Abstract
Microorganisms can be versatile in their interactions with each other, being variously beneficial, neutral or antagonistic in their effect. Although this versatility has been observed among many microorganisms and in many environments, little is known regarding the mechanisms leading to these changes in behavior. In the present work, we analyzed the mechanism by which the soil bacterium Pseudomonas fluorescens BBc6R8 shifts from stimulating the growth of the ectomycorrhizal fungus Laccaria bicolor S238N to killing the fungus. We show that among the three secondary metabolites produced by the bacterial strain—the siderophores enantio-pyochelin and pyoverdine, and the biosurfactant viscosin—the siderophores are mainly responsible for the antagonistic activity of the bacterium under iron-limited conditions. While the bacterial strain continues to produce beneficial factors, their effects are overridden by the action of their siderophores. This antagonistic activity of the strain P. fluorescens BBC6R8 in iron-depleted environments is not restricted to its influence on L. bicolor, since it was also seen to inhibit the growth of the actinomycete Streptomyces ambofaciens ATCC23877. We show that the strain P. fluorescens BBc6R8 uses different strategies to acquire iron, depending on certain biotic and abiotic factors.
Keywords: mutualism, antagonism, mycorrhiza helper bacteria, iron, siderophores, viscosin
Pseudomonas fluorescens produces several secondary metabolites in its interactions with soil fungi and bacteria, and shifts from being mutualistic to antagonist under iron-limited conditions.
INTRODUCTION
Historically, microorganisms have been classified according to their effects on other organisms as pathogens, antagonists, parasites, mutualistics and commensals. While some microorganisms, such as biotrophic pathogens and symbionts, have a strict and specific lifestyle, most are versatile in their behaviors and can vary from antagonistic to neutral or beneficial and vice versa depending on various biotic and abiotic parameters. Opportunistic pathogens are often commensal microorganisms that take advantage of the weaknesses of their host defenses. For example, the opportunistic human pathogen Candida albicans is a commensal microorganism, commonly found as part of the normal flora of the digestive track (Jenkinson and Douglas 2002). Similarly, saprophytic fungi such as Alternaria or Fusarium that normally live on dead organic matter can become pathogenic when they encounter debilitated, stressed or senescent plants (Prell and Day 2001). Most documented cases of opportunistic infections are related to deficiencies of the immune system of the host, whether a plant or animal (Whipps 2001; Berg, Eberl and Hartmann 2005). Less is known regarding the potential shift of behavior of mutualistic microorganisms during their lifetime. In the present work, we address this point by focusing on the interaction between an ectomycorrhizal (EcM) fungus and a mycorrhiza helper bacterium.
EcM fungi establish a symbiotic relationship with trees in which they provide water and inorganic nutrients for their host in exchange for sugars (Smith and Read 2008). These exchanges take place in ectomycorrhiza, a mixed organ composed of root and fungal tissues. EcM symbiosis is widespread across the world, with most trees from temperate and boreal forests having been found to evidence its working. EcM symbiosis is not restricted to the mutualistic association between symbiotic fungi and plant roots, since mycorrhizal root tips are colonized by a complex microbiome with which plant roots and EcM fungi interact (Deveau 2016). Within this complex microbiome, some bacteria, called mycorrhiza helper bacteria (MHB), can be beneficial in stimulating the formation or function of mycorrhiza (Frey-Klett, Garbaye and Tarkka 2007; Deveau and Labbé 2016). A number of genera of both Gram-negative and Gram-positive bacteria constitute the range of MHB. Mechanisms of action differ between strains and can involve spore germination, or stimulation of pre-symbiotic growth, plant defenses or short root production (Frey-Klett, Garbaye and Tarkka 2007). MHB have aroused the interest of the scientific community due to their potential to improve health and growth of tree seedlings (Garbaye 1994). However, despite several significant successes of field inoculations of MHB to improve mycorrhiza formation and tree growth (Garbaye and Bowen 1989; Poole et al.2001; Founoune et al.2002; Frey-Klett, Garbaye and Tarkka 2007; Zhao et al.2014), no MHB is currently routinely used to inoculate tree nurseries, and none has been commercialized yet. This is partly due to the seeming adaptive nature of these strains where, for reasons largely unknown, their beneficial effects are not always observed. Thus, they can be good models to study the mechanisms by which a beneficial strain can shift to being either harmless or harmful towards other organisms. In our own work, we applied the EcM fungus Laccaria bicolor S238N and the MHB strain Pseudomonas fluorescens BBc6R8 as model organisms.
Pseudomonas fluorescens BBc6R8 is an MHB strain that has been isolated from a sporocarp of L. bicolor and which promotes the establishment of EcM symbiosis between L. bicolor and Douglas fir (Frey-Klett, Pierrat and Garbaye 1997). Its MHB effect is found in its ability to stimulate the survival and the pre-symbiotic growth of L. bicolor S238N mycelium under unfavorable growth conditions, notably thanks to nutrient complementation (Brulé et al.2001; Deveau et al.2010). This MHB strain also appears to benefit from the interaction with the fungus, since its survival in the soil is increased in the presence of the EcM fungus (Deveau et al.2010). However, the helper effect of the strain P. fluorescens BBc6R8 is lost when the bacterial strain is inoculated at elevated concentrations (Frey-Klett et al.1999). The objective of the present work was to decipher the mechanism by which P. fluorescens BBc6R8 shifts from having a beneficial influence towards an antagonist one. In addition, we analyzed the specificity of this antagonistic activity through the analysis of the interaction between P. fluorescens BBc6R8 and the soil bacterium Streptomyces ambofaciens ATCC23877.
MATERIALS AND METHODS
Fungal–bacterial and bacterial–bacterial confrontation bioassays
All strains used in this work are listed in Table 1. The EcM basidiomycete L. bicolor S238N was maintained on Pachlewski agar medium P5 (Di Battista et al.1996) at 25°C for 3 weeks. Pseudomonas fluorescens BBc6R8 WT and the mutants ΔviscB, Tn5::foxA, 7B3 and 7C3 were stored at –80°C in LB Medium (Sambrook, Fritsch and Maniatis 1989) with 20% glycerol added and kanamycin (50 μg mL−1) when appropriate. To prepare the bacterial inoculum for the in vitro bioassays with L. bicolor, the bacterial strains were first grown on 10% TSA plates (3 g L−1 Tryptic Soy Broth from Difco and 15 g L−1 of agar) supplemented with kanamycin when appropriate at 25°C for 65 h. The bioassay was prepared as described in Deveau et al. (2007), except that King's B (KB) agar medium (20 g L−1 proteose peptone #3 from Difco, 1.5 g L−1 K2HPO4, 15 g L−1 of agar, supplied after autoclaving with 10 mL L−1 100% glycerol and 6 mL L−1 MgSO4 1M) was used instead of P20Th- for iron starvation conditions. Briefly, four droplets of bacterial solution of a single strain (OD600 nm 0.7 in sterile water) were distributed at 1.2 cm from the center of a fungal plug. Control treatments in which bacteria were replaced by sterile water droplets were performed. Plates were incubated in the dark at 10°C to reproduce soil temperature. The diameter of fungal colonies was measured every 5–8 days. For each treatment, seven replicates were performed. The full experiment was performed four times independently.
Table 1.
List of strains used in this study.
| Strains | Characteristics | References |
|---|---|---|
| L. bicolor S238N | EcM basidiomycete fungus | Deveau et al. (2007) |
| S. ambofaciens ATCC23877 | Reference strain isolated from soil | Pinnert-Sindico (1954) |
| P. fluorescens BBc6R8 | Mycorrhiza helper bacterium isolated from sporocarps of L. bicolor S238N. Wild-type strain | Frey-Klett, Pierrat and Garbaye (1997) |
| P. fluorescens BBc6R8 Tn5::foxA | Mutant derived from P. fluorescens BBc6R8, Tn5 insertion in foxA | Galet et al. (2015) |
| P. fluorescens BBc6R8 7B3 | Mutant derived from P. fluorescens BBc6R8 by Tn5 insertion | This study |
| P. fluorescens BBc6R8 7C3 | Mutant derived from P. fluorescens BBc6R8 by Tn5 insertion | This study |
| P. fluorescens BBc6R8 ΔviscB-(P448) ΔviscB-(P450) | Knock-out isogenic mutants of gene viscB derived from P. fluorescens BBc6R8 | This study |
| P. tolaasii LMG 2342T | White line agar assay indicator strain | Wong and Preece (1979) |
| P. reactans LMG5329 | WLIP producer | Wong and Preece (1979) |
| P. fluorescens SBW25 | Viscosin producer | de Bruijn et al. (2007) |
The S. ambofaciens ATCC23877–P. fluorescens BBc6R8 interaction was analyzed using the following setup. Five-microliter drops of S. ambofaciens (109 spores mL−1) and of P. fluorescens BBc6R8 (109 cfu mL−1) were deposited 4 mm apart on KB agar plates and were incubated at 28°C for 24–72h. Control experiments with P. fluorescens BBc6R8 or S. ambofaciens alone were performed for each set of incubation conditions. The full experiment was performed three times independently.
Transposon mutagenesis of P. fluorescens BBc6R8 and screen of the library for the loss of siderophore production
Transposon mutagenesis was carried using the transposon mini Tn5::gfp::lux cloned into pGP704 (Fones et al.2010). The transposon was introduced into P. fluorescens BBc6R8 by triparental mating using the helper plasmid pRK2013. The resultant bacterial mixture was spread onto M9 agar containing 50 μg mL−1 kanamycin. Resulting colonies were inoculated into 100 μL of KB broth in 42 96-well plates and incubated at 28°C overnight. A total of 75 μL of 50% (v/v) glycerol were then added to each well, and plates were stored at –80°C as a mutant library.
Mutants for siderophore production were screened by replication of the Tn5 bacterial library on KB plates. After 48 h of growth at 28°C, plates were observed under UV light, and colonies that did not produce fluorescence were selected.
Direct mutagenesis of P. fluorescens BBc6R8 viscB gene
Since viscB is a very large gene (12.9 kb), partial as opposed to full deletion of the gene 2562998405 (locus MHB_001840; Deveau et al.2014) was carried out (between positions 146 and 7113), with a stop codon introduced at the site of the deletion. The deletion was created using the two-step allelic exchange strategy described by Zhang and Rainey (2007) and previously used to generate T3SS secretion mutants in P. fluorescens BBc6R8 (Cusano et al.2011). Flanking regions were cloned into the suicide integration vector pUIC3 using the In-fusion BioBrick assembly system (Clontech, TakaraBio Europe, Saint Germain en Laye, France). Cycloserine enrichment was used to enrich strains that had lost the chromosomally integrated pUIC3-based vector. Strains were grown overnight in 100 mL LB broth; 500 μL of the overnight culture were inoculated into 30 mL pre-warmed LB broth and cultivated at 28°C with shaking (200 rpm) for 2 h. Tetracycline (Tc) was added at the final concentration of 10 μg mL−1 to inhibit the growth of cells that had lost pUIC3. After growth for 3 h, cycloserine was added at 3 mg mL−1 and growth was continued for another 3 h (during this step the growing Tc-resistant cells were killed). The cells were then washed in sterile water, diluted and inoculated onto M9 plus X-Gal plates. Double recombinants were verified by loss of lacZY activity, loss of Tc resistance, PCR screening for recombinants in which the deletion had occurred and finally sequencing across the deleted region. Two confirmed mutants, designated ΔviscB-(P448) and ΔviscB-(P450), were chosen for use in subsequent assays to control for possible non-specific changes (such as point mutations elsewhere in the genome) which could potentially have been introduced during the mutagenesis procedure and affected bacterial fitness.
Extraction, isolation and identification of enantio-pyochelin and desferrioxamine E
Pseudomonas fluorescens BBc6R8 and S. ambofaciens ATCC23877 were grown on separate KB agar plates at 26°C. After 72 h of incubation, cells and agar media were recovered and transferred to glass scintillation vials. One milliliter of ethyl acetate/methanol/formic acid (65:35:0.1, v/v) were added to the samples. Samples were then sonicated for 10 min. The extraction procedure was repeated three times. The resulting crude extracts were centrifuged at 10 000 rpm for 2 min. The supernatants were recovered and dried in vacuo. Samples were then resuspended in 100 μL of methanol for further analysis. Samples were directly infused into the mass spectrometer using a Triversa nanomate-electrospray ionization source (Advion Biosystems, Ithaca, NY, USA) coupled to a 6.42 T Thermo LTQ-FT-ICR mass spectrometer (Thermo-Electron Corporation, San Jose, CA, USA). FT-MS and ion trap MS/MS spectra were acquired using Tune Plus software version 1.0 and Xcalibur software version 1.4 SR1 (Thermo-Electron Corporation). The instrument was tuned on m/z 816, the 15+ charge state of cytochrome C.
Extraction, isolation and identification of pyoverdine
Pseudomonas fluorescens BBc6R8 was grown in 1.5 L of Davis Minimum Broth without dextrose (Becton, Dickinson and Company), supplemented with 20 mmol glycerol (DMBgly) with shaking (130 rpm) at 28°C for 90 h. Cells were removed by centrifugation (20 min, 4200 g, 4°C). The pH of the supernatant was adjusted to 6, Amberlite XAD-4 adsorbent resin (50 g/L) was added and the mixture was agitated for 4 h at 70 rpm at 28°C. The XAD-4 resin was then passed through a fritted funnel, and the bound metabolites were eluted with MeOH/H2O (50:50) under agitation, for 4 h. The combined eluates were dried using a rotary evaporator, resuspended in 1 mL of deionized water and a second chromatography step was performed on an open column of Sephadex LH-20 (15 g). After loading, the column was rinsed with deionized water and colored bands were systematically collected. Four fractions were obtained, lyophilized and subsequently analyzed by LC/MS. The LC/MS system consisted of an Agilent HPLC 1100 Series system coupled to an AB Sciex 3200 Q TRAP mass spectrometer. The HPLC separation was carried out on reversed phase using a linear gradient of 2:98–100:0 MeOH/H2O over a period of 30 min, followed by isocratic elution with 100% MeOH for 30 min (Macherey-Nagel Nucleodur PolarTec column; 250 × 4.6 m; particle size 5 μm; flow rate 1 mL min−1; UV-DAD monitoring).
Extraction, isolation and identification of viscosin
Pseudomonas fluorescens BBc6R8 was grown with shaking (140 rpm) at 26°C in six 5 L Erlenmeyer flasks, each containing 1.5 L of DMBgly. After 72 h of incubation, cell and supernatant fractions of the cultures were separated by centrifugation at 4000 rpm for 10 min. The supernatant was acidified to pH 3 with HCl and extracted twice with ethyl acetate (1:1, v/v). The resulting crude extract was fractionated by vacuum liquid chromatography over reversed-phase silica gel (Macherey-Nagel Polygoprep 50–60) using a stepwise gradient of methanol–water and finally dichloromethane to give six fractions. 1H NMR profiling of these fractions indicated the 100% MeOH fraction to be of further interest because its 1H NMR spectrum exhibited three clusters of resonances typical of peptides, i.e. downfield amide signals, α- proton resonances and upfield side chain protons. RP-HPLC separation of this fraction using a Waters HPLC system (consisting of a 600 pump, a 996 photodiode array detector, a Rheodyne 7161 injector and a Perkin Elmer Series 200 vacuum degasser) and gradient elution from 80%–100% MeOH in H2O (0.05% TFA) over 45 min, followed by isocratic elution at 100% MeOH for an additional 10 min (column: Phenomenex Luna C18(2)-100A, 10 × 250 mm, 5-μm column in combination with a Phenomenex SecurityGuard Luna-C18(2) 10 × 10 mm pre-column; 2.0 mL min−1 flow rate; UV monitoring at 215 nm) gave viscosin in a semi pure form. The compound was subsequently purified twice by RP-HPLC; first by employing a Macherey-Nagel Nucleosil 120–5, 4 × 250 mm column (1.0 mL min−1, 40 min gradient elution from 60% to 100% MeOH-H2O (0.05% TFA) followed by a 20-min hold) and second by using a Waters SymmetryShield RP18, 4.6 × 250 mm column (1.0 mL/min, 40 min gradient elution from 50% to 100% ACN-H2O (0.05% TFA) followed by a 20-min hold) to yield 13.5 mg of pure viscosin: ESI-TOF-MS: [M+H]+ = 1126.7, [M+Na]+ = 1148.7; HR-ESI-TOF-MS: [M+Na]+ = 1148.6798 m/z consistent with the molecular formula C54H95N9O16Na. HR-ESI-TOF-MS spectra were recorded on a Bruker Daltonic's microTOF-Q instrument. NMR experiments were carried out on a Bruker Avance 300 DPX and on a Bruker DRX-250 spectrometer, respectively. Spectra were referenced to the residual solvent signal of d4-MeOH with resonances at δH/C 3.35/49.0.
White-line-in-agar test
The white line test was performed according to the method of Wong and Preece (1979). Pseudomonas fluorescens BBc6R8 was streaked on KB medium at 1 cm distance next to P. tolaasii LMG 2342T and the formation of a white precipitate in the agar was evaluated after 24–72 h growth at 26°C. Pseudomonas reactans LMG 5329 (WLIP producer) and P. fluorescens SBW25 (viscosin producer) (de Bruijn et al.2007) were included as positive and negative controls, respectively. Assays were carried out in duplicate and repeated twice.
MALDI imaging mass spectrometry
The experiment was performed as previously described (Phelan et al.2014). Briefly, a region of agar including the microorganisms grown alone or in interaction was excised with a razor blade and laid upon a MALDI MSP 96 anchor target plate (Bruker Daltonics). A photograph was taken of the MALDI plate holding the agar samples. Universal MALDI matrix (Sigma-Aldrich) was applied on top of the samples using a 53-μm sieve, and samples were dried at 37°C overnight. A second photograph was taken of the MALDI plate. All microbial colonies were subjected to MALDI-TOF MS in positive reflectron mode using a 400–500 μm spatial resolution in XY by a Bruker Daltonics Microflex. Data were analyzed using FlexImaging 3.0.
Statistical analyses
Statistical analyses were performed using the R software. Normality of datasets was tested using the Shapiro–Wilk test followed by appropriate tests depending on the normality of data.
RESULTS
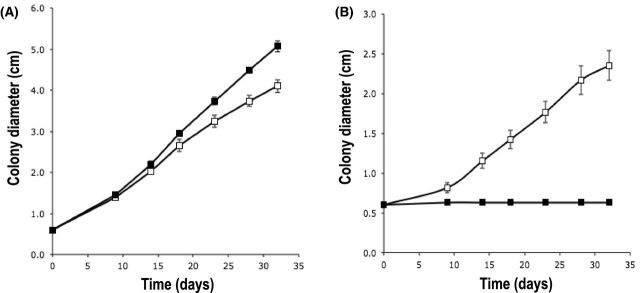
Pseudomonas fluorescens BBc6R8 stimulates L. bicolor S238N radial growth in vitro on P20Th- medium but inhibits it on KB.
As previously reported (Deveau et al.2007), the ability of P. fluorescens BBc6R8 to stimulate the growth of L. bicolor S238N (Brulé et al.2001) can be reproduced in vitro when the two microorganisms are co-cultivated on P20Th- medium (Fig. 1A). However, the growth of L. bicolor was completely inhibited by the strain BBc6R8 when P20Th- medium was replaced by the iron-limited KB medium in a similar set up (Fig. 1B).
Figure 1.
Effect of P. fluorescens BBc6R8 on the radial growth of L. bicolor S238N on P20Th- (A) and KB (B) agar media. Open squares: L. bicolor S238N alone, filled squares: L. bicolor S238N co-cultivated with P. fluorescens BBc6R8. Each point is the mean value of seven biological replicates ±SD. The experiment was reproduced three times independently and similar results were obtained each time.
Identification of the secondary metabolites produced by strain BBc6R8 through genome mining and imaging mass spectrometry
In order to identify the metabolites potentially involved in this inhibitory effect, the genome sequence of strain P. fluorescens BBc6R8 (Deveau et al.2014) was explored to identify gene clusters involved in the production of secondary metabolites. None of the genes commonly responsible for the production of secondary metabolites involved in Pseudomonas antagonism towards fungi and other eukaryotes, such as 2,4-diacetylphloroglucinol (DAPG), hydrogen cyanide (HCN), pyrrolnitrin, pyoluteorin or rhizoxins (Loper et al.2008, 2012; Gross and Loper 2009), was found in the genome of strain BBc6R8. However, complete gene clusters coding for the non-ribosomal peptide synthetases responsible for the biosynthesis of the two siderophores enantio-pyochelin (Youard et al.2007) and a pyoverdine, and for the production of a cyclic lipopeptide, were found (Table 1, Supporting Information). In addition, a truncated gene cluster coding for enzymes involved in the production of mangotoxin was found, but it is expected to be non-functional due to the absence of the corresponding mbo-like operon (Carrión et al.2012). The production of the two siderophores was confirmed by mass spectrometry. The MS2 profile of the ion with a precursor mass of m/z 325 [M+H]+ completely matched with the fragmentation profile of enantio-pyochelin (Fig. 1, Supporting Information). The capability of strain BBc6R8 to produce a pyoverdine is in line with the bioinformatic analysis and was already proven in previous studies by UV- and MS-based experiments with the purified compound (Gamalero et al.2003). At that time, a mass of 1305.7 Da was determined using ESI-MS. In this study, we detected this pyoverdine as a minor metabolite along with a suite of four major pyoverdine derivatives, ranging in size from 1276.5 to 1306.6 Da (Fig. 2A, Supporting Information). Empirically, pyoverdines of one strain differ in the nature of the dicarboxylic acid side chain. So far glutamic acid, α-ketoglutaric acid, succinic acid (amide) and malic acid (amide) were found (Budzikiewicz 2004) and the relative amounts of the various side chains of a pyoverdine depend on the cultivation time, iron concentration and pH of the culture broth (Schäfer, Tarak and Budzikiewicz 1991). Therefore, the occurrence of the compounds can be readily attributed to the presence of the succinic acid amide (Pyo-Suca, m/z 1277.7 [M+H]+), succinic acid (Pyo-Suc, m/z 1278.7 [M+H]+), malic acid (Pyo-Mal, m/z 1295.8 [M+H]+), α-ketoglutaric acid (Pyo-Kgl, m/z 1306.6 [M+H]+) and glutamic acid (Pyo-Glu, m/z 1307.7 [M+H]+) congeners (Fig. 2A, Supporting Information). All pyoverdine derivatives showed the pyoverdine-characteristic UV-absorption maxima at 365 and 380 nm (Fig. 2B, Supporting Information), which result from their common fluorescent chromophore (Abdallah 1991) and indicated in addition that they were detected as deferrated free pyoverdines. Lastly, the structure elucidation of the lipopeptide was obtained by NMR and MS analyses. The purified lipopeptide gave an [M+Na]+ peak at m/z 1148.6798 in the high-resolution mass spectrum (Fig. 3, Supporting Information), appropriate for a molecular formula of C54H95N9O16 which corresponded according to a database search to the structures of either viscosin or WLIP. These two structures represent epimers, i.e. the only structural difference between viscosin and WLIP is the D versus L configuration of one leucine residue. The white-line-agar assay was used to discriminate between the two lipopeptides. The absence of a white line in this assay when BBc6R8 was grown alongside P. tolaasii supports the deduction that the lipopeptide of BBc6R8 is viscosin (Fig. 4, Supporting Information) because WLIP is known to produce a white precipitate in combination with tolaasin, whereas viscosin does not (Wong and Preece 1979). Furthermore, the 1D-NMR experiments confirmed the overall lipopeptidic nature of the isolated compound (Fig. 5, Supporting Information), and demonstrated that the observed chemical shift values were in good agreement with the reported 1H NMR data for viscosin (Laycock et al.1991).
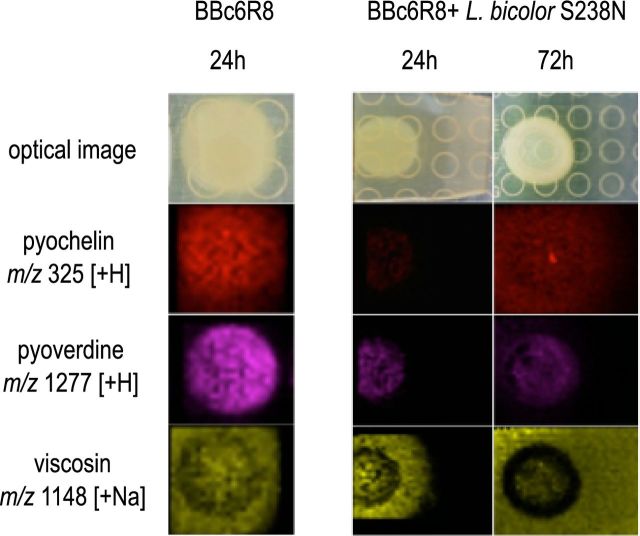
Next, imaging mass spectrometry (IMS) was used to follow the production of enantio-pyochelin, pyoverdine and viscosin during the interaction between L. bicolor S238N and P. fluorescens BBc6R8 on P20Th- and KB medium. None of the three compounds was detected when the microorganisms were cultured alone or together on P20Th- medium (data not shown). In contrast, the three compounds were produced by the bacterial strain on KB medium when the bacterium was cultivated alone or in the presence of L. bicolor S238N (Fig. 2). Both enantio-pyochelin and viscosin were seen diffusing from the bacterial colonies into the external medium as soon as 24 h post-inoculation, and reached the fungal colonies at 72 h.
Figure 2.
Microbial IMS images of secondary metabolites produced by P. fluorescens BBc6R8 grown alone (left images) and side by side with L. bicolor S238N (right images) on KB medium. Laccaria bicolor S238N was grown for 3 weeks on KB medium to allow hyphal development before the inoculation of P. fluorescens BBc6R8. Optical images are displayed in the top row, all other images are overlays of falsely colored m/z distributions over optical images.
Laccaria bicolor S238N is not sensitive to viscosin but to siderophores
Viscosin and other similar cyclic lipopetides have been reported to have antibiotic and antifungal activities (Raaijmakers et al.2010). To determine whether viscosin could be responsible for the inhibitory effect of BBc6R8, two knock-out isogenic mutants were constructed by deletion of the viscB gene encoding for a non-ribosomal peptide synthetase. The non-polar deletions were checked by PCR, and the inability of the mutants to produce viscosin was confirmed by LC-MS (data not shown). No difference in the inhibition of the growth of L. bicolor S238N was observed between the WT bacterial strain and the viscB mutants (Fig. 6, Supporting Information). In addition, to test whether viscosin can have an impact on L. bicolor S238N growth in conditions where no other secondary metabolite is produced, purified viscosin was added to P20Th- prior inoculation with L. bicolor S238N. Two concentrations were tested: one corresponding to the concentration measured in the extracellular medium of pure cultures of P. fluorescens BBc6R8 (1.5 mg L−1) and a 10-fold higher concentration close to that produced by the soil biocontrol strain P. fluorescens SBW25 (de Bruijn et al.2007). Strain SBW25 is phylogenetically closely related to strain BBc6R8 (Jun et al.2016), and has been shown to induce lysis of zoospores of the oomycete Phytophtora infestans through the production of viscosin. A slight inhibition of the radial growth of L. bicolor S238N was measured (–12%, P < 0.01 Kruskal–Wallis test) at the highest concentration, but not at the level that is produced by BBc6R8 (Table 2). Thus, we conclude that L. bicolor S238N is very slightly sensitive to viscosin, and that the lipopeptide is not responsible for the growth inhibition observed on KB medium in the presence of P. fluorescens BBc6R8.
Table 2.
Effect of viscosin on the radial growth of L. bicolor S238N after 5 and 10 days of co-culture. Each value corresponds to the mean diameter of six biological replicates ±SD. ‘a’ indicates values which are statistically significantly different according to a Kruskal–Wallis test (P < 0.01) followed by a Mann–Whitney post-hoc test. dpi: days post-inoculation.
| Control | Viscosin | Viscosin | |
|---|---|---|---|
| (cm) | 1.5 mg L−1 (cm) | 15 mg L−1 (cm) | |
| 5 dpi | 1.7 ± 0.08 | 1.6 ± 0.06 | 1.3 ± 0.08a |
| 10 dpi | 2.8 ± 0.06 | 2.8 ± 0.11 | 2.5 ± 0.1a |
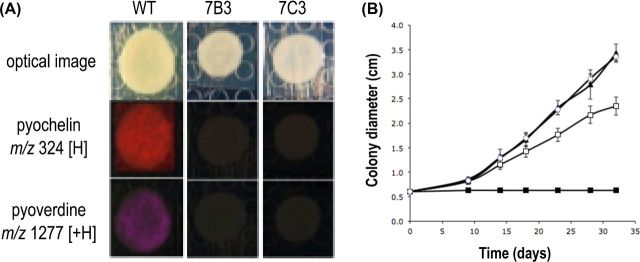
In the second step, we tested whether the production of siderophores by P. fluorescens BBc6R8 could inhibit the growth of the EcM fungus. To do so, we isolated two clones from a Tn5 mutant library of strain BBc6R8 that did not produce fluorescence under UV excitation. Neither pyoverdine nor enantio-pyochelin could be detected by IMS when the two mutants were grown on KB medium (Fig. 3A). The effect of the two clones on the growth of L. bicolor S238N was tested in vitro on iron-deficient and non-deficient media. Mutants 7B3 and 7C3 harbored the same phenotype as the wild type when confronted with L. bicolor S238N on P20Th- medium (data not shown). In contrast, none of the mutants inhibited the growth of L. bicolor S238N on KB medium, suggesting that competition for iron is responsible for the antagonistic effect of BBc6R8 (Fig. 3B). Unfortunately, we were unable to identify the position of the Tn5 insertion in the genome sequences of the mutant 7B3 and 7C3, for unknown technical reasons. Thus, we cannot exclude the possibility that mutants 7B3 and 7C3 have lost other functions in addition to siderophore production. Therefore, we tested whether competition for iron was responsible for the growth inhibition observed on KB medium by supplementing KB medium with iron(III) chloride (20 mg L−1) and by monitoring the radial growth of L. bicolor cultivated alone or together with BBc6R8 WT. If competition for iron is involved in the inhibition of growth of L. bicolor in the presence of P. fluorescens BBc6R8, then the addition of iron should suppress this inhibition. Under these conditions, BBc6R8 no longer inhibited the radial growth of L. bicolor S238N, but rather stimulated it (+16% after 9 days of incubation, t-test P < 0.05). Altogether, our data suggest that the production of siderophores is very likely to be responsible for the inhibitory effect of BBc6R8. In addition, our data suggest that P. fluorescens BBc6R8 also produces beneficial compounds on KB medium, but that their beneficial effect is masked by the antagonistic activity of the siderophores.
Figure 3.
Suppression of pyochelin and pyoverdine production in P. fluorescens BBc6R8 and its effects on the interaction with L. bicolor S238N. (A) IMS images of secondary metabolites produced by P. fluorescens BBc6R8 WT and by the mutants 7B3 and 7C3 grown on KB medium for 24 h. Similar results were obtained at 72 h and 7 days. Optical images are displayed in the top row, all other images are overlays of falsely colored m/z distributions over optical images. (B) Effects of P. fluorescens BBc6R8 WT and of the mutants 7B3 and 7C3 on the radial growth of L. bicolor S238N on KB agar medium. Open squares: L. bicolor S238N alone, closed squares: L. bicolor S238N co-cultivated with P. fluorescens BBc6R8 WT, closed triangles: L. bicolor S238N co-cultivated with 7B3, open circles: L. bicolor S238N co-cultivated with 7C3. Each point is the mean value of seven biological replicates ±SD. The experiment was reproduced three times independently and similar results were obtained each time.
Siderophores of P. fluorescens BBc6R8 also inhibit the growth of S. ambofaciens ATCC23877 on KB medium
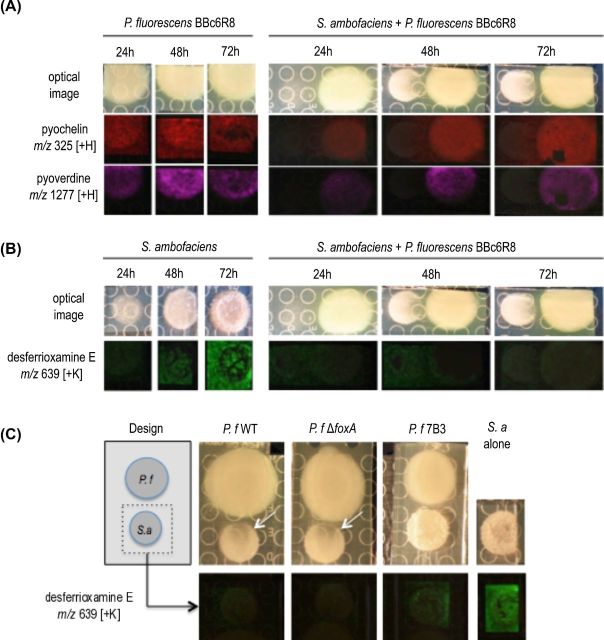
We demonstrated in a previous study that P. fluorescens BBc6R8 ceases production of pyoverdine and enantio-pyochelin when desferrioxamine and coelichelin, two siderophores produced by the actinomycete S. ambofaciens, are present in the environment (Galet et al.2015). Instead of using its own siderophores to scavenge iron, P. fluorescens BBc6R8 captures S. ambofaciens siderophores thanks to the receptor FOXA (Galet et al.2015). This observation was made when the two microorganisms were cultivated side by side on 26A agar medium (Galet et al.2015). However, when we replaced 26A agar medium by KB agar medium using the same co-cultivation assay, P. fluorescens BBc6R8 continued to produce enantio-pyochelin and pyoverdine in the presence of S. ambofaciens as indicated by IMS (Fig. 4A). Streptomyces ambofaciens also produced a siderophore—desferrioxamine E—when grown alone on KB medium (Fig. 4B). The identity of desferrioxamine E was confirmed by tandem mass spectrometry (Fig. 7, Supporting Information). A very low amount of desferrioxamine E was detected when the two microorganisms were co-cultured on KB medium (Fig. 4B). This reduction of desferrioxamine E production was independent of siderophore piracy by FOXA and of the siderophore production by P. fluorescens BBc6R8, since both mutants Tn5::foxA and 7B3 induced the same phenotype as that of the WT strain (Fig. 4C). In addition to its effect on siderophore production, P. fluorescens BBc6R8 inhibited the development of S. ambofaciens colonies (Fig. 4A, C). This inhibition was not due to S. ambofaciens siderophore piracy by P. fluorescens BBc6R8, since the Tn5::foxA mutant similarly inhibited the growth of S. ambofaciens (Fig. 4C). In contrast, mutant 7B3 and 7C3 that lost the ability to produce siderophores did not seem to inhibit the growth of S. ambofaciens, suggesting that P. fluorescens siderophores are also responsible, at least in part, for the inhibition of the growth of S. ambofaciens on KB medium (Fig. 4C).
Figure 4.
Microbial IMS images of secondary metabolites produced by P. fluorescens BBc6R8 (A) and S. ambofaciens ATCC23857 (B) when cultured alone or side by side on KB medium. Optical images are displayed in the top row, all other images are overlays of falsely colored m/z distributions over optical images. (C) Production of desferrioxamine E by S. ambofaciens ATCC23857 when cultured alone or side by side with P. fluorescens BBc6R8 WT or mutants Tn5::foxA or 7B3. The white arrows on the optical images point at the inhibition zone on the colony of S. ambofaciens.
DISCUSSION
MHB have aroused interest due to their potential application in improving seedling growth and survival in tree nurseries (Frey-Klett, Garbaye and Tarkka 2007; Deveau and Labbé 2016), and to reduce the use of fertilizers in intensive tree culture systems such as short rotation coppices or Poplar culture (Zhao et al.2014). However, the transition from laboratory to field use has not yet been successfully proven, partly because of the changeable effects of MHB. Their application requires an understanding of their mechanisms of action and a delineation of the precise conditions in which these bacteria exert their beneficial activities. Indeed, as most MHB are non-obligate mutualists, the outcome of the interaction with EcM fungi may be strongly influenced by external parameters. Here, we show that the beneficial activity of the MHB strain P. fluorescens BBc6R8 on the growth of the EcM fungus L. bicolor S238N in vitro is lost in iron starvation conditions. Determinants responsible for the stimulatory activity are most probably still produced during iron starvation, since mutants for siderophore synthesis recovered their ability to stimulate the growth of the fungus in vitro. However, these beneficial determinants would be nullified by the deleterious impact of competition for iron. A similar pattern has been described for the MHB strain Paenibacillus sp. EJP73: the bacterial strain produces a mixture of compounds among which some inhibit the growth of the EcM fungus Lactarius rufus, while others are stimulatory (Aspray et al.2013). The set of inhibitory compounds produced by the strain BBc6R8 is likely to be reduced, since genes linked to the synthesis of antibiotic compounds classically produced by P. fluorescens strains (e.g. DAPG, HCN, pyrrolnitrin, pyoluteorin, rhizoxins; Gross and Loper 2009) were absent from the genome of the MHB strain. In addition, the biosurfactant viscosin produced by the MHB strain BBc6R8 does not appear to be involved in its antagonistic activity, in contrast to other Pseudomonas strains (Laycock et al.1991; de Bruijn et al.2007; Alsohim et al.2014). In this regard, the MHB strain BBc6R8 is very close to the plant growth-promoting rhizobacteria (PGPR) WGS358, 374 and 417 that also lack genes for antibiotic synthesis and rely solely on their high competiveness for iron to exert their PGPR activity (Berendsen et al.2015).
The high efficiency of strain BBc6R8 in the capture of iron strongly contrasts with the limited ability of the fungus to achieve the same. Laccaria bicolor produces several siderophores of the hydroxamate family in the presence of low iron levels, the main representative being linear fusigen (Haselwandter et al.2013). However, the quantity produced by the fungus is very low, since linear fusigen, as well as the two other siderophores produced by the fungus, ferricrocin and triacetylfusarinine, cannot be detected by LC-UV methods, and high-sensitivity LC-MS is required to detect them (Haselwandter et al.2013). In accordance with this observation, we were not able to detect any of the L. bicolor-derived siderophores on KB medium by MALDI-IMS when grown alone or in the presence of P. fluorescens BBc6R8 (data not shown). In addition, Haselwandter et al. (2013) reported that the release of siderophores by L. bicolor into the medium is greatly delayed, only occurring after several weeks of growth in low-iron medium. This contrasts strongly with the behavior of P. fluorescens BBc6R8, which releases massive quantities of the high-affinity siderophore pyoverdine a few hours after encountering low-iron conditions (Galet et al.2015). In the present case, we also observed an important production and diffusion of the low affinity siderophore enantio-pyochelin. In addition to the production of siderophores, the bacterium possesses a wide range of receptors to uptake xenosiderophores, among which hydroxamate siderophores are included (Galet et al.2015). However, this potential piracy of fungal siderophores, if it exists, would probably play a minor role in the antagonistic activity of BBc6R8, as pyoverdine mutants recovered their stimulatory ability. Nevertheless, it is likely that most of the iron present is taken up by the bacterium, and that the fungus cannot obtain enough to sustain its own growth. Iron is essential to the growth of the majority of microorganisms due to its involvement in a wide variety of housekeeping processes, such as respiration or DNA synthesis. Very few organisms can live in the absence of bioavailable iron by substituting iron with other transition metals such as cobalt, manganese or copper. Despite its relatively high abundance in soil, iron is generally poorly bioavailable to organisms, existing mainly in its ferric form which is insoluble in water and highly inaccessible to biological ligands (Kosman 2003). To overcome this problem, many microorganisms produce siderophores. Fe3+-siderophore complexes are then taken up by cells through specific membrane receptors. Competition for iron is often a component of antagonistic interactions between fungi and fluorescent pseudomonads (Cornelis and Matthijs 2002). It is one of the well-known mechanisms of biocontrol of plant root pathogenic fungi (Haas and Défago 2005). Pyoverdines can sequester iron from the rhizosphere and render it unavailable for the nutrition of rhizospheric fungi producing less potent siderophores. Our data suggest that this antagonism may not be restricted to phytopathogenic fungi, but that it could also affect plant mutualistic fungi. Whether the antagonism described here in vitro also occurs in natural soil remains to be tested. However, previous analyses regarding the availability of iron to P. fluorescens in the rhizosphere and in soil indicated that other P. fluorescens strains actively transcribe genes involved in siderophore production within hours after inoculation in the soil (Loper and Henkels 1997). We observed that mutants 7B3 and 7C3 poorly survived in soil compared to the WT (data not shown), thus suggesting that P. fluorescens also produces siderophores in soil. Frey-Klett et al. (1999) showed that high inocula of P. fluorescens BBc6R8 inhibited mycorrhiza formation, and they hypothesized that high bacterial density may lead to harmful competition. Competition for iron may be part of the antagonistic interaction occurring at high bacterial density in soil.
The high efficiency of P. fluorescens in scavenging iron does not only specifically affect the EcM fungus L. bicolor S238N but also the actinomycete bacterium S. ambofaciens. Therefore, we can expect that it has a broad influence in natural environments. Interestingly, P. fluorescens did not use the same strategy to obtain iron when it was co-cultured with S. ambofaciens, depending on the composition of the medium. In one case, P. fluorescens BBc6R8 massively produced its two siderophores, while in another instance, it stopped producing its own siderophores and captured the siderophores produced by S. ambofaciens thanks to its receptor for xenosiderophore FOXA (Galet et al.2015). The behavior of S. ambofaciens in the presence of P. fluorescens also altered depending on the growth conditions: the actinomycete strongly reduced the production of the siderophores ferrioxamine in the presence of P. fluorescens on KB medium. The physiological consequences of such a reduction in the biosynthesis of desferrioxamine may not be limited to the acquisition of iron, since desferrioxamine production has been also linked to morphogenesis in Streptomyces (Lambert et al.2014). The two media mainly differed in their source of sugar (glucose versus glycerol) and peptides (tryptone versus peptone). Whether these changes in behavior are related to nutrient quality or to other factors remains to be investigated. Nevertheless, this indicates that we cannot predict the behavior of bacterial strains in natural environments on the basis of their genomic contents, since a single strain has numerous strategies to solve a single issue, such as in the well-studied case of iron starvation, and the factors that determine which strategy is used await elucidation. This issue is not restricted to the environment, since the human opportunistic pathogen P. aeruginosa also uses multiple pathways to acquire iron during the infection of lungs (Konings et al.2013) and adapts its strategies to acquire iron depending on the type of infection at hand (Cornelis and Dingemans 2013). Therefore, despite our extensive knowledge on the mechanisms of iron scavenging by microorganisms, a lot remains to be discovered regarding the regulation of the process in situ.
Supplementary Material
Acknowledgments
We are grateful to Julia Paterson (Gross Lab) for proofreading the manuscript. AD would like to thank Amina Bouslimani, Don Nguyen, Dimitri Floros, Tal Luzzatto and Mauricio Caraballo Rodriguez (Dorrestein Lab, UCSD) for their help with MALDI-IMS and LC-MS analyses, and Pieter Burlinson (INRA-Nancy) for his help for the construction of Tn5 mutant library.
SUPPLEMENTARY DATA
FUNDING
This work was supported by the French National Research Agency through the Laboratory of Excellence ARBRE (ANR-11- LABX-0002-01) and by the Genomic Science Program (project ‘Plant-Microbe Interactions’), US Department of Energy, Office of Science, Biological and Environmental Research under the contract DE6AC05-00OR22725. Funding of the work in the H.G. laboratory by the Deutsche Forschungsgemeinschaft (GR 2673/2-1) within the frame of the Research Unit FOR854 is gratefully acknowledged. HG and SM would like to gratefully acknowledge the generous contribution of the Alexander von Humboldt Foundation, which provided financial support for conducting this research (Georg Forster Fellowship awarded to SM). We further acknowledge Bruker and NIH Grant GMS10RR029121 for the support of the shared instrumentation infrastructure that enabled this work
Conflict of interest. None declared.
REFERENCES
- Abdallah M. Handbook of Microbial Iron Chelates. Boca Raton: CRC Press; 1991. Pyoverdins and pseudobactins; pp. 139–53. [Google Scholar]
- Alsohim AS, Taylor TB, Barrett GA, et al. The biosurfactant viscosin produced by Pseudomonas fluorescens SBW25 aids spreading motility and plant growth promotion. Environ Microbiol. 2014;16:2267–81. doi: 10.1111/1462-2920.12469. [DOI] [PubMed] [Google Scholar]
- Aspray TJ, Jones EE, Davies MW, et al. Increased hyphal branching and growth of ectomycorrhizal fungus Lactarius rufus by the helper bacterium Paenibacillus sp. Mycorrhiza. 2013;23:403–10. doi: 10.1007/s00572-013-0483-1. [DOI] [PubMed] [Google Scholar]
- Berendsen RL, van Verk MC, Stringlis IA, et al. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16:539. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7:1673–85. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- Brulé C, Frey-Klett P, Pierrat JC, et al. Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol Biochem. 2001;33:1683–94. [Google Scholar]
- Budzikiewicz H. Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non-fluorescent Pseudomonas spp.) In: Herz W, Falk H, Kirby G, editors. Progress in the Chemistry of Organic Natural Products. Vol. 87. Vienna: Springer; 2004. pp. 81–237. [DOI] [PubMed] [Google Scholar]
- Carrión VJ, Arrebola E, Cazorla FM, et al. The mbo operon is specific and essential for biosynthesis of mangotoxin in Pseudomonas syringae. PLoS One. 2012;7:e36709. doi: 10.1371/journal.pone.0036709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P, Matthijs S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ Microbiol. 2002;4:787–98. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- Cusano AM, Burlinson P, Deveau A, et al. Pseudomonas fluorescens BBc6R8 type III secretion mutants no longer promote ectomycorrhizal symbiosis. Environ Microbiol Rep. 2011;3:203–10. doi: 10.1111/j.1758-2229.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- de Bruijn I, de Kock MJ, Yang M, et al. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol. 2007;63:417–28. doi: 10.1111/j.1365-2958.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- Deveau A. How does the tree root microbiome assemble? Influence of ectomycorrhizal species on Pinus sylvestris root bacterial communities. Environ Microbiol. 2016 doi: 10.1111/1462-2920.13214. [DOI] [PubMed] [Google Scholar]
- Deveau A, Brulé C, Palin B, et al. Role of fungal trehalose and bacterial thiamine in the improved survival and growth of the ectomycorrhizal fungus Laccaria bicolor S238N and the helper bacterium Pseudomonas fluorescens BBc6R8. Environ Microbiol Rep. 2010;2:560–8. doi: 10.1111/j.1758-2229.2010.00145.x. [DOI] [PubMed] [Google Scholar]
- Deveau A, Gross H, Morin E, et al. Genome sequence of the mycorrhizal helper bacterium Pseudomonas fluorescens BBc6R8. Genome Announc. 2014;2:e01152–13. doi: 10.1128/genomeA.01152-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau A, Labbé J. Mycorrhiza helper bacteria. In: Martin F, editor. Molecular Mycorrhizal Symbiosis. Oxford: Wiley-Blackwell; 2016. in press. [Google Scholar]
- Deveau A, Palin B, Delaruelle C, et al. The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol. 2007;175:743–55. doi: 10.1111/j.1469-8137.2007.02148.x. [DOI] [PubMed] [Google Scholar]
- Di Battista C, Selosse M-A, Bouchard D, et al. Variations in symbiotic efficiency, phenotypic charachters and ploidy level among different isolates of the ectomycorrhizal basidiomycete Laccaria bicolor strain S238. Mycol Res. 1996;100:1315–24. [Google Scholar]
- Fones H, Davis CA, Rico A, et al. Metal hyperaccumulation armors plants against disease. PLoS Pathog. 2010;6:e1001093. doi: 10.1371/journal.ppat.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founoune H, Duponnois R, Bâ AM, et al. Mycorrhiza helper bacteria stimulate ectomycorrhizal symbiosis of Accaria holoserica with the Pisolithus albus. New Phytol. 2002;153:81–9. [Google Scholar]
- Frey-Klett P, Churin JL, Pierrat JC, et al. Dose effect in the dual inoculation of an ectomycorrhizal fungus and a mycorrhiza helper bacterium in two forest nurseries. Soil Biol Biochem. 1999;31:1555–62. [Google Scholar]
- Frey-Klett P, Garbaye J, Tarkka M. The mycorrhiza helper bacteria revisited. New Phytol. 2007;176:22–36. doi: 10.1111/j.1469-8137.2007.02191.x. [DOI] [PubMed] [Google Scholar]
- Frey-Klett P, Pierrat JC, Garbaye J. Location and survival of mycorrhiza helper Pseudomonasfluorescens during establishment of ectomycorrhizal symbiosis between Laccaria bicolor and Douglas Fir. Appl Environ Microb. 1997;63:139–44. doi: 10.1128/aem.63.1.139-144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galet J, Deveau A, Hôtel L, et al. Pseudomonas fluorescens pirates both ferrioxamine and ferricoelichelin siderophores from Streptomyces ambofaciens. Appl Environ Microb. 2015;81:3132–41. doi: 10.1128/AEM.03520-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E, Fracchia L, Cavaletto M, et al. Characterization of functional traits of two fluorescent pseudomonads isolated from basidiomes of ectomycorrhizal fungi. Soil Biol Biochem. 2003;35:55–65. [Google Scholar]
- Garbaye J. Mycorrhiza helper bacteria: a new dimension to the mycorrhizal symbiosis. New Phytol. 1994;128:197–210. doi: 10.1111/j.1469-8137.1994.tb04003.x. [DOI] [PubMed] [Google Scholar]
- Garbaye J, Bowen G. Stimulation of ectomycorrhizal infection of Pinus radiata by some microorganisms associated with the mantle of ectomycorrhizas. New Phytol. 1989;112:383–8. [Google Scholar]
- Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–46. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- Haselwandter K, Häninger G, Ganzera M, et al. Linear fusigen as the major hydroxamate siderophore of the ectomycorrhizal Basidiomycota Laccaria laccata and Laccaria bicolor. Biometals. 2013;26:969–79. doi: 10.1007/s10534-013-9673-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Douglas LJ. Interactions between Candida species and bacteria in mixed infections. In: Brogden K, Guthmiller J, editors. Polymicrobial Diseases. Washington, D.C.: ASM Press; 2002. pp. 357–74. [PubMed] [Google Scholar]
- Jun S, Wassenaar TM, Nookaew I, et al. A Diversity of Pseudomonas genomes, including populus -associated isolates, as revealed by comparative genome analysis. Appl Environ Microb. 2016;82:375–83. doi: 10.1128/AEM.02612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings AF, Martin LW, Sharples KJ, et al. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun. 2013;81:2697–704. doi: 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185–97. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- Lambert S, Traxler MF, Craig M, et al. Altered desferrioxamine-mediated iron utilization is a common trait of bald mutants of Streptomyces coelicolor. Metallomics. 2014;6:1390–9. doi: 10.1039/c4mt00068d. [DOI] [PubMed] [Google Scholar]
- Laycock M V, Hildebrand PD, Thibault P, et al. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. J Agric Food Chem. 1991;39:483–9. [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Henkels MD. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl Environ Microb. 1997;63:99–105. doi: 10.1128/aem.63.1.99-105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Henkels MD, Shaffer BT, et al. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl Environ Microb. 2008;74:3085–93. doi: 10.1128/AEM.02848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan VV, Moree WJ, Aguilar J, et al. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J Bacteriol. 2014;196:1683–93. doi: 10.1128/JB.01258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnert-Sindico. Une nouvelle espece de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n sp caracteres culturaux Ann Inst Pasteur. 1954;87:702–7. [PubMed] [Google Scholar]
- Poole EJ, Bending GD, Whipps JM, et al. Bacteria associated with Pinussylvestris-Lactarius rufus ectomycorrhizas and their effects on mycorrhiza formation in vitro. New Phytol. 2001;151:743–51. doi: 10.1046/j.0028-646x.2001.00219.x. [DOI] [PubMed] [Google Scholar]
- Prell HH, Day P. Plant-Fungal Pathogen Interaction: A Classical and Molecular View. Berlin Heidelberg: Springer; 2001. [Google Scholar]
- Raaijmakers JM, De Bruijn I, Nybroe O, et al. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–62. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Maboratory Press; 1989. [Google Scholar]
- Schäfer H, Tarak K, Budzikiewicz H. Zur genese der amidisch an den chromophor von pyoverdinen gebundenen dicarbonsäuren. Z Naturforsch. 1991;46c:398–406. [Google Scholar]
- Smith S, Read D. Mycorrhizal Symbiosis. 3rd edn. London: Academic Press; 2008. [Google Scholar]
- Whipps JM. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- Wong WC, Preece TF. Identification of Pseudomonas tolaasi: the white line in agar and mushroom tissue block rapid pitting tests. J Appl Bacteriol. 1979;47:401–7. [Google Scholar]
- Youard ZA, Mislin GLA, Majcherczyk PA, et al. Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J Biol Chem. 2007;282:35546–53. doi: 10.1074/jbc.M707039200. [DOI] [PubMed] [Google Scholar]
- Zhang X-X, Rainey PB. Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25. Genetics. 2007;176:2165–76. doi: 10.1534/genetics.107.075713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wu XQ, Ye JR, et al. Isolation and characterization of a mycorrhiza helper bacterium from rhizosphere soils of poplar stands. Biol Fertil Soils. 2014;50:593–601. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.