Abstract
In this paper, we describe the microbial composition and their predictive metabolic profile in the sea urchin Lytechinus variegatus gut ecosystem along with samples from its habitat by using NextGen amplicon sequencing and downstream bioinformatics analyses. The microbial communities of the gut tissue revealed a near-exclusive abundance of Campylobacteraceae, whereas the pharynx tissue consisted of Tenericutes, followed by Gamma-, Alpha- and Epsilonproteobacteria at approximately equal capacities. The gut digesta and egested fecal pellets exhibited a microbial profile comprised of Gammaproteobacteria, mainly Vibrio, and Bacteroidetes. Both the seagrass and surrounding sea water revealed Alpha- and Betaproteobacteria. Bray–Curtis distances of microbial communities indicated a clustering profile with low intrasample variation. Predictive metagenomics performed on the microbial communities revealed that the gut tissue had high relative abundances of metabolisms assigned to the KEGG-Level-2 designation of energy metabolisms compared to the gut digesta, which had higher carbohydrate, amino acid and lipid metabolisms. Overall, the results of this study elaborate the spatial distribution of microbial communities in the gut ecosystem of L. variegatus, and specifically a selective attribute for Campylobacteraceae in the gut tissue. Also, the predictive functional significance of bacterial communities in uniquely compartmentalized gut ecosystems of L. variegatus has been described.
Keywords: 16S rRNA, Gulf of Mexico, bioinformatics, microbiota, next-generation sequencing, PICRUSt
This study describes the distribution of microbiota, and their predicted functional attributes, in the gut ecosystem of sea urchin, Lytechinus variegatus, from its natural habitat of Gulf of Mexico.
INTRODUCTION
The sea urchin, Lytechinus variegatus, inhabits the eastern coast of the United States, ranging through the Gulf of Mexico from NC, USA to the northern coast of Brazil (Hendler et al.1995; Watts, McClintock and Lawrence 2013). This species is typically found in shallow nearshore meadows of turtlegrass Thalassia testudinum, upon which they graze and ingest the leaves, including the associated epibionts and microbiota (Moore et al.1963; Zieman 1982; Beddingfield and McClintock 2000). Although seasonal variations occur, seagrass leaves consist of carbohydrates (45%–60% dry wt., with a majority being insoluble at 35%–45% dry wt.), proteins (10%–15% dry wt.) and low levels of lipids (<5% dry wt.) (Zieman 1982; Pradheeba et al.2011). Once consumed by the sea urchin, ingested materials (ingesta) will receive a mucosal contribution from the pharynx, and envelop the ingesta in the form of a spherical pellet, herein referred to as gut digesta. This unique digestive feature demonstrates a physical compartmentalization of the ingesta from the surface of the gut tissue, and is considered to be an advantageous digestive strategy for this animal (Brooks and Wessel 2003; Ziegler et al.2010). Importantly, gut digesta formation is accompanied by an apparent microbial enrichment that is distinctively different from the microbial community of the gut tissue (Hakim et al.2015). With this distribution of microbial communities between the gut digesta and the gut tissue, an allocation of defined metabolic profiles would be expected, enriched with those metabolic genes represented by the heightened microbial taxa. Additionally, pellets representing the gut digesta remain intact even after egestion (egested fecal pellets) (Sauchyn, Lauzon-Guay and Scheibling 2011; Holland 2013), and are predicted to undergo microbial-driven molecular transitions (Sauchyn, Lauzon-Guay and Scheibling 2011; Hakim et al.2015). These egested fecal pellets have been acknowledged as an enriched source of nutrient to organisms at various trophic levels in the hydrosphere (Johannes and Satomi 1966; Koike, Mukai and Nojima 1987; Sauchyn, Lauzon-Guay and Scheibling 2011), and therefore sea urchin grazing of marine seagrass and other available sea vegetation has been identified as a major factor in nutrient cycling within benthic marine communities (Eklöf et al.2008; Miyata 2010).
Distribution of microbial communities in such a unique and compartmentalized ecosystem suggests either (i) a selective attribute of the host to promote the growth of key microbial members, (ii) a microbial life strategy selecting the host gut environment as conducive to growth and division or (iii) a complex combination of both circumstances (Bäckhed et al.2005; Shade and Handelsman 2012). Recently, the gut microbial community of the sea urchin L. variegatus cultured in the laboratory and fed with a formulated diet was described using next generation sequencing (NextGen) technology and bioinformatics analyses (Hakim et al.2015). This study established a baseline bacterial profile in the gut lumen, gut digesta and egested fecal pellets, including the feed and culture environment. Additionally, the study showed that although a diverse microbial community exists in their feed and surrounding culture environment, a select group of microbial taxa is differentially enriched in the gut lumen and gut digesta (Hakim et al.2015). Given the importance of the sea urchin gut microbiota to the host as it pertains to digestive physiology, as well as the nutritional benefit provided by bacteria enveloped in the egested fecal pellets to the ecosystem they inhabit, it is imperative to map the microbial profiles of the gut as they occur in nature, including comparisons of the microbial communities of the marine environment to the sea urchin's own microbiota. These comparisons would define the selectiveness of the sea urchin gut ecosystems, if such selection exists, for preferred bacterial taxa in their natural habitat. The role of microbiota in digestion of the ingested food within the gut lumen and gut digesta has not been confirmed in sea urchins. However, based on the suggested role of gut microbiota in other organisms, it is likely that there is an intimate association with the digestion process (Guerinot and Patriquin 1981; Nelson et al.2010). With the advent of bioinformatics tools (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States, PICRUSt v.1.0.0) which utilize the 16S rRNA NextGen sequencing data, it is possible to predict the functional attributes of microbiota residing in various components of the sea urchin digestive system (Langille et al.2013).
In this study, we collected fresh specimens of the sea urchin L. variegatus from shallow-water seagrass beds located in the northern Gulf of Mexico. We have identified the microbiota occurring in the lumen of the gut and pharynx, the gut digesta and the egested fecal pellets, as well as the surrounding sea water and natural seagrass (turtlegrass, T. testudinum) with high taxonomic coverage using a culture-independent NextGen Illumina MiSeq sequencing technology and bioinformatics tools. In addition, we have used the PICRUSt v.1.0.0 on 16S rRNA gene sequence datasets and determined the predictive functional profile of microbial communities in the naturally occurring sea urchin gut microbiome.
MATERIALS AND METHODS
Collection of Lytechinus variegatus and sample collection
Lytechinus variegatus (n = 3) were collected in October 2014 from within 1 m2 of each other in the Saint Joseph Bay Aquatic Preserve, Florida (29.80°N 85.36°W), placed in a clean plastic cooler containing sea water collected from the same location, and transported to the laboratory at the University of Alabama at Birmingham. Oceanic water conditions were recorded as 20 ± 2°C, with a pH of 7.8 ± 0.2 and salinity of 28 ± 1 ppt. Leaves of the seagrass Thalassia testudinum were harvested by excision at the urchin collection site and placed in plastic bags for further microbiota extraction. Fresh sea water samples were collected within the top 1 m of the collection site and placed in sterile containers. Three adult sea urchins were dissected for the study (UR1 d = 55 mm, wet wt. = 31.59 g; UR2 d = 55 mm, wet wt. = 38.58 g; and UR3 d = 60 mm, wet wt. = 46.77 g). Tissue extraction and environmental sample preparation for NextGen began 7 ± 1 h following collection. Prior to dissection, the sea urchins were placed in individual containers containing sea water from the sample site. From these containers, egested fecal pellets were collected upon their release, to ensure that the released fecal pellets (egesta) were appropriately collected from each sea urchin, without the contamination of another sea urchin's fecal pellets. The gut tissue and pharynx tissue were collected as described in Hakim et al. (2015). The gut digesta was voided and collected from the gut tissue by gentle shaking in autoclaved (121°C for 20 min at 103.42 kPa) sea water. The microbiota of the sea water (water) (n = 3) from Saint Joseph Bay Aquatic Preserve, Florida, was vacuum-filtered through Millipore 0.22 μm filtration paper (EMD Millipore Corporation, Danvers, MA, USA). The seagrass collected from the area of sea urchin grazing were minced using a sterile scalpel.
Metacommunity DNA purification and generation of 16S rRNA amplicon library
Microbial genomic DNA was isolated using the Fecal DNA isolation kit from Zymo Research (Irvine, CA, USA; catalog no. D6010) according to the manufacturer's instructions. An amplicon library was prepared from metacommunity DNA, using unique bar-coded oligonucleotide primers through PCR to amplify the hyper variable region 4 (V4) of the 16S rRNA gene (Kozich et al.2013; Kumar et al.2014). Oligonucleotide primers were adapted from the standard protocols of the Earth Microbiome Project (www.earthmicrobiome.org; Caporaso et al.2011, 2012), and were as follows: forward primer (515F) V4: 5′-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTG-CCAGCMGCCGCGGTAA-3′; and reverse primer modified from 806R to include uniquely bar-coded 5′ region and adaptor sequence V4: 5′-CAAGAGAAGACGGCATAC-GAGATNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT-3′ (Eurofins Genomics, Inc., Huntsville, AL, USA) (Kumar et al.2014). PCR amplification was set up as follows: 10 μL of 5× Reaction Buffer; 1.5 μL (200 μM) of each of the dNTPs; 2 μL (1.5 μM) of each oligonucleotide primer solution; 1.5 μL (5 U) of the LongAmp® enzyme kit (New England Biolabs, Ipswich, MA, USA; catalog no. E5200S); 30 μL (2–5 ng μL–1) of the template DNA; and 3 μL of sterile H2O for a total reaction volume of 50 μL. The PCR cycling parameters were as follows: initial denaturation at 94°C for 1 min; 32 cycles of amplification with each cycle consisting of 94°C for 30 s, 50°C for 1 min, 65°C for 1 min; followed by final extension of 65°C for 3 min and a final hold at 4°C. The resultant PCR reaction was electrophoresed on a 1.0% (w/v) Tris-borate-EDTA/agarose gel, and the PCR product (∼380 bp predicted product size) was visualized using UV illumination. The amplified DNA band was excised and purified from the agarose matrix using QIAquick Gel Extraction Kit according to the manufacturer's instructions (Qiagen Inc., Venlo, Limburg; catalog no. 28704).
NextGen sequencing by Illumina and bioinformatics
In preparation for NextGen sequencing, PicoGreen dye (Life Technologies, Grand Island, NY, USA) was used to quantitate the samples, which were adjusted to a concentration of 4 nM (Kumar et al.2014). To sequence the PCR products, the single lane flowcell NextGen sequencing Illumina MiSeq platform (Kozich et al.2013; Kumar et al.2014) was used, incorporating the 250 bp paired-end kits from Illumina specific to the V4 region of the 16S rRNA gene. Raw sequence data were demultiplexed and converted to FASTQ format (Cock et al.2010), evaluated for quality and filtered using the FASTX toolkit (Gordon and Hannon 2010). The overlapping regions of the paired-end reads (∼245 bp) were merged using the ‘fastq_mergepairs’ module of USEARCH (Edgar 2010). Read pairs with <50 bp overlap and/or over 20 mismatching nucleotides were discarded, and chimeras were removed using ‘identify_chimeric_seqs.py’ module of USEARCH (Edgar 2010). Read quality was assessed before and after filtering using FASTQC (Andrews 2010). All NextGen raw sequence data files have been deposited to NCBI SRA for public access (the accession number is SRP076869).
The following steps were performed using the Quantitative Insights Into Microbial Ecology microbiome analysis package (QIIME, v1.8.0) (Lozupone et al.2007; Caporaso et al.2010; Navas-Molina et al.2013; Kumar et al.2014). First, sequences were grouped into operational taxonomic units (OTUs) at a similarity threshold of 97% using UCLUST (Edgar 2010), and taxonomic assignments (to the species level when possible) were achieved through the Ribosomal Database Project (RDP) classifier (Wang et al.2007), trained using the Greengenes (v13.8) 16S rRNA database (DeSantis et al.2006; McDonald et al.2011), at a 60% confidence threshold (Wang et al.2007). OTUs observed to be <0.0005% abundant were removed. To summarize taxa abundance at different hierarchical levels (phylum, class, order, family and genus), biological replicates in the resultant OTU table were grouped according to analysis of similarity (ANOSIM) of the weighted (R = 0.7251; P = 0.001) Unifrac distances calculated between each sample (Lozupone and Knight 2005; Hamady, Lozupone and Knight 2010), conducted at 999 permutations using QIIME (v1.8.0). The top 100 most resolved taxa from this this data were used to construct stacked column bar charts using Microsoft Excel (Microsoft, Seattle, WA, USA). Alpha diversity was estimated using the observed OTUs, Shannon (Shannon 1948; Hill et al.2003; Marcon et al.2014), and Simpson (Simpson 1949; Hill et al.2003) diversity was measured using QIIME (v1.8.0).
To construct the multidimensional-scaling (MDS) plots (Kruskal and Wish 1978; Clarke 1993; Clarke and Gorley 2001), the quality assessed unfiltered OTU table was used, and all samples were rarefied to the median OTU value (87 225) to account for variation in read depth (QIIME, v1.8.0) (Gotelli and Colwell 2011). Those samples with OTUs totaling less than the median value were maintained at their original value, and included in the Beta-diversity analysis (de Cárcer et al.2011). Beta diversity was visualized using PRIMER-6 software (Primer-E Ltd, Plymouth Marine Laboratory, Plymouth UK, v6.1.2), by standardizing and square root-transforming the subsampled OTUs from each sample (Clarke and Gorley 2001), and MDS plots were generated according to the Bray–Curtis similarity values (Bray and Curtis 1957). These similarity values were also used to construct the complete-linkage hierarchical clustering dendrogram (Krebs 1999; Clarke and Gorley 2001; Dawyndt, De Meyer and De Baets 2005) used in the heatmap, to show triplicate sample clustering according to the Bray–Curtis values (Bray and Curtis 1957). A heatmap was constructed by merging sample replicates, followed by filtering out those taxa with <1% (<0.01) from the top 100 OTUs, using the ‘heatmap.2’ function of the R package (available at http://CRAN.R-project.org/package=gplots). The dendrograms were created using complete-linkage clustering of the compositional data using the Bray–Curtis values in the R package. An additional dendrogram to show intrasample variation in the heatmap was created using Bray–Curtis similarity and complete-linkage clustering using PRIMER-6 (v6.1.2) software.
PICRUSt (v1.0.0) and STAMP (v2.1.3) analysis for predictive metagenomics
PICRUSt v1.0.0 (Langille et al.2013) was used to determine the predictive metagenomes of the microbial populations from each sample. PICRUSt (v1.0.0) extrapolates known metabolic characteristics based on the associated phylogeny from the sequence data achieved after NextGen sequencing of the variable V4 region of the 16S rRNA gene (Langille et al.2013; Lang, Eisen and Zivkovic 2014). To achieve a predicted metagenome of each sample, the FASTA file created after quality assessment, chimeric trimming and filtering of sequences (<0.0005%) was used. OTUs corresponding to the sequence file were closed-referenced picked against the Greengenes (v13.5) database at a 97% identity, as suggested in PICRUSt (v1.0.0). The resultant OTU table was then supplemented with de novo OTUs present at >100 in any sample, as determined by open-reference OTU picking against the Greengenes (v13.5) database. The resultant OTU table was normalized, and metagenomes were predicted by referencing the assigned Greengenes Ids to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) Database (Kanehisa and Goto, 2000; Kanehisa et al.2014), using the ‘predict_metagenomes.py’ module of PICRUSt (v1.0.0). The predicted metagenomes were collapsed into hierarchical categories (KEGG-Level-2 and 3), and the relative abundances of the gut digesta (n = 3) and the gut tissue (n = 3) metagenome categories were calculated and graphed using Microsoft Excel software (Microsoft). The KEGG-Level-2 functional categories were used for two-group box-plot analysis in Statistical Analysis of Metagenomic Profiles (STAMP v2.1.3) (Parks et al.2014). Variance was calculated using a two-sided Welch's t-test, which does not assume equal variance (Welch 1938), along with the Benjamini–Hochberg false discovery rate (FDR) (Benjamini and Hochberg 1995) statistic for multiple test corrections (Parks et al.2014; Zhao et al.2015). Confidence intervals were set to 95% (0.95). The P-value for the total variance between the two groups is listed in the box plots. The P-value for a significant difference between the mean relative abundances of each group was recognized as P < 0.05.
RESULTS
Total Illumina sequence reads, quality trimming and OTU designation
NextGen sequencing of the V4 region of the 16S rRNA gene using the Illumina MiSeq platform resulted in a total of 1761 403 raw sequence reads from the 18 samples of the study (sea urchin samples n = 3; water n = 3; seagrass n = 3) (Table 1). Quality assessment and chimeric trimming produced 1362 092 sequence reads, and removal of those unique sequences accounting for <0.0005% resulted in a total of 1294 253 reads. Of these reads, the gut tissues (260 551 reads) expressed a combined total of 467 OTUs, and the pharynx tissue (238 729 reads), a total of 1893 OTUs. The gut digesta samples (302 287 reads) displayed a total of 6740 OTUs, and the egested fecal pellet samples (351 170 reads), a total of 10 172 OTUs. Of the environmental contributions to the gastrointestinal microbiota, the sea water (11 508 reads) produced 914 total OTUs, and the seagrass (130 008 reads) produced 2271. Rarefaction curves reached or approached a plateau, indicating sufficient sampling depth (data not shown).
Table 1.
Sample statistics corresponding to the 18 samples of the study, determined after NextGen sequencing of the V4 variable region of the 16S rRNA gene using the Illumina MiSeq platform. Included are the raw sequences prior to analysis, followed by the quality assessed sequence read counts and the resultant unique observations (unfiltered). This is followed by the filtered count (in which those sequences occurring at <0.0005% were removed), as well as the resultant unique observations. The Shannon and Simpson diversity values corresponding to each sample are also included. For Shannon diversity, a value much higher than 0 would be considered more diverse, and for Simpson diversity, a value closer to 1 would be considered more diverse.
| Raw | Quality | Unique observations | Filtered | Unique observations | |||
|---|---|---|---|---|---|---|---|
| Sample | sequences | assessment | (unfiltered) | reads | (filtered) | Shannon | Simpson |
| Egested fecal pellet 1 | 204 150 | 149 503 | 16 030 | 133 208 | 3431 | 8.33 | 0.97 |
| Egested fecal pellet 2 | 158 186 | 126 394 | 10 306 | 117 384 | 3367 | 8.14 | 0.97 |
| Egested fecal pellet 3 | 138 390 | 108 855 | 9538 | 100 578 | 3374 | 8.46 | 0.98 |
| Gut digesta 1 | 132 132 | 105 608 | 7550 | 99 468 | 2554 | 6.94 | 0.95 |
| Gut digesta 2 | 143 149 | 118 643 | 5712 | 114 053 | 2004 | 5.76 | 0.93 |
| Gut digesta 3 | 113 415 | 92 589 | 5131 | 88 766 | 2182 | 7.20 | 0.96 |
| Gut tissue 1 | 89 881 | 74 363 | 1205 | 73 264 | 184 | 0.83 | 0.19 |
| Gut tissue 2 | 125 274 | 108 088 | 1627 | 106 493 | 173 | 0.52 | 0.09 |
| Gut tissue 3 | 95 653 | 81 861 | 1089 | 80 794 | 110 | 0.60 | 0.12 |
| Pharynx tissue 1 | 140 831 | 104 981 | 3416 | 101 762 | 667 | 4.14 | 0.67 |
| Pharynx tissue 2 | 94 593 | 68 311 | 2637 | 65 931 | 672 | 4.18 | 0.76 |
| Pharynx tissue 3 | 99 972 | 72 323 | 1678 | 71 036 | 554 | 2.21 | 0.38 |
| Seagrass 1 | 230 | 175 | 70 | 128 | 41 | 5.66 | 0.97 |
| Seagrass 2 | 696 | 525 | 222 | 470 | 177 | 7.23 | 0.99 |
| Seagrass 3 | 208 197 | 137 462 | 7006 | 129 410 | 2053 | 6.87 | 0.88 |
| Water 1 | 494 | 228 | 104 | 195 | 76 | 6.01 | 0.97 |
| Water 2 | 12 175 | 9271 | 1026 | 8621 | 568 | 5.83 | 0.91 |
| Water 3 | 3985 | 2912 | 439 | 2692 | 270 | 5.73 | 0.92 |
| Total 18 | 1761 403 | 1362 092 | 74 786 | 1294 253 | 22 457 |
Microbial diversity across different samples
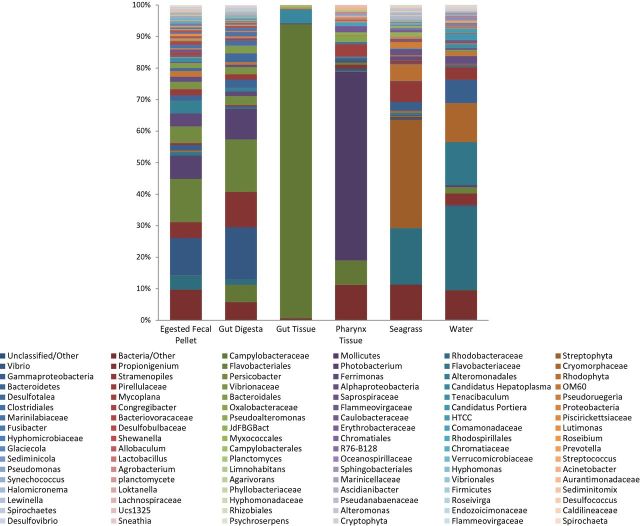
Relative abundances of the top 100 resolved taxa determined through the RDP classifier using Greengenes (v13.8) at a 60% confidence threshold are elaborated in Fig. 1. Biological replicates were merged on the basis of ANOSIM of the weighted UniFrac distances (R = 0.7251; P = 0.001). From the gut tissues collected from the three sea urchins, microorganisms belonging to phylum Proteobacteria constituted the highest relative abundance. At the class level, Epsilonproteobacteria accounted for the highest abundance, revealing family Campylobacteraceae to be heightened (93%). In the gut tissue, genus level taxa assignments could not be achieved using QIIME (v1.8.0). Subsequent BLAST (Altschul et al.1990; Morgulis et al.2008) search of the overrepresented sequence (253 bp) corresponding to family Campylobacteraceae, and occurring across all samples, revealed uncultured Arcobacter sp. (identity: 91%, E-value: 1.82e–87), Arcobacter bivalviorum (identity: 91%, E-value: 2.00e–89), Sulfuricurvum sp. (identity: 90%, E-value: 4.00e–86) and other uncultured bacterium clone (identity: 90%, E-value: 2.00e–89). Additionally, Candidatus Hepatoplasma (∼5%), of phylum Tenericutes, were observed. The pharynx tissue revealed a high abundance of Tenericutes (∼60%), showing class Mollicutes to be the most resolved taxa. Proteobacteria were also observed, with class Alpha-, Beta-, Epsilon- and Gammaproteobacteria showing as prevalent (Fig. 1).
Figure 1.
Stacked column bar graph of the top 100 most resolved taxa (to the genus level where possible) across all samples are presented. Replicates (n = 3) were merged, and OTUs were left untrimmed. Proteobacteria was found to be considerably abundant across all samples, as well as Bacteroidetes in the gut digesta, egested fecal pellets and water samples. The seagrass contained a high abundance of Cyanobacteria, and the pharynx tissue was dominated by class Mollicutes of phylum Tenericutes. Family Campylobacteraceae was determined to be the most abundant taxa in the gut tissue. In the gut digesta and egested fecal pellets, Vibrio, Propionigenium, and Flavobacteriales, and Photobacterium were most abundant. Relative abundances were calculated in QIIME (v1.8.0), and graphed using Microsoft Excel software (Microsoft).
The gut digesta and egested fecal pellets of the sea urchins revealed similar microbial profiles, and were dominated by phyla Proteobacteria (∼50%) with class Gammaproteobacteria appearing as heightened, followed by Alpha-, Delta-, and Epsilonproteobacteria at approximately equal prevalence. The heightened genera were Vibrio, Photobacterium, Propionigenium and Ferrimonas. Bacteroidetes were also observed, with the classes Flavobacteria, Cytophagia and Bacteroidia represented. Persicobacter and Tenacibaculum were the observed genera from Bacteroidetes.
From the surrounding environment in the Gulf of Mexico, the grazed upon seagrass revealed a high abundance of Cyanobacteria, followed by Proteobacteria and Bacteroidetes. Positive identification from within Cyanobacteria revealed order Streptophyta to be the most represented taxa, though resolution past the order level could not be achieved. However, the occurrence of this order was insignificant in the gut microbial community, and therefore their role in digestive process, if any, is unknown. Both the seagrass and the water samples revealed phylum Proteobacteria, particularly Rhodobacteraceae (class Alphaproteobacteria). The water samples displayed phylum Bacteroidetes, with a fairly high relative abundance of class Flavobacteria (∼25%), with family Flavobacteraceae and Cryomorphaceae represented. Observed taxa at the genus level of all samples are elaborated in Supplementary Table 1 (Supporting Information).
Statistical analysis
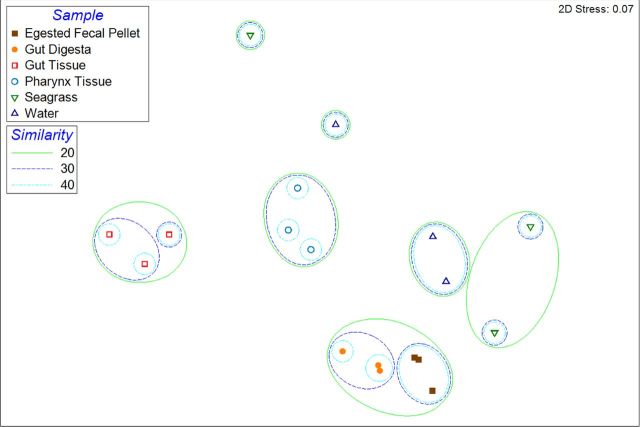
The Shannon diversity and Simpson diversity indices were calculated for the 18 samples of the study, revealing the gut tissue to have the least diversity of observed OTUs (Table 1). The pharynx tissue followed with a moderate diversity, and a high diversity was observed to be associated with the gut digesta and egested fecal pellets. The water and seagrass samples also displayed a high microbial diversity. MDS plot analysis revealed observable clustering of biological replicates from the sea urchin (Fig. 2). Three gut tissue samples clustered together at 20% Bray–Curtis similarity, as did the pharynx tissue samples. The gut digesta and fecal pellet samples also clustered together. The seagrass and water displayed more intrasample variability, and did not cluster as predictably as the other sea urchin samples. Filtering for heatmap analysis (<1% of the top 100 most resolvable taxa) resulted in 42 taxa (Fig. 3). This analysis revealed the gut tissue to be unique in microbial composition, separate from the other samples of the study, with a minor similarity to the pharynx tissue as depicted through complete-linkage hierarchical clustering (Fig. 3). The OTU data of the gut digesta and egested fecal pellets were shown to have a similar microbial profile, and the sea water and seagrass showed a moderate similarity through complete-linkage (Fig. 3). Dendrogram analysis of sample replicates using Bray–Curtis distances through complete-linkage hierarchical clustering supported both ANOSIM as well as the sample type grouping for the heatmap analysis.
Figure 2.
2D multidimensional-scaling (MDS) graph produced by PRIMER-6 (v6.1.2) using subsampled OTU data generated through QIIME (v1.8.0). Overlay of clusters were generated according to Bray–Curtis similarity, followed by complete-linkage clustering with similarity thresholds set at 10% intervals from 20%–40%. The samples obtained from the sea urchin microbiome showed distinct clustering patterns, with low intrasample (n = 3) variation. Similarity = Bray–Curtis similarity (scaled to 100).
Figure 3.
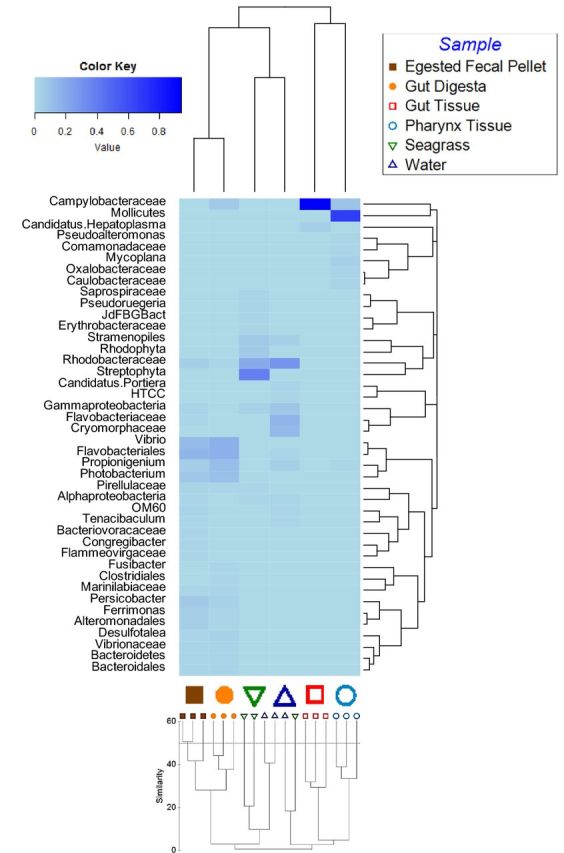
Heatmap generated using OTU data of the top most resolved taxa as determined using QIIME (v1.8.0), and filtered to 42 taxa by including only those taxa representing >1% of the total abundance. The rows correspond to bacterial taxa, and the columns represent the six different sample types (merged replicates) of this study. Dendrograms were created using complete-linkage hierarchical clustering of the compositional data. Heatmap was generated using the ‘heatmap.2’ function in R package (available at http://CRAN.R-project.org/package=gplots). Below is a complete-linkage (furthest neighbor) hierarchical clustering graph produced by PRIMER-6 (v6.1.2) using subsampled OTU data generated through QIIME (v1.8.0). Replicates corresponding to the sea urchin gut system possessed a low intrasample variation, and the environmental sample replicates showed slight intrasample variation. Similarity index is depicted on the left of the graph below, and similarity to a value of 60 is shown. Similarity = Bray–Curtis similarity (scaled to 100).
Predicted metagenomes based on PICRUSt (v1.0.0)
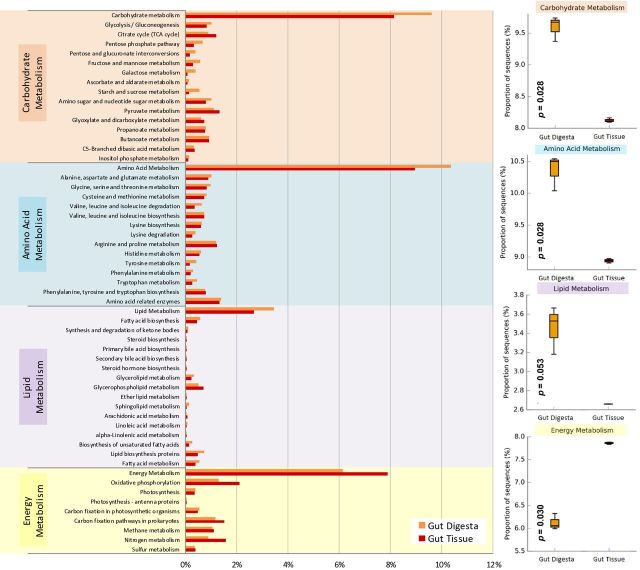
The microbial populations of those samples likely influencing the digestive processes occurring in the gut of the sea urchin were subjected to PICRUSt (v1.0.0) analysis to determine the metabolic processes predicted to be occurring in grouped populations, and compare those processes across compartmentalized microbial populations using the Welch's t-test of two groups (Fig. 4). The KEGG-Level-2 categories considered were as follows: carbohydrate metabolism, amino acid metabolism, lipid metabolism and energy metabolism. The groups compared included the gut digesta and gut tissues. The gut digesta revealed a heightened relative abundance of the KEGG-Level-2 categories corresponding to carbohydrate (9.59%; P = 0.028) and amino acid (10.39%; P = 0.028) metabolisms, as well as a marginally heightened abundance of lipid (3.45%; P = 0.053) metabolisms compared to the gut tissue. Conversely, the gut tissue displayed high abundances of pathways related to energy (7.87%; P = 0.030) metabolism (Fig. 4). Investigations of the KEGG-Level-3 subcategories from carbohydrate metabolism assigned to the gut digesta showed various sugar metabolic processes, such as the pentose phosphate pathway, pentose glucoronate interconversion pathway and glycolysis. Starch, sucrose, fructose, mannose, galactose and amino acid sugar metabolisms were also pronounced as compared to the gut tissue. KEGG-Level-3 categories corresponding to amino acid metabolism showed the gut digesta to exceed the gut tissue in the degradation or biosynthesis of multiple amino acids (alanine, aspartate, glutamate, glycine, serine, threonine, cysteine, methionine, valine, leucine, isoleucine, lysine, histidine, tyrosine, phenylalanine and tryptophan). KEGG-Level-3 categories of lipid metabolisms also showed the gut digesta to contain pathways related to the biosynthesis of fatty acids, unsaturated fatty acids and glycerolipids, among others. From the gut tissue, KEGG-Level-3 pathways of energy metabolisms revealed relative abundances that surpassed the gut digesta in oxidative phosphorylation, prokaryotic carbon fixation, and methane, nitrogen and sulfur metabolisms. An elaboration of STAMP (v2.1.3) statistics, including the mean relative abundances of each Level-2 and 3 KEGG metagenomics category, as well as the other KEGG processes identified using PICRUSt (v1.0.0), are listed in Supplementary Table 2 (Supporting Information).
Figure 4.
PICRUSt (v1.0.0) analysis of predicted metagenomes generated by using the 16S rRNA gene data of the gut tissue (n = 3) and gut digesta (n = 3) samples. OTUs were picked using closed-reference picking, as suggested by PICRUSt (v1.0.0), and merged with open-reference picked de novo OTUs (occurring at >100 per sample) which included each OTU's respective representative Greengenes ID. KEGG pathways were assigned (KO IDs) using the ‘predict_metagenomes.py’ module, and collapsed into hierarchical KEGG pathways (KEGG-Level-2 and 3). (A) The mean relative abundance of KEGG-Level-2 metadata categories are listed along with the associated KEGG-Level-3 pathways. (B) Box plots of the KEGG-Level-2 category of carbohydrate metabolism, amino acid metabolism, lipid metabolism and energy metabolism were generated using STAMP (v2.1.3) analytical software according to two-group statistics, using a two-sided Welch's t-test (not assuming equal variance) along with Benjamini–Hochberg FDR. Confidence intervals were selected as 95% (i.e.: 0.95), and P-value of each KEGG-Level-2 two-group analyses is listed in the respective box plot. Relative abundance data were graphed using Microsoft Excel software (Microsoft), and box plots generated using STAMP (v2.1.3).
DISCUSSION
Previous studies on the sea urchin digestive tract microbiota have described a distinct and pervasive bacterial profile that will colonize the apparent gut epithelium tissue, recognizably different from the bacterial community of the ingested feed, marine environment and pelleted gut digesta and egested fecal pellets (Guerinot and Patriquin 1981; Meziti et al.2007; Lawrence, Lawrence and Watts 2013). The microbial profile of the laboratory-cultured sea urchin Lytechinus variegatus, studied previously by our lab, revealed a near-exclusive occurrence of the order Campylobacterales associated with the gut tissue, of which oligotyping depicted Arcobacter sp., Sulfuricurvum sp. and Arcobacter bivalviorum (each with BLAST identities ≥90%) (Hakim et al.2015). The aforementioned microbial distribution was corroborated in the naturally occurring sea urchins of the current study, as the highly abundant representative sequences were assigned to Campylobacteraceae (93%) (Fig. 1), and also determined to be Arcobacter sp., Sulfuricurvum sp. and A. bivalviorum. ANOSIM and subsequent OTU Bray–Curtis MDS cluster analysis revealed the microbial community of each gut tissue sample (n = 3) to cluster together (Fig. 2), away from all other samples of the study. These results support a distinct gut tissue-associated bacterial profile, as reported in previous studies (Guerinot and Patriquin 1981; Meziti et al.2007). This is to note that the sample replicates used in this study exhibited a sufficient number of sequences to generate a reliable taxonomical identification of microbial communities, as well as their predictive functional attributes, which was supported by the statistical analyses of the current study (Table 1; Fig. 2). Previously, three or less replicate samples were reported in NextGen sequencing analyses comparing microbial composition in various ecosystems showing adequate power that was verified by relevant statistical methods (Hong et al.2015; Manzari et al.2015; Sha et al.2016). Since the heightened taxa of the gut tissue in this study showed a high BLAST similarity to Arcobacter, it is important to note that phylotypes related to Arcobacter have been observed in other marine invertebrates, such as the Chilean oyster Tiostrea chilensis (Romero et al.2002), shrimp Rimicaris exoculata (Durand et al.2010) and the hydrothermal vent-dwelling gastropod Alviniconcha aff. Hessleri (Suzuki et al.2005). To a much lesser extent, members of the representative class (Epsilonproteobacteria) have been observed in the cecum of mice (Gu et al.2013), and in humans (Eppinger et al.2004; Larsen and Dai 2015), in which both pathogenic and non-pathogenic roles have been described. However, it is unlikely that the Campylobacteraceae taxa observed in this study are antagonistic to the sea urchin L. variegatus, considering the near-dominant and consistent presence of these bacteria signifying host-selection (Hakim et al.2015), and the described non-detrimental association of this taxa described in the aforementioned marine invertebrates. The pharynx tissue also revealed a discernable microbial composition from the other sample types, and was found to consist of a high prevalence of Mollicutes (60%), belonging to phylum Tenericutes (Figs 1 and 2). Members of Mollicutes have previously been observed in the stomach of the eastern oyster, Crassostrea virginica (King et al.2012), as well as the terrestrial isopod, Porcellio scaber (Wang et al.2004). The unique group-specific clustering of the microbial communities of the gut and pharynx tissues of L. variegatus was further evidenced through heatmap analysis (Fig. 3).
The microbiota of the gut digesta and egested fecal pellets revealed Vibrio to be noticeably abundant in the naturally occurring sea urchin. Interestingly, Vibrio was not found to be significantly associated with the gut and pharynx tissues (<1%) (Fig. 1). This taxon has previously been identified in various sea urchins (Unkles 1977; Guerinot et al.1982), particularly L. variegatus (Nelson et al.2010; Hakim et al.2015), which have been described to play a role in nitrogen fixation, as well as protein assimilation in gonadal tissues (Fong and Mann 1980; Guerinot et al.1982). Certain Vibrio spp. are considered stable and common associates of many marine invertebrates, and have been observed in copepod species from the Gulf of Maine (Moisander, Sexton and Daley 2015), various crabs (Carcinus maenas and Hemigrapsus sanguineus), mussels (Mytilus edulis) and zooplankton (Preheim et al.2011). The representative class (Gammaproteobacteria) has also been observed in a number of marine and freshwater fish (Roeselers et al.2011), with studies showing significant abundances of Vibrio in zebrafish (Danio rerio) (Roeselers et al.2011), the abalone (Haliotis discus hannai) (Tanaka et al.2004) and the farmed marine turbot fish (Scophthalmus maximus) (Xing et al.2013). Additionally, in this study, Photobacterium of family Vibrionaceae were found to be abundant in the gut digesta and egested fecal pellets. Members of this genus, specifically Photobacterium lipolyticum, have been identified in the herbivorous sea urchin Paracentrotus lividus (Meziti et al.2007; Yeruham et al.2015) and other marine organisms (Gomez-Gil et al.2011), with a potential role in lipolytic activity (Seo et al.2005; Yoon et al.2005). Lastly, Flavobacterales, commonly found in the marine environment (Smith et al.2013), were the likely source of high abundance in the gut digesta and egested fecal pellets in the naturally occurring sea urchins of this study (Fig. 1).
Considering the nutritive profile of seagrass, and the limitations of the innate gut digestive enzymes of the sea urchin, it has been proposed that the bacteria of the gastrointestinal tract contribute to the digestion of complex sugars and cellulose (Lasker and Giese 1954; García-Tello and Baya 1973; Unkles 1977; Becker et al.2009), as well as the metabolism or synthesis of necessary biomolecules for protein and lipid incorporation (Tysskt et al.1961; Fong and Mann 1980; Lawrence, Lawrence and Watts 2006; Arafa et al.2012). The gastrointestinal tract of this animal represents a physical compartmentalization of the gut digesta from the gut tissue, and are each accompanied by uniquely dissimilar microbial profiles. This would indicate disparate functional profiles along with those divergent microbial populations, suggesting an allocation of microbial metabolisms and digestive responsibilities occurring in the compartmentalized gut ecosystem of this animal. By using PICRUSt (Fig. 4), a bioinformatics program that has shown considerable efficacy in the predictions of metabolic functions of microbial communities (Langille et al.2013), we were able to predict carbohydrate metabolic pathways, seeming to be essential for the digestion of starch and cellulose from turtlegrass consumed within its natural habitat. This is supported by a previous study that reported bacterial digestion of complex carbohydrates from alginate within the sea urchin gut (Sawabe et al.1995; Nelson et al.2010), identifying members of Vibrio—a genus that appeared heightened in the gut digesta revealed in this study (Fig. 1). PICRUSt analysis also showed protein and lipid metabolisms to be elevated in the gut digesta as compared to the gut tissue (Fig. 4). The reliance on gut microbial communities for protein metabolisms in the sea urchin has been previously addressed by Fong and Mann (1980), who demonstrated that suppressed microbial growth in Strongylocentrotus droebachiensis by antibiotic treatment resulted in a significantly reduced incorporation of essential amino acids into the gonadal tissue. Also, from the current study, the observation of pathways related to lipid metabolism in the gut digesta is likely to be conducted by the microbial community, and such metabolisms have been supported by previous studies of the microbial involvement in lipid and fatty acid biosynthesis (Leo and Parker, 1966) in sea urchins Psammechinus miliaris (Cook et al.2000), Paracentrotus lividus (Arafa et al.2012) and other marine invertebrates (Phillips 1984).
Conversely, the bacteria of the gut tissue surpassed the gut digesta in the KEGG-Level-2 category of energy metabolism, which encompasses the subcategories of oxidative phosphorylation, carbon fixation, methane, nitrogen and sulfur metabolisms. Such metabolisms denote interdependency between the bacterial members associated with the gut tissue and other microbial communities of the sea urchin gut ecosystem (such as the gut digesta) (Fischbach and Sonnenburg 2011). Bacteria colonizing the mucosal layer of the gastrointestinal tract in humans and other animals have been implicated in a myriad of roles in the development, maintenance and homeostatic preservation of the digestive tract, often outcompeting transient microbiota, as a product of a co-evolution of host and microbe (Rakoff-Nahoum et al.2004; Tlaskalová-Hogenová et al.2011; Schluter and Foster 2012; Wu and Wu 2012). Considering the near-exclusive abundance of Campylobacteraceae in the gut tissue, and the substantial relative abundance of energy metabolisms performed by this community, it appears that a mutualistic relationship may be occurring between the sea urchin L. variegatus and its selected gut mucosal resident, though the mechanisms of selection and the supposed benefit, if one exists, are unclear at this time.
In summary, the results of this study have revealed the gut microbial communities, with the highest taxonomic coverage, in the sea urchin L. variegatus from their natural habitat of the Gulf of Mexico. An enrichment of Campylobacteraceae (93%) was observed in the gut lumen, and is similar to that observed in L. variegatus held in culture and fed formulated diets. Given the near-exclusive abundance of Campylobacteraceae in the gut tissue, one can predict that the high energy metabolism observed by PICRUSt (v1.0.0) analysis is attributed by the members of this taxonomic group. In contrast, OTUs assigned to Vibrio, Photobacterium, Propionigenium and Flavobacteria were found to occur with high abundance in the gut digesta and egested fecal pellets. The innate digestive enzymes capable of processing complex carbohydrates, proteins and lipids from diet (turtlegrass) are scarcely reported in sea urchins, and consequently, studies have implicated the bacteria in the gut ecosystem in executing such necessary metabolic processes (Leo and Parker 1966; Phillips 1984; Schlosser et al.2005; Arafa et al.2012). This indicates the bacterial community within the mucosally enveloped ingesta of the naturally occurring sea urchin likely to be involved, as determined by PICRUSt (v1.0.0), in the metabolism of carbohydrates, amino acids and lipids. Further metagenomics analyses would help substantiate the metabolic attributes of the gut microbiota in benefitting health and digestive physiology of the animal, as well as the community in their inhabited ecosystem.
Supplementary Material
Acknowledgments
We would like to thank Dr Peter Eipers of the Department of Cell, Developmental and Integrative Biology, and Dr Micheal Crowley of the Heflin Center for Genomics Sciences at the University of Alabama at Birmingham (UAB), for their assistance in NextGen sequencing for this study. We would also like to thank UAB Institutional Animal Care and Use Committee (IACUC), and the Biology Department at UAB for logistics and graduate tuition and stipend support.
SUPPLEMENTARY DATA
FUNDING
We thank the UAB Institutional Animal Care and Use Committee for their support in this research. Also, we thank Edward Partridge, MD (Comprehensive Cancer Center; grant P30AR050948), Robert Kimberly, MD (Center for Clinical Translational Science; grant UL1TR000165), Michael Saag, MD (Center for AIDS Research; grant 5P30AI027767) and David Allison, PhD (UAB Nutrition Obesity Research Center (NORC); grant NIH P30DK056336) for providing support with the center grants and core facilities for this work. This work was supported by the Microbiome Resource at the UAB, School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767) and Center for Clinical Translational Science (UL1TR001417). Animal husbandry was supported in part by the Aquatic Animal Research Core, part of the UAB NORC (NIH P30DK056336).
Conflict of interest. None declared.
REFERENCES
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews SF. FASTQC. A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (5 July 2016, date last accessed)
- Arafa S, Chouaibi M, Sadok S, et al. The influence of season on the gonad index and biochemical composition of the sea urchin Paracentrotus lividus from the Golf of Tunis. Sci World J. 2012;2012:1–8. doi: 10.1100/2012/815935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Becker PT, Samadi S, Zbinden M, et al. First insights into the gut microflora associated with an echinoid from wood falls environments. Cah Biol Mar. 2009;50:343. [Google Scholar]
- Beddingfield SD, McClintock JB. Demographic characteristics of Lytechinus variegatus (Echinoidea:Echinodermata) from three habitats in a North Florida Bay, Gulf of Mexico. Mar Ecol. 2000;21:17–40. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–49. [Google Scholar]
- Brooks JM, Wessel GM. A diversity of yolk protein dynamics and function. Recent Dev Cell Res. 2003;1:1–30. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci USA. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–24. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR. Non‐parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–43. [Google Scholar]
- Clarke KR, Gorley RN. Primer V5 (Plymouth Routines in Multivariate Ecological Research): User Manual/tutorial. Plymouth, UK: Primer-E; 2001. [Google Scholar]
- Cock PJ, Fields CJ, Goto N, et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–71. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EJ, Bell MV, Black KD, et al. Fatty acid compositions of gonadal material and diets of the sea urchin, Psammechinus miliaris: trophic and nutritional implications. J Exp Mar Biol. 2000;255:261–74. doi: 10.1016/s0022-0981(00)00301-4. [DOI] [PubMed] [Google Scholar]
- Dawyndt P, De Meyer H, De Baets B. The complete linkage clustering algorithm revisited. Soft Comput. 2005;9:385–92. [Google Scholar]
- de Cárcer DA, Denman SE, McSweeney C, et al. Evaluation of subsampling-based normalization strategies for tagged high-throughput sequencing data sets from gut microbiomes. Appl Environ Microb. 2011;77:8795–8. doi: 10.1128/AEM.05491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand L, Zbinden M, Cueff-Gauchard V, et al. Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol Ecol. 2010;71:291–303. doi: 10.1111/j.1574-6941.2009.00806.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eklöf J, De la Torre-Castro M, Gullström M, et al. Sea urchin overgrazing of seagrasses: a review of current knowledge on causes, consequences, and management. Estuar Coast Shelf S. 2008;79:569–80. [Google Scholar]
- Eppinger M, Baar C, Raddatz G, et al. Comparative analysis of four Campylobacterales. Nat Rev Microbiol. 2004;2:872–85. doi: 10.1038/nrmicro1024. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–47. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong W, Mann K. Role of gut flora in the transfer of amino acids through a marine food chain. Can J Fish Aquat Sci. 1980;37:88–96. [Google Scholar]
- García-Tello P, Baya A. Acerca de la posible función de bacterias agaroliticas aisladas del erizo blanco (Loxechinus albus (Mol.)) Mus Nac Hist Nac Pub Oc. 1973;15:3–8. [Google Scholar]
- Gomez-Gil B, Roque A, Rotllant G, et al. Photobacterium swingsii sp. nov., isolated from marine organisms. Int J Syst Evol Micr. 2011;61:315–9. doi: 10.1099/ijs.0.019687-0. [DOI] [PubMed] [Google Scholar]
- Gordon A, Hannon G. Fastx-toolkit. FASTQ/A short-reads pre-processing tools. 2010. http://hannonlab.cshl.edu/fastx_toolkit/ (21 March 2016, date last accessed)
- Gotelli NJ, Colwell RK. Estimating species richness. In: Magurran AE, McGill BJ, editors. Biological Diversity: Frontiers in Measurement and Assessment. Oxford: Oxford University Press; 2011. pp. 39–54. [Google Scholar]
- Gu S, Chen D, Zhang JN, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. 2013;8:e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot M, Patriquin D. N2-fixing vibrios isolated from the gastrointestinal tract of sea urchins. Can J Microbiol. 1981;27:311–7. doi: 10.1139/m81-048. [DOI] [PubMed] [Google Scholar]
- Guerinot M, West P, Lee J, et al. Vibrio diazotrophicus sp. nov., a marine nitrogen-fixing bacterium. Int J Syst Bacteriol. 1982;32:350–7. [Google Scholar]
- Hakim JA, Koo H, Dennis LN, et al. An abundance of Epsilonproteobacteria revealed in the gut microbiome of the laboratory cultured sea urchin, Lytechinus variegatus. Front Microbiol. 2015;6:1047. doi: 10.3389/fmicb.2015.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendler G, Kier PM, Miller JE, et al. Sea Stars, Sea Urchins, and Allies: Echinoderms of Florida and the Caribbean. Vol. 390. Washington, DC: Smithsonian Institution Press; 1995. [Google Scholar]
- Hill TC, Walsh KA, Harris JA, et al. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- Holland ND. Digestive System. In: Lawrence JM, editor. Sea Urchins: Biology and Ecology. Vol. 38. UK: Elsevier; 2013. p. 119. [Google Scholar]
- Hong C, Si Y, Xing Y, et al. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ Sci Pollut R. 2015;22:10788–99. doi: 10.1007/s11356-015-4186-3. [DOI] [PubMed] [Google Scholar]
- Johannes R, Satomi M. Composition and nutritive value of fecal pellets of a marine crustacean. Limnol Oceanogr. 1966;11:191–7. [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Judd C, Kuske CR, et al. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS One. 2012;7:e51475. doi: 10.1371/journal.pone.0051475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike I, Mukai H, Nojima S. The role of the sea urchin, Tripneustes gratilla (Linnaeus), in decomposition and nutrient cycling in a tropical seagrass bed. Ecol Res. 1987;2:19–29. [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microb. 2013;79:5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CJ. Ecological Methodology. Vol. 620. CA, USA: Benjamin/Cummings Menlo Park; 1999. [Google Scholar]
- Kruskal J, Wish M. Quantitative Applications in the Social Sciences: Multidimensional Scaling. Vol. 11. Beverly Hills, CA: Sage; 1978. [Google Scholar]
- Kumar R, Eipers P, Little RB, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet. 2014;82:18.8.1–29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Eisen JA, Zivkovic AM. The microbes we eat: abundance and taxonomy of microbes consumed in a day's worth of meals for three diet types. PeerJ. 2014;2:e659. doi: 10.7717/peerj.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PE, Dai Y. Metabolome of human gut microbiome is predictive of host dysbiosis. GigaScience. 2015;4:1–16. doi: 10.1186/s13742-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker R, Giese AC. Nutrition of the sea urchin, Strongylocentrotus purpuratus. Biol Bull. 1954;106:328–40. [Google Scholar]
- Lawrence J, Lawrence A, Watts S. Feeding, digestion, and digestibility of sea urchins. In: Lawrence JM, editor. Sea Urchins: Biology and Ecology. Vol. 38. UK: Elsevier; 2013. pp. 135–54. [Google Scholar]
- Lawrence JM, Lawrence AL, Watts SA. Feeding, digestion, and digestibility. In: Lawrence JM, editor. Edible Sea Urchins: Biology and Ecology. Vol. 37. UK: Elsevier; 2006. p. 135. [Google Scholar]
- Leo RF, Parker PL. Branched-chain fatty acids in sediments. Science. 1966;152:649–50. doi: 10.1126/science.152.3722.649. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb. 2007;73:1576–85. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzari C, Fosso B, Marzano M, et al. The influence of invasive jellyfish blooms on the aquatic microbiome in a coastal lagoon (Varano, SE Italy) detected by an Illumina-based deep sequencing strategy. Biol Invasions. 2015;17:923–40. [Google Scholar]
- Marcon E, Scotti I, Hérault B, et al. Generalization of the partitioning of Shannon diversity. PLoS One. 2014;9:e90289. doi: 10.1371/journal.pone.0090289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziti A, Kormas KA, Pancucci-Papadopoulou M-A, et al. Bacterial phylotypes associated with the digestive tract of the sea urchin Paracentrotus lividus and the ascidian Microcosmus sp. Russ J Mar Biol. 2007;33:84–91. [Google Scholar]
- Miyata T. Reducing overgrazing by sea urchins by market development. Bull Fish Res Agen. 2010;32:103–7. [Google Scholar]
- Moisander PH, Sexton AD, Daley MC. Stable associations masked by temporal variability in the marine copepod microbiome. PLoS One. 2015;10:e0138967. doi: 10.1371/journal.pone.0138967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HB, Jutare T, Bauer J, et al. The biology of Lytechinus variegatus. B Mar Sci. 1963;13:23–53. [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, et al. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–64. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Method Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson L, Blair B, Murdock C, et al. Molecular Analysis of gut microflora in captive‐raised sea urchins (Lytechinus variegatus) J World Aquacult Soc. 2010;41:807–15. [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips NW. Role of different microbes and substrates as potential suppliers of specific, essential nutrients to marine detritivores. B Mar Sci. 1984;35:283–98. [Google Scholar]
- Pradheeba M, Dilipan E, Nobi E, et al. Evaluation of seagrasses for their nutritional value. IJMS. 2011;40:105. [Google Scholar]
- Preheim SP, Boucher Y, Wildschutte H, et al. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol. 2011;13:265–75. doi: 10.1111/j.1462-2920.2010.02328.x. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J, Garcia-Varela M, Laclette JP, et al. Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis) Microbial Ecol. 2002;44:365–71. doi: 10.1007/s00248-002-1063-7. [DOI] [PubMed] [Google Scholar]
- Sauchyn LK, Lauzon-Guay J-S, Scheibling RE. Sea urchin fecal production and accumulation in a rocky subtidal ecosystem. Aquat Biol. 2011;13:215–23. [Google Scholar]
- Sawabe T, Oda Y, Shiomi Y, et al. Alginate degradation by bacteria isolated from the gut of sea urchins and abalones. Microb Ecol. 1995;30:193–202. doi: 10.1007/BF00172574. [DOI] [PubMed] [Google Scholar]
- Schlosser SC, Lupatsch I, Lawrence JM, et al. Protein and energy digestibility and gonad development of the European sea urchin Paracentrotus lividus (Lamarck) fed algal and prepared diets during spring and fall. Aquacult Res. 2005;36:972–82. [Google Scholar]
- Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HJ, Bae SS, Lee J-H, et al. Photobacterium frigidiphilum sp. nov., a psychrophilic, lipolytic bacterium isolated from deep-sea sediments of Edison Seamount. Int J Syst Evol Micr. 2005;55:1661–6. doi: 10.1099/ijs.0.63338-0. [DOI] [PubMed] [Google Scholar]
- Sha Y, Liu M, Wang B, et al. Gut bacterial diversity of farmed sea cucumbers Apostichopus japonicus with different growth rates. Microbiology. 2016;85:109–15. [Google Scholar]
- Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. 2012;14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. [Google Scholar]
- Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Smith MW, Allen LZ, Allen AE, et al. Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol. 2013;4:120. doi: 10.3389/fmicb.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Sasaki T, Suzuki M, et al. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl Environ Microb. 2005;71:5440–50. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Ootsubo M, Sawabe T, et al. Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques. Aquaculture. 2004;241:453–63. [Google Scholar]
- Tlaskalová-Hogenová H, Štěpánková R, Kozáková H, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–20. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tysskt C, Mailloux M, Brisou J, et al. Sur la Microflore Normale de L'oursin Violet Des Cotes Algéroises (Paracentrotus lividus, Ltmk) Archives de l'Institut Pasteur d'Algérie. 1961;39:271. [PubMed] [Google Scholar]
- Unkles S. Bacterial flora of the sea urchin Echinus esculentus. Appl Environ Microb. 1977;34:347–50. doi: 10.1128/aem.34.4.347-350.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Stingl U, Anton-Erxleben F, et al. “Candidatus Hepatoplasma crinochetorum,” a new, stalk-forming lineage of Mollicutes colonizing the midgut glands of a terrestrial isopod. Appl Environ Microb. 2004;70:6166–72. doi: 10.1128/AEM.70.10.6166-6172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts SA, McClintock JB, Lawrence JM. Lytechinus. In: Lawrence JM, editor. Sea Urchins: Biology and Ecology. Vol. 38. UK: Elsevier; 2013. pp. 475–86. [Google Scholar]
- Welch BL. The significance of the difference between two means when the population variances are unequal. Biometrika. 1938;29:350–62. [Google Scholar]
- Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Hou Z, Yuan J, et al. Taxonomic and functional metagenomic profiling of gastrointestinal tract microbiome of the farmed adult turbot (Scophthalmus maximus) FEMS Microbiol Ecol. 2013;86:432–43. doi: 10.1111/1574-6941.12174. [DOI] [PubMed] [Google Scholar]
- Yeruham E, Rilov G, Shpigel M, et al. Collapse of the echinoid Paracentrotus lividus populations in the Eastern Mediterranean—result of climate change? Sci Rep. 2015;5:1–6. doi: 10.1038/srep13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Lee J-K, Kim Y-O, et al. Photobacterium lipolyticum sp. nov., a bacterium with lipolytic activity isolated from the Yellow Sea in Korea. Int J Syst Evol Micr. 2005;55:335–9. doi: 10.1099/ijs.0.63215-0. [DOI] [PubMed] [Google Scholar]
- Zhao W, Wang Y, Liu S, et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PloS One. 2015;10:1–13. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Mooi R, Rolet G, et al. Origin and evolutionary plasticity of the gastric caecum in sea urchins (Echinodermata: Echinoidea) BMC Evol Biol. 2010;10:1–32. doi: 10.1186/1471-2148-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieman JC. Ecology of the seagrasses of South Florida: a community profile. Technical report. University of Virginia Deptartment of Environmental Sciences; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.