (MSI), and the BRAFV600E mutation. Furthermore, ZNF331 methylation was strongly associated with CpG island methylator phenotype (CIMP) and MLH1 methylation (Table 1). Wang et al. [5] reported a similar methylation frequency of ZNF331 in colorectal cancer (67%; 98/146). However, in contrast to our data Wang et al. did not find associations between methylated ZNF331 and BRAF mutation, CIMP nor MLH1 methylation, which may be explained by differences in sample size (Wang et al., n = 146; current study, n = 423), marker panels to define CIMP, method to identify methylation, age (median age Wang et al. 60; current study 72), and/or ethnicity (Wang et al.: Asian; current study: Caucasian).
Table 1.
Associations between ZNF331 methylation and clinical and molecular features
| Total | ZNF331 unmethylated | ZNF331 methylated | P value | |
|---|---|---|---|---|
| n | n (%) | n (%) | ||
| No. of patients | 423 | 122 (29) | 301 (71) | |
| Gender | 0.165 | |||
| Male | 213 | 68 (32) | 145 (68) | |
| Female | 210 | 54 (26) | 156 (74) | |
| Age | 0.074 | |||
| < 60 | 70 | 26 (37) | 44 (63) | |
| 60–74 | 178 | 55 (31) | 123 (69) | |
| ≥ 75 | 175 | 41 (23) | 134 (77) | |
| Stage | 0.683 | |||
| I | 79 | 20 (25) | 59 (75) | |
| II | 169 | 51 (30) | 118 (70) | |
| III | 118 | 32 (27) | 86 (73) | |
| IV | 56 | 19 (34) | 37 (66) | |
| Localization | < 0.001 | |||
| Right colon | 167 | 27 (16) | 140 (84) | |
| Left colon | 130 | 47 (36) | 83 (64) | |
| Rectum | 121 | 46 (38) | 75 (62) | |
| MSI status | < 0.001 | |||
| MSS | 325 | 111 (34) | 214 (66) | |
| MSI | 89 | 8 (9) | 81 (91) | |
| BRAF | < 0.001 | |||
| BRAF wt | 356 | 120 (34) | 236 (66) | |
| BRAF mut | 67 | 2 (3) | 65 (97) | |
| CIMP | < 0.001 | |||
| CIMP− | 355 | 121 (34%) | 234 (66) | |
| CIMP+ | 65 | 0 (0) | 65 (100) | |
| MLH1 methylation | < 0.001 | |||
| MLH1 unmeth | 360 | 117 (32.5) | 243 (67.5) | |
| MLH1 meth | 60 | 4 (7) | 56 (93) | |
| Series | 0.439 | |||
| Oslo 3 | 59 | 14 (24) | 45 (76) | |
| Oslo 2 | 364 | 108 (30) | 256 (70) |
Meth methylated, mut mutation, No. number, unmeth unmethylated, wt wild type
Wang et al. [5] further reported that patients with ZNF331 promoter methylation had a worse prognosis than patients with unmethylated promoters. Our results were in accordance with their study, although statistical significance was not reached in the multivariate Cox regression model adjusting for age and stage (HR = 1.44 (0.97–2.14), P = 0.069; Table 2). The univariate model is presented in Fig. 1 (P = 0.143).
Table 2.
Multivariate Cox proportional hazard analysis with overall survival as endpoint
| Patients, n | Multivariate HR (95% CI) | P value | |
|---|---|---|---|
| Age | |||
| < 60 | 70 | 1.00 (ref) | |
| 60–74 | 176 | 1.70 (0.91–3.18) | 0.099 |
| ≥ 75 | 173 | 3.42 (1.84–6.34) | < 0.001 |
| Stage | |||
| I | 78 | 1.00 (ref) | |
| II | 168 | 1.24 (0.66–2.34) | 0.498 |
| III | 117 | 2.32 (1.24–4.34) | 0.009 |
| IV | 56 | 11.10 (5.91–20.85) | < 0.001 |
| ZNF331 methylation | |||
| ZNF331 unmeth | 121 | 1.00 (ref) | |
| ZNF331 meth | 298 | 1.44 (0.97–2.14) | 0.069 |
Variables not selected by the backward likelihood method to be included in the final model: series, gender, CIMP-, MSI-, and BRAF mutation status
Meth methylated, unmeth unmethylated
Fig. 1.
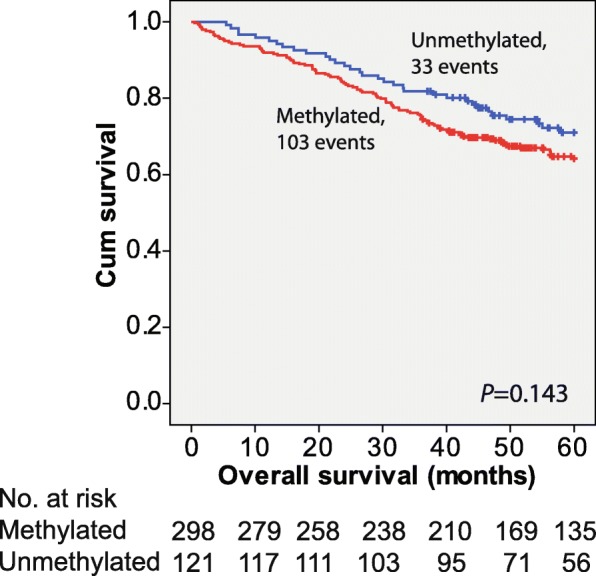
Effect of ZNF331 promoter methylation on overall survival modeled by the Kaplan-Meier method and compared using the log-rank test
In conclusion, in an extended series of colorectal cancer samples, we have showed the potential of promoter methylation of ZNF331 as a biomarker for colorectal cancer detection. We have further provided data indicating a trend towards poorer prognosis for patients with ZNF331 methylation.
Material and methods
Colorectal cancer tissue samples
This study included 423 colorectal cancer tissue samples. Fifty-nine of the samples were obtained from several different hospitals in the southeast region of Norway in the period 1987–1989 (Oslo 3 series; described in [6]), and 364 of the samples were obtained from patients undergoing surgical resection at the Oslo University Hospital–Aker from 2005 to 2011 (Oslo 2 series; described in [7, 8]). Survival data was available for 419 patients (Oslo 3, n = 59; Oslo 2, n = 360).
Bisulfite treatment and quantitative methylation-specific PCR (qMSP)
DNA from cancer tissue samples were bisulfite treated using the EpiTect Bisulfite Kit (Qiagen), and the samples were purified using the QIAcube (Qiagen).
Quantitative methylation-specific PCR (qMSP) was used to analyze the methylation of the ZNF331 promoter (NM_018555), with primers and probe sequences as reported earlier [4]. The method was performed as previously described [4, 9], with the ALU-C4 element as a normalization control [10]. As described in ref. [4], samples with percent methylated reference (PMR) values ≥ 1 were considered methylated. Information about MSI, CIMP, MLH1 methylation, and BRAF mutation status were available from previous studies [11, 12].
Statistical analyses
Associations between ZNF331 methylation and clinicopathological data were analyzed by Pearson chi-square or Fisher’s exact tests. For all analyses, patients were divided into three age groups (< 60 years, 60–74 years, and ≥ 75 years). Breakpoints were chosen as previously described [11]. Overall survival was used as endpoint in the survival analyses and was calculated from time of surgery until death of any cause. Cases were censored at last follow-up. The univariate effect of ZNF331 on survival was modeled by the Kaplan-Meier method and compared using the log-rank test. A multivariate Cox’s proportional hazard model was generated by a stepwise selection procedure (backward likelihood model) in order to identify a subset of relevant predictor variables from the set of available clinicopathological data (series, age, stage, gender, CIMP-, MSI-, BRAF-, and ZNF331 methylation status). Hazard ratios (HRs) and 95% confidence intervals (CIs) were derived from the model, and significance of the parameters was assessed using Wald’s test. To evaluate the assumption of proportionality, a chi-square test was performed. A P value < 0.05 was considered statistically significant. The analyses were performed using IBM SPSS Statistics 21 and R version 3.4.1.
Acknowledgments
Funding
This work was supported by grants from the South-Eastern Norway Regional Health Authority (project number 2016071 to G.E. Lind, funding HM Vedeld as a postdoc).
Availability of data and materials
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Abbreviations
- CIMP
CpG island methylator phenotype
Authors’ contributions
GEL contributed to the conception and design. HMV, AN, and RAL contributed to the acquisition of data. HMV, AN, RAL, and GEL contributed to the analyses and interpretation of the data. HMV contributed to the drafting of the manuscript. All authors were involved in the revision of the manuscript and have approved the final version.
Ethics approval and consent to participate
The research biobanks have been registered according to national legislation (numbers 2781 and 236-2005-16141). The study is part of a project approved by the Regional Committee (REC) for Medical and Health Research Ethics (numbers 1.2005.1629 and S-09282c 2009/4958).
Competing interests
RAL and GEL are inventors of a US provisional patent application filed in 2011, describing methylation of ZNF331 and five additional genes as biomarkers for detection of gastrointestinal cancers (61/451,198, INVEN-31899/US-1/PRO). The rest of the authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: the current state and clinical perspectives. Semin Cancer Biol. 2017; 10.1016/j.semcancer.2017.12.004. [DOI] [PMC free article] [PubMed]
- 2.Lothe RA, Ahmed D, Andresen K et al. Methods and biomarkers for detection of gastrointestinal cancers. US provisional application filed 61/451,198, INVEN-31899/US-1/PRO 2011.
- 3.Yu J, Liang QY, Wang J, et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2012;32:307–317. doi: 10.1038/onc.2012.54. [DOI] [PubMed] [Google Scholar]
- 4.Vedeld HM, Andresen K, Eilertsen IA, et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, He T, Herman JG, et al. Methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin Epigenetics. 2017;9:115. doi: 10.1186/s13148-017-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 7.Sveen A, Agesen TH, Nesbakken A, et al. ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res. 2012;18:6001–6010. doi: 10.1158/1078-0432.CCR-11-3302. [DOI] [PubMed] [Google Scholar]
- 8.Berg M, Danielsen SA, Ahlquist T, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind GE, Danielsen SA, Ahlquist T, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. MolCancer. 2011;10:85. doi: 10.1186/1476-4598-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vedeld HM, Merok M, Jeanmougin M, et al. CpG island methylator phenotype identifies high risk patients among microsatellite stable BRAF mutated colorectal cancers. Int J Cancer. 2017;141:967–976. doi: 10.1002/ijc.30796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merok MA, Ahlquist T, Royrvik EC, et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol. 2013;24:1274–1282. doi: 10.1093/annonc/mds614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.


