Abstract
UDP-xylose is a sugar donor required for the synthesis of diverse and important glycan structures in animals, plants, fungi, and bacteria. Xylose-containing glycans are particularly abundant in plants and in the polysaccharide capsule that is the major virulence factor of the pathogenic fungus Cryptococcus neoformans. Biosynthesis of UDP-xylose is mediated by UDP-glucuronic acid decarboxylase, which converts UDP-glucuronic acid to UDP-xylose. Although this enzymatic activity was described over 40 years ago it has never been fully purified, and the gene encoding it has not been identified. We used homology to a bacterial gene, hypothesized to encode a related function, to identify a cryptococcal sequence as putatively encoding a UDP-glucuronic acid decarboxylase. A soluble 47-kDa protein derived from bacteria expressing the C. neoformans gene catalyzed conversion of UDP-glucuronic acid to UDP-xylose, as confirmed by NMR analysis. NADH, UDP, and UDP-xylose inhibit the activity. Close homologs of the cryptococcal gene, which we termed UXS1, appear in genome sequence data from organisms ranging from bacteria to humans.
UDP-xylose (UDP-Xyl) is an important nucleotide sugar required for the synthesis of numerous glycoconjugates. Examples include proteoglycans, abundant molecules in the extracellular matrix and on the cell surface of animal cells; plant polysaccharides such as xyloglucan and xylan; and the capsular polysaccharides of certain fungi. We have examined synthesis of this compound in the pathogenic fungus Cryptococcus neoformans.
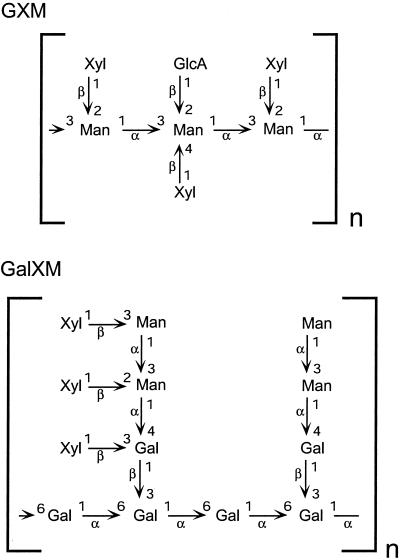
C. neoformans is responsible for serious disease in the setting of immune compromise. A distinguishing feature of this organism is its extensive polysaccharide capsule, which is absolutely required for virulence (1). Capsular polysaccharides are shed copiously into the environment and mediate multiple processes by which this microbe inhibits the host immune response (2–4). The most abundant component of the capsule is termed glucuronoxylomannan, a polymer that reaches sizes of roughly one million daltons and is composed of a mannose backbone substituted with glucuronic acid and xylose (Fig. 1; ref. 5). The ratio of xylose/glucuronic acid/mannose ranges from 1:1:3 to 4:1:3, depending on the strain, and this variation is likely to be responsible for differences between cryptococcal serotypes that have distinct host range and growth characteristics (6). The remainder of the capsule is composed of a smaller galactoxylomannan (GalXM, Fig. 1); this polymer consists of a galactose backbone with trimer side chains of galactose and mannose. Each GalXM side chain bears between one and three xylose residues (7).
Figure 1.
Structures of cryptococcal capsular polysaccharides. The structures of C. neoformans glucuronoxylomannan (GXM) (serotype B) and galactoxylomannan (GalXM) are shown. Not shown are the 6-O-acetyl groups that modify up to 10% of mannose residues in GXM. Component monosaccharides are d-pyranoses.
We are investigating cryptococcal glycan formation, with particular focus on the capsular polysaccharides because of their unique structures and role in virulence. Our interests include the glycosyltransferase-catalyzed reactions that transfer sugars from activated nucleotide sugar precursors to acceptor polysaccharides during capsule synthesis and the upstream metabolic pathways that provide the appropriate precursors (8). The pathway of UDP-Xyl synthesis is of particular interest in C. neoformans, because of the abundant and strain-specific modification of glucuronoxylomannan and galactoxylomannan with xylose. It is not known whether UDP-Xyl is the immediate donor of xylose to these polysaccharides or whether another donor compound is interposed. Studies by White and colleagues (9) suggested that lipid-linked xylose is formed in vitro by cryptococcal membranes, but whether this compound plays a role in capsular polysaccharide synthesis was not determined (9). Xylose also occurs in a novel family of glycophosphosphingolipids that is found in cryptococcus but not in mammals.§
Two enzymes are required for the synthesis of UDP-Xyl from UDP-glucose (UDP-Glc). First, UDP-Glc dehydrogenase catalyzes the dehydrogenation of its substrate to form UDP-glucuronic acid (UDP-GlcA); this is then acted on by UDP-GlcA decarboxylase to produce UDP-Xyl. We have used homology to known enzymes in other systems to clone a functional C. neoformans UDP-Glc dehydrogenase (M.B.-P. and T.L.D., unpublished data). In this article we focus on the second step in synthesis.
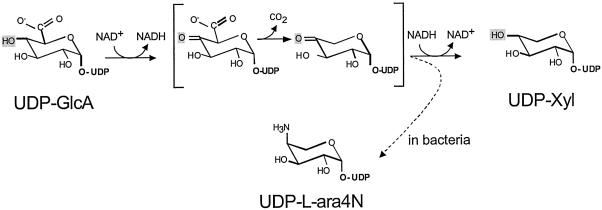
Decarboxylation of UDP-GlcA to form UDP-Xyl was first demonstrated in plant extracts (11). Although early studies of this enzyme indicated that it generated a mixture of UDP-d-xylose and UDP-l-arabinose, it was later shown that this was caused by a contaminating UDP-l-arabinose-4-epimerase; the UDP-GlcA decarboxylase itself produces only UDP-Xyl (12). This activity was partially purified from soluble fractions from both wheat germ (12–14) and a nonpathogenic species of cryptococcus, C. laurentii (15). The mechanism of decarboxylation has been suggested to proceed by several steps (ref. 12; Fig. 2). First, UDP-GlcA is oxidized to UDP-4-keto-GlcA. This β-keto acid may be readily decarboxylated, generating an intermediate UDP-4-keto pentose; this product may then be reduced stereospecifically to UDP-Xyl.
Figure 2.
Model for the mechanism of UDP-GlcA decarboxylase, based on refs. 12 and 23. The compounds in brackets are 4-keto intermediates. Depending on the system the same intermediates could give rise to end products of UDP-Xyl or UDP-aminoarabinose (dashed arrow, in bacteria). The substituent at the 4-position is shaded in each structure.
To identify the UDP-GlcA decarboxylase in C. neoformans, we exploited information derived from bacterial systems. Studies in Salmonella typhimurium have identified two loci, pmrE and pmrF, which are required for addition of l-4-aminoarabinose to lipid A (16). PmrE encodes a UDP-Glc dehydrogenase. The UDP-GlcA synthesized by this enzyme could serve as a precursor to UDP-aminoarabinose (16–18), via initial reactions similar to those that convert UDP-GlcA to UDP-Xyl in eukaryotic systems (Fig. 2). PmrF is located in a putative operon with several genes whose sequences suggest roles in sugar metabolism; Gunn et al. (16) have proposed that these genes also may participate in aminoarabinose addition. Extending this model, the groups of Raetz and Morona (17, 18) independently hypothesized a pathway for UDP-aminoarabinose synthesis based on sequence analysis of the genes in the pmrF cluster. They suggested that the protein product of one sequence, ORF3, might catalyze oxidation of the 4-position of UDP-GlcA. This could lead to decarboxylation of the resulting UDP-4-keto sugar and subsequent transamination to form UDP-aminoarabinose (Fig. 2, dashed arrow).
We reasoned that a protein that catalyzes oxidation of UDP-GlcA en route to UDP-Xyl might bear structural similarity to one that performs the same function during UDP-aminoarabinose synthesis. Based on this hypothesis we cloned a cDNA from C. neoformans that encodes a polypeptide homologous to that predicted by the bacterial ORF3. Below we describe functional analysis and characterization of the UDP-GlcA decarboxylase encoded by this C. neoformans gene, which we have named UXS1 for its role in UDP-Xyl synthesis. These studies demonstrate a UDP-GlcA decarboxylase from cryptococcus, help validate the model for bacterial aminoarabinose synthesis, and provide sequence information on a class of enzymes important across biological kingdoms.
Experimental Procedures
Cloning the Cryptococcal Homolog of Bacterial orf3.
The sequence of ORF3 from the pmrF gene cluster in S. typhimurium (GenBank accession no. AF036677) was used to search publicly available C. neoformans genomic sequences (November 14, 2000; http://sequence-www.stanford.edu/group/C.neoformans/index.html). A 165-bp sequence in the database (contained in read 967214E04.x1) was identified as part of a potential homolog and used to obtain the corresponding cryptococcal gene by reverse transcriptase–PCR. Briefly, total RNA was isolated from C. neoformans strain Cap67, reverse-transcribed by using an oligo(dT) primer, and used as a template for rapid amplification of cDNA ends (RACE) analysis using CLONTECH Marathon reagents according to the manufacturer's instructions. Gene-specific primers for 5′ and 3′ RACE reactions were DC-3 (5′-ATCAAGTCGTGGACGTATTGGAAGGATCGGGTCTG-3′) and DC-2 (5′-AGGGCAAGCGTGTCGCTGAGACCTTGACC-3′), respectively. RACE reaction products were sequenced and assembled into a 1,360-bp contiguous region containing initiation and stop codons bounding an ORF homologous to ORF3. A NotI-modified 3′primer (DC-4, 5′-GCGGCCGCTAGATGGACACCTCTCGGATTCGGG-3′) and an NdeI-modified 5′ primer (DC-5, 5′-CATATGAGCTCCGAGAAGCCTGAGACTATCACCC-3′) were used to amplify the full-length ORF, and the resulting reverse transcriptase–PCR product was cloned into the expression vector pET-28a (Novagen) to form plasmid pDC22.6. DNA sequence of the coding region was confirmed by sequencing (Nucleic Acid Chemistry Laboratory, Washington University School of Medicine) and submitted to GenBank (accession no. AF385328). This sequence exactly matched portions of a 1,932-bp genomic sequence in the C. neoformans database, which also included six introns ranging in size from 50 to 438 bp. Primers DC-4 and DC-5 also were used to amplify the complete gene from C. neoformans strain B4500.
Protein Expression and Assay.
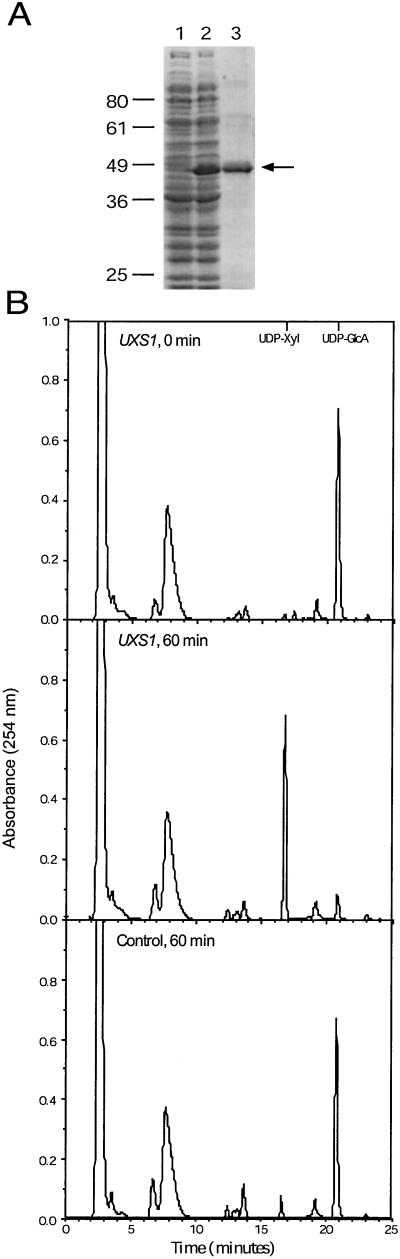
For assay of expressed protein, Escherichia coli strain BL21 (DE3)pLysS cells (Stratagene), carrying either pDC22.6 (with C. neoformans UXS1) or the pET-28a vector alone, were grown to OD600 = 0.5 and then induced for 3 h at 30°C with 1 mM isopropyl β-d-thiogalactoside. Cells were collected, washed with dH2O, and stored at −80°C. Cell pellets were resuspended in 20 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.8/10% glycerol/1 mM EDTA/1 mM DTT/0.5 mM PMSF) and broken in a French press (1,100 psi). Particulate matter was sedimented (Sorvall SS34 rotor, 13,000 rpm, 30 min, 4°C), and the supernatant fraction was stored in aliquots at −20°C. For purification, bacterial protein was thawed, bound to Ni-NTA agarose (Qiagen, Valencia, CA) in 50 mM NaH2PO4, 300 mM NaCl, 1 mM DTT, pH 8, and the column was washed with the binding buffer containing 10 mM imidazole. Protein was eluted with binding buffer containing 250 mM imidazole, typically with 50% yield and ≈30-fold enrichment to a specific activity of at least 150 nmol UDP-Xyl min−1⋅mg−1 in a standard assay (below). SDS/PAGE of the starting material and eluted fraction are shown below (see Fig. 4A).
Figure 4.
Expression and assay of Uxs1. (A) SDS/PAGE analysis of bacterial proteins stained with Coomassie blue. Lane 1, soluble protein from bacteria containing vector alone; lane 2, soluble protein from bacteria expressing Uxs1; lane 3, Uxs1 protein purified by nickel chromatography. The migration positions of standards (in kDa) are shown at left, and the arrow indicates Uxs1. (B) HPLC profiles of standard assays performed for the times indicated using soluble protein from bacteria carrying either a plasmid containing UXS1 or a control plasmid. As indicated at the top the UDP-xylose standard elutes just before 17 min (at 0.21 M KH2PO4) and UDP-GlcA just before 21 min (at 0.28 M KH2PO4). The large peak near 8 min is NAD+.
Standard UDP-GlcA decarboxylase assays (50 μl) were performed at 23°C for 15 min and contained final concentrations of 40 mM Tris⋅HCl (pH 7.4), 1 mM NAD+, 1 mM UDP-GlcA, and 2–10 mg/ml total bacterial protein. Reactions were stopped by the addition of 50 μl of phenol/chloroform (1:1, vol/vol), vortex-mixed, and subjected to centrifugation (microfuge, 16,000 × g, 5 min, room temperature). The upper (aqueous) phase was reserved, and the lower phase was re-extracted with 80 μl dH2O. The two aqueous phases were pooled and analyzed by HPLC (Waters) on a 250 × 4.6 mm quaternary amine-silica gel anion exchange column (Hypersil 5 μ SAX resin; Phenomenox, Torrance, CA) run at 1.5 ml/min. After injection the column was washed for 5 min with buffer A (5 mM phosphate buffer, pH 3.2) and then eluted with a 20-min linear gradient from 0 to 60% buffer B (0.6 M KH2PO4) as described (19).
Product Analysis.
One hundred-microliter UDP-GlcA decarboxylase assays were performed by using protein prepared from bacteria carrying either pDC22.6 or the vector alone. Assay products were analyzed by HPLC on a SAX column developed with ammonium formate, and the reaction product was collected. This sample was lyophilized, dissolved in water, relyophilized several times, and then exchanged twice with 99.96% D2O. Proton NMR data were collected at 25°C on Varian Inova 500-MHz and 600-MHz spectrometers.
Results and Discussion
To identify potential UDP-GlcA decarboxylase sequences, we first examined the sequence of the ≈74-kDA polypeptide encoded by S. typhimurium ORF3 from the pmrF gene cluster. This sequence showed homology to two groups of polypeptides: the N-terminal portion to prokaryotic methionyl-tRNA formyltransferases and the C-terminal region to a family of putative dTDP-glucose 4–6 dehydratases. Based on this homology we hypothesized that the C-terminal domain of the protein encoded by ORF3 was most likely related to a nucleotide sugar decarboxylase, and we used this sequence to do a blast search (20) of the partially completed C. neoformans genome sequence (November 14, 2000; http://sequence-www.stanford.edu/group/C.neoformans/index.html; ref. 21). One sequence demonstrated homology and was used to obtain a complete ORF (Fig. 3) from cryptococcal cDNA as described in Experimental Procedures. The C. neoformans ORF contained a putative nicotinamide adenosine dinucleotide (NAD+) binding motif (22) near the N terminus (Fig. 3, *). This GXGXXG motif also is found in dTDP-glucose 4–6 dehydratases and other proteins that bind NAD+; its presence in the cryptococcal sequence is consistent with the requirement for an electron acceptor in the first step of the decarboxylation reaction (Fig. 2).
Figure 3.
Comparison of C. neoformans Uxs1 to S. typhimurium Orf3. Identical amino acids are shaded dark gray and similar residues are shaded light gray. The NAD+ binding motif of Uxs1 is underscored with * and is aligned with a similar motif in the bacterial sequence that falls in a well-conserved region. The N-terminal 200 residues of Orf3 are not shown.
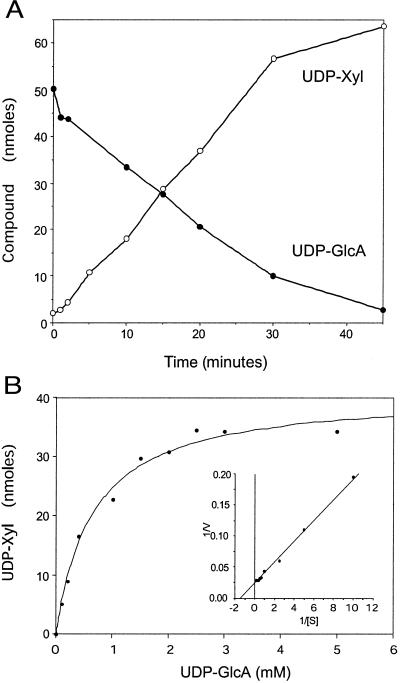
To test for UDP-GlcA-decarboxylase activity, we expressed the cryptococcal coding sequence in bacteria and assayed soluble protein for the ability to convert UDP-GlcA to UDP-Xyl. Fig. 4A shows protein extracts from induced bacteria harboring either a vector control or the UXS1 expression plasmid, analyzed by SDS/PAGE. UXS1 encodes a protein about ≈47 kDa in size, in agreement with the size predicted from the amino acid sequence (46.5 kDa), and mass spectrometry of tryptic fragments (not shown) confirmed identity. As shown in Fig. 4B, protein from bacteria expressing the cryptococcal gene, but not from control cells, converted UDP-GlcA to a product that comigrated with a UDP-Xyl standard. This conversion depended on time (Fig. 5A) and the amount of protein (not shown), and no product was formed when the bacterial protein was heat-inactivated (not shown).
Figure 5.
Characterization of product formation. (A) Time course. Standard Uxs1 reactions (Experimental Procedures) were carried out for the indicated times, and the amounts of reactant and product were analyzed. ●, UDP-GlcA; ○, UDP-Xyl. (B) Effect of UDP-GlcA concentration. Reactions (15 min) were performed without added NAD+ and in the presence of the indicated concentrations of UDP-GlcA. (Inset) A reciprocal plot of the data.
Replacement of the UDP-GlcA substrate with UDP-Glc or UDP-galacturonic acid yielded no product, indicating specificity of the reaction for UDP-GlcA. Kinetic studies (Fig. 5B) indicated that the activity has a Km of ≈0.7 mM for UDP-GlcA, similar to values determined in previous studies of partially purified enzyme preparations from C. laurentii (1 mM) and wheat germ (0.18–0.53 mM; refs. 12 and 13). This value is within the range of reported intracellular concentrations of UDP-GlcA. Under the conditions tested the Vmax was 0.8 μmol UDP-Xyl min−1⋅mg−1 for nickel column-purified enzyme.
The expressed protein was active at temperatures from 23°C to 42°C, with highest activity at 37°C, consistent with efficient function within a mammalian host. The enzyme demonstrated good activity over a broad pH range (6.5–8.5), with 7.5 as the optimum pH. Metal ions were not required, with over 90% of control activity retained in the presence of 5 mM EDTA, and there was slight inhibition by the presence of 10 mM MgCl2, CaCl2, or MnCl2. The decarboxylase was stable at −70°C for at least several months, and activity was reduced by less than 10% by the inclusion in assay mixtures of 50 mM DTT or 5% glycerol and by less than 20% by the presence of 20% glycerol or 100 mM NaCl.
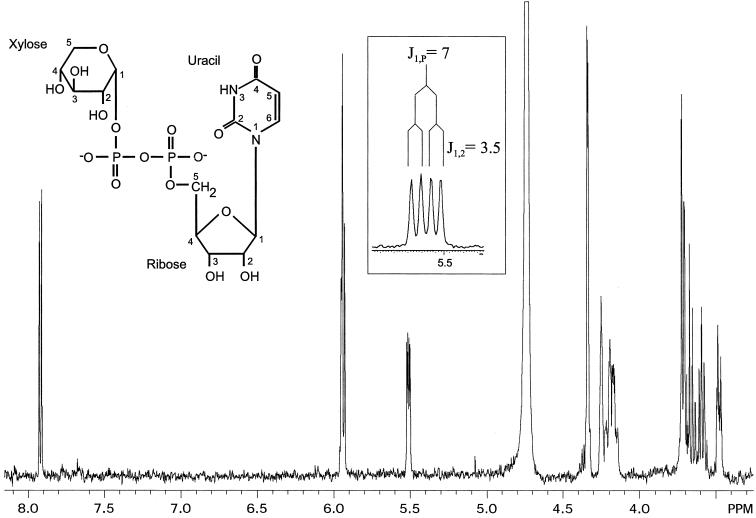
Substitution of UDP-[14C]GlcA for UDP-GlcA in standard reactions yielded a radiolabeled product that was analyzed by TLC. The compound comigrated with a UDP-Xyl standard in two solvent systems, and acid hydrolysis of the material resulted in a radiolabeled species that comigrated with a xylose standard (not shown). To confirm identity we used NMR to compare the product of a nonradiolabeled reaction to a UDP-xylose standard. The one-dimensional proton spectrum of the compound produced by Uxs1 (Fig. 6 and Table 1) matched that of a UDP-Xyl standard and indicated 95% purity of the assay product. A two-dimensional proton-proton correlated spectroscopy spectrum confirmed the expected assignments (not shown). The α-anomeric configuration of the xylose was confirmed by the 3JH1,H2 coupling constant of 3.5 Hz (Table 1), and the 3J3,4 coupling constant of 8.9 Hz confirmed the assignment of xylose as opposed to arabinose (3J3,4 = 3.4 Hz), demonstrating stereo-specific reduction of the 4-keto intermediate. This product analysis demonstrated that the expressed protein encodes a UDP-GlcA decarboxylase capable of synthesizing UDP-Xyl (E.C. designation 4.1.1.35), and we named the cryptococcal gene UXS1, for its participation in UDP-Xyl synthesis. We have similarly termed the cryptococcal gene encoding a UDP-Glc dehydrogenase UXS2 (M.B.-P. and T.L.D., unpublished work; GenBank accession no. AF405548).
Figure 6.
Product analysis. The one-dimensional proton NMR spectrum of the product of the Uxs1 assay is shown. (Left Inset) The structure of UDP-Xyl. (Boxed Inset) An expanded region of the axis just above 5.5 ppm to show the xylose H1 peak splitting.
Table 1.
Chemical shifts of the Uxs1 assay product in D2O at 25°C (in ppm), referenced to internal acetone at δ2.218 relative to 3-(trimethylsilyl)-1-propanesulfonic acid
| H1 | H2 | H3 | H4 | H5, 5′ | H6 | |
|---|---|---|---|---|---|---|
| Xylose | 5.517 | 3.483 | 3.679 | 3.592 | 3.72 | |
| 3J1,23.5 | 3J2,38.9 | 3J3,48.9 | ||||
| 3J1,P7.0 | 3J2,P3.5 | |||||
| Ribose | 5.938 | 4.34 | 4.255 | 4.34 | 4.15–4.22 | |
| Uracil | 5.956 | 7.927 |
Scalar coupling constants (J, in Hz) for some internal residues are also indicated. Identical results were obtained with standard UDP-Xyl.
The first step in the conversion of UDP-GlcA to UDP-Xyl, oxidation to UDP-4-keto-GlcA, is believed to require NAD+. After decarboxylation of this intermediate, the product may be reduced stereo-specifically by NADH to yield UDP-Xyl (Fig. 2). Consistent with this mechanism, previous studies of UDP-GlcA decarboxylase activity from C. laurentii indicated an absolute requirement for exogenous NAD+ (23). However, studies of the enzyme from wheat germ showed neither activation by NAD+ nor inhibition by NADH (12). Further examination of preparations from wheat germ suggested that in fact NAD+ is tightly bound to the enzyme, resisting even the action of NADase (12). Lack of NADH dissociation also explains the observed retention of tritium label at C-4 throughout a reaction postulated to proceed via a 4-keto intermediate (12, 23). The C. neoformans enzyme converted UDP-GlcA to UDP-Xyl in the absence of exogenous NAD+, although activity was 2-fold higher in the presence of 1 mM NAD+ (Table 2, compare standard reaction and reaction without NAD+). As shown in Table 2, the enzyme was inhibited by UDP and NADH, but was not affected by NADP+ and only slightly inhibited by NADPH. In contrast to earlier studies on crude enzyme from C. neoformans (24), UDP-Xyl was only moderately inhibitory, with an effect similar to that of UDP-Glc.
Table 2.
Inhibition of the standard Uxs1 assay
| Condition | Concentration, mM | % of control |
|---|---|---|
| Standard assay | — | 100 |
| − NAD+ | — | 49 |
| + NADH | 1 | 34 |
| + NADH | 5 | 4 |
| + NADP+ | 1 | 91 |
| + NADPH | 1 | 74 |
| + UDP | 1 | 29 |
| + UDP-Xyl | 1 | 91 |
| + UDP-Xyl | 5 | 58 |
| + UDP-Glc | 1 | 97 |
| + UDP-Glc | 5 | 68 |
Standard enzyme assays (15 min; 23°C; 40 mM M Tris⋅HCl, pH 7.4; 1 mM NAD+; 1 mM UDP-GlcA; 2 mg/ml bacterial protein) were performed in the absence of NAD+ (− NAD+) or in the presence of the indicated compound. Data are shown as per cent activity compared to a standard assay. Each value is the mean of triplicate assays performed at least twice.
Previous literature on UDP-GlcA decarboxylase activities has indicated that enzymes from different sources vary in terms of subcellular localization. Plant and fungal enzymes are soluble (12, 23) while the activity is associated with a particulate fraction in both cultured chondrocytes and hen oviduct (25, 26). Consistent with these data, the expressed cryptococcal enzyme is found primarily in a soluble fraction, and the sequence has no regions suggestive of membrane spanning domains.
Our interest in examining the UDP-GlcA decarboxylase from cryptococcus stems from considerations of both biochemistry and potential therapeutics. Future experiments should examine the polysaccharides produced in cryptococcal cells in which UXS1 has been disrupted, which may help elucidate pathways of capsular polysaccharide formation. Because the capsule is required for cryptococcal virulence it is likely that altered capsule synthesis caused by reduced decarboxylase activity will lead to reduced virulence of the disrupted strains. If this is the case, compounds that inhibit this enzyme have potential as therapeutic agents. The difference in subcellular localization between fungal and mammalian enzymes suggests that identifying selective inhibitors may be possible.
Beyond cryptococcus, UDP-GlcA decarboxylases are found in numerous organisms, as suggested by the importance of xylose in plant and animal biology and strengthened by the closely related sequences identified by blast searches with the predicted translation product of UXS1. Close homologs of C. neoformans Uxs1 (typically >50% identical over 300 residues) were found in Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, mouse, and human (20). As expected from the absence of xylose synthesis in Saccharomyces cerevisiae, no close homolog was found in searches of the yeast genome.
In plants xylose polymers may comprise up to 25% of dry weight (27), so production of UDP-Xyl is a crucial function. Additionally, xylose addition is one feature that differentiates plant glycoproteins from those of mammals and may be responsible for their immunogenicity. In animals xylose links the glycosaminoglycans heparan sulfate and chondroitin sulfate to the serine residues of proteoglycan core proteins. Proteoglycans are central to physiological processes from hemostasis to cell migration and development (28–30) and to pathologic processes such as microbial adherence and invasion (10); the availability of UDP-GlcA decarboxylase sequence will facilitate investigation of these events. The cloning and expression of UDP-GlcA decarboxylase from C. neoformans therefore should contribute to a broad array of investigations, ranging from plant biology, protein expression, and fungal pathogenesis to mammalian cell biology.
Acknowledgments
We thank David Feingold for encouraging us to pursue studies of this enzyme. We are especially grateful to Jeff Esko for the suggestion that we consider the orf3 sequence in taking a molecular approach to this project, an idea that stemmed from conversations with Chris Raetz and Steve Breazeale, and for constructive comments on the manuscript. We appreciate helpful discussions with Chris Raetz, Eduardo Groisman, Steve Levery, Robert Cherniak, and members of the Doering laboratory. We thank John Glushka for performing NMR analysis, Jeremi D. Johnson and Ron Orlando for MS analysis, and Hong Liu for preparing Fig. 3. We also gratefully acknowledge the funding support that the Burroughs Wellcome Fund and the National Institutes of Health (Grant AI47087) have provided to the C. neoformans Genome Project at the Stanford Genome Technology Center. This work was supported by grants to T.L.D. from the Burroughs Wellcome Fund (Junior Investigator Award in Molecular Pathogenic Mycology), Neose Corporation (GRANT Award), the Andrew and Virginia Craig Faculty Research Fund, and National Institutes of Health (Grant AI49173) and by startup funds provided to M.B.-P. by the University of Georgia.
Note Added in Proof.
The sequence of a UDP-GlcA-carboxylyase from Pisum sativum has been deposited in GenBank by Kobayashi et al. (accession no. BAB40967). Orthologs from Aribidopsis thaliana (Bar-Peled, accession nos. AAK70880, AAK70881, and AAK70882) and Mus musculus (Olson and Esko, accession no. AAK85410) have also recently been deposited.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF385328).
De Souza Gutierrez, A. L., Heise, N., Wait, R., Jones, C., Previato, J. O. & Mendonca-Previato, L. (1999) Glycobiology, 9, 1114 (abstr.).
References
- 1.Chang Y C, Kwon-Chung K J. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, DC: Am. Soc. Microbiol.; 1998. [Google Scholar]
- 3.Buchanan K L, Murphy J W. Emerg Infect Dis. 1998;4:71–83. doi: 10.3201/eid0401.980109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues M L, Alviano C S, Travassos L R. Microbes Infection. 1999;1:293–301. doi: 10.1016/s1286-4579(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 5.Cherniak R, Sundstrom J B. Infect Immun. 1994;62:1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherniak R, Valafar H, Morris L C, Valafar F. Clin Diagn Lab Immunol. 1998;5:146–159. doi: 10.1128/cdli.5.2.146-159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnav V V, Bacon B E, O'Neill M O, Cherniak R. Carbohydr Res. 1998;306:315–330. doi: 10.1016/s0008-6215(97)10058-1. [DOI] [PubMed] [Google Scholar]
- 8.Doering T L. Trends Microbiol. 2000;8:547–553. doi: 10.1016/s0966-842x(00)01890-4. [DOI] [PubMed] [Google Scholar]
- 9.White C W, Cherniak R, Jacobson E S. J Med Vet Mycol. 1990;28:289–301. doi: 10.1080/02681219080000381. [DOI] [PubMed] [Google Scholar]
- 10.Rostand K S, Esko J D. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feingold D S, Neufeld E F, Hassid W Z. J Biol Chem. 1960;235:910–913. [PubMed] [Google Scholar]
- 12.Ankel H, Feingold D S. Biochemistry. 1965;4:2468–2475. [Google Scholar]
- 13.John K V, Schutzbach J S, Ankel H. J Biol Chem. 1977;252:8013–8017. [PubMed] [Google Scholar]
- 14.Kyossev Z N, Drake R R, Kyosseva S V, Elbein A D. Eur J Biochem. 1995;228:109–112. [PubMed] [Google Scholar]
- 15.Ankel H, Ankel E, Feingold D S. Biochemistry. 1966;5:1864–1869. doi: 10.1021/bi00870a012. [DOI] [PubMed] [Google Scholar]
- 16.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Lin S, Cotter R J, Raetz C R H. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 18.Baker S J, Gunn J S, Morona R. Microbiology. 1999;145:367–368. doi: 10.1099/13500872-145-2-367. [DOI] [PubMed] [Google Scholar]
- 19.Bar-Peled M, Lewinsohn E, Fluhr R, Gressel J. J Biol Chem. 1991;266:20953–20959. [PubMed] [Google Scholar]
- 20.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitman J, Casadevall A, Lodge J K, Perfect J R. Mycopathologia. 1999;148:1–7. doi: 10.1023/a:1007136602930. [DOI] [PubMed] [Google Scholar]
- 22.Wierenga R K, Terpstra P, Hol W G. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 23.Ankel H, Feingold D S. Biochemistry. 1966;5:182–189. doi: 10.1021/bi00865a024. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson E S, Payne W R. J Bacteriol. 1982;152:932–934. doi: 10.1128/jb.152.2.932-934.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bdolah A, Feingold D S. Biochem Biophys Res Commun. 1965;21:543–546. doi: 10.1016/0006-291x(65)90519-x. [DOI] [PubMed] [Google Scholar]
- 26.John K V, Schwartz N B, Ankel H. J Biol Chem. 1977;252:6707–6710. [PubMed] [Google Scholar]
- 27.Zablackis E, Huang J, Muller B, Darvill A G, Albersheim P. Plant Physiol. 1995;107:1129–1138. doi: 10.1104/pp.107.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esko J D. Curr Opin Cell Biol. 1991;3:805–816. doi: 10.1016/0955-0674(91)90054-3. [DOI] [PubMed] [Google Scholar]
- 29.Park P W, Reizes O, Bernfield M. J Biol Chem. 2000;275:29923–29926. doi: 10.1074/jbc.R000008200. [DOI] [PubMed] [Google Scholar]
- 30.Turnbull J, Powell A, Guimond S. Trends Cell Biol. 2001;11:75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]