Abstract
Kinase gene fusions are important drivers of oncogenic transformation and can be inhibited with targeted therapies. Clinical grade diagnostics using RNA sequencing to detect gene rearrangements in solid tumors are limited, and the few that are available require prior knowledge of fusion break points. To address this, we have analytically validated a targeted RNA sequencing assay (OSU-SpARKFuse) for fusion detection that interrogates complete transcripts from 93 kinase and transcription factor genes. From a total of 74 positive and 36 negative control samples, OSU-SpARKFuse had 93.3% sensitivity and 100% specificity for fusion detection. Assessment of repeatability and reproducibility revealed 96.3% and 94.4% concordance between intrarun and interrun technical replicates, respectively. Application of this assay on prospective patient samples uncovered OLFM4 as a novel RET fusion partner in a small-bowel cancer and led to the discovery of a KLK2-FGFR2 fusion in a patient with prostate cancer who subsequently underwent treatment with a pan–fibroblast growth factor receptor inhibitor. Beyond fusion detection, OSU-SpARKFuse has built-in capabilities for discovery research, including gene expression analysis, detection of single-nucleotide variants, and identification of alternative splicing events.
The cancer-driving chromosomal rearrangement BCR-ABL1 was initially described >50 years ago in chronic myeloid leukemia.1, 2 Since that time, technological advancements have enabled the discovery of gene fusions in many solid tumors, including carcinomas of the thyroid, salivary gland, prostate, lung, breast, head and neck, brain, skin, gastrointestinal tract, and kidney.3 In particular, the advent of next-generation sequencing (NGS) has radically changed the landscape of gene fusions in cancer, with 90% of the nearly 10,000 gene fusions that have been reported being identified by sequencing approaches.4 Methods using both DNA sequencing (DNAseq) and RNA sequencing (RNAseq) have been used to identify rearrangements in tumor specimens; however, DNA approaches do not distinguish expressed gene fusions likely representing driver events from nonexpressed passenger fusion events.5, 6, 7 Therefore, RNAseq data have been extensively used to identify chimeric transcripts in diverse solid tumors.8, 9 Examination of transcriptome data from 4300 primary tumor samples representing 13 tumor types in the Cancer Genome Atlas revealed in-frame protein kinase fusions to be present in >7% of samples.9 Importantly, druggable kinase fusions that involve ALK, ROS1, RET, NTRKs, and FGFRs were detected at a frequency of 1% to 9% in individual cancer types examined.
Despite its broad potential, whole transcriptome sequencing is not ideal for routine clinical grade testing for individual patients because of the massive scale, cost, and prolonged turnaround times. Several research groups have used a targeted approach, similar to whole exome capture, to focus on genes of interest in the transcriptome. An initial study using the leukemia cell line K-562 found successful enrichment of select cancer-related transcripts using complementary oligonucleotide probes, including a significant increase in the number of sequenced reads identifying the BCR-ABL1 kinase fusion.10 Subsequent commercial and research grade applications have been developed, including a capture kit to sequence the human RNA kinome, sequencing of noncoding RNAs, and whole exome capture of the transcriptome.11, 12, 13, 14 Although targeted RNAseq is a valuable research tool, this method has not yet been applied for clinical grade testing in solid tumor specimens. Several groups have used other RNAseq strategies to detect gene rearrangements in clinical tumor specimens; however, these methods are limited by a need for prior knowledge of fusion break points and directionality.15, 16 Recently, a targeted capture method using both DNAseq and RNAseq was validated for identification of base substitutions, copy number alterations (CNAs), and genomic rearrangements, however this assay has limited utility as it is currently only available for use on samples derived from hematological malignancies.17 Clinical reporting of gene fusions in solid tumors is paramount to patient care because rearrangements that involve ALK and RET are proven targets for therapy in non–small-cell lung cancer.18, 19, 20, 21 In addition, RET, NTRK1, and BRAF rearrangements have been successfully targeted in the clinic and are therefore viable targets in patients with appropriate molecular profiles.22, 23, 24
We describe the analytic validation of a targeted RNAseq assay to detect gene fusions that involve 93 kinase and transcription factor (TF) genes in solid tumor specimens performed in our Clinical Laboratory Improvement Amendments–certified laboratory. A cohort of 110 positive and negative control samples was used for determining assay sensitivity and specificity. We generated serial dilutions of positive control cell lines that contained nine unique gene fusions to examine detection limits for the assay and also to assess intrarun repeatability and interrun reproducibility. Application of the assay on 95 prospective patient specimens revealed novel fusion partners for both RET and FGFR2 and additionally identified a well-characterized resistance mutation in a patient with leukemia and a MET exon skipping event in a lung cancer sample. Implementation of targeted RNAseq in clinical laboratories will help expand the knowledge base of gene fusions in solid tumors and has the potential to directly affect patient care by detecting therapeutically actionable targets.
Materials and Methods
Probe Design
For our 93 target kinase/TF genes (Supplemental Table S1), all reference sequence (RefSeq) transcripts were identified using the University of California, Santa Cruz (UCSC) Genome Browser. Nonoverlapping 5′biotinylated 120-mer probes were designed for each RefSeq transcript and were allowed to cross exon-exon junctions. For exons <120 bp, an additional probe was designed that was centered over the exon and extended into adjacent intronic regions. Probes that covered the same positions and splice junctions were removed using custom scripts; however, two pairs of duplicate probes were inadvertently included in the final design. A total of 3143 probes were selected for the kinase genes and TFs. An additional 149 nonoverlapping probes were designed to target four 5000-bp genomic regions (chr2:176550000-176555000, chr3:83270000-83275000, chr6:99275000-99280000, and chr12:28125000-28130000) to assess DNA contamination. These regions were selected based on high complexity and lack of gaps in the reference sequence. Probes that overlapped with low complexity regions (as defined by RepeatMasker) were removed from the final design. Ten control transcripts from the External RNA Controls Consortium (ERCC) were targeted with a total of 73 nonoverlapping probes. Nine housekeeping genes were selected based on their relative abundance in 14 Cancer Genome Atlas RNAseq data sets, representing a variety of cancer types. The selected control genes had similar expression rates as targeted kinase genes. Nonoverlapping 120mer probes were selected as described above, with additional probes added for exons <120 bp. In all, 157 probes were selected for the nine control genes. To assess the overall specificity of our probe design, we performed a BLAST search against the human transcriptome. Most probes (3212 of 3522) had only one result. Of the remaining 310 probes, 222 are control probes intended to target genomic DNA or ERCC sequences not present in the human transcriptome. A total of 88 probes that targeted kinase/TF genes had off-target binding sites within the transcriptome; however, these probes were included with the understanding that some minimal off-target capture might occur. A list of all probe sequences can be found in Supplemental Table S2.
RNA Extraction and Quality Control Assessment
RNA was extracted from cell lines and fresh-frozen tissues using the miRNeasy Kit (Qiagen, Valencia, CA) per the manufacturer's protocol. RNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissues using the miRNAeasy FFPE kit (Qiagen). Sample quality was assessed using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and TapeStation 2200 (Agilent, Santa Clara, CA).
rRNA Depletion, cDNA Synthesis, Library Preparation and Amplification, Targeted Capture, and Illumina Sequencing
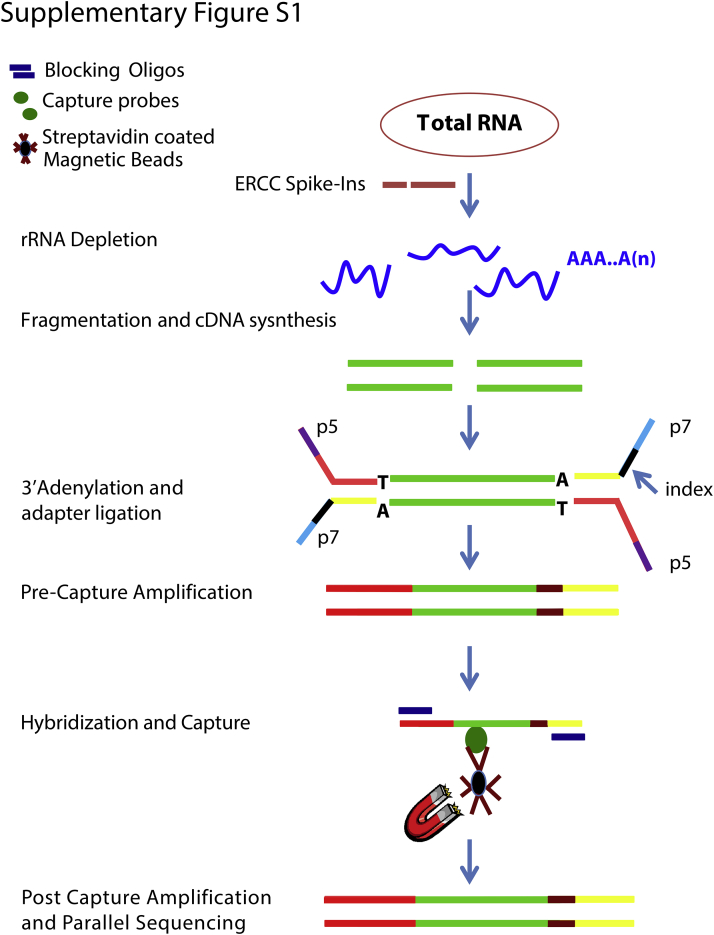
Ambion ERCC Spike-In Mix (Thermo Fisher Scientific) was added to total RNA (250 ng). Ribo-Zero (Illumina, San Diego, CA) rRNA depletion was performed followed by chemical fragmentation, cDNA synthesis, A-tailing, and ligation of unique sequencing indexes using Illumina TruSeq Stranded Total RNA Library Kit. Fresh-frozen tissue and cell line RNA was fragmented for 8 minutes. All FFPE RNA and degraded RNA with a percentage of RNA fragments >200 nucleotides (DV200) ≤30% skipped fragmentation and proceeded to first strand cDNA synthesis. Adapter ligated cDNA was then PCR amplified for a total of 15 cycles. Libraries were quantitated using Qubit dsDNA HS Kit (Invitrogen, Carlsbad, CA) and quality assessed with TapeStation 2200 (Agilent) as per the manufacturer's protocol. Four libraries were then pooled at 125 ng each for a total of 500 ng. Cot-1 DNA (Sigma-Aldrich, St. Louis, MO), universal blocking oligonucleotides (IDT, Coralville, IA), and adapter-specific blocking oligonucleotides (IDT) were added to the pooled libraries and dried in a SpeedVac. The dried mixture was then resuspended in NimbleGen 2× Hybridization Buffer and Hybridization Component A (Roche, Madison, WI) as per IDT xGen Lockdown Probes Protocol (IDT) and hybridized for 16 to 18 hours with OSU-SpARKFuse custom probes (IDT). Streptavidin DynaBeads (Invitrogen) were used for capture and wash was performed using NimbleGen Hybridization and Wash Kit (Roche). Final hybridized product was amplified using KAPA Hifi HotStart Ready Mix (KAPA Biosystems, Wilmington, MA) and Illumina sequencing primers (Sigma-Aldrich) for a total of 12 cycles. Final library quantification was performed using Qubit dsDNA HS kit (Life Technologies) and TapeStation 2200 (Agilent). Paired-end 2 × 100-bp sequencing was performed on the Illumina MiSeq Desktop Sequencer using the MiSeq Reagent 300-cycle Kit v2. In brief, pooled captured libraries were denatured, diluted to 10 to 20 pmol/L and loaded on a flow cell.
Cell Culture
H2228 (CRL-3935; ATCC, Manassas, VA), HCC-78 (ACC 563; Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany), lymphoblastoid cell lines (Coriell, Camden, NJ; generously donated by Dr. Michael Snyder, Stanford University, Palo Alto, CA), MEG-01 (CRl-2021; ATCC), and EOL-1 (ACC386; DSMZ) were cultured in RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 2 mmol/L l-glutamine (Sigma-Aldrich) and 10% fetal bovine serum (FBS) (Sigma-Aldrich). NALM-1 (DSMZ) was cultured in RPMI 1640 medium supplemented with 2 mmol/L l-glutamine and 15% FBS. KG1a (CCl-243; ATCC), HL60 (CCL-240; ATCC), and K-562 (CCL-243; ATCC) were cultured in Iscove's modified Dulbecco's medium (Sigma-Aldrich) supplemented with 20% and 10% FBS, respectively. HEK-293FT (R700-07; Thermo Fisher Scientific) was cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 0.1 mmol/L MEM Non-Essential Amino Acids (NEAA) (Sigma Aldrich), 6 mmol/L l-glutamine, 1 mmol/L MEM Sodium Pyruvate (Life Technologies), and 10% FBS. SW780 (ATCC, CRL-2169) was cultured in Leibovitz's L-15 Medium (Sigma Aldrich) with 10% FBS. SUP-B15 (ATCC, CRL-1929) was cultured in Iscove's modified Dulbecco's medium containing 4 mmol/L l-glutamine and 1.5 g/L sodium bicarbonate (Sigma Aldrich) and supplemented with 80% 0.05 mmol/L 2-mercaptoethanol (Sigma Aldrich), and 20% FBS. LC-2 (RIKEN BioResource Center, Japan, RCB0440) was cultured in a 1:1 mixture of RPMI 1640 and HAMS F12 (Cellgro, Manassas, VA) supplemented with 25 mmol/L HEPES (Sigma Aldrich) and 15% FBS. RT4 (ATCC, HTB-2) was cultured in McCoy's 5a Medium with 10% FBS. VCaP (ATCC, CRl-2876) was cultured in Dulbecco's modified Eagle's medium with 10% FBS and 50 mg of Normacin (InvivoGen, San Diego, CA). TC-71 cells were kindly gifted by Dr. Beth Lawlor (University of Michigan, Ann Arbor, MI) and cultured with RPMI 1640 medium supplemented with 5 mmol/L l-glutamine and 10% FBS. All cells were cultured at 37°C and 5% CO2. Fusion-containing cell lines were authenticated based on the presence of previously described unique fusions.25, 26 HapMap cell lines were internally authenticated by comparing single-nucleotide polymorphism (SNP) array data (ftp://ftp.ncbi.nlm.nih.gov/hapmap/genotypes/2010-08_phaseII+III/forward) to in house–derived SNP data from custom exon sequencing, with the exception of GM12978 for which no SNP array data were available.
HEK293 FT Cell Transfection
Eighteen pLVX-IRES-Puro vectors that expressed different gene fusions were kindly supplied by ARIAD Pharmaceuticals (Cambridge, MA) (Supplemental Table S2). HEK293FT cells were plated 24 hours before transfection at a density of 105 cells per well in a 6-well clear bottom tissue culture plate (Corning, Corning, NY) in complete growth medium supplemented with 10% FBS. Plasmid DNA (2.5 μg) was added to 250 μL of Opti-MEM1 (Invitrogen) then mixed with 7.5 μL of TransIT-LT1 Reagent (Mirus, Madison, WI). The TransIT LT/DNA complex was added to cells and incubated for 48 hours at 37°C, 5% CO2. Cells were then collected, and RNA extraction was performed.
RT-PCR, Sanger Sequencing, and FISH
cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD) from 1 μg of total RNA. cDNA was PCR amplified with fusion specific primers (IDT). RefSeq accession numbers for transcripts involved in the indicated fusions can be found at https://www.ncbi.nlm.nih.gov/refseq. Amplified product was purified with PureLink PCR purification kit (Invitrogen) and sequenced at The Ohio State University Comprehensive Cancer Center Genomics Shared Resource (Columbus, OH).
The RET break-apart fluorescence in situ hybridization (FISH) assay was conducted with the Vysis LSI RET (Tel) SpectrumRed and Vysis LSI RET (Cen) SpectrumGreen probes from Abbott Molecular (Chicago, IL) as previously described with minor modifications.27 At least 50 tumor cells were scored, and 3′ and 5′ signals physically separated by ≥1 signal diameter were considered split. Specimens were considered positive for RET rearrangement if ≥15% of the cells had split signals, single 3′ signals (red), or single 5′ signals (green).
Validation Specimens
Validation specimens used in this study included 18 HEK transfected cell lines, 13 fusion-positive cell lines, one fusion-negative cell line, 19 lymphoblastoid cell lines, 15 fusion-positive xenograft FFPE specimens purchased from Crown Bioscience Inc. (Santa Clara, CA), 10 FFPE tumor samples generously donated by Dr. Dara Aisner (University of Colorado, Denver), one fusion-positive and 17 fusion-negative lung FFPEs from The Ohio State University Tissue Archive Services (collected under institutional review board–approved study OSU-15030, Novel Molecular Diagnostics for Cancer: Clinical Validation and Discovery, Ohio State University, Columbus, OH), and 16 fresh-frozen or FFPE samples (collected under institutional review board–approved study OSU-13053, Precision Cancer Medicine for Advance Cancer Through High-Throughput Sequencing, Ohio State University, Columbus OH). The percentage of tumor content for all FFPE and fresh-frozen tissues was estimated by a board-certified pathologist.
Fusion Detection, Gene Expression Analysis, and Single-Nucleotide Variant Calling
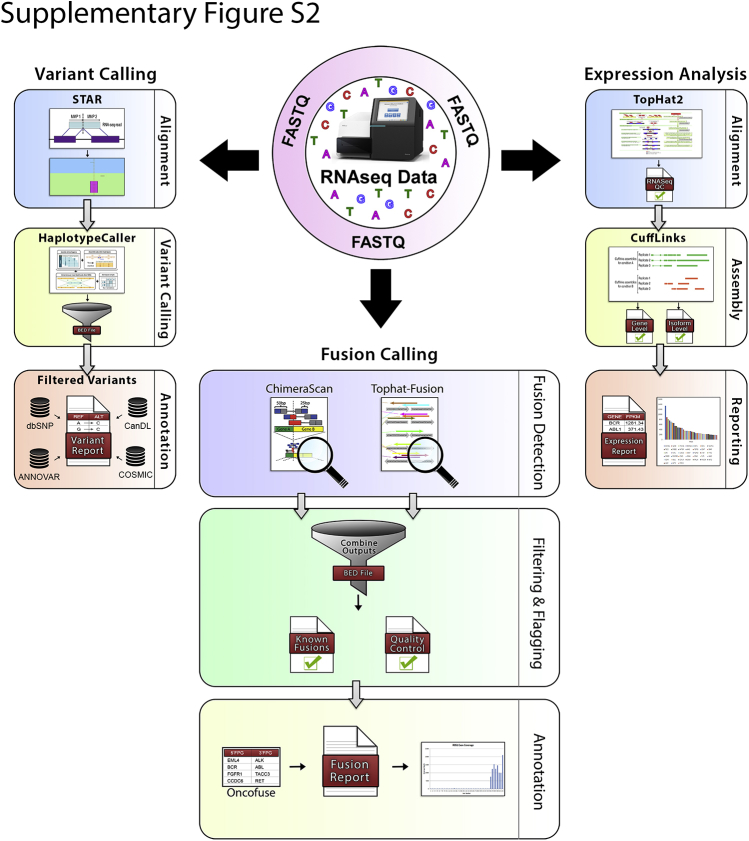
FASTQ files generated by the Illumina MiSeq were analyzed by our custom RNAseq pipeline that contained three major components: fusion calling, gene expression, and variant calling. Fusions were called using two callers: ChimeraScan version 0.4.5 and TopHat-Fusion (tophat-2.0.10.Linux_x86_64).28, 29 Bowtie was used for alignment in both fusion callers (ChimeraScan version 0.12.7 and TopHat-Fusion version 1.1.1), and resulting BAM files were used to identify translocation events. To rescue expected fusions that were filtered out by TopHat-Fusion, the TopHat-Fusion postscript was manually changed. The following modifications were made: candidate fusions were required to have at least 10 bases covered on either side of the break point (default required at least 16 bases covered on either side), a read filter was turned off that filtered out fusions with high expression of wild-type transcripts, and a filter was removed that used the uniformity of read distribution around a break point (because only one gene in a fusion is typically targeted, read distributions are not expected to be similar on both sides of the break point).
Clinically relevant and known fusions were curated using literature review and the Archer Quiver Fusion Database (ArcherDX, Boulder, CO; http://archerdx.com/software/quiver), which collates gene fusions from six publicly available data sources [Catalogue of Somatic Mutations in Cancer (COSMIC), ChimerDB, TICdb, Mitleman, Chromosomal Rearrangements in Diseases (dbCRiD), and Chimeric Transcripts and RNA-Seq (ChiTars)]. Additional fusions detected by OSU-SpARKFuse that underwent subsequent validation were also added to our internal database. All fusions included in this database can be found in Supplemental Table S3. Oncofuse version 1.0.9b2 was used to annotate fusions, including domain information.30 Mean coverage, exonic coverage, and rRNA percentage were calculated from the BAM generated by TopHat, using custom Python scripts (Github; https://github.com/OSU-SRLab/SpARKFuseValidation).
To calculate gene expression, TopHat2 version 2.0.10 (with parameters -p 6, --library-type fr-firststrand) was used for aligning the FASTQ files to a modified human reference genome UCSC build hg19 assembly which included the ERCC genes.31 A UCSC gene annotation file in GTF format was also supplied during the alignment. Gene expression for known genes was calculated as FPKM (fragments per kilobase of transcript per million mapped reads) using CuffLinks version 2.1.1 from the Tuxedo suite, whereas the gene annotation file from UCSC was provided to keep the gene format consistent throughout the pipeline.32 The aligned BAM file from TopHat2 was assessed by RNASeqQC version v1.1.7 to generate alignment metrics.33 Picard Tools version 1.84 (Broad Institute, Cambridge, MA; https://broadinstitute.github.io/picard) CollectInsertSizeMetrics and a custom python script were used to calculate library insert sizes from the down-sampled SAM files.
To call single-nucleotide variants, FASTQ files were initially aligned using STAR version 2.4.0.34 The Picard Tools utility AddOrReplaceReadGroups was used to modify read group information, which included adding the library information, sequencing platform, and sample name. MarkDuplicates was used to mark the read duplicates in the BAM file without removing them. Indel realignment was performed using utilities of the Genome Analysis Toolkit (GATK) package version 3.3-0 (Broad Institute) in the following order: SplitNCigarReads, RealignerTargetCreator, and IndelRealigner.35 Mate information was fixed using Picard's FixMateInformation.jar function. Base quality score recalibration was performed at this step using BaseRecalibrator from GATK. HaplotypeCaller (with parameters -dontUseSoftClippedBases -stand_call_conf 20 -stand_emit_conf 20) was used to call variants and indels from the raw data.35 For filtering the variants, VariantFiltration option (with parameters -window 35 -cluster 3 -filterName FS -filter FS >30.0 -filterName QD -filter QD <2.0) from GATK was used. The filtered variants and indels were annotated for gene information using custom scripts. All variant calling steps were performed on targeted regions only. All custom python scripts are hosted on Github.
Study Approval
All studies that involved humans were approved by The Ohio State University Institutional Review Board. For study OSU-13053 (2013C0152), informed consent was obtained after the nature and possible consequences of the study were explained. No consent was required for study OSU-15030 (2015C0021).
Results
Clinical Targeted RNAseq Assay for Gene Fusion Detection in Solid Tumors
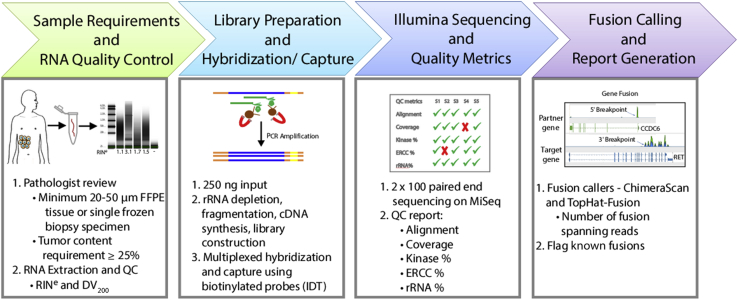
We validated a targeted RNAseq assay termed OSU-SpARKFuse (Ohio State University-Spanning Actionable RNA Kinase Fusions) for clinical use on both FFPE and fresh-frozen tissue to detect gene fusion events in solid tumors (Figure 1 and Supplemental Figure S1). After tumor content estimation by a pathologist, specimens underwent RNA extraction followed by quality control assessment, including RNA concentration, RNA integrity number equivalent (RINe) and DV200.36 Because of the highly degraded nature of FFPE RNA, an rRNA depletion strategy to enrich mRNA species from 250 ng of total RNA input was used. Before this depletion, control RNA fragments from the ERCC were spiked in to serve as positive controls for library preparation and analysis.37 RNAs were subjected to cDNA synthesis, library preparation, and multiplexed hybridization capture using custom probes that targeted 93 kinase/TF genes, nine control housekeeping genes, four control genomic DNA regions, and 10 control ERCC transcripts (Supplemental Table S1). Final captured libraries were sequenced on the Illumina MiSeq platform with 2 × 100-bp read length. Sequencing data were analyzed using a custom pipeline integrating quality control assessment with two fusion callers (TopHat-Fusion and ChimeraScan) to maximize assay sensitivity (Supplemental Figure S2).28, 29 Fusions called by one or both fusion callers were sorted based on clinical significance and level of support (number of fusion spanning reads). Because of the high false-positive rates, a custom clinical filter was implemented whereby previously published or verified fusions were flagged (Fusion Detection, Gene Expression Analysis, and Single-Nucleotide Variant Calling),25, 26 and only these were considered as true-positive results, pending sufficient supporting evidence (Supplemental Table S3). For prospective application, identification of novel fusion partners or break points requires further investigation before clinical reporting, which includes manual inspection of fusion-spanning reads and/or confirmation via RT-PCR and Sanger sequencing.
Figure 1.
OSU-SpARKFuse workflow. After tumor content estimation, RNA is extracted from routine clinical specimens. A total of 250 ng of RNA is used for library construction, including rRNA depletion, cDNA synthesis, and ligation of unique indexed adapters. cDNA libraries are hybridized and captured with 3522 custom probes and sequenced on the Illumina MiSeq. FASTQ files are processed with a customized in-house pipeline to generate alignment metrics and accurately call gene fusions. High-confidence fusion calls are reported. DV200, percentage of RNA fragments >200 nucleotides; ERCC, External RNA Controls Consortium; QC, quality control; RINe, RNA integrity number equivalent.
Target Enrichment and OSU-SpARKFuse Performance
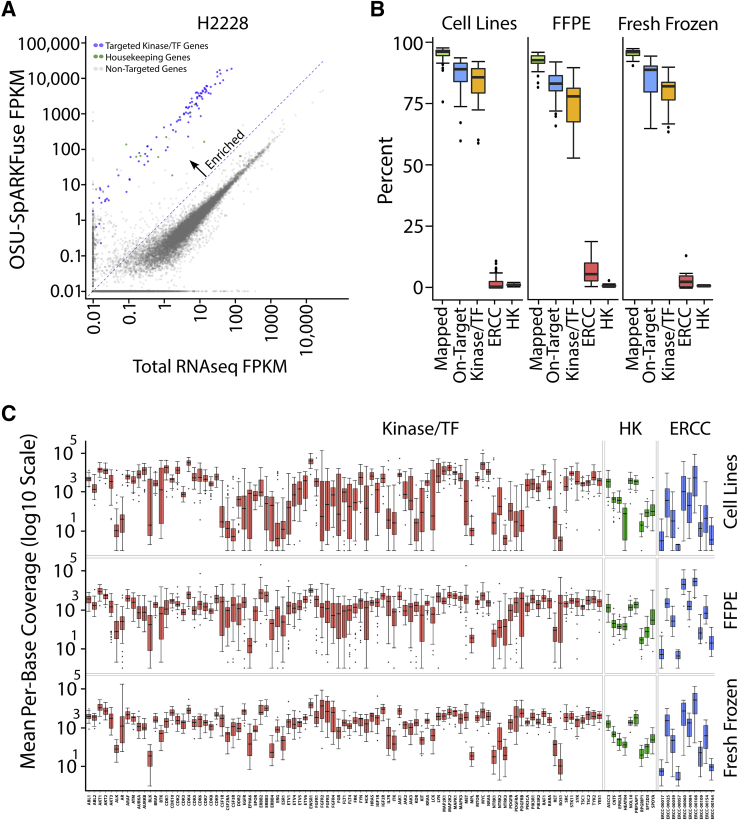
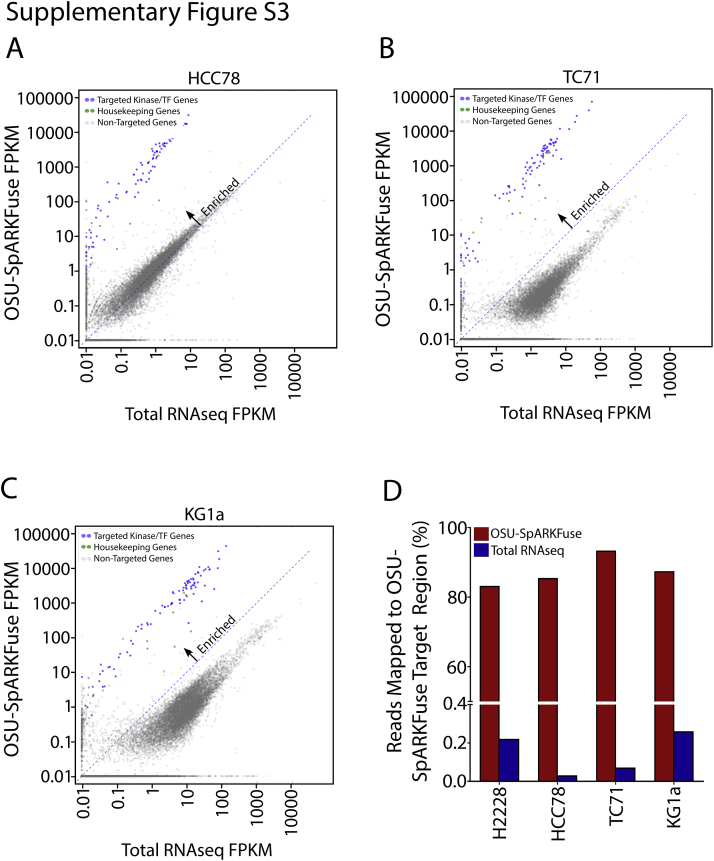
Enrichment of targeted transcripts was evaluated by comparing normalized gene expression from OSU-SpARKFuse and total RNAseq data on four cancer cell lines with well-characterized rearrangements: H2228 (EML4-ALK), TC-71 (EWSR1-FLI1), HCC-78 (SLC34A2-ROS1), and KG1a (FGFR1OP2-FGFR1).38, 39, 40, 41 A mean FPKM value was calculated for all targeted genes, which revealed enrichment of kinase and housekeeping genes in OSU-SpARKFuse compared with traditional transcriptome sequencing (Figure 2A and Supplemental Figure S3, A–C). Using total RNAseq, <0.3% of reads mapped to our targeted regions, whereas >80% of reads mapped to targeted regions using OSU-SpARKFuse (Supplemental Figure S3D).
Figure 2.
Target enrichment and performance of OSU-SpARKFuse on 110 validation samples. A: Comparison of gene expression [measured as fragments per kilobase of transcript per million mapped reads (FPKM)] in total RNA sequencing (RNAseq) data versus OSU-SpARKFuse data in the H2228 cell line. B: Percentage of reads aligning to the HG19 transcriptome (mapped), OSU-SpARKFuse target regions (on-target), targeted kinase/transcription factor (TF) genes, External RNA Controls Consortium (ERCC) transcripts, and housekeeping (HK) genes in cell lines (51 samples), formalin-fixed, paraffin-embedded (FFPE) tissues (43 samples), and fresh-frozen tissues (16 samples). C: Distribution of mean per-base coverage for 93 kinase/TF genes, nine housekeeping genes, and 10 ERCC transcripts in cell lines, FFPE tissues, and fresh-frozen tissues. Outliers are plotted as individual dots. Kinase/TF genes, housekeeping genes, and ERCC vtranscripts on the y axis are listed in Supplemental Table S1. n = 51 cell line samples (C); n = 43 FFPE tissue samples (C); n = 16 fresh-frozen tissue samples (C).
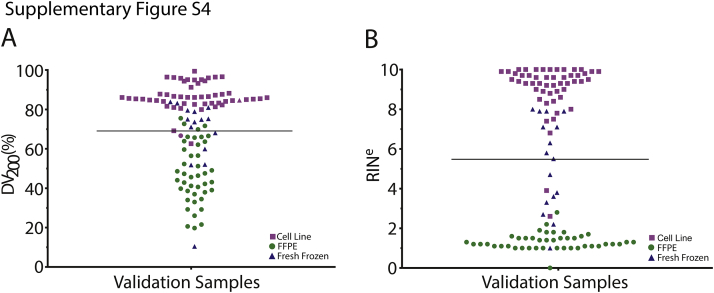
Performance of OSU-SpARKFuse was examined on 74 positive and 36 negative control specimens used for assay validation, including 51 cell line samples, 43 FFPE tissues, and 16 fresh-frozen tissues (Supplemental Table S4). To pass our quality control thresholds, these samples were required to have a minimum of 2 × 106 kinase/TF reads that constituted at least 50% of total sequencing reads. Samples with <2 × 106 kinase/TF reads did not pass and were not considered for validation purposes. Similarly, samples that contained >2 × 106 kinase/TF reads that constituted <50% of total reads did not pass and again were not considered for validation purposes. RNA derived from this cohort varied significantly in terms of quality, with RINe and DV200 values ranging from 0 to 10 and 10% to 99%, respectively (Supplemental Figure S4). As expected, most RNA derived from FFPE tissues had lower RINe and DV200 values compared with cell line and fresh-frozen tissue RNA.
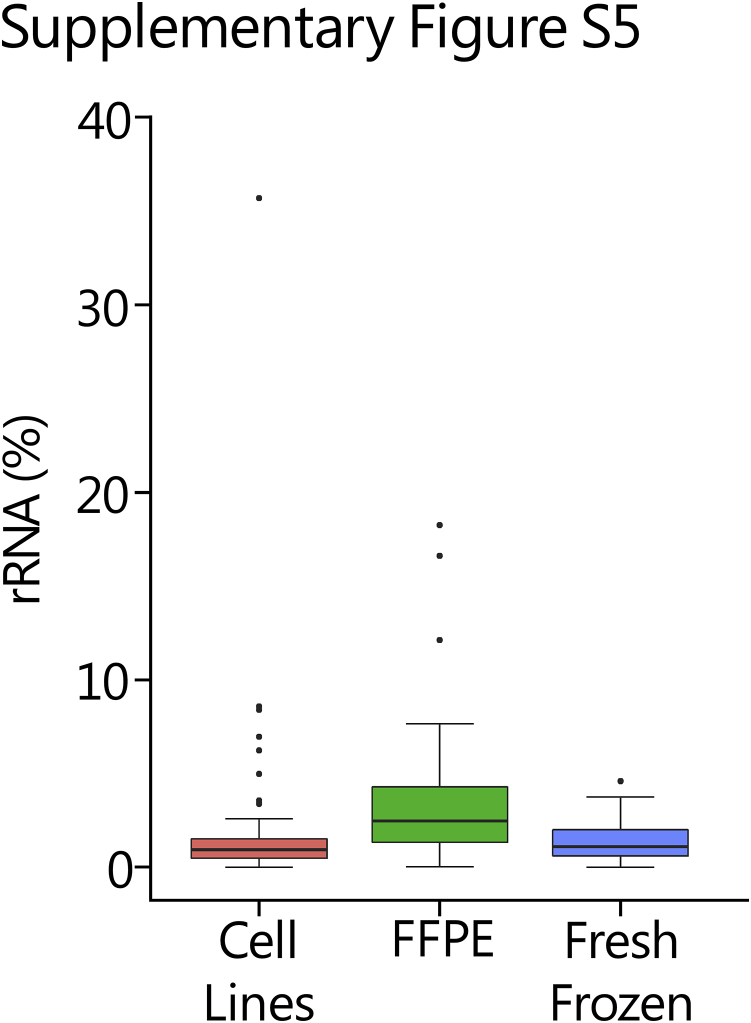
Of a mean 10.5 million reads per sample, only 2.7% of these reads mapped to rRNA regions [2.32%, 3.65%, and 1.53% in cell line (n = 51), FFPE (n = 43), and fresh-frozen (n = 16) tissues, respectively], indicating efficient removal of ribosomal transcripts (Supplemental Figure S5). Capture efficiency was assessed for each sample type by determining the proportion of sequencing reads mapping to our targeted regions of interest, including kinase/TF genes, ERCCs, and housekeeping genes (Figure 2B). Kinase/TF reads constituted a mean of 84.56%, 74.72%, and 79.57% of total reads in cell line, FFPE, and fresh frozen samples, respectively. Interestingly, targeted ERCC transcripts comprised nearly 7% of total reads in FFPE samples, which was quintuple or double the amount present in cell line and fresh-frozen samples. This bias in ERCC reads is likely attributable to the low quality of RNA present in FFPE samples. The mean percentage of reads supporting housekeeping genes was 1.15% and as expected was more consistent among the three sample types (1.35%, 0.98%, and 0.92% in cell line, FFPE, and fresh-frozen samples, respectively).
To examine sequencing depth, the mean per base coverage of each targeted gene in cell line, FFPE, and fresh frozen samples (Figure 2C) were each calculated. The mean per base coverage for all targeted bases in the three sample types was 4849×, 3078×, and 3808×, respectively. Although considerable variation in gene expression across our validation samples was expected, per-base coverage of targeted genes in this cohort was investigated to determine whether any genomic regions consistently failed to be captured and/or sequenced across all samples. Of the 256,148 kinase/TF bases targeted by OSU-SpARKFuse, all had at least 10× coverage within the 110 validation samples, indicating efficacy of the assay in capturing the desired targets. Similar analysis was performed for ERCC transcripts and housekeeping genes, which had only 0.74% of ERCC transcript bases and zero housekeeping transcript bases with <10× coverage, again indicating efficient capture of these targeted transcripts.
Sensitivity and Specificity of OSU-SpARKFuse
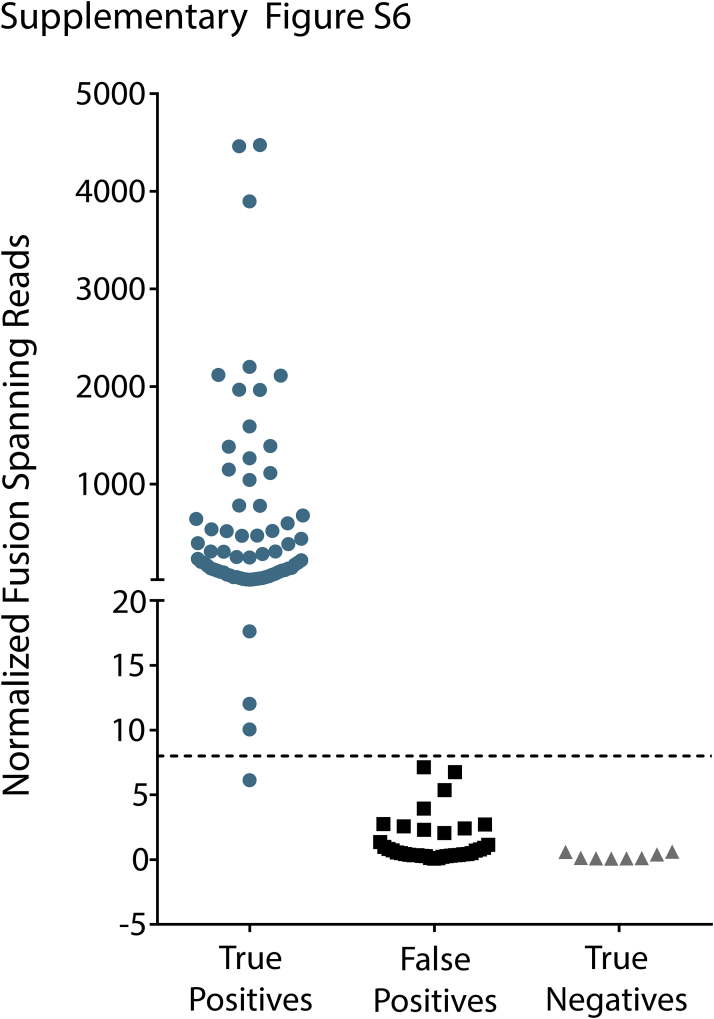
To determine the accuracy of OSU-SpARKFuse, we examined fusion calls from both TopHat-Fusion and ChimeraScan in our 110-sample validation cohort. Fusions called by either tool were considered in the analysis. Importantly, we used our clinical filter, and only fusions that passed this filter were considered when determining the sensitivity and specificity of the assay. For all fusion calls, the number of fusion-spanning reads was normalized per 1 × 106 reads that mapped to targeted kinase/TF genes [normalized fusion-spanning reads (NFSRs)]. A total of 75 true-positive fusion events that involved 18 unique gene targets were assessed from 74 unique positive control samples, consisting of 31 cell lines, 27 FFPE samples, and 16 fresh-frozen samples (Supplemental Table S4). Samples with rearrangements that involved diverse genes were selected to examine performance of the assay over a range of genomic loci and fusions with variable expression. The cell line cohort included 13 cancer cell lines with previously published and validated gene fusions (Fusion Detection, Gene Expression Analysis, and Single-Nucleotide Variant Calling),25, 26 as well as 18 fusion constructs transfected into HEK293FT cells. Gene rearrangements in FFPE and fresh frozen samples were previously determined using standard methods, including FISH, DNA intron sequencing (NGS), and Sanger sequencing. Because we only expected to detect 75 true-positive fusion events in our positive control samples, additional unanticipated fusions found in these samples were considered false-positive results, and this information was used to establish an NFSR cutoff for high-confidence fusion calls. Of these 74 samples, 25 had NFSR values >0 for a false-positive fusion; however, this number did not exceed 8 NFSRs (Supplemental Figure S6). Using this as our threshold for high-confidence fusion calls, 70 of our 75 true-positive fusion events were correctly identified, revealing an overall sensitivity of 93.3% (95% CI, 84.47%–95.52%). Of the false-negative event, four had zero fusion-spanning reads, and one fell below our NFSR threshold with a value of 6.12 (Supplemental Figure S6). All samples that failed to call the correct expected fusion were derived from poor-quality FFPE tissues, indicating a lower sensitivity of OSU-SpARKFuse on this particular sample type (81.5%). The utility of combining two fusion callers was made apparent because only 61 and 58 fusions were correctly identified using TopHat-Fusion (95% CI, 70.33%–89.06%) and ChimeraScan (95% CI, 65.94%–85.88%) alone, respectively.
To examine specificity, OSU-SpARKFuse was applied to an independent negative control cohort composed of 19 well-characterized cell lines from the 1000 Genomes Project (HapMap cell lines) not expected to contain gene fusions and an additional 16 FFPE lung cancer samples negative for ALK, RET, and ROS1 fusions based on FISH from the pathology archives at The Ohio State University (Supplemental Table S4).42 In addition, the acute myelogenous leukemia cell line HL60 was used, which was previously reported to contain a fusion that involved NSMCE2 and BF104016, neither of which are targeted by OSU-SpARKFuse.43 Of these 36 samples, seven had NFSR values >0 for a false-positive fusion; however, the highest NFSR observed in these samples was 1.42, and therefore no high confidence fusions were called in these 36 samples, establishing a specificity of 100% (95% CI, 87.99%–100%) (Supplemental Figure S6).
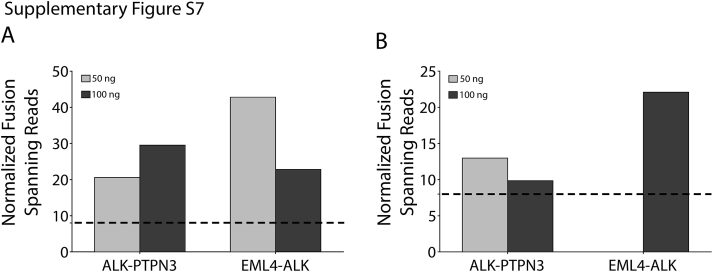
RNA Input, Quality Assessment, and Fusion Detection Limits
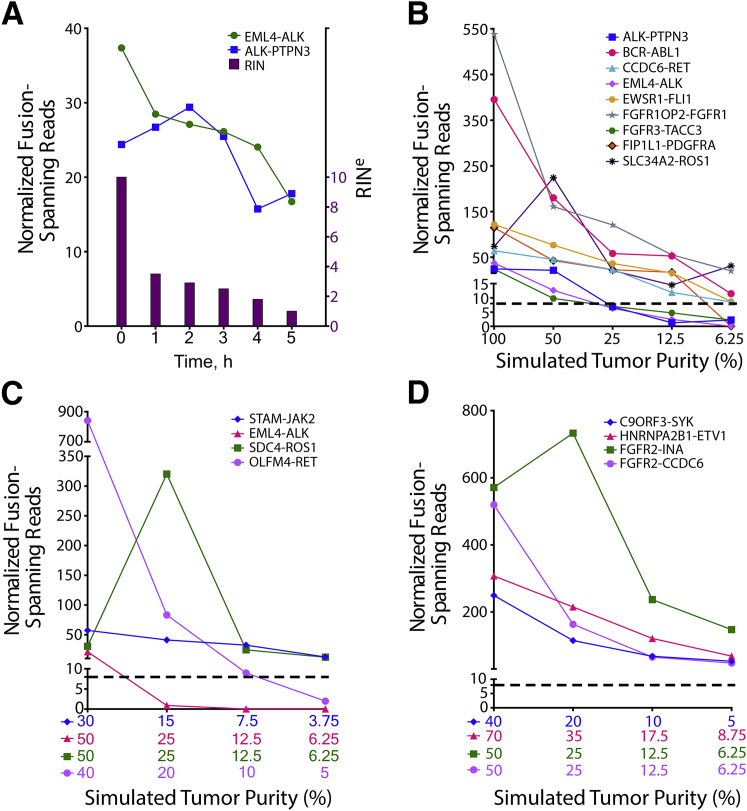
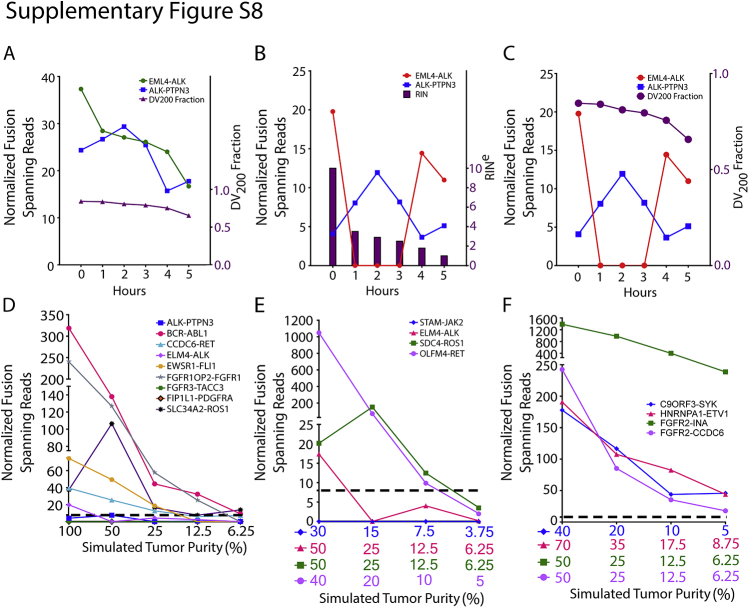
Although 250 ng of RNA was attainable from our 110 validation samples, OSU-SpARKFuse was capable of detecting known fusions when the input limit was challenged with 50 and 100 ng of RNA from the H2228 cell line, indicating that a lower sample input may be used with this assay (Supplemental Figure S7). To determine limitations for fusion detection in highly degraded samples, we artificially degraded RNA from this same cell line. After heat treatment at 90°C for 1 to 5 hours, a decrease was observed in both the RINe and DV200 values at increasing time points (RINe, 10–1; DV200, 85%–65%) (Figure 3A and Supplemental Figure S8A). Although a relatively steady decrease in the number of NFSRs for EML4-ALK and ALK-PTPN3 was observed as the heating time was increased, both fusions remained detectable above our 8-NFSR threshold (Figure 3Aand Supplemental Figure S8, A–C).
Figure 3.
Fusion detection in degraded and diluted fusion-positive samples. A: RNA from H2228 was incubated at 90°C for 0 to 5 hours, and RNA integrity number equivalent (RINe) was determined (purple bars). Normalized fusion-spanning reads derived from TopHat-Fusion are plotted at each time point for EML4-ALK and ALK-PTPN3 fusions. RNA from eight fusion-positive cell lines (B), four fusion-positive FFPE tissues (C), and four fusion-positive fresh frozen tissues (D) was serially diluted to simulate the indicated tumor purities. Dashed lines indicate threshold for high-confidence fusion calls.
To examine the lower detection limit of OSU-SpARKFuse, serial dilutions of RNA were generated to simulate various tumor purity levels. Eight cell lines with nine mutually exclusive gene fusions (ALK-PTPN3, BCR-ABL1, CCDC6-RET, EML4-ALK, EWSR1-FLI1, FGFR1OP2-FGFR1, FGFR3-TACC3, FIP1L1-PDGFRA, and SLC34A2-ROS1) were sequentially mixed to generate four samples with fusions present at 50% (mix A, B, C, and D), two samples with fusions present at 25% (mix AB and mix CD), and one sample with fusions present at 12.5% (mix ABCD) (Supplemental Table S5). To generate a final dilution with fusions present at 6.25%, equal volumes of RNA from mix ABCD and the fusion-negative cell line GM12878 were combined. The NFSR values were again calculated for each fusion at all dilutions using both TopHat-Fusion (Figure 3B) and ChimeraScan (Supplemental Figure S8D). In contrast to DNAseq in which detection limits rely strictly on the prevalence of cancer cells present in the tumor sample, additional variation must be considered when dealing with RNAseq data, including expression of fusion genes, which can be highly variable. TopHat-Fusion derived NFSR values that supported the nine fusions present in the cell lines used for mixes ranged from 24 to 537 in undiluted samples, indicating a wide range of expression levels. At 50% simulated tumor purity, all fusion events were detected above our NFSR cutoff of 8 (Figure 3B). However, on further dilution of these fusions to 25%, 12.5%, and 6.25%, NFSRs for ALK-PTPN3, EML4-ALK and FGFR3-TACC3 fell below this threshold.
To ascertain whether the detection limit of OSU-SpARKFuse was similar in clinical samples, we generated serial dilutions of RNA from four positive control FFPE (STAM-JAK2, EML4-ALK, SDC4-ROS1, and OLFM4-RET) and fresh-frozen samples (C9ORF3-SYK, HNRNPA2B1-ETV1, FGFR2-INA, and FGFR2-CCDC6) (Supplemental Tables S6 and S7). Again, significant variability was observed in the undiluted expression of these fusions, with TopHat-Fusion–derived NFSR values ranging from 21 to 840 in FFPE samples and 248 to 571 in fresh-frozen samples (Figure 3, C and D, and Supplemental Figure S8, E and F). In contrast to cell lines, clinical samples are inherently diluted by surrounding normal cells; therefore, tumor purity values are unique to each sample. The FFPE tissues used for serial dilutions had initial tumor purities in the range of 30% to 50% and on subsequent dilution were simulated to the range of 3.75% to 6.25% tumor cells (Figure 3C). Both the STAM-JAK2 and SDC4-ROS1 fusions were detectable above our high-confidence NFSR cutoff at these lowest dilutions. Similarly, serially diluted fresh-frozen tissues had initial tumor purities in the range of 40% to 70%, and all fusion events were detected above our NFSR cutoff of 8 in final dilution samples, with simulated tumor purities ranging from 5% to 8.75% (Figure 3D).
Repeatability and Reproducibility of OSU-SpARKFuse
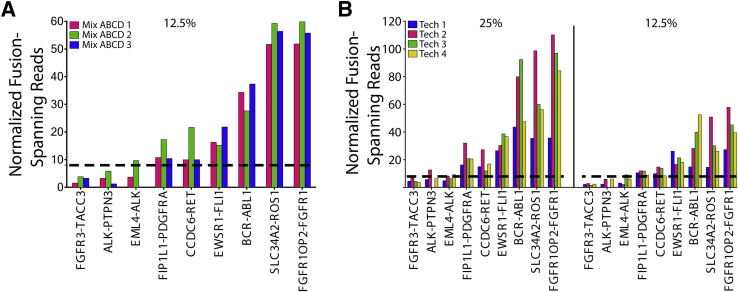
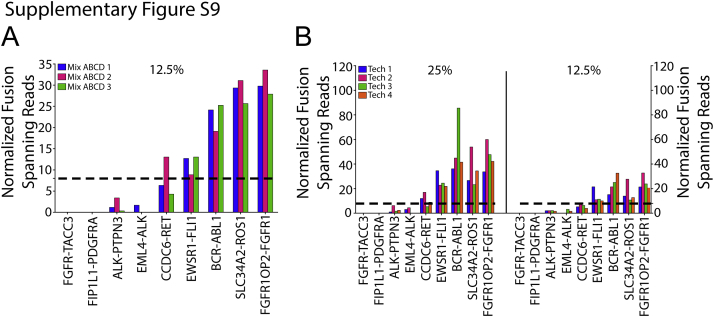
The repeatability of OSU-SpARKFuse was examined using our 12.5% dilution (mix ABCD) that contained nine gene fusions to maximize data points for comparison. Three technical replicates were prepared and sequenced in parallel by the same technician, and NFSR values were calculated using both TopHat-Fusion (Figure 4A) and ChimeraScan (Supplemental Figure S9A). Six of the nine known fusions were uniformly called across the replicates. Although NFSRs for FGFR3-TACC3, ALK-PTPN3, and EML4-ALK did not meet our established threshold of 8 at the 12.5% dilution, this observation was consistent for these particular fusions. To examine overall concordance, we determined whether the reads for a particular fusion were above or below our high-confidence threshold of 8 NFSR. Using these criteria, we determined the overall concordance among the three technical replicates to be 96.3% (26/27 concordant calls). Reproducibility of OSU-SpARKFuse was tested using our 25% and 12.5% cell line dilutions again using TopHat-Fusion (Figure 4B) and ChimeraScan (Supplemental Figure S9B). Four independent technicians prepared and sequenced libraries from the same starting RNA on two different MiSeq instruments. Similar to our observations from the reproducibility experiment, an overall concordance of 94.4% was achieved (68/72 concordant calls).
Figure 4.
Intrarun repeatability and interrun reproducibility of OSU-SpARKFuse. A: RNA isolated from 12.5% cell line dilutions was prepared and sequenced in the same run by the same technician for a total of three replicates. Normalized fusion-spanning reads derived from TopHat-Fusion are plotted. B: RNA isolated from 25% and 12.5% cell line mixes was prepared by four different technicians and sequenced on two different MiSeq instruments. Normalized fusion-spanning reads derived from TopHat-Fusion are plotted. Dashed lines indicate threshold for high confidence fusion calls.
Detection of Novel RET and FGFR2 Fusion Partners
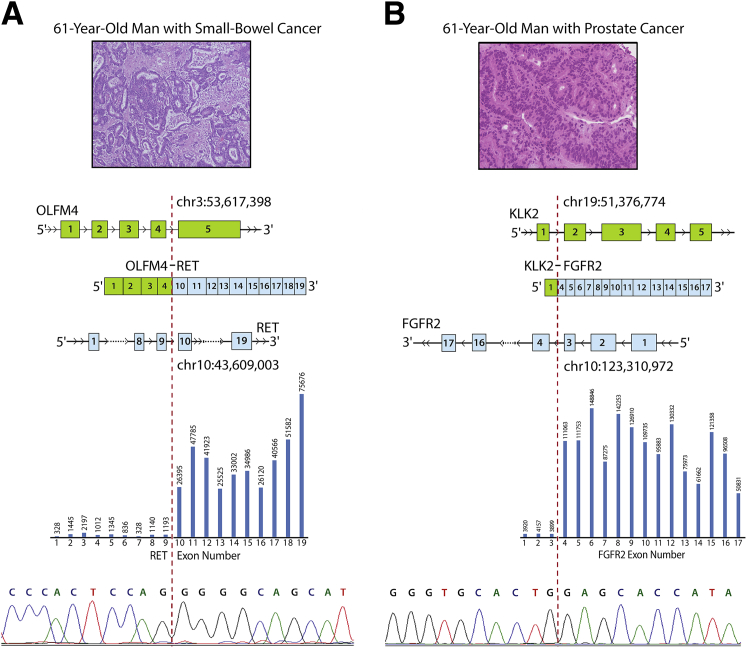
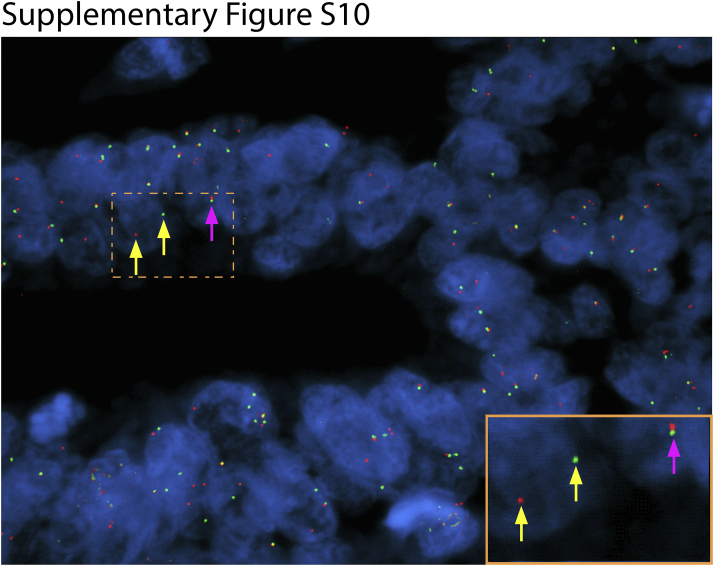
Because of the unbiased design of OSU-SpARKFuse, the potential for discovery of novel gene fusions and partners is limited only by our target genes of interest. This assay was used to assess gene fusion status in 95 tissue samples from patients with advanced cancer at The Ohio State University as part of a clinical tumor sequencing study (OSU-13053). Fusions that involved RET are present in 1% to 2% of non–small cell lung carcinomas and recently were identified in colorectal cancer at a frequency of 0.2%.44, 45 Using OSU-SpARKFuse, we discovered OLFM4 to be a novel fusion partner of RET in a 61-year-old patient with metastatic small-bowel cancer (Figure 5A). Both TopHat-Fusion and ChimeraScan detected this rearrangement with 840 and 1048 NFSRs, respectively. In this chimeric transcript, exons 1 to 4 of OLFM4 are fused to exons 10 to 20 of RET to generate an in-frame gene that contained the coiled coil domain of OLFM4 and the entire kinase domain of RET (Figure 5A). Analysis of exon-level coverage of RET clearly distinguished exons not involved in the fusion transcript (1 to 9) from exons that were included (10 to 20) because read depth increased nearly 36-fold in these latter exons (Figure 5A). This previously undescribed fusion was confirmed by RT-PCR and subsequent Sanger sequencing using custom primers that spanned the break point and FISH using break-apart probes for RET (Figure 5A, Table 1, and Supplemental Figure S10).
Figure 5.
Detection of novel clinically actionable fusions. A: A novel fusion that involves exons 1 to 4 of OLFM4 and exons 10 to 19 of RET was detected in a formalin-fixed, paraffin-embedded sample from a 61-year-old man with small-bowel cancer. B: A novel fusion that involves exon 1 of KLK2 and 4 to 17 of FGFR2 was detected in a fresh frozen biopsy sample from a 61-year-old man with prostate cancer. Top: Representative hematoxylin and eosin images from a Whipple resection (A) and a liver biopsy (B). Middle: Schematic of fusion gene with indicated exonic break points. Bar graph represents average exon level read depth for indicated RET exons (https://www.ncbi.nlm.nih.gov/refseq; accession number NM_020630) (A) and FGFR2 exons (https://www.ncbi.nlm.nih.gov/refseq; accession number NM_001144913.1) (B). Bottom: chromatogram trace of OLFM4-RET (A) and KLK2-FGFR2 (B) fusion transcripts. Dashed lines indicate break point.
Table 1.
Primer Sequences Used for Sanger Sequencing
| Gene | Sequence | Expected Amplicon Size, bp |
|---|---|---|
| OLFM4_exon4_F | 5′-TGGCTCTGAAGACCAAGCTG-3′ | 201 |
| RET_exon10_R | 5′-CCTCCTCAGGGAAGCAGTTG-3′ | |
| KLK2_exon1_F | 5′-CATGTGGGACCTGGTTCTCT-3′ | 194 |
| FGFR2_exon4_R | 5′-CCTGCTTAAACTCCTTCCCG-3′ | |
| KLK2_exon2_F | 5′-ATCCAGTCTCGGATTGTGGG-3′ | 293 |
| FGFR2_exon4_R | 5′-CCTGCTTAAACTCCTTCCCG-3′ |
Indicated primer pairs were used to generate amplicons for Sanger sequencing of novel fusions identified using OSU-SpARKFuse.
F, forward direction; R, reverse direction.
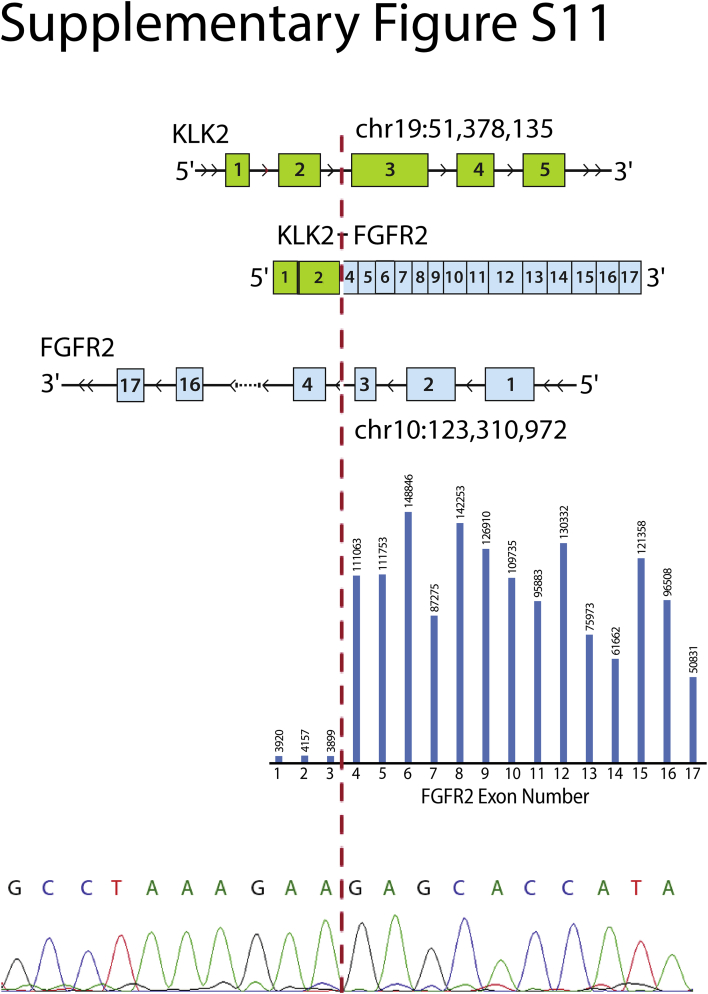
OSU-SpARKFuse also identified a novel gene fusion in a 61-year-old patient with metastatic prostate cancer that involved KLK2 and FGFR2. Interestingly, two separate break points for this fusion were observed, one including exon 1 of KLK2 and the other including both exons 1 and 2 of KLK2 (Figure 5B and Supplemental Figure S11). The dominant fusion involved exon 1 and had 2200 NFSRs (by ChimeraScan). Similar to observations made with respect to exon bias in RET, an approximate 27-fold increase in read depth for FGFR2 exons involved in the fusion transcript was seen (exons 1 to 3 versus 4 to 17) (Figure 5B). KLK2 expression is unique to prostate tissue and correlates with increased cellular proliferation and decreased apoptosis in castrate-resistant prostate cancer specimens.46 Several reports have identified gene fusions that involve KLK2 and the transcription factors ETV1 and ETV4 in prostate cancer samples.47, 48 Expression of KLK2 is regulated by androgen receptor, which likely drives expression of FGFR2 and explains the activation mechanism of this fusion gene.49 Custom primers that target both the exon 1 and exon 2 break points were designed, and both fusions were confirmed by RT-PCR and Sanger sequencing (Figure 5B, Table 1, and Supplemental Figure S11). Discovery of this chimeric transcript provided eligibility for this patient to receive a novel FGFR inhibitor as part of a basket clinical trial at The Ohio State University.
Gene Expression Analysis, SNP Calling, and Alternative Splicing Events
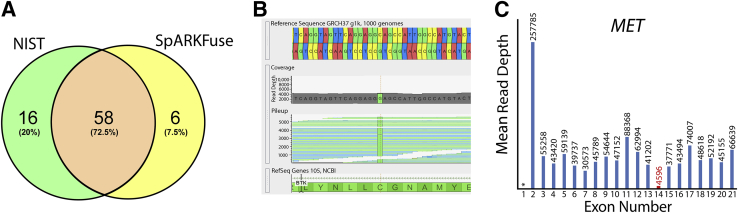
Although OSU-SpARKFuse was designed for detection of clinically relevant gene fusions, additional capabilities were built into our custom pipeline to enable discovery research. One obvious application of RNAseq data is gene expression analysis. To address this, a mean gene expression value (FPKM) was included for all targeted kinase/TF genes. In addition, variant calling was enabled using the GATK. To gauge the accuracy of identifying SNPs from RNAseq data, we used the HapMap cell line GM12878 that was extensively characterized by the National Institute of Standards and Technology (NIST) and has publically available data for high-confidence SNP calls based on various DNAseq methods.50 Raw SNP calls derived from OSU-SpARKFuse and NIST were filtered through a common target regions file, and concordance was determined (Figure 6A). A total of 96 SNP positions were identified; however, 16 occurred in locations that were not expressed at the RNA level and were therefore excluded from the analysis. Of the remaining 80 SNPs, 58 (72.5%) were detected by both NIST and OSU-SpARKFuse. An additional 16 SNPs were only identified by NIST, whereas six SNPs were called exclusively by OSU-SpARKFuse. A closer examination of the 16 SNPs missed by OSU-SpARKFuse revealed 11 of these positions to have <10× sequencing coverage, which likely contributes to why these calls were missed. The six additional SNPs only called by OSU-SpARKFuse were regarded as false-positive results, with maximum coverage of 18 and minimum coverage of two at these positions. Variant calling was applied on a sample from a 66-year-old man with chronic lymphocytic leukemia that progressed to Richter transformation after treatment with the Bruton tyrosine kinase inhibitor ibrutinib for 16 months. OSU-SpARKFuse identified a C481S mutation in Bruton tyrosine kinase at the binding site of ibrutinib that promotes drug resistance (Figure 6B).51 Although it can provide important information, one obvious limitation of variant calling exclusively from RNAseq data is that variants from nonexpressed or low-expressed genes will be missed; therefore, we would not be confident relying solely on OSU-SpARKFuse for variant calling.
Figure 6.
Potential clinical applications of OSU-SpARKFuse. A: Venn diagram representing concordance of variant calls from OSU-SpARKFuse and high-confidence variant calls from National Institute of Standards and Technology (NIST) for the GM12878 cell line. B: Genome Browser screen shot depicting C to G bp substitution, resulting in a C481S mutation in a patient with chronic lymphocytic leukemia resistant to treatment with ibrutinib. C: Mean exon-level read depth for indicated MET exons (https://www.ncbi.nlm.nih.gov/refseq; accession number NM_001127500). Red text indicates skipped exon. Asterisk represents untranslated region not covered by OSU-SpARKFuse probes.
In addition to gene level expression, our custom pipeline also calculates sequencing depth on a per-exon level for all transcripts of our targeted kinase/TF genes. This level of resolution can be used to identify exon imbalance events that support the presence of gene fusions as described previously (Figure 5) but may also indicate alternative splicing. Recently, comprehensive DNAseq of splice site alterations at MET exon 14 revealed 126 distinct variants that could potentially lead to exon skipping and subsequent MET activation.52 However, not all these variants will result in the same level of exon 14 skipping; therefore, having a semiquantitative detection method is advantageous. Using OSU-SpARKFuse, we were able to identify a MET exon 14 skip event in a sample from a 79-year-old man with metastatic lung adenocarcinoma (Figure 6C). In this case, we observed a nearly 10-fold decrease in the mean read depth of exon 14 compared with the 5′ and 3′ adjacent exons.
Discussion
Although RNAseq has been routinely applied as a research tool for the discovery of gene fusions in cancer, an unbiased clinical grade RNAseq assay capable of detecting both known and novel gene fusions in solid tumors has not been developed for patient care.10, 11, 12, 14, 53, 54 We describe the extensive analytical validation of a targeted RNAseq assay termed OSU-SpARKFuse and establish both the accuracy (sensitivity, specificity) and precision (reproducibility, repeatability) of this assay for detecting clinically actionable gene fusions that involve kinases and canonical TFs. In addition to fulfilling an unmet clinical need, OSU-SpARKFuse also enables discovery research and opens doors for future clinical applications that involve the cancer transcriptome, including exon skipping, resistance mutations, and alternative splicing (Figure 6).51, 52, 55 Using a cohort of 110 positive and negative control validation specimens, we found the successful performance of OSU-SpARKFuse on diverse sample types, including cell lines, FFPE tissues, and fresh-frozen tissues that varied widely in RNA quality (RINe and DV200). Samples were required to have a minimum of 2 × 106 kinase/TF reads that constituted 50% of total sequencing reads to be considered for gene fusion calling. In addition to being highly sensitive for fusion detection (93.3%), a critical advantage of OSU-SpARKFuse over other methods is that prior knowledge of intronic/exonic break points or fusion partners is not required, as evidenced by discovery of novel fusions that involve RET and FGFR2 oncogenes (Figure 5). OSU-SpARKFuse is also suitable for real-time patient testing with a turnaround of approximately 5 days, including RNA extraction, library construction, hybridization and capture, sequencing, and analysis.
OSU-SpARKFuse has immediate clinical implications for the care of patients with cancer by detecting therapeutically actionable gene fusions. With the recent discovery of activating gene fusions that involve kinases, such as ALK, FGFRs, RET, ROS1, and NTRKs, multiple opportunities for treatment with kinase inhibitors have emerged in clinical trials. A recent review summarized 35 different trials that involved gene fusions in epithelial cancers.3 In early-phase studies for ALK fusion-positive lung cancer, patients were screened using FISH.20 However, FISH is costly to develop for custom detection of multiple gene fusions and can exhaust small tumor samples acquired through needle biopsies from patients. Thus, development of assays that can cost effectively detect fusions across multiple genes from small tumor samples is essential. OSU-SpARKFuse fulfills this need and facilitates identification of patients with gene fusions who can then be eligible for novel targeted therapies, such as FGFR inhibitors (Figure 6) or NTRK inhibitors.56
Depending on the desired application, other sequencing approaches may be used for gene fusion detection, although each has distinct advantages and disadvantages. Several groups have published on multiplex amplicon approaches that specifically target fusions across known break points.15, 57, 58 The main advantages of such amplicon approaches include lower-input requirements, potentially increased sensitivity attributable to extensive amplification, shorter technical time for the assay, and reduced complexity for data analysis. However, amplicon approaches are severely limited in terms of discovery because they can only amplify known fusion break points with known partners. Without prior knowledge of these potential partners, amplicon-based technologies are unable to detect rearrangements that involve novel partners that may have clinical relevance. Another less biased strategy is anchored multiplex PCR, which requires knowledge of only one end of a target region for enrichment, can be used with a lower input, and has a quick turnaround time.16 However, information regarding specific exons involved in fusions and directionality are required for assay design, which limits the ability of this method to detect fusions that involve novel break points. In addition, upper limits exist with respect to the amount of content that can be targeted. Methods that use DNAseq have also been applied for fusion detection and have the advantage of identifying the genomic break point, which is particularly useful for the discovery of novel gene fusions. However, because of the cumulative size of exons, DNAseq approaches are considerably more expensive. For example, to detect equivalent fusions that involve 93 transcripts captured by OSU-SpARKFuse, a DNA intron capture design would exceed 10,000,000 bp (40-fold larger than the approximately 250,000 bp in SpARKFuse). Thus, an RNAseq-based design is smaller, suitable for desktop sequencers, and 40 times less expensive. Furthermore, DNA strategies lack the expanded potential of RNAseq for detection of exon skipping and alternative splicing.
The chief advantage of targeted RNAseq compared with other RNA-based approaches is the unbiased detection of any fusion partner in any direction. In addition, this method opens the door for further applications of the cancer transcriptome, including identification of resistance mutations, exon skipping, and splice variants. Despite these advantages of targeted RNAseq, there are limitations to its application. First, RNA quality will likely be a challenge for older archival specimens compared with DNA. Most FFPE specimens we and others have evaluated for assay development are <5 years old. Thus, retrospective projects on older specimens may not be ideal compared with DNA approaches. Second, detection limits for targeted RNAseq may not be as sensitive as amplicon-based approaches for lower-expressing fusions; therefore, application of this assay is restricted to specimens that contain a minimum of 25% tumor content. Data from our limit of detection experiments, as well as routine use of OSU-SpARKFuse have led us to identify EML4-ALK as one such low-expressing fusion transcript; therefore, detection of this fusion event below our current NFSR threshold of 8 may warrant further investigation. Despite this limitation, OSU-SpARKFuse was able to correctly detect fusions in many samples with tumor fractions as low as 4% to 9% (Figure 3, B–D). Third, targeted RNAseq requires a priori selection of transcripts for investigation and thus may miss opportunities to identify novel transcripts that can be detected with whole transcriptome sequencing. However, whole transcriptome approaches are more expensive and are currently difficult to scale practically for real-time patient care. Finally, analysis of RNAseq data for a clinical genetics laboratory requires dedicated personnel for bioinformatics analysis and quality review. On the other hand, targeted RNAseq can identify multiple classes of alterations, which may reduce the number of validated assays required for comprehensive genomic analysis of patient samples. This cost savings may help compensate for additional bioinformatics personnel.
Looking ahead to the era of precision medicine for cancer care, diagnostics based on DNAseq and RNAseq are likely to be complementary rather than mutually exclusive tools. Genomic alterations can be corroborated at the transcriptome level with respect to expression or loss of expression for given variants. Although variant calling exclusively from RNAseq data is not recommended, ranking variants detected by DNAseq according to expression level may be one way in which these two technologies could complement one another. Along these same lines, correlation of transcriptome signatures with genomic alterations may enable more accurate prediction of response to certain targeted therapies before treatment. In addition, clinical grade targeted RNAseq has tremendous potential for application using liquid biopsy approaches, which could include testing of tumor exosome RNA derived from peripheral blood samples.59 Liquid biopsy approaches enable simultaneous real-time sequencing of tumor RNAs while patients are receiving novel therapies. Such integrative tissue and liquid biopsy strategies may lead to the identification of RNA biomarkers that predict whether a patient is responding or developing resistance to a therapy. Lastly, targeted RNAseq can be scaled and optimized for additional transcripts of interest and can be used for broader applications in patients with solid tumors.
Acknowledgments
We thank Jenny Badillo for her administrative support to our team, the Ohio Supercomputer Center for providing disk space and processing capacity to run our analyses, the Comprehensive Cancer Center (The Ohio State University Wexner Medical Center) for their administrative support, especially The Ohio State University Comprehensive Cancer Center Genomics Shared Resource and Tissue Archive Services, Dr. Michael Snyder (Stanford University, Palo Alto, CA) for lymphoblastoid cell lines, and Dr. Beth Lawlor (University of Michigan, Ann Arbor, MI) for TC-71 cells.
J.W.R. supervised the project, analyzed and interpreted data, and wrote the manuscript; D.M. designed and conducted experiments, analyzed and interpreted data, and wrote the manuscript; J.M. analyzed data and wrote of the manuscript; M.R.W., A.S., and M.R. contributed to reproducibility experiments; H.P. and K.R.N. contributed to in vitro transfection experiments; M.V.-G. developed and performed FISH experiments; E.A.K., E.L., E.Z., and E.S. analyzed data; J.G. provided gene fusion constructs; N.N., A.G.F., and J.C. reviewed pathology on tissue specimens; K.D.D. and D.L.A. provided clinical positive control specimens; L.Y. provided statistical support; S.R. conceived the study, supervised the project, and wrote the manuscript.
Footnotes
Supported by American Cancer Society grant MRSG-12-194-01-TBG (S.R.), the Prostate Cancer Foundation, National Human Genome Research Institute grant UM1HG006508-01A1, National Cancer Institute grant UH2 CA202971-01, Fore Cancer Research, the American Lung Association, Pelotonia, and a Roessler research scholarship from the Ohio State University College of Medicine (E.H.L.).
J.W.R., D.M., and J.M. contributed equally to this work.
Disclosures: An immediate family member of S.R. owns stock in Johnson and Johnson.
Current address of J.C., GenomOncology, Cleveland, OH.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2017.05.006.
Supplemental Data
Supplemental Figure S1.
The OSU-SpARKFuse assay schematic. External RNA Controls Consortium (ERCC) control RNA is added to total RNA isolated from clinical specimens. After rRNA depletion, samples undergo chemical fragmentation, cDNA synthesis, A-tailing, adapter ligation, and PCR amplification. Pooled cDNA libraries are hybridized to biotinylated custom probes in the presence of blocking oligos and captured using streptavidin-coated magnetic beads. Final libraries undergo a second round of PCR amplification and subsequent sequencing.
Supplemental Figure S2.
The OSU-SpARKFuse pipeline schematic. RNA sequencing (RNAseq) data are analyzed using variant calling, expression analysis, and fusion calling modules. Raw FASTQ files generated by the sequencing instrument (MiSeq) are processed through each module with the specified alignment tool. For variant calling, FASTQ files are aligned using STAR, and Genome Analysis Toolkit's HaplotypeCaller is used to nominate single-nucleotide variants and indels. The output is filtered for in-target regions and annotated using ANNOVAR, Catalogue of Somatic Mutations in Cancer (COSMIC), Cancer Data Log (CanDL), and Single Nucleotide Polymorphism database (dbSNP). For expression analysis, FASTQ files are aligned using TopHat2, and Cufflinks is used to calculate gene and isoform level expression (as fragments per kilobase per million mapped reads). For fusion calling, tool-specific versions of Bowtie are used for alignment, and then fusion calling algorithms are applied. NFSRs are determined, and the output is filtered for in-target fusions. Quality control metrics are determined, known fusions present in our internal database are flagged, and protein domains are annotated using Oncofuse.
Supplemental Figure S3.
OSU-SpARKFuse target enrichment. A–C: Comparison of gene expression [measured as fragments per kilobase per million mapped reads (FPKM)] in total RNA sequencing (RNAseq) data versus OSU-SpARKFuse data in HCC78 (A), TC71 (B), and KG1a (C) cell lines. D: Percentage of sequencing reads mapped to OSU-SpARKFuse target regions from total RNAseq and OSU-SpARKFuse data.
Supplemental Figure S4.
RNA quality in analytical validation cohort. Distribution of percentage of RNA fragments >200 nucleotides (DV200) (A) and RNA integrity number equivalent (RINe) (B) values for RNA derived from cell lines (51 samples), formalin-fixed, paraffin-embedded (FFPE) tissues (43 samples), and fresh-frozen tissues (16 samples). Line indicates mean for all 110 samples.
Supplemental Figure S5.
rRNA depletion in analytical validation cohort. Percentage of rRNA reads detected in cell lines (51 samples), formalin-fixed, paraffin-embedded (FFPE) tissues (43 samples), and fresh-frozen tissues (16 samples). Outliers are plotted as individual dots.
Supplemental Figure S6.
Threshold determination for high-confidence fusion calls. Normalized fusion spanning read counts were determined for 75 fusion events from 74 positive control samples (true positives), for additional unexpected fusion events from these samples (false positives), and for fusion events present in 36 negative control samples (true negatives). Dashed line indicates threshold for high-confidence fusion calls.
Supplemental Figure S7.
Fusion detection in low-input samples. RNA (50 and 100 ng) from the H2228 cell line was used as input for OSU-SpARKFuse. Normalized fusion spanning reads derived from TopHat-Fusion (A) and ChimeraScan (B) are plotted for ALK-PTPN3 and EML4-ALK. Dashed lines indicate threshold for high-confidence fusion calls.
Supplemental Figure S8.
Fusion detection in degraded and diluted fusion-positive samples. RNA from H2228 was incubated at 90°C for 0 to 5 hours, and percentage of RNA fragments >200 nucleotides (DV200) (A and C) and RNA integrity number equivalent (RINe) (B) values were determined (purple lines and bars). Normalized fusion-spanning reads derived from TopHat-Fusion (A) and ChimeraScan (B and C) are plotted at each time point for EML4-ALK and ALK-PTPN3 fusions. RNA from eight fusion-positive cell lines (D), four fusion-positive formalin-fixed, paraffin-embedded tissues (E), and four fusion-positive fresh-frozen tissues (F) was serially diluted to simulate the indicated tumor purities. Normalized fusion-spanning reads derived from ChimeraScan are plotted. Dashed lines indicate threshold for high-confidence fusion calls.
Supplemental Figure S9.
Intrarun repeatability and interrun reproducibility of OSU-SpARKFuse. A: RNA isolated from 12.5% cell line dilutions was prepared and sequenced in the same run by the same technician for a total of three replicates. Normalized fusion-spanning reads derived from ChimeraScan are plotted. Dotted line indicates threshold for high-confidence fusion calls. B: RNA isolated from 25% and 12.5% cell line mixes was prepared by four different technicians and sequenced on two different MiSeq instruments. Normalized fusion-spanning reads derived from ChimeraScan are plotted. Dashed lines indicates threshold for high-confidence fusion calls.
Supplemental Figure S10.
Florescence in situ hybridization (FISH) to detect novel RET fusion. Representative image of RET break-apart FISH on formalin-fixed, paraffin-embedded small-bowel cancer specimen. Inset shows the boxed area at higher magnification. Red, RET 3′ signal; green, RET 5′ signal; blue, DAPI. Yellow arrows represent break-apart RET signal. Pink arrows represent intact RET signal. Original magnification, ×100 (main image).
Supplemental Figure S11.
Detection of novel clinically actionable fusion. A second novel fusion break point was identified that involved exons 1 to 2 of KLK2 and 4 to 17 of FGFR2 in a fresh-frozen biopsy sample from a 61-year-old man with prostate cancer. Top: Schematic of fusion gene with indicated exonic break points. Middle: Bar graph represents average exon level read depth for indicated FGFR2 exons (https://www.ncbi.nlm.nih.gov/refseq; accession number NM_001144913.1). Bottom: Chromatogram trace of KLK2-FGFR2 fusion transcript. Dashed line indicates break point.
References
- 1.Nowell P.C., Hungerford D.A. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 2.Nowell P.C., Hungerford D.A. Chromosome studies in human leukemia, II: chronic granulocytic leukemia. J Natl Cancer Inst. 1961;27:1013–1035. [PubMed] [Google Scholar]
- 3.Kumar-Sinha C., Kalyana-Sundaram S., Chinnaiyan A.M. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129. doi: 10.1186/s13073-015-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertens F., Johansson B., Fioretos T., Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 5.Maher C.A., Kumar-Sinha C., Cao X., Kalyana-Sundaram S., Han B., Jing X., Sam L., Barrette T., Palanisamy N., Chinnaiyan A.M. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maher C.A., Palanisamy N., Brenner J.C., Cao X., Kalyana-Sundaram S., Luo S., Khrebtukova I., Barrette T.R., Grasso C., Yu J., Lonigro R.J., Schroth G., Kumar-Sinha C., Chinnaiyan A.M. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens P.J., McBride D.J., Lin M.L., Varela I., Pleasance E.D., Simpson J.T., Stebbings L.A., Leroy C., Edkins S., Mudie L.J., Greenman C.D., Jia M., Latimer C., Teague J.W., Lau K.W., Burton J., Quail M.A., Swerdlow H., Churcher C., Natrajan R., Sieuwerts A.M., Martens J.W., Silver D.P., Langerod A., Russnes H.E., Foekens J.A., Reis-Filho J.S., van 't Veer L., Richardson A.L., Borresen-Dale A.L., Campbell P.J., Futreal P.A., Stratton M.R. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stransky N., Cerami E., Schalm S., Kim J.L., Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin J.Z., Berger M.F., Adiconis X., Rogov P., Melnikov A., Fennell T., Nusbaum C., Garraway L.A., Gnirke A. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabanski C.R., Magrini V., Griffith M., Griffith O.L., McGrath S., Zhang J., Walker J., Ly A., Demeter R., Fulton R.S., Pong W.W., Gutmann D.H., Govindan R., Mardis E.R., Maher C.A. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J Mol Diagn. 2014;16:440–451. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieslik M., Chugh R., Wu Y.M., Wu M., Brennan C., Lonigro R., Su F., Wang R., Siddiqui J., Mehra R., Cao X., Lucas D., Chinnaiyan A.M., Robinson D. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015;25:1372–1381. doi: 10.1101/gr.189621.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark M.B., Mercer T.R., Bussotti G., Leonardi T., Haynes K.R., Crawford J., Brunck M.E., Cao K.A., Thomas G.P., Chen W.Y., Taft R.J., Nielsen L.K., Enright A.J., Mattick J.S., Dinger M.E. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods. 2015;12:339–342. doi: 10.1038/nmeth.3321. [DOI] [PubMed] [Google Scholar]
- 14.Majewski I.J., Mittempergher L., Davidson N.M., Bosma A., Willems S.M., Horlings H.M., de Rink I., Greger L., Hooijer G.K., Peters D., Nederlof P.M., Hofland I., de Jong J., Wesseling J., Kluin R.J., Brugman W., Kerkhoven R., Nieboer F., Roepman P., Broeks A., Muley T.R., Jassem J., Niklinski J., van Zandwijk N., Brazma A., Oshlack A., van den Heuvel M., Bernards R. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230:270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 15.Beadling C., Wald A.I., Warrick A., Neff T.L., Zhong S., Nikiforov Y.E., Corless C.L., Nikiforova M.N. A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing. J Mol Diagn. 2016;18:165–175. doi: 10.1016/j.jmoldx.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J., Pao W., Engelman J.A., Iafrate A.J., Le L.P. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 17.He J., Abdel-Wahab O., Nahas M.K., Wang K., Rampal R.K., Intlekofer A.M. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergethon K., Shaw A.T., Ou S.H., Katayama R., Lovly C.M., McDonald N.T., Massion P.P., Siwak-Tapp C., Gonzalez A., Fang R., Mark E.J., Batten J.M., Chen H., Wilner K.D., Kwak E.L., Clark J.W., Carbone D.P., Ji H., Engelman J.A., Mino-Kenudson M., Pao W., Iafrate A.J. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drilon A., Wang L., Hasanovic A., Suehara Y., Lipson D., Stephens P., Ross J., Miller V., Ginsberg M., Zakowski M.F., Kris M.G., Ladanyi M., Rizvi N. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Janne P.A., Costa D.B., Varella-Garcia M., Kim W.H., Lynch T.J., Fidias P., Stubbs H., Engelman J.A., Sequist L.V., Tan W., Gandhi L., Mino-Kenudson M., Wei G.C., Shreeve S.M., Ratain M.J., Settleman J., Christensen J.G., Haber D.A., Wilner K., Salgia R., Shapiro G.I., Clark J.W., Iafrate A.J. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw A.T., Ou S.H., Bang Y.J., Camidge D.R., Solomon B.J., Salgia R., Riely G.J., Varella-Garcia M., Shapiro G.I., Costa D.B., Doebele R.C., Le L.P., Zheng Z., Tan W., Stephenson P., Shreeve S.M., Tye L.M., Christensen J.G., Wilner K.D., Clark J.W., Iafrate A.J. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doebele R.C., Davis L.E., Vaishnavi A., Le A.T., Estrada-Bernal A., Keysar S., Jimeno A., Varella-Garcia M., Aisner D.L., Li Y., Stephens P.J., Morosini D., Tuch B.B., Fernandes M., Nanda N., Low J.A. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015;5:1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farago A.F., Le L.P., Zheng Z., Muzikansky A., Drilon A., Patel M., Bauer T.M., Liu S.V., Ou S.H., Jackman D., Costa D.B., Multani P.S., Li G.G., Hornby Z., Chow-Maneval E., Luo D., Lim J.E., Iafrate A.J., Shaw A.T. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol. 2015;10:1670–1674. doi: 10.1097/01.JTO.0000473485.38553.f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbiah V., Westin S.N., Wang K., Araujo D., Wang W.L., Miller V.A., Ross J.S., Stephens P.J., Palmer G.A., Ali S.M. Targeted therapy by combined inhibition of the RAF and mTOR kinases in malignant spindle cell neoplasm harboring the KIAA1549-BRAF fusion protein. J Hematol Oncol. 2014;7:8. doi: 10.1186/1756-8722-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes S.A., Beare D., Boutselakis H., Bamford S., Bindal N., Tate J., Cole C.G., Ward S., Dawson E., Ponting L., Stefancsik R., Harsha B., Kok C.Y., Jia M., Jubb H., Sondka Z., Thompson S., De T., Campbell P.J. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies K.D., Le A.T., Theodoro M.F., Skokan M.C., Aisner D.L., Berge E.M., Terracciano L.M., Cappuzzo F., Incarbone M., Roncalli M., Alloisio M., Santoro A., Camidge D.R., Varella-Garcia M., Doebele R.C. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18:4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer M.K., Chinnaiyan A.M., Maher C.A. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D., Salzberg S.L. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shugay M., Ortiz de Mendibil I., Vizmanos J.L., Novo F.J. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29:2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 31.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11:11.0.1–11.0.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu T.L., Goss V.L., Reeves C., Popova L., Nardone J., Macneill J., Walters D.K., Wang Y., Rush J., Comb M.J., Druker B.J., Polakiewicz R.D. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108:4202–4204. doi: 10.1182/blood-2006-06-026666. [DOI] [PubMed] [Google Scholar]
- 39.Jung Y., Kim P., Jung Y., Keum J., Kim S.N., Choi Y.S., Do I.G., Lee J., Choi S.J., Kim S., Lee J.E., Kim J., Lee S., Kim J. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–597. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- 40.Koivunen J.P., Mermel C., Zejnullahu K., Murphy C., Lifshits E., Holmes A.J., Choi H.G., Kim J., Chiang D., Thomas R., Lee J., Richards W.G., Sugarbaker D.J., Ducko C., Lindeman N., Marcoux J.P., Engelman J.A., Gray N.S., Lee C., Meyerson M., Janne P.A. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 42.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 43.Chinen Y., Sakamoto N., Nagoshi H., Taki T., Maegawa S., Tatekawa S., Tsukamoto T., Mizutani S., Shimura Y., Yamamoto-Sugitani M., Kobayashi T., Matsumoto Y., Horiike S., Kuroda J., Taniwaki M. 8q24 amplified segments involve novel fusion genes between NSMCE2 and long noncoding RNAs in acute myelogenous leukemia. J Hematol Oncol. 2014;7:68. doi: 10.1186/s13045-014-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Rolle A.F., Klempner S.J., Garrett C.R., Seery T., Sanford E.M., Balasubramanian S., Ross J.S., Stephens P.J., Miller V.A., Ali S.M., Chiu V.K. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget. 2015;6:28929–28937. doi: 10.18632/oncotarget.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulligan L.M. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer. 2014;14:173–186. doi: 10.1038/nrc3680. [DOI] [PubMed] [Google Scholar]
- 46.Shang Z., Niu Y., Cai Q., Chen J., Tian J., Yeh S., Lai K.P., Chang C. Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumour Biol. 2014;35:1881–1890. doi: 10.1007/s13277-013-1253-6. [DOI] [PubMed] [Google Scholar]
- 47.Hermans K.G., Bressers A.A., van der Korput H.A., Dits N.F., Jenster G., Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 48.Pflueger D., Terry S., Sboner A., Habegger L., Esgueva R., Lin P.C., Svensson M.A., Kitabayashi N., Moss B.J., MacDonald T.Y., Cao X., Barrette T., Tewari A.K., Chee M.S., Chinnaiyan A.M., Rickman D.S., Demichelis F., Gerstein M.B., Rubin M.A. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G., Jones S.J., Marra M.A., Sadar M.D. Identification of genes targeted by the androgen and PKA signaling pathways in prostate cancer cells. Oncogene. 2006;25:7311–7323. doi: 10.1038/sj.onc.1209715. [DOI] [PubMed] [Google Scholar]
- 50.Zook J.M., Chapman B., Wang J., Mittelman D., Hofmann O., Hide W., Salit M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 51.Woyach J.A., Furman R.R., Liu T.M., Ozer H.G., Zapatka M., Ruppert A.S., Xue L., Li D.H., Steggerda S.M., Versele M., Dave S.S., Zhang J., Yilmaz A.S., Jaglowski S.M., Blum K.A., Lozanski A., Lozanski G., James D.F., Barrientos J.C., Lichter P., Stilgenbauer S., Buggy J.J., Chang B.Y., Johnson A.J., Byrd J.C. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frampton G.M., Ali S.M., Rosenzweig M., Chmielecki J., Lu X., Bauer T.M. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 53.Qadir M.A., Zhan S.H., Kwok B., Bruestle J., Drees B., Popescu O.E., Sorensen P.H. ChildSeq-RNA: a next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16:361–370. doi: 10.1016/j.jmoldx.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Scolnick J.A., Dimon M., Wang I.C., Huelga S.C., Amorese D.A. An efficient method for identifying gene fusions by targeted RNA sequencing from fresh frozen and FFPE samples. PLoS One. 2015;10:e0128916. doi: 10.1371/journal.pone.0128916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C., Chen Y., Mohammad T.A., Chen Y., Fedor H.L., Lotan T.L., Zheng Q., De Marzo A.M., Isaacs J.T., Isaacs W.B., Nadal R., Paller C.J., Denmeade S.R., Carducci M.A., Eisenberger M.A., Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo M., Misale S., Wei G., Siravegna G., Crisafulli G., Lazzari L., Corti G., Rospo G., Novara L., Mussolin B., Bartolini A., Cam N., Patel R., Yan S., Shoemaker R., Wild R., Di Nicolantonio F., Bianchi A.S., Li G., Siena S., Bardelli A. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016;6:36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 57.Pfarr N., Stenzinger A., Penzel R., Warth A., Dienemann H., Schirmacher P., Weichert W., Endris V. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosomes Cancer. 2016;55:30–44. doi: 10.1002/gcc.22297. [DOI] [PubMed] [Google Scholar]
- 58.Takeda M., Sakai K., Terashima M., Kaneda H., Hayashi H., Tanaka K., Okamoto K., Takahama T., Yoshida T., Iwasa T., Shimizu T., Nonagase Y., Kudo K., Tomida S., Mitsudomi T., Saigo K., Ito A., Nakagawa K., Nishio K. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol. 2015;26:2477–2482. doi: 10.1093/annonc/mdv475. [DOI] [PubMed] [Google Scholar]
- 59.San Lucas F.A., Allenson K., Bernard V., Castillo J., Kim D.U., Ellis K., Ehli E.A., Davies G.E., Petersen J.L., Li D., Wolff R., Katz M., Varadhachary G., Wistuba I., Maitra A., Alvarez H. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016;27:635–641. doi: 10.1093/annonc/mdv604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.