Abstract
The bacterium Chlamydia trachomatis is the etiological agent of the most common sexually transmitted infection in North America and Europe. Medical complications resulting from genital C. trachomatis infections arise predominantly in women where the initial infections often remain asymptomatic and thus unrecognized. Untreated asymptomatic infections in women can ascend into the upper genital tract and establish persistence, ultimately resulting in extensive scarring of the reproductive organs, pelvic inflammatory disease, infertility and ectopic pregnancies. Previously resolved C. trachomatis infections fail to provide protective immune memory, and no effective vaccine against C. trachomatis is currently available. Critical determinants of the pathogenesis and immunogenicity of genital C. trachomatis infections are cell-autonomous immune responses. Cell-autonomous immunity describes the ability of an individual host cell to launch intrinsic immune circuits that execute the detection, containment and elimination of cell-invading pathogens. As an obligate intracellular pathogen C. trachomatis is constantly under attack by cell-intrinsic host defenses. Accordingly, C. trachomatis evolved to subvert and co-opt cell-autonomous immune pathways. This review will provide a critical summary of our current understanding of cell-autonomous immunity to C. trachomatis and its role in shaping host resistance, inflammation and adaptive immunity to genital C. trachomatis infections.
Keywords: Chlamydia trachomatis, TLR, inflammasome, interferon-inducible GTPases, indole-dioxygenase, STING
This review describes the tug of war between cell-autonomous immunity and bacterial counter-immunity that determines the final outcome of an infection with the obligate intracellular pathogen Chlamydia trachomatis.
AN INTRODUCTION TO CHLAMYDIA INFECTION BIOLOGY
Several species of the genus Chlamydia cause human diseases (Belland, Ojcius and Byrne 2004). Most human Chlamydia infections are caused by the human-adapted pathogens Chlamydia trachomatis and C. pneumoniae. These infections are common throughout the world with up to 80% of some adult populations testing seropositive for Chlamydia (Kuo et al. 1995; Choi et al. 1998). Chlamydia pneumoniae is responsible for both asymptomatic and acute pulmonary infections which have been associated with the development or exacerbation of persistent respiratory ailments such as asthma and chronic obstructive pulmonary disease (Blasi et al. 1993; Kuo et al. 1995; Hahn et al. 2012; Patel et al. 2012; Shimada, Crother and Arditi 2012). Chlamydia trachomatis infections are also frequent and occur in approximately 100 million individuals annually worldwide, causing significant morbidity in the human population. Distinct C. trachomatis subspecies or strains are adapted to different human cell types and tissues and accordingly associate with distinct disease manifestations (Stephens et al. 2009). Strains within serovars A to C infect conjunctival epithelial cells and cause endemic ocular infection. If left untreated, repeated infections can result in trichiasis (in-turning of the eye lashes), leading to corneal abrasions, corneal scarring, opacification and ultimately blindness. Strains within serovars D to K infect genital tract epithelial cells. Recurring or persistent genital C. trachomatis infections can cause urethritis, pelvic inflammatory disease (PV), infertility or lead to neonatal infections (Haggerty et al. 2010; Mylonas 2012). Lastly, the lymphogranuloma venereum (LGV) strains L1, L2 and L3 not only infect epithelial cells but also infect macrophages and spread systemically through lymph nodes. A detailed description of the pathogenesis of both ocular and genital infections was presented in a number of comprehensive recent reviews (Darville and Hiltke 2010; Abdelsamed, Peters and Byrne 2013; Taylor et al. 2014; Hafner 2015). Our review will specifically focus on cell-autonomous immune responses that underlie the pathogenesis of genital C. trachomatis infections. Because C. trachomatis is a highly adapted human pathogen with reduced virulence in the mouse (Coers, Starnbach and Howard 2009), many investigators have opted to use the closely related rodent-adapted pathogen C. muridarum to perform murine infection experiments. Important scientific insights have been gleaned from rodent studies using either C. trachomatis or C. muridarum as the infectious agent, and our review will encompass findings obtained from either model.
All Chlamydia species are obligate intracellular pathogens that replicate within an intracellular vacuolar compartment known as an ‘inclusion’. Chlamydia trachomatis enters host cells in its infectious form known as the elementary body (EB). Following invasion, EBs differentiate into the replicative reticulate body (RB) form, which can undergo binary fission within the confines of the expanding inclusion (Abdelrahman and Belland 2005). At mid stage of the developmental cycle, RBs begin to differentiate back into EBs, which can exit the spent host cell either through lysis or vacuolar extrusion (Hybiske and Stephens 2007). Following host cell exit, antibody-mediated immunity can restrict the dissemination of C. muridarum and thereby prevent systemic infections in mouse models (Li and McSorley 2013). However, antibody-mediated immunity is largely dispensable for the clearance of C. muridarum at the primary infection site and similarly fails to provide protective immunity (Morrison et al. 2000; Li and McSorley 2013, 2015). Instead, clearance of infections and protective immunity is dependent on Chlamydia-specific T cells. Mucosal Chlamydia infections in mice evoke both CD4+ and CD8+ T-cell responses. Yet, while depletion of CD8+ T cells fails to compromise immune protection against genital infections with either C. trachomatis or the rodent-adapted strain C. muridarum, CD4+ T cells are both necessary and sufficient to confer protection (Starnbach, Bevan and Lampe 1994, 1995; Morrison, Feilzer and Tumas 1995; Lampe et al. 1998; Morrison et al. 2000; Coers et al. 2011; Gondek et al. 2012; Nogueira et al. 2015). Critical for the anti-Chlamydia response of CD4+ T cells is their ability to secrete the proinflammatory cytokine interferon-γ (IFNγ) (Johansson et al. 1997; Perry et al. 1999; Roan et al. 2006; Gondek, Roan and Starnbach 2009). Priming of host cells with IFNγ induces a network of cell-autonomous defense modules (Boehm et al. 1997). Here, we will review IFNγ-dependent and –independent cell-autonomous host defense pathways that mediate resistance to Chlamydia and how these responses impact the pathogenesis of genital Chlamydia infections. Additionally, we will summarize and evaluate the literature on innate immune sensing pathways that modulate cell-autonomous host defense to Chlamydia, impact the activation of adaptive immune responses and control inflammation.
AN OVERVIEW OF CELL-AUTONOMOUS IMMUNITY AND ITS IMPORTANCE TO CHLAMYDIA TRACHOMATIS PATHOGENESIS
Vertebrates maintain a sophisticated network of specialized immune cells that includes innate lymphoid cells, professional antigen presenting cells, B cells and T cells. This network of professional immune cells—essential for host protection against the onslaught of myriads of potential infectious agents—is traditionally equated with the immune system. However, more often than not professional immune cells exert their function through the secretion of proinflammatory cytokines that bind to their cognate receptors on the surface of non-immune cells and thereby instruct these non-immune cells to enter a state of heightened resistance towards pathogens. This cytokine-induced state of heightened resistance is due to the cytokine-induced expression of various cell-autonomous immune pathways (Randow, MacMicking and James 2013; Pilla-Moffett et al. 2016)
The term ‘cell-autonomous immunity’ is not to be confused with ‘cellular immunity’. While cellular immunity refers to immune functions of professional immune cells that operate independently of antibodies, cell-autonomous immunity describes the capacity of individual cells of virtually all cell lineages to independently defend themselves against infections. Functionally, cell-autonomous immune responses can be placed into two categories: detection of pathogens by pattern recognition receptors (PRRs) and execution of antimicrobial effector functions. Pathogen recognition by PRRs is the first and essential step in cell-autonomous host defense, as it enables the host cell to detect the presence and precise location of a pathogen. Once localized, the pathogen can be captured and delivered into terminal lysosomes. Here, the host cell bombards the ‘microbial prisoner’ with an array of antimicrobial molecules that are active within acidified lysosomes (Randow, MacMicking and James 2013). While microbial destruction within lysosomes appears to be the default cell-autonomous defense pathway, intracellular pathogens can also be contained within multilamellar autophagosome-like structures or simply be expelled from the infected host cell (McCormack et al. 2013; Randow, MacMicking and James 2013; Miao et al. 2015; Selleck et al. 2015). Cytokines, and IFNs in particular, are often required to induce or enhance cell-autonomous immunity against C. trachomatis and many other intracellular pathogens (Pilla-Moffett et al. 2016). However, some cell types including ‘non-immune cells with barrier functions’, such as keratinocytes or mucosal epithelial cells, appear to have subsets of cell-autonomous defense programs switched on at all times (Bernard and Gallo 2011; Hooper 2015; McCormack et al. 2015).
Cell-autonomous host defense is central to the pathogenesis of C. trachomatis infections. Infected cells can sense the presence of C. trachomatis through several PRRs (Fig. 1) and the importance of PRRs in shaping the outcome of C. trachomatis infections is becoming increasingly evident (Darville and Hiltke 2010; Hafner 2015). PRR stimulation promotes the recruitment of professional immune cells to the site of infection. Therefore, PRR activation creates a more proinflammatory microenvironment surrounding C. trachomatis-infected cells and thereby induces cell-autonomous host defense in both the infected cell and non-infected bystander cells (Darville and Hiltke 2010; Hafner 2015). We will discuss the various types of cell-autonomous defense pathways that have been shown to mediate bactericidal or bacteriostatic activities against C. trachomatis and C. muridarum. We will further explore the role of these cell-autonomous defense pathways in host resistance, inflammation and infection-induced pathology and, if known, highlight bacterial counter-immune mechanisms that allow Chlamydia to block or even co-opt cell-autonomous immunity.
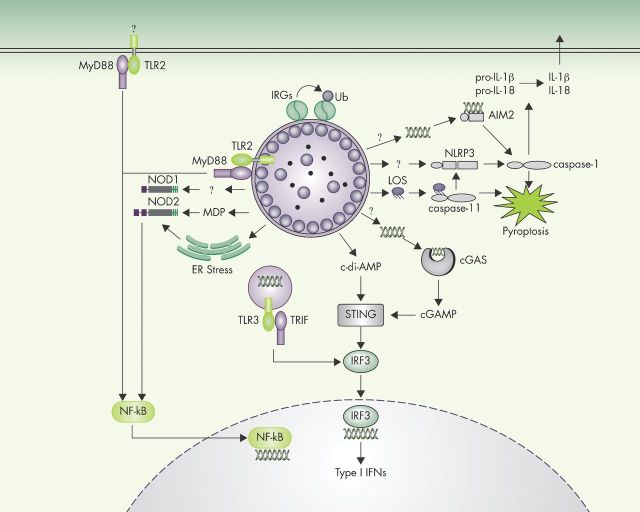
Figure 1.
Innate immune sensors of C. trachomatis infection. A wide variety of sensors collectively detect C. trachomatis infections and activate immune response pathways. Following the detection of unknown C. trachomatis-derived ligands in the extracellular or vacuolar milieu, TLR2 and TLR3 induce the transcription of cytokine-encoding genes. The cytosolic sensor STING directly detects the presence of bacterial cyclic di-AMP or—indirectly through cGAS—the presence of DNA in the host cell cytosol. STING activation leads to IRF3-dependent induction of type I IFN production. Cytosolic NOD1 and NOD2 detect bacterial cell-wall components, for example, muramyl dipeptide, and Chlamydia-induced ER stress to induce NF-κB activation and transcription of proinflammatory cytokines. Three distinct cytosolic surveillance pathways can induce inflammasome activation in response to C. trachomatis: AIM2, NLRP3 and caspase-11. Activated inflammasomes execute pyroptotic cell death, and IL-1β/IL-18 secretion. IRGs detect C. trachomatis inclusions in mouse cells and recruit host factors, promoting inclusion ubiquitination and ultimately leading to inclusion rupture.
The conflict between cell-autonomous host defense and bacterial counter-immunity is a reflection of the continuous evolutionary arms race between Chlamydia species and their hosts. As a result of this and similar host–pathogen conflicts, cell-autonomous host defense strategies continue to diverge in different host species (Daugherty and Malik 2012). Accordingly, distinct Chlamydia species evolved disparate virulence strategies to cope with cell-autonomous defenses unique to their preferred host species. For example, C. trachomatis has adapted to IFNγ-inducible human- but not rodent-specific cell-autonomous responses and thereby further solidified its tropism for the human host (Caldwell et al. 2003; Nelson et al. 2005; Bernstein-Hanley et al. 2006a,b; Coers, Starnbach and Howard 2009). The evolutionary conflict between human immunity and C. trachomatis counter-immunity is ongoing, as mirrored in the genetic diversity of C. trachomatis strains and some recent observations indicating that human genetic variation affecting cell-autonomous immunity can alter susceptibility to C. trachomatis-associated diseases (Abdelsamed, Peters and Byrne 2013).
INNATE IMMUNE SENSORS OF CHLAMYDIA TRACHOMATIS INFECTIONS
Toll-like receptors
Toll-like receptors (TLRs) comprise a large family of type I transmembrane receptor proteins that recognize structurally conserved pathogen-associated molecular patterns (PAMPs) as well as some endogenous ligands released in the context of infection or cellular damage. TLRs form both homo- and heterodimers and each dimer displays different ligand specificities (Kawai and Akira 2011). TLR4 and myeloid differentiation factor 2 (MD-2) form receptor complexes that detect the presence of lipopolysaccharide (LPS), also known as endotoxin (Kawai and Akira 2011). Although C. trachomatis is a Gram-negative bacterium and expresses an LPS-like lipooligosaccharide (LOS), C. trachomatis infections fail to stimulate robust TLR4-dependent responses (Ingalls et al. 1995; Nguyen et al. 2011). The lack of TLR4 activation by C. trachomatis is a result of the specific structure of Chlamydia LOS. The immune-stimulatory activities of LPS or LOS are determined by its core structural component, lipid A. The precise molecular configuration of lipid A differs amongst distinct bacterial species correlating with its immunogenicity; the number of acyl chains and chain lengths of a given lipid A molecule determine the efficiency with which the TLR4-MD-2-LPS complex is formed (Kawai and Akira 2011; Scior, Alexander and Zaehringer 2013). Whereas strong activators of TLR4 signaling such as Salmonella or Escherichia coli lipid A contain six acyl chains between 12 and 14 carbons in length, Chlamydia lipid A is penta-acylated with non-hydroxylated fatty acid chains of up to 20 carbons in length rendering it a poor ligand for MD-2 (Brade et al. 1997; Rund et al. 1999; Heine et al. 2007; Scior, Alexander and Zaehringer 2013). Accordingly, purified Chlamydia LOS is 100-fold less potent than Salmonella LPS at inducing TLR4/MD-2-dependent cytokine responses and TLR4-deficient mice clear genital Chlamydia infections with kinetics similar to wild-type mice (Ingalls et al. 1995; Darville et al. 2003). Moreover, an unbiased forward genetics approach in mice failed to detect linkage between susceptibility to C. trachomatis infections and the Tlr4 loss-of-function allele present in C3H/HeJ inbred mice (Bernstein-Hanley et al. 2006a,b). Furthermore, human studies with patient cohorts consisting predominantly of white women revealed no significant association between infection rates and polymorphisms in TLR4 or its coreceptor CD14, or between tubal factor infertility and TLR4 or CD14 variants (Morre et al. 2003; Ouburg et al. 2005; Wang et al. 2005; Verweij et al. 2016). A single study examining a small cohort of African American women found an association between the TLR4 rs1727911 CC genotype and cervical or endometrial C. trachomatis infections, specifically amongst patients with BV (Taylor et al. 2012), suggesting that TLR4 variants could exacerbate C. trachomatis-induced sequelae. However, the latter study has yet to be confirmed using a validation cohort. All of these studies together indicate that C. trachomatis infections are unlikely to directly induce robust TLR4 activation in mice or humans. Future studies will need to address whether TLR4 signaling could play a role in the pathogenesis of C. trachomatis genital infections, possibly by altering the composition of the vaginal microbiome or susceptibility to BV (Wiesenfeld et al. 2003; Yoshimura et al. 2009).
While Chlamydia LOS is a poor agonist for TLR4, C. trachomatis and C. muridarum infections were shown to induce robust secretion of proinflammatory cytokines including tumor necrosis factor α (TNFα), thus suggesting that Chlamydia infections stimulate TLRs other than TLR4 (Williams et al. 1989; Ingalls et al. 1995; Prebeck et al. 2003). Subsequent studies revealed that C. trachomatis and C. muridarum infections trigger TLR2-dependent secretion of interleukin 6 (IL-6), IL-8 and TNFα (Darville et al. 2003; Derbigny, Kerr and Johnson 2005; O'Connell et al. 2006). TLR2 was shown to colocalize with inclusion membranes (O'Connell et al. 2006) and therefore signaling may originate from the inclusion membrane itself or, alternatively, from the plasma membrane (Fig. 1).
Several Chlamydia-derived molecules were proposed to act as TLR2 agonists including heat shock protein 60 (hsp60), the chlamydial major outer membrane protein and a lipoprotein named macrophage infectivity potentiator (Mip) (Vabulas et al. 2001; Bulut et al. 2002; Bas et al. 2008; Massari et al. 2013). While various structurally distinct PAMPs are implicated as TLR2 agonists, direct binding to TLR2 has only been demonstrated for a small number of molecules including amphiphilic lipopeptides or lipoproteins and the fungal cell-wall component zymosan (Lien et al. 1999; Sato et al. 2003; Jin et al. 2007; Kang et al. 2009). The crystal structure of TLR1-TLR2 heterodimer bound to diacylated lipopeptide and TLR2-TLR6 heterodimer bound to triacylated lipopeptide confirmed that lipopeptides act as bona fide TLR2 ligands, which activate TLR2 signaling at picomolar concentrations (Muhlradt et al. 1997; Jin et al. 2007; Kang et al. 2009). Some previously considered agonists, such as peptidoglycan (PG), fail to induce TLR2 activation when chemically synthesized, suggesting that contaminations with lipopeptides could account for the induction of TLR2 signaling by a number of substances including PG-derived structures purified from bacteria (Zahringer et al. 2008).
TLR2 activation is dramatically diminished when cells are exposed to plasmid-cured Chlamydia strains, suggesting that plasmid-encoded genes either control the synthesis or the processing of TLR2-activating molecules (O'Connell et al. 2007, 2011). The plasmid-encoded protein Pgp4 acts as a transcriptional regulator and controls the expression of multiple chromosomal genes including glgA, which encodes a glycogen synthase (Song et al. 2013). Accordingly, plasmid-cured Chlamydia strains display a defect in glycogen synthesis and glycogen accumulation (Matsumoto et al. 1998; O'Connell and Nicks 2006; O'Connell et al. 2011). More recently, it was demonstrated that enzymatically synthesized glycogen activates TLR2 signaling, opening up the intriguing possibility that Chlamydia-derived glycogen functions as the primary trigger for infection-induced TLR2 activation (Kakutani et al. 2012). In contrast to the defect in glycogen accumulation plasmid-cured Chlamydia strains express wild-type levels of the lipoprotein Mip, all but excluding Mip as the driver of Chlamydia-induced TLR2 activation (O'Connell et al. 2011). Using recently developed genetic approaches, loss-of-function mutations in pgp4, mip and other chlamydial genes could be used to define the Chlamydia-derived TLR2 agonist. How glycogen or other TLR agonists become available to their immune sensors during the course of an infection is currently unknown and will need to be addressed in the future.
Further confirming a role for TLR2 as a sensor for Chlamydia infections, Darville and colleagues found that the secretion of the cytokines TNFα, IL-6 and CXCL2 was decreased in the genital mucosa of C. muridarum-infected Tlr2−/− mice relative to wild-type mice. In spite of the diminished cytokine levels, TLR2 deficiency unexpectedly failed to change the kinetics by which C. muridarum infections were cleared in the rodent host. Instead, Tlr2−/− mice displayed a marked reduction in infection-induced oviduct and mesosalpinx pathologies, suggesting a specific role for TLR2 signaling in the development of Chlamydia-induced sequelae but a lesser role in immune clearance (Darville et al. 2003). Similarly, plasmid-cured C. muridarum strains that fail to activate TLR2 signaling cause substantially reduced pathology relative to infections with plasmid-bearing wild-type strains (O'Connell et al. 2007). Collectively, these observations establish an important role for TLR2 signaling in Chlamydia infection-associated pathologies.
In humans, TLR2 is highly expressed in the fallopian tube and the cervix suggesting that TLR2 sensing of C. trachomatis could also drive pathologies in human genital infections (Pioli et al. 2004). Indeed, a moderate association between TLR2 haplotypes and the clinical course of C. trachomatis infections has been observed (Verweij et al. 2016). On the other hand, comparable disease manifestations were observed in rhesus macaques inoculated with either wild-type or with plasmid-cured C. trachomatis, the latter strain lacking TLR2 activating properties (Qu et al. 2015). These data indicate that TLR2 signaling plays a lesser role in the pathogenesis of C. trachomatis infections in primates than it does in rodents, and furthermore, that additional PRRs other than TLR2 are likely to be involved in Chlamydia-induced immunopathologies.
A strong candidate to act as the key TLR2-induced cytokine promoting oviduct pathology in mice is TNFα; similar to Tlr2−/− mice, TNFα−/− mice are also protected against pathologies induced by C. muridarum infections while resolving C. muridarum infections with kinetics comparable to wild-type mice (Murthy et al. 2011). These data suggest that TNFα is necessary to induce sequelae in the mouse model but do not exclude the involvement of other TLR2-induced cytokines in the pathogenesis of Chlamydia infections. Regardless of the specific mechanism by which TLR2 activation leads to development of immunopathologies, these observations demonstrate that immune clearance of genital Chlamydia infections can be uncoupled from sequelae-inducing immune responses, an observation with important implications for vaccine development (Stary et al. 2015).
Whereas TLR2 activation triggers Myd88-dependent NF-kB signaling, engagement of endosomal TLR3 predominantly triggers the production of IFNβ (Kawai and Akira 2010). Chlamydia infections induce IFNβ secretion in a number of cell types including murine oviduct epithelial cells (Xia et al. 2003; Lad et al. 2005; Derbigny et al. 2007) and it was shown that epithelial cells deficient in TLR3 expression showed a marked, albeit not complete reduction in Chlamydia-induced IFNβ secretion (Derbigny et al. 2012). TLR3 recognizes double-stranded RNA (dsRNA), a molecular pattern first associated with viral infections (Kawai and Akira 2010). It was later demonstrated that bacterial dsRNA also activates TLR3 signaling (Kawashima et al. 2013), suggesting that Chlamydia-derived dsRNA may act as a TLR3 agonist in epithelial and possibly also in dendritic cells (Fig. 1). Considering the inherently anti-inflammatory nature of TLR3-driven IFNβ production (Kawai and Akira 2010), a potential role for TLR3 signaling in dampening immunity to Chlamydia needs to be explored further in the future.
Stimulator of interferon genes
Signaling via TLR3 is not the only mechanism by which Chlamydia infections induce type I IFN production. A second PRR that senses the presence of Chlamydia and induces the secretion of IFNβ is stimulator of interferon genes (STING) (Prantner, Darville and Nagarajan 2010). The importance of STING as an inducer of IFNβ production in response to Chlamydia infections was demonstrated in multiple cell lines (Prantner, Darville and Nagarajan 2010; Barker et al. 2013). For example, human embryonic kidney (HEK) 293T cells are inherently deficient for STING expression and only respond to C. trachomatis with IFNβ secretion when STING is ectopically expressed. Similarly, STING-deficient mouse embryonic fibroblasts fail to secrete IFNβ in response to C. trachomatis infections (Barker et al. 2013).
Several studies have demonstrated that STING dimerization leads to the activation of the TANK-binding kinase 1 resulting in the phosphorylation of interferon regulatory transcription factor 3 (IRF3) and IRF3-dependent transcription of type I IFN-encoding genes (Barber 2014). Two pathways result in STING dimerization and activation: in the first pathway, STING acts as a direct sensor of bacteria-derived cyclic dinucleotides and in the second pathway, STING acts as an indirect sensor of cytosolic double-stranded DNA (dsDNA) (Sun et al. 2013; Wu et al. 2013). Both pathways have been implicated in the activation of STING by Chlamydia infections (Barker et al. 2013; Zhang et al. 2014).
Cyclic di-GMP and cyclic di-AMP are bacterial second messenger molecules that are expressed at varying concentrations (0.1–10 μM) inside a bacterial cell (Ryan, Tolker-Nielsen and Dow 2012). It was demonstrated in vitro that one cyclic di-GMP molecule forms a complex with the C-terminal domains of two STING molecules, thereby promoting STING dimerization and initiating STING signaling. While the structural data convincingly demonstrate direct binding of cyclic di-GMP to STING, the measured binding affinity of cyclic di-GMP to STING was relatively low compared to other ligand–PRRs interactions (Ouyang et al. 2012; Yin et al. 2012). Accordingly, it was found that STING activation by cyclic dinucleotides under physiological conditions requires the helicase DDX41, which binds cyclic dinucleotides and then forms a complex with STING (Parvatiyar et al. 2012). STING can also be activated by cyclic-GMP-AMP (cGAMP) (Sun et al. 2013; Wu et al. 2013). The production of cGAMP mediated the host enzyme cGAMP synthase (cGAS) in response to the presence of cytosolic dsDNA (Sun et al. 2013). Following dimerization induced by either bacterial cyclic dinucleotides or by host cGAMP, STING rapidly traffics from the endoplasmic reticulum (ER) through to the Golgi to perinuclear endosomes where it promotes the phosphorylation and activation of IRF3 (Barber 2014).
Cyclic di-AMP is a nucleic acid metabolite that contributes to cell-wall homeostasis in Gram-positive bacteria (Corrigan et al. 2011). Chlamydia trachomatis was identified as the first Gram-negative bacteria to also produce cyclic di-AMP but its physiological function in C. trachomatis is currently unknown (Barker et al. 2013). Intrabacterial concentrations of cyclic di-AMP rapidly increase as replicating RBs transform back into infectious EBs and this increase in cyclic di-AMP levels correlates with an increase in IFNβ production (Barker et al. 2013). Exogenously supplied cyclic di-AMP or C. trachomatis infections induce IFNβ production in HEK 293T cells that ectopically express murine STING but lack the dsDNA sensor cGAS, suggesting that chlamydial cyclic di-AMP acts directly as a STING agonist (Barker et al. 2013). However, coexpression of STING and cGAS in HEK 293T cells elicits greater IFNβ production in response to Chlamydia infections than expression of STING alone (Zhang et al. 2014). Therefore, Chlamydia infections appear to induce STING activation not only through the production of cyclic di-AMP but also through an infection-dependent increase in cytosolic dsDNA levels (Fig. 1). This model is also supported by studies demonstrating that cGAS expression was required for robust IFNβ production in Chlamydia-infected human and murine oviductal epithelial cell lines (Zhang et al. 2014). Failure to efficiently degrade cytosolic dsDNA due to the absence of the exonuclease TREX1 exacerbated IFNβ production in response to Chlamydia-infected cells, thus further supporting a model in which cytosolic dsDNA contributes to Chlamydia-induced type I IFN production (Zhang et al. 2014). Unanswered so far remain the questions of whether the inflammatory dsDNA is bacterial or host derived and how dsDNA leaks either from the bacteria and their surrounding inclusion space or from the host cell nucleus into the host cell cytosol.
While both cyclic di-AMP and dsDNA seem to promote STING activation in Chlamydia-infected cells, which one of the two pathways is predominantly activated may depend on the infected cell type, the host species or the individual host haplotype. Indeed, it was reported that common SNPs in the human STING gene significantly alter the ability of STING protein to respond to agonists, thus skewing the binding preference of STING either towards cGAMP or cyclic dinucleotides (Yi et al. 2013). It is therefore plausible that STING allelic variants could influence the magnitude or duration of the type I IFN response during chlamydial infections.
How relevant is STING activation in the pathogenesis of C. trachomatis infections? Mouse fibroblasts deficient for STING signaling allow for exacerbated chlamydial growth, revealing a potential function for STING in cell-autonomous immunity to C. trachomatis (Barker et al. 2013). It was proposed that STING mediates its anti-Chlamydia activity through autocrine cell activation by STING-induced type I IFNs, but this model has not yet been experimentally confirmed. While the type I IFN receptor IFNAR induces cell-autonomous immunity in tissue culture models, IFNAR paradoxically promotes C. trachomatis survival in vivo (Nagarajan et al. 2008). Therefore, STING activation and the resulting production of type I IFNs could potentially promote C. trachomatis survival in the genital tract. Predicting the in vivo function of STING is further complicated by the possibility that STING-mediated IRF3 activation could provide protection against immunopathologies independently of IFNAR signaling (Prantner et al. 2011). In order to clearly define how STING-mediated responses affect the pathogenesis of Chlamydia infections, additional in vivo studies are needed.
Nucleotide-binding oligomerization domain-containing proteins
Activation of the cytosolic PRRs NOD1 and NOD2 drives NF-κB-dependent expression of proinflammatory cytokines such as IL-6. Recently, it was demonstrated that activation of NOD2 by either single-stranded RNA (ssRNA) or N-glycosylated muramyl dipeptide could also induce type I IFN production (Pandey et al. 2009; Sabbah et al. 2009; Caruso et al. 2014). Whereas activation of NOD2 by ssRNA induces type I IFN production in an IRF3- and MAVS-dependent manner (Caruso et al. 2014), mycobacteria-derived muramyl dipeptides trigger type I IFN secretion in an IRF5-dependent manner (Pandey et al. 2009). In line with a role for nucleotide-binding oligomerization domain (NOD) proteins as initiators of type I IFN responses, it was reported that maximal induction of IFNβ expression during chlamydial infection requires NOD1 (Prantner, Darville and Nagarajan 2010). A separate study reported NOD1-mediated NF-κB signaling in C. muridarum-infected fibroblasts, which resulted in the secretion of the cytokine IL-6 and the chemokine CXCL2 (Welter-Stahl et al. 2006). However, the chlamydial agonist responsible for NOD1 activation or the downstream signaling events were not described in these studies.
Both NOD1 and NOD2 are known to recognize structures derived from PG, a polymer imbedded in bacterial cell walls. NOD1 recognizes the PG dipeptide D-gamma-glutamyl-meso-diaminopimelic acid (iE-DAP), which is made by all Gram-negative and a few Gram-positive bacterial species. NOD2, on the other hand, binds murmayl dipeptide, a common building block of PG in all bacterial phyla (Caruso et al. 2014). While the presence of PG in the cell wall of Chlamydiae had been controversial for half a century, recent work by Maurelli and colleagues unequivocally showed that Chlamydiae produce detectable amounts of PG (Liechti et al. 2014; Packiam et al. 2015). The same group demonstrated that Chlamydia-derived muramyl peptides activated NOD2 signaling and the canonical NF-κB pathway (Packiam et al. 2015). Although these observations establish that chlamydial PG acts as NOD2 agonist, it remains unclear whether chlamydial PG is the chief activator of NOD1/2 signaling in response to Chlamydia infections. As an alternative model, Chlamydia infection-induced ER stress could be the dominant factor driving NOD1/2 signaling. This model is based on a recent report demonstrating that an inhibitor of the ER stress response kinase IRE1α blocked IL-6 secretion in C. muridarum-infected human HeLa cells (Keestra-Gounder et al. 2016).
Collectively, these observations establish that Chlamydia can induce NOD signaling. The importance of NOD signaling in the pathogenesis of genital C. trachomatis infections is less clear. A polymorphic variant in human NOD1 associates with decreased rates of genital C. trachomatis infections in women but increased incidence of tubal factor infertility (Brankovic et al. 2015), suggesting a functional role for NOD1 in human Chlamydia infections. In mice, on the other hand, clearance rates of experimental C. muridarum infections were unaffected by the absence of NOD1 (Welter-Stahl et al. 2006). The immune response of Chlamydia-infected NOD2-deficient animals has yet to be reported. Considering that chlamydial PG has been confirmed as a NOD2 agonist, a more careful examination of the role of NOD2 in anti-Chlamydia immunity is warranted.
Inflammasomes
Inflammasomes are critical for host defense against a broad spectrum of card-carrying pathogens but also provide essential host resistance against environmental bacteria that are non-pathogenic in immunocompetent hosts (Guo, Callaway and Ting 2015; Maltez et al. 2015). While the absence of inflammasome function results in increased susceptibility to infections, dysregulation of inflammasomes has been linked to a host of autoinflammatory and autoimmune diseases (Guo, Callaway and Ting 2015). Inflammasomes are multiprotein oligomers that form in response to intracellular PAMPs or host-derived damage-associated molecular patterns (DAMPs). The canonical inflammasome consists of the cysteine protease caspase-1, an upstream sensor protein and in most cases the adapter protein apoptosis-associated speck-like protein containing a carboxy-terminal CARD (ASC) (Latz, Xiao and Stutz 2013). The non-canonical inflammasome is less well characterized and is defined by the presence of caspase-11. Cytosolic PAMPs or DAMPs trigger the assembly of either type of inflammasome and can induce a proinflammatory cell death pathway known as pyroptosis. Canonical inflammasomes additionally mediate the proteolytic processing of the cytokines IL-1β and IL-18. Processing of pro-IL-1β and pro-IL-18 by caspase-1 transforms these cytokine proforms into their active forms that can be delivered into the extracellular milieu by a poorly characterized secretion pathway (Dupont et al. 2011). Chlamydia infections in a variety of epithelial cell lines as well as in monocytes induce caspase-1-dependent IL-1β and IL-18 secretion (Lu, Shen and Brunham 2000; Abdul-Sater et al. 2010). Caspase-1 activation and the associated cytokine release occur as a result of the detection of Chlamydia by at least three separate cytosolic surveillance pathways (Fig. 1): a cytosolic dsDNA-sensing pathway that requires absent in melanoma 2 (AIM2), direct activation of NLR family pyrin domain containing 3 (NLRP3) by unknown ligand(s) and indirect activation of NLRP3 via the non-canonical caspase-11-containing inflammasome (Abdul-Sater et al. 2009, 2010; Finethy et al. 2015).
Initial studies attributed Chlamydia-induced caspase-1 activity to the NLRP3 inflammasome. These studies found that in unprimed cells interference with NLRP3 expression resulted in a corresponding drop in IL-1β secretion (Abdul-Sater et al. 2009, 2010). In naive cells, Chlamydia induces NLRP3 activation in a manner dependent on both chlamydial protein synthesis and the activity of type III secretion system (T3SS) (Lu, Shen and Brunham 2000; Abdul-Sater et al. 2010). It was hypothesized that a secreted chlamydial protein induced K+ efflux leading to consequential generation of radical oxygen species (ROS), a known inducer of NLRP3 activity. Thus far, no such chlamydial protein has been identified. Alternatively, T3SS may be required for cell priming rather than for direct NLRP3 activation. In support of the latter model, priming with LPS was shown to obviate the need for chlamydial growth or T3SS function to induce IL-1β secretion in response to Chlamydia infections (Prantner et al. 2009).
While no Chlamydia-derived molecule has so far been identified as a direct agonist for NLRP3, one potential candidate is cyclic di-AMP, which can induce K+ efflux and NLRP3 activation through an unknown mechanism independent of STING (Abdul-Sater et al. 2013). Furthermore, the ability of cyclic di-AMP to activate STING during a Chlamydia infection suggests that cyclic di-AMP is available in sufficient quantities to activate NLRP3. Alternatively, NLRP3 activation may occur in response to infection-associated cellular stress rather than a microbe-derived agonist, consistent with the putative role for NLRP3 as a promiscuous sensor of infection-induced DAMPs (Latz, Xiao and Stutz 2013). As will be discussed in more detail later, Chlamydia infections also activate caspase-11, which in turn induces NLRP3 function and thus is responsible for some but not all NLRP3-mediated secretion of IL-1β and IL-18 (Finethy et al. 2015). These data hence imply the existence of at least two NLRP3 activation pathways in Chlamydia-infected cells: a caspase-11-dependent and a caspase-11-independent pathway. The precise nature of the caspase-11-independent inducer of NLRP3 activation in Chlamydia-infected cells remains to be determined.
In IFNγ- or LPS-primed murine bone marrow-derived macrophages (BMDMs), complete loss of NLRP3 expression results in a partial loss of IL-1β and IL-18 secretion (Nagarajan et al. 2012; Finethy et al. 2015). Loss of AIM2 expression also results in a moderate reduction in IL-1β and IL-18 production in primed BMDMs (Finethy et al. 2015). These observations suggest that NLRP3 and AIM2 inflammasomes function independently of one another and have partly redundant functions in primed BMDMs. Accordingly, concurrent loss of NLRP3 and AIM2 gene function leads to the complete abrogation of Chlamydia-induced IL-1β and IL-18 secretion, indicating that NRLP3 and AIM2 are the sole canonical inflammasomes activated by C. muridarum or C. trachomatis in BMDMs (Finethy et al. 2015).
AIM2 functions as PRRs for cytosolic dsDNA (Burckstummer et al. 2009; Fernandes-Alnemri et al. 2009; Hornung et al. 2009; Roberts et al. 2009). As with cGAS, the source of the inflammatory dsDNA activating AIM2 is unknown. Similarly, very little is known about the mechanism by which AIM2 is regulated. In the absence of an external priming signal, Chlamydia induces an inflammasome response primarily through NLRP3, demonstrating that AIM2 requires an IFN-mediated second signal in order to detect Chlamydia infections (Finethy et al. 2015). The nature of this second signal is currently unknown but may involve an IFN-inducible host response that results in the release of either host or bacterial dsDNA into the host cell cytosol.
As already mentioned, Chlamydia infections prompt the activation of the non-canonical caspase-11 inflammasome in BMDMs, triggering NLRP3-dependent IL-1β and IL-18 secretion. Activated caspase-11 also initiates pyroptosis in BMDMs in an NLRP3- and caspase-1-independent manner (Finethy et al. 2015). Findings made in regard to the function of caspase-11 in murine BMDMs could be of significance for understanding immunity to C. trachomatis in humans. Similar to their murine caspase-11 homolog, human caspase-4 and caspase-5 are activated by Gram-negative infections (Knodler et al. 2014; Shi et al. 2014). Recent studies with purified mouse caspase-11 and human caspase-4 protein revealed that both the murine and human protein display similar ligand specificities for penta- and hexa-acylated forms of LPS (Shi et al. 2014). Penta-acylated LOS derived from Chlamydia is therefore a strong candidate to function as a caspase-11 or caspase-4 ligand. Future experiments will need to test whether purified Chlamydia LOS functions as a direct ligand of murine caspase-11 and/or human caspase-4/ −5. Additionally, the question of how chlamydial LOS becomes available to cytosolic sensors needs to be addressed. One possibility is that host-directed inclusion lysis coupled to bacteriolysis could liberate LOS making it available for sensing in the host cytosol. However, in murine BMDMs both C. muridarum and C. trachomatis induce robust caspase-11 activation despite the ability of C. muridarum to interfere with host-mediated inclusion lysis (Finethy et al. 2015; Haldar et al. 2015). Alternatively, LOS molecules or small outer membrane vesicles released from individual bacteria (Vanaja et al. 2016) could accumulate inside inclusions and ‘leak’ into the host cytosol ‘accidently’.
Although the activation of caspase-11—as well as AIM2—in response to Chlamydia has only been described in BMDMs so far, these sensors might also function in the epithelial cell-autonomous immune response to Chlamydia. Caspase-11 and AIM2 were shown to be expressed and function in epithelial cells (Knodler et al. 2014; Hu et al. 2015). The relative in vivo contribution of caspase-11 and AIM2 to Chlamydia-induced inflammasome activation in urogenital epithelial cells is unknown. However, even if these pathways were inactive in epithelial cells, it would not exclude a role for these proteins in cell-autonomous immunity to Chlamydia. While C. trachomatis predominantly infects epithelial cells, macrophage immunity is not irrelevant to the course of infection. LGV serovars L1, L2, and L3 are able to infect macrophages thereby encouraging systemic infection (Mabey and Peeling 2002). Thus, while speculative, it is possible that pyroptotic responses in macrophages could diminish C. trachomatis LGV dissemination beyond the site of infection.
As discussed, priming of host cells with IFNs prior to infection promotes rapid and potent inflammasome responses. In part, this is due to the ability of IFNs to induce expression of inflammasome components such as caspase-11 (Schauvliege et al. 2002). However, IFN-induced expression of these components alone is not sufficient to explain the enhanced response of IFN-primed cells, and it recently became apparent that IFN priming induces additional cofactors that increase the sensitivity for inflammasome activation (Broz et al. 2012). One such group of cofactors constitutes guanylate-binding proteins (GBPs), a family of IFN-inducible GTPases (Pilla-Moffett et al. 2016). Studies performed in murine BMDMs found that loss of GBP expression led to a reduction in pyroptosis as well as IL-1β and IL-18 secretion in response to both cytosolic and vacuolar bacterial pathogens (Meunier et al. 2014, 2015; Pilla et al. 2014; Man et al. 2015). Recently, we demonstrated that GBP-deficient BMMs also displayed a muted inflammasome response when infected with either C. trachomatis or C. muridarum (Finethy et al. 2015).
GBPs have been proposed to assist inflammasome activation through three distinct mechanisms by mediating (i) lysis of pathogen-containing vacuoles (PVs) and/or (ii) bacteriolysis thereby inducing the release of PAMPs into the cytosol or by (iii) directly promoting formation of the inflammasome complex. In support of the first mechanism, GBPs have been shown to target to and to facilitate the rupture of PVs formed by the protozoan pathogen Toxoplasma gondii (Yamamoto et al. 2012). GBPs also associate with C. trachomatis inclusions formed in mouse fibroblasts (Haldar et al. 2013, 2014), and possibly in human cells, although the latter observation remains controversial and requires further investigation (Tietzel, El-Haibi and Carabeo 2009; Al-Zeer et al. 2013; Johnston et al. 2016). In spite of the association of GBPs with C. trachomatis inclusions, the function of GBPs in Chlamydia-induced inflammasome activation is most likely unrelated to PV lysis; we found that C. muridarum precludes GBP localization to the inclusion membrane through an unidentified mechanism, and failed to observe a link between GBPs and inclusion rupture (Finethy et al. 2015; Haldar et al. 2015). Therefore, in this instance, GBPs are likely to function independently of PV lysis.
Recently, it was demonstrated that GBPs could promote inflammasome activation during infection with the cytosolic bacterium Francisella novicida. These studies showed that GBPs could colocalize with cytosolic bacteria and mediate bacteriolysis (Man et al. 2015; Meunier et al. 2015). While it is plausible that GBPs could lyse exposed Chlamydiae following inclusion rupture, no association of GBPs with expelled Chlamydia was observed (Finethy et al. 2015). The available data therefore favor a model wherein the GBPs function outside the host–pathogen interface in promoting inflammasome activation in response to Chlamydia. This model is in agreement with a study that reported how murine GBPs could exacerbate caspase-11-dependent pyroptosis in response to purified LPS delivered into the host cytosol, indicating that GBPs act downstream of the release of PAMPs by intracellular pathogens (Pilla et al. 2014). Another study reported that the GBP family member GBP5 promotes NLRP3-ASC oligomerization in response to a subset of canonical NLRP3 agonists (Shenoy et al. 2012), although this report has been challenged (Meunier et al. 2014; Man et al. 2015). Thus, the function of GBPs outside of direct PV or pathogen destruction remains unclear and future experiments will be necessary to determine the mechanism of GBP-dependent inflammasome regulation. Moreover, the importance of GBPs in controlling inflammation in response to Chlamydia infections in vivo still needs to be addressed.
Immunity-related GTPases
In addition to GBPs, a second family of IFN-inducible GTPases, the so-called immunity-related GTPases (IRGs), plays an important role in the biology of Chlamydia infections. IFN-inducible GTPases belong to the dynamin superfamily and have emerged as essential mediators of host resistance to a broad spectrum of pathogens including viruses and bacteria. Members of IRG protein family provide cell-autonomous immunity to infections with a number of intracellular bacterial and protozoan pathogens (Pilla-Moffett et al. 2016). In accord with their role in host resistance, allelic variants of the human IRGM locus are associated with altered susceptibility to bacterial infections (Intemann et al. 2009; Yang et al. 2014). Naturally occurring genetic variations in murine IRG genes are associated with altered susceptibility to C. trachomatis infections (Bernstein-Hanley et al. 2006a,b) and the IRG resistance system has emerged as the single most potent IFNγ-inducible defense system active against C. trachomatis in the mouse (Nelson et al. 2005; Coers et al. 2008, 2011).
The IRG resistance systems in mice and humans are different from one another in a number of ways starting with the number of IRG genes encoded by the respective murine and human genomes: whereas mouse genomes typically harbor about 20 IRG genes, the human genome contains only one ubiquitously expressed IRG gene (Bekpen et al. 2005, 2009; Coers, Starnbach and Howard 2009). All IRG proteins encoded by the mouse genome can be placed into two subfamilies defined by sequence and functional differences: the regulatory IRGM and the ‘effector’ GKS proteins (Bekpen et al. 2005; Hunn et al. 2008). The effector GKS proteins are defined by the presence of a canonical GxxxxGKS motif in the P-loop of the GTP-binding site. IFN priming of mouse cells strongly induces expression of IRGM and GKS proteins. In uninfected cells, GKS proteins are found predominantly in the GDP-bound form and reside in the cytosol or associate transiently with cell organelles (Hunn et al. 2008). In infected cells, GKS proteins translocate and bind to C. trachomatis inclusions in their active, GTP-bound form (Coers et al. 2008; Haldar et al. 2013).
In contrast to GKS proteins, IRGM proteins contain a non-canonical GxxxxGMS P-loop sequence and are therefore sometimes referred to as ‘GMS proteins’ (Bekpen et al. 2005; Hunn et al. 2008). Following their induction by IFNs, murine GDP-bound IRGM proteins stably reside on most if not all endomembranes and organelles including theER, mitochondria and lipid droplets (LDs). Here, the regulatory IRGM proteins prevent off-target activation of GKS proteins and thereby guard self-structures such as LDs against GKS binding (Haldar et al. 2013). In contrast to endomembranes and organelles, inclusions are largely devoid of IRGM proteins and therefore permissive for the recognition by GKS proteins (Fig. 2). The absence of IRGM proteins from inclusion membranes provides a ‘missing-self’ signal resulting in the transition of GKS proteins into a GTP-bound active state in which GKS proteins can stably attach to inclusion membranes (Coers 2013; Haldar et al. 2013). This process is further supported by the ATG8 conjugation system (Al-Zeer et al. 2009; Haldar et al. 2014). Once loaded with GKS proteins, inclusions become decorated with a ubiquitin coat and ultimately undergo host-mediated rupture by a poorly characterized process (Haldar et al. 2015), as discussed in more detail below. Therefore, the critical function of GKS proteins is to direct antimicrobial effector pathways to their intended ‘non-self target’, the inclusion (Fig. 1). In how far PRRs other than GKS proteins fulfill similar functions in cell-autonomous immunity to C. trachomatis in human cells remains to be determined.
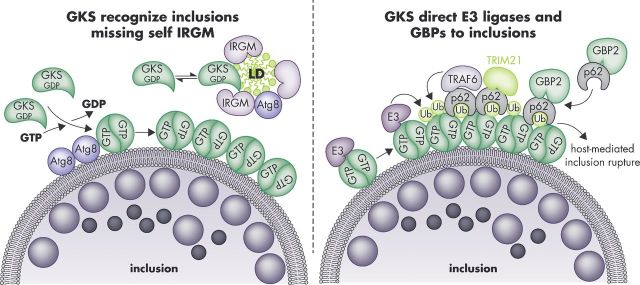
Figure 2.
Targeting of GKS proteins and GBPs to C. trachomatis inclusions in mouse cells. The GKS class of IRG proteins is guided towards inclusion membranes through a missing-self principle. The IRGM proteins Irgm1 and Irgm3 reside on ‘self’ membranes and organelles such as LDs and block GKS protein activation at these sites. The absence of these IRGM proteins from inclusions enables GTP-bound GKS dimers to form and associate with inclusion membranes. This association of GKS proteins with inclusions is further enhanced by the presence of lipidated Atg8 proteins at inclusion membranes (left panel). Inclusion-bound GKS proteins promote the recruitment of ‘pioneering’ ubiquitin E3 ligases (E3) and p62-interacting E3 ligases (TRAF6, TRIM21), which promote the decoration of inclusions with ubiquitin. Potential ubiquitination substrates are the GKS proteins themselves. The ubiquitin-binding protein p62 escorts GBP2 to inclusions (right panel). Additional p62-independent mechanisms of GBP recruitment exist (not shown). GKS-decorated inclusions rupture in a p62-dependent manner.
PATHWAYS OF CELL-AUTONOMOUS DEFENSE AGAINST CHLAMYDIA TRACHOMATIS
Ubiquitination of inclusions and associated defense pathways
Ubiquitin is a small (8.5 kDa) protein—expressed in virtually all cells of the human body—that can be covalently attached to other proteins; this process is known as ubiquitination. Ubiquitin can be attached to other proteins as a single molecule (monoubiquitination) or as multiple chain-linked molecules (polyubiquitination). Ubiquitination regulates many cellular processes that include receptor internalization and proteolysis. The attachment of polyubiquitin chains to large intracellular structures such as protein aggresomes or damaged mitochondria promotes the degradation of these structures inside autolysosomes (Komander and Rape 2012; Klionsky et al. 2016). Intracellular pathogens can also become ubiquitin decorated, resulting in their capture and degradation inside autolysosomes, or their containment with autophagosome-like structures (Boyle and Randow 2013). We have recently demonstrated that ubiquitin systems promote the rupture of inclusions in mouse fibroblasts (Haldar et al. 2015), adding to the list of antimicrobial responses that are ubiquitin dependent.
The coating of intracellular pathogens with ubiquitin is a conserved defense mechanism found in host organisms as diverse as fruit flies and humans (Boyle and Randow 2013; Manzanillo et al. 2013). The ubiquitin E3 ligases Parkin and LRSAM1 were previously shown to be required for the ubiquitination of intracellular Mycobacterium tuberculosis and Salmonella enterica in unprimed mouse and human cells (Huett et al. 2012; Manzanillo et al. 2013). Inclusions formed by C. trachomatis can also become ubiquitin decorated in mouse cells after priming with IFNs. This IFN-dependent pathway uses a set of E3 ligases that is distinct from the previously reported Parkin and LRSAM1 and instead depends in part on the E3 ligases TRAF6 and Trim21. The ubiquitination of C. trachomatis inclusions in mouse cells is absolutely dependent on the murine IRG resistance system. The data support a model according to which GKS proteins recruit several ubiquitin E3 ligases including the aforementioned TRAF6 and Trim21 to inclusions resulting in the deposition of K48- and K63-linked polyubiquitin and potentially other ubiquitin species at the inclusion membrane (Haldar et al. 2015). The ubiquitination substrates are currently unknown but may include GKS proteins themselves (Traver et al. 2011). The ubiquitin coat surrounding GKS-decorated inclusions recruits ubiquitin-binding proteins including p62. The latter protein escorts members of the GBP protein family to ubiquitinated inclusions, and GBPs are likely to enlist additional antimicrobial host factors (Haldar et al. 2015). As a result, ubiquitinated inclusions become endowed with a diverse set of host defense proteins, ultimately resulting in inclusion rupture and spillage of the inclusion's bacterial contents into the host cell cytosol. While the molecular mechanisms underlying inclusion lysis are not yet defined, ubiquitin systems appear to be essential for the execution of this novel host defense pathway (Haldar et al. 2015). The fate of C. trachomatis RBs and EBs ejected from inclusions into the host cytosol is also unclear but may involve capture and degradation through a xenophagic pathway (Al-Zeer et al. 2009).
While C. trachomatis is highly susceptible to IRG-mediated cell-autonomous immunity in murine cells, closely related C. muridarum is resistant to cell-intrinsic defense pathways orchestrated by IRGs. This resistance is founded in the ability of rodent-adapted C. muridarum to actively block the accumulation of GKS proteins at its surrounding inclusion membranes (Coers et al. 2008). The mechanism by which C. muridarum interferes with GKS function and the bacterial virulence factors responsible for immune evasion are currently unknown. Because C. trachomatis and C. muridarum are highly syntenic and display high genomic diversity only within a region of the bacterial chromosome termed the plasticity zone (PZ), genes within the C. muridarum PZ including the toxin-encoding gene tc0438 were attractive candidates to be involved in IRG evasion (Nelson et al. 2005). However, a careful mutational analysis of C. muridarum PZ genes including tc0438 strongly argued against a role for PZ-encoded factors in IRG evasion (Rajaram et al. 2015).
Because GKS proteins solicit ubiquitin systems to the inclusion, C. muridarum resists ubiquitination of its inclusion by interfering with GKS function (Haldar et al. 2015). This difference between C. trachomatis and C. muridarum may have far-reaching implications for the study of adaptive immunity to Chlamydia infections, as it was recently shown that ubiquitin tagging of Toxoplasma gondii PVs affects antigen presentation and CD8+ T-cell activation (Lee et al. 2015). Future work will need to address whether human cells are equipped with the cell-intrinsic capacity to label inclusions with ubiquitin. Because human cells lack GKS proteins entirely (Bekpen et al. 2005), the attachment of ubiquitin tags to inclusions in human cells would require a convergent but GKS-independent system. Such a human-specific ubiquitination system may indeed exist, as it was recently shown that T. gondii PVs become ubiquitin-decorated in IFNγ-primed human epithelial cells (Coers and Haldar 2015; Selleck et al. 2015).
The enzyme indole-2,3-dioxygenase provides nutritional immunity in human cells
In light of the importance of IRG-mediated immunity for controlling C. trachomatis infection in mice, it may seem surprising that an IFNγ-inducible IRG system appears to be lacking in humans. As mentioned before, mice express about 20 separate IRG proteins upon IFNγ stimulation but human epithelial cells express only a single IRG protein known as IRGM (Bekpen et al. 2005). Human IRGM expression, however, is not induced by IFNγ, but instead is constitutively driven off a retroviral element, albeit at low levels. In comparison to its mouse orthologs, the human IRGM protein is severely truncated at both the N- and C-termini implying some degree of functional divergence between mouse and human IRGM proteins (Bekpen et al. 2005, 2009). Although human IRGM is a functional protein involved in the regulation of autophagy (Singh et al. 2006, 2010; Chauhan, Mandell and Deretic 2015), these findings suggest that human and mouse IRGs are sufficiently distinct to dramatically differ in their ability to restrict growth of C. trachomatis and, further, that human and mouse cells may apply divergent strategies to contain or eliminate intracellular infections with C. trachomatis.
Cell-autonomous host defense to Chlamydia is indeed substantially different between mouse and human cells. This concept is supported by the finding that IFNγ-primed human but not mouse cells restrict Chlamydia growth through the induction of indole-2,3-dioxygenase (IDO)-mediated tryptophan depletion. Because C. trachomatis is a natural tryptophan auxotroph, tryptophan depletion by IDO restricts intracellular growth of C. trachomatis (Byrne, Lehmann and Landry 1986; Nelson et al. 2005; Roshick et al. 2006). However, it is less clear whether IDO-mediated nutritional immunity ultimately benefits the human host or the bacterial intruder.
Chlamydia trachomatis is highly adapted to the human IDO pathway. In response to IDO-induced tryptophan starvation, C. trachomatis undergoes dramatic physiological and morphological changes, and enters a state of persistence (Beatty, Byrne and Morrison 1993; Kane et al. 1999; MacKenzie et al. 2007; Wyrick 2010). As C. trachomatis enters persistence, the bacterium upregulates a partial trp operon enabling it to consume exogenous indole—most likely originating from the genital microbiome—and to produce enough tryptophan to survive in the persistent state (Caldwell et al. 2003; Wyrick 2010; Aiyar et al. 2014). Therefore, C. trachomatis is able to enter, survive and endure in the persistent state for as long as host cells remain in the IFNγ-primed state characterized by diminished intracellular tryptophan stores. As we will discuss later in more detail, this persistent state may play an important role in the establishment of chronic C. trachomatis infections (Fig. 3).
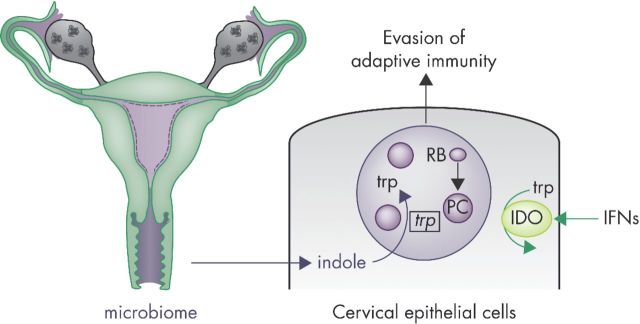
Figure 3.
Chlamydia trachomatis responds to changes in the tryptophan metabolism of IFNγ-primed epithelial cells of the female genital tract. Chlamydia trachomatis colonizes columnar epithelial cells of the endocervix following infection of the female genital tract. Infections induce the expression of lymphocyte-derived IFNγ, which primes human epithelial cells to induce the expression of the tryptophan-catabolizing enzyme IDO. Depletion of intracellular tryptophan stores by IDO instructs C. trachomatis RBs to undergo a dramatic morphological transformation into ‘persister cells’ (PCs). PCs halt replication and drastically change their cell physiology. While still an untested hypothesis, these substantial changes may allow PCs to evade adaptive immune responses. As one of their adaptations to IFNγ priming, PCs upregulate expression of the trp operon and thereby activate an indole-scavenging pathway. Consuming indole—most likely derived from the vaginal mircobiome—enables C. trachomatis PCs to survive tryptophan-spent conditions within its host cell.
This interaction between host IDO and the C. trachomatis trp operon is unique to human infections and absent from the interaction between the murine host and its host-adapted pathogen C. muridarum. Mouse genital epithelial cells express little to no IDO, even when primed with IFNγ, and accordingly C. muridarum experienced no selective pressure to acquire the ability to generate tryptophan de novo (Nelson et al. 2005; Roshick et al. 2006; Coers, Starnbach and Howard 2009; Abdelsamed, Peters and Byrne 2013). Instead, C. muridarum evolved to cope with IRG responses unique to the rodent host. Therefore, the ability of C. muridarum to counteract murine IRG immunity and the ability of C. trachomatis to co-op the human IDO response beautifully illustrate how two closely related Chlamydia species are specifically adapted to their respective host species. These findings also highlight the importance of studying specific adaptations of C. trachomatis to its human host.
Nitric oxide synthase mediates macrophage immunity to Chlamydia trachomatis infections
Chlamydia trachomatis predominantly infects epithelial cells. Vaginal infections with serovars D-K can spread from columnar epithelial cells of the endocervix to the fallopian tubes or the ovaries but typically remain localized to the genital tract. The LGV serovars L1, L2 and L3 on the other hand are invasive and can cause systemic infections by spreading through the lymphatic drainage system (Belland, Ojcius and Byrne 2004). While all genital C. trachomatis strains are adapted to grow and survive inside epithelial cells, only LGV strains are also adapted to survive inside macrophages (Mabey and Peeling 2002; Belland, Ojcius and Byrne 2004). Survival of C. trachomatis LGV strains inside macrophages has been linked to dissemination of infection, chronic infections and tolerance towards antibiotic treatment (Ramsey et al. 2001a; de Vries et al. 2009). Therefore, macrophages appear to be equipped with additional cell-autonomous defense mechanisms that are lacking from epithelial cells. These macrophage-specific defense mechanisms are presumably more effective against non-LGV than LGV strains.
One critical mediator of macrophage immunity against Chlamydia infections is nitric oxide synthase 2 (NOS2). It has been demonstrated that NOS2 restricts the dissemination of genital C. muridarum infections and also limits infection-induced pathology in mice (Ramsey et al. 2001a). NOS2 synthesizes nitric oxide (NO), a noxious gas with both antimicrobial and immunemodulatory functions. In combination with ROS, NO forms reactive nitrogen species (RNS), which react with proteins and thereby directly damage bacteria engulfed within NOS2-positive phagosomes (Bogdan 2015). NOS2-dependent killing of Chlamydia inside macrophages also requires the function of the lysosomal enzyme cathepsin B, which acts upsteam of RNS production but may also act synergistically with RNS in Chlamydia killing (Rajaram and Nelson 2015). The increased dissemination rates observed in NOS2-deficient mice could therefore be a direct consequence of diminished killing of Chlamydia inside NOS2-deficient macrophages.
In addition to murine macrophages, IFNγ-primed murine epithelial cells also restrict growth of C. trachomatis in a NO-dependent manner (Igietseme et al. 1996, 1997). The importance of NO in cell-autonomous immunity to C. trachomatis in human cells is less clear. Infections with C. trachomatis result in undetectable to moderate induction of NO production in a variety of human cells types, and pharmacological inhibition of NOS2 function has no significant effect on C. trachomatis replication in human cells (Roshick et al. 2006; Agrawal et al. 2011). The limited role for NOS2 in human cell-autonomous resistance to C. trachomatis could be due to the ability of C. trachomatis to inhibit the NOS2 pathways in human cells: in human mesenchymal stem cells, C. trachomatis infections induce polyamine synthesis and thereby suppress NOS2 expression and NO production. Blocking the polyamine pathway through the ablation of ornithine decarboxylase function results in an increase in NO production and enhanced restriction of C. trachomatis growth in mesenchymal stem cells (Abu-Lubad, Meyer and Al-Zeer 2014). Whether C. trachomatis can block NO production in other human cell types including macrophages remains to be investigated.
While defects in the bactericidal activity of macrophages could boost inflammation in NOS2-deficient mice due to an increase in bacterial burden, NOS2 deficiency has also been shown to affect inflammation independently of its impact on bacterial burden (Mishra et al. 2013). In addition to its antimicrobial properties, NOS2-produced NO can also regulate macrophage function through the activation of the cyclic GMP-dependent pathway, or through protein S-nitrosylation, a reversible post-translational modification of protein sulfhydryl groups (Bogdan 2015). Specifically, it was shown that IFN priming of macrophages inhibits NLRP3 inflammasome assembly through NOS2-dependent S-nitrosylation of NLRP3 (Hernandez-Cuellar et al. 2012; Mishra et al. 2013). The inactivation of NLRP3 by this pathway plays an import role in limiting the destructive, tissue-damaging effects of chronic M. tuberculosis infections (Mishra et al. 2013). It is conceivable that NOS2-mediated inhibition of NLRP3 assembly could be similarly indispensable in restricting the damaging effects of NLRP3 activation during Chlamydia infections and that the increased immunopathologies observed in C. muridarum-infected NOS2-deficient mice are largely the result of NLRP3 hyperactivation. This intriguing hypothesis postulates that the increased incidence of infection-induced pathologies in C. muridarum-infected NOS2-deficient mice could be reversed by the simultaneous deletion of NLRP3. Future studies need to delineate the relative contribution of NOS2 to anti-Chlamydia host defense in macrophages or other cell types and the importance of NOS2 as an anti-inflammatory factor during C. trachomatis infections.
CELL-AUTONOMOUS IMMUNE RESPONSES REGULATE INFLAMMATION AND ADAPTIVE IMMUNITY TO CHLAMYDIA TRACHOMATIS
The role of inflammasomes in the pathogenesis of Chlamydia-associated disease
As just discussed for NOS2-deficient mice, the activation or inhibition of inflammasomes may dramatically alter the course of Chlamydia infections. This prompts the question as to how inflammasomes regulate immune functions triggered by Chlamydia infections. Following the sensing of microbial PAMPs or infection-induced DAMPs, activated inflammasomes can induce pyroptotic cell death and the release of IL-1α/β and IL-18 (Latz, Xiao and Stutz 2013). The ability of IL-1 and IL-18 receptor signaling to shape downstream immune responses implicates inflammasomes as key regulators of chlamydial disease outcome.
Recent studies have identified IL-1 receptor (IL-1R) signaling as a determinant of Chlamydia infection-associated disease. Mice-lacking IL-1R expression exhibit increased bacterial shedding during the course of urogenital C. muridarum infections compared to wild-type mice. Conversely, mice deficient for the endogenous IL-1R antagonist clear infections more rapidly than wild-type mice. These results highlight a role for IL-1R signaling in controlling pathogen clearance. Importantly, loss of IL-1R signaling additionally leads to decreased incidence of Chlamydia-induced sequelae (Nagarajan et al. 2012). This suggests that while IL-1R-dependent mechanisms promote clearance, they also result in collateral tissue damage. This is consistent with the prevalent model suggesting that host-driven inflammation is responsible for chlamydial disease.
IL-1R signaling can be activated by either IL-1α or IL-1β. Mice deficient for IL-1β develop higher chlamydial burden and less pathology similar to IL-1R-deficient mice, suggesting that IL-1β-initiated signaling drives the observed IL-1R-dependent phenotypes (Prantner et al. 2009). However, these observations do not exclude a role for IL-1α in the pathogenesis of chlamydial disease, as synergistic activity of IL-1α and IL-1β may be responsible for the full extent of the observed immunopathologies.
How does IL-1R signaling dictate the course of infection and disease outcome? IL-1R signaling both shapes T-cell responses and controls innate immune cell recruitment. Specifically, loss of IL-1R is associated with decreased infiltration of CD45+ Ly6G+ F4/80− polymorphonuclear cells (PMNs) into the uterus throughout the course of genital C. muridarum infections (Nagarajan et al. 2012). The decreased recruitment of PMNs likely contributes to the diminished histopathology observed in IL-1R-deficient mice. This model is supported by the observations that neutropenic mice infected with C. muridarum exhibited decreased inflammatory damage while clearing infections at normal rates (Lee et al. 2010). Nonetheless, whether IL-1R signaling contributes to disease solely through the recruitment of PMNs remains unclear. The reported cytopathic effects of IL-1R signaling on epithelial cells could additionally contribute to Chlamydia-induced pathologies; it was shown that human ex vivo fallopian tube organ cultures displayed fewer markers of tissue damage following C. trachomatis infections when treated with exogenous IL-1R antagonist (Hvid et al. 2007). Conceivably, IL-1R signaling could exert its effects on tissue inflammation in a dual manner by promoting the recruitment of PMNs to the site of infection while also directly acting on epithelial cells. The recruited PMNs themselves could act as source of additional IL-1 cytokine release, ultimately resulting in a feed-forward mechanism and enhanced tissue destruction (Lapointe et al. 2010). Together, these results support a model in which PMNs are required for the induction of IL-1R-dependent immunopathologies but dispensable for IL-1R-dependent immune clearance of Chlamydia infections.
Whereas IL-1R-mediated immunopathologies involve PMNs, the impact of IL-1R signaling on infection burden could be due to changes in the CD4+ T-cell response. IL-1α and IL-1β have both been reported to directly enhance antigen-dependent CD4+ T-cell expansion and differentiation (Ben-Sasson et al. 2009). In one C. muridarum infection study, loss of IL-1R resulted in decreased CD4+ cell numbers at 20 days post-infection (dpi) in oviducts. However, similar changes in CD4+ T-cell numbers were not observed at 10 dpi, when differences in bacterial burden became first apparent (Nagarajan et al. 2012). Therefore, changes in absolute CD4+ T-cell numbers cannot fully explain the role of IL-1R signaling in promoting clearance of C. muridarum infections. A non-mutually exclusive mode of action for shaping the adaptive immune response is the ability of IL-1R signaling to affect T-cell polarization. Inflammasome-dependent IL-1α/β production can favor a Th17-skewed response in a variety of contexts (Acosta-Rodriguez et al. 2007; Rao, Tracey and Pober 2007). IL-17 receptor-deficient mice exhibit a suppressed Th1 response to genital C. muridarum infections accompanied by a decrease in local IFNγ secretion (Scurlock et al. 2011). Therefore, changes in the polarization of the T-cell responses provide an attractive framework to account for the role of IL-1R signaling in the clearance of Chlamydia infections. However, alternative mechanisms should also be considered in future studies.
The adaptor protein ASC is required for canonical inflammasome assembly and inflammasome-dependent IL-1β production (Latz, Xiao and Stutz 2013). Similar to IL-1R deficiency, loss of ASC expression results in increased burden in C. muridarum-infected mice. This phenotype is not evident until ∼10 dpi, suggesting that observed changes in bacterial burden result from a diminished adaptive immune response. In contrast to IL-1R signaling deficiencies, however, loss of ASC expression has no impact on Chlamydia-induced pathologies (Nagarajan et al. 2012).
Why do mice deficient in either ASC or in IL-1R display distinct phenotypes? While ASC is required for the release of IL-1 through the activity of canonical inflammasomes, secreted IL-1 can also originate from other sources. For example, neutrophil-derived serine proteases are capable of processing IL-1β, thus facilitating IL-1β secretion (Netea et al. 2010). Some observations suggest that neutrophils could supply IL-1β independently of ASC during Chlamydia infections in vivo: mice lacking ASC exhibit only a partial decrease in IL-1β levels accompanied by an increase in neutrophil infiltration (Nagarajan et al. 2012). Therefore, increased neutrophil-derived IL-1β secretion in ASC-deficient mice could be compensating for the defect in inflammasome-dependent IL-1β production in the same animals.
Another factor that could account for the increased susceptibility of ASC-deficient mice is IL-18. IL-18 is a macrophage-derived cytokine that signals through the IL-18 receptor and activates T-cell immunity (Sedimbi, Hagglof and Karlsson 2013). ASC-containing canonical inflammasomes produce IL-18 in response to Chlamydia infections (Finethy et al. 2015) and IL-18 is produced in vivo in an ASC-dependent manner in C. muridarum-infected mice (Nagarajan et al. 2012). However, the importance of IL-18 in the pathogenesis of Chlamydia infections has not yet been explored. An attractive and testable model evokes synergistic activities between IL-1 and IL-18 cytokines in shaping the adaptive immune response essential for sterilizing immunity.
While an atypical caspase-8-dependent pathway also exists, caspase-1 is essential for pro-IL-1β and pro-IL18 processing in most contexts (Monie and Bryant 2015). Mice deficient for both caspase-1 and caspase-11 exhibit decreased genital tract pathologies accompanied by no change in bacterial burden during the course of the infection (Cheng et al. 2008). The difference between Casp1/Casp11 double knockout and ASC-deficient mice in resolving Chlamydia infections is unexpected considering that both capase-1 and ASC are required for processing and secretion of IL-1β and IL-18 in Chlamydia-infected macrophages (Finethy et al. 2015). Confounding the interpretation of the in vivo data is the lack of knowledge regarding the contribution of caspase-11 to the outcome of a chlamydial infection in vivo. While caspase-1 and ASC are required for IL-1β and IL-18 production in Chlamydia-infected macrophages, these proteins are dispensable for caspase-11 activity, which is sufficient for the induction of pyroptosis and the associated release of IL-1α (Kayagaki et al. 2011; Finethy et al. 2015). In order to solve the caspase-1 conundrum, and to fully understand the role of canonical and non-canonical inflammasomes in dictating chlamydial immunity and disease progression, future studies need to examine the in vivo function of caspase-1 and caspase-11 individually.
On the role of host IDO in the establishment of persistent Chlamydia trachomatis infections
Immunopathologies are associated with persistent or recurring C. trachomatis infections in humans, and likely occur as a consequence of the continuous stimulation of immune sensors such as TLR2 or inflammasomes. Therefore, the prevention of persistent C. trachomatis infections could help to substantially reduce medical complications associated with genital C. trachomatis infections. Currently, it is not understood how C. trachomatis is able to establish long-lasting infections in a subset of patients. However, as hypothesized previously (Caldwell et al. 2003; Nelson et al. 2005; Coers, Starnbach and Howard 2009; Aiyar et al. 2014), the unique adaptation of genital C. trachomatis strains to the human IDO-driven immune response could be playing an important part in the establishment of chronic C. trachomatis infections within the environment of the female genital tract.
In order to cause persistent infections, a microbe such as Chlamydia must be able to prevent clearance by the adaptive branch of the immune system. Two simple models may explain how C. trachomatis can survive the adaptive immune response. In the first model, bacterial replication and the clearance of C. trachomatis by the adaptive immune response occur at similar rates, thus creating a stalemate. In the second model, C. trachomatis exists in two subpopulations: a replicating form that is most abundant during the acute phase of the infection but susceptible to the adaptive immune response and a non-replicating form (‘persister’) that is either immunologically invisible or resistant to the adaptive immune response. According to the latter model, activation of the adaptive immune response would result in the removal of the more abundant, replicating forms of C. trachomatis. The drastic reduction in bacterial burden would be a contributing factor to the contraction of the adaptive immune response. Once host immunity wanes, non-replicating persister cells could revert to the replicating form and expand until an increase in microbial numbers triggers a renewed immune response. In support of this model, so called ‘dormant’, ‘quiescent’ or ‘persistent’ bacterial subpopulations have been described for several pathogenic bacterial species that cause chronic infections including C. trachomatis (Lewis 2010; Wyrick 2010).
The existence of a persistent form of C. trachomatis has been extensively demonstrated in tissue culture model, but there is as yet no in vivo model that recapitulates the epidemiology and clinical data of C. trachomatis infections. However, key aspects of persistence can be replicated in human tissue culture cells. Cultured human epithelial cells can be infected with EBs, which differentiate into larger, metabolically active and replicating RBs. After several rounds of replication inside the inclusion, RBs differentiate back into EBs, which are then released from the spent host cell and able to infect neighboring cells. This developmental cycle is disrupted when C. trachomatis is exposed to environmental stresses like IDO-induced tryptophan starvation. In response to tryptophan starvation, the natural tryptophan auxotroph C. trachomatis transforms from an active, replicating, state into a persistent form (Wyrick 2010). Growth of these organisms slows dramatically, yet they endure as persister cells until tryptophan becomes available again
Chlamydia trachomatis persister cells can survive in the presence of limiting amounts of tryptophan. Severe tryptophan starvation, however, is ultimately bactericidal to C. trachomatis (Leonhardt et al. 2007). Therefore, in adaptation to the human IDO response, genital strains of C. trachomatis express a tryptophan synthase that can use exogenous indole for the synthesis of tryptophan (Caldwell et al. 2003). The ability of C. trachomatis to salvage exogenous indole may provide C. trachomatis with a lifeline to endure IDO-mediated tryptophan starvation in vivo. Because neither human cells nor C. trachomatis produces indole, the genital microbiota is the only possible source of indole in vivo. However, the most abundant member of the genital microbiota, Lactobacillus, is not an indole producer. Therefore, when the population of Lactobacillus collapses, as it does during BV, indole-producing bacteria including Prevotella species, Bacteriodes species and Escherichia coli have an opportunity to expand in numbers (Onderdonk, Delaney and Fichorova 2016), possibly leading to increased vaginal indole levels. In support of this hypothesis, positive correlations between BV and rates of C. trachomatis infections have been reported (Wiesenfeld et al. 2003; Yoshimura et al. 2009). Differences in the vaginal microbiome—either inherent to the individual or induced by BV—may also help explain why some women are at greater risk to develop persistent infections than others. To further substantiate or refute the hypothesis that C. trachomatis co-opts the human cell-autonomous IDO response to establish persistent infections, future studies need to examine how genital indole concentrations and vaginal microbiome dynamics correlate with C. trachomatis infections in human patients. Further, the development of experimental animal models to examine the relationship between IDO, the genital microbiome and the course of C. trachomatis infections would provide a major advance in our ability to dissect and ultimately understand C. trachomatis induced diseases of the genital tract.
CONCLUSIONS AND FUTURE DIRECTIONS
Progress in understanding chronic Chlamydia trachomatis infection has been severely hampered by the lack of small animal models that recapitulate disease as it manifests itself in humans. Unlike humans, laboratory mice rapidly clear experimentally induced genital infections with C. trachomatis and do not develop significant disease phenotypes (Perry et al. 1999). If it were possible to model elements of human C. trachomatis pathogenesis in mice—with a mouse model of chronic human infection—it would accelerate the study of this disease and the development of therapies to combat it. Our understanding of the immune mechanisms that prevent the establishment of chronic C. trachomatis infections in mice has advanced to a point where we can try to apply this knowledge to create a mouse model of chronic genital infection with C. trachomatis. As a first step towards this goal, mice need to be stripped of the murine IFNγ-induced IRG response that is absent from humans but rapidly clears C. trachomatis from mice, as already described (Coers et al. 2011). As a second step, an inducible human IDO response needs to be inserted into epithelial cells of the murine genital tract, so that IDO-mediated induction of C. trachomatis aberrant forms can be achieved in vivo. This engineered mouse would provide a tool to begin to address the questions of whether and how the expression of IDO in genital epithelial cells and the establishment of chronic infections leads to the development of infection-associated pathologies, hinders the development of an adaptive, sterilizing immune response to C. trachomatis or alters the immune response to other pathogens or self-antigens. Using gnotobiotic mouse technology and controlled association of mice with defined microbial communities would further test the idea that the composition of the microbiota of the genital tract is a critical factor in modulating the course of an infection with C. trachomatis. This concept is based on the previous observations that C. trachomatis can survive IDO-mediated tryptophan starvation in the presence of indole and that the only possible source of indole are other microbes.
The interplay between the human IDO response and the indole-scavenging pathway employed by C. trachomatis persister cells exemplifies the importance of cell-autonomous immunity and microbial adaptations to these cell-intrinsic host defense pathways in defining the outcome of C. trachomatis infections. We can expect that many equally important interactions between C. trachomatis and the cell-autonomous defense system of its human host must exist, many of which may still be unknown. For example, a recent report identified perforin-2 as a novel effector molecule that provides cell-autonomous immunity to C. trachomatis infections in macrophages (Fields et al. 2013). Perforin-2 contains a membrane attack complex/perforin domain also found in complement protein C9 and has pore-forming properties (McCormack et al. 2015). Whether and how perforin-2 inserts bactericidal pores into Chlamydia is unknown, but it seems likely that C. trachomatis would have evolved counter-immune strategies to circumvent such a powerful host defense system. The unexplored role of ubiquitin systems in host defense against C. trachomatis infections provides another example of a poorly characterized relationship between C. trachomatis and cell-autonomous immunity. We recently showed that murine cells could label inclusions with ubiquitin and further demonstrated that the rodent-adapted pathogen C. muridarum blocks inclusion ubiquitination in mouse cells (Haldar et al. 2015). Whether a functionally orthologous ubiquitination pathway directed at Chlamydia exists in human cells and whether C. trachomatis can evade ubiquitination of its inclusion in human cells remain to be explored. Investigating the ‘battle over ubiquitin’ between C. trachoamatis and its human host is timely, as the ubiquitination of PVs has been linked to antigen presentation and CD8 + T-cell expansion (Lee et al. 2015). Addressing these types of questions will dramatically advance our understanding of cell-autonomous immunity to C. trachomatis, will reveal novel bacterial virulence strategies and will open up avenues towards the development of innovative treatment strategies and vaccine designs.
Acknowledgments
The authors would like to thank Danielle Pillla-Moffett and Chris Moffett for their assistance in generating some of the artwork for this manuscript.
FUNDING
This work was supported by a National Science Foundation predoctoral award (to RF) and by the National Institute Health grant R01AI103197 (to JC).
Conflict of interest. None declared.
REFERENCES
- Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–59. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Abdelsamed H, Peters J, Byrne GI. Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol. 2013;8:1129–46. doi: 10.2217/fmb.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Sater AA, Koo E, Hacker G, et al. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem. 2009;284:26789–96. doi: 10.1074/jbc.M109.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Sater AA, Said-Sadier N, Padilla EV, et al. Chlamydial infection of monocytes stimulates IL-1beta secretion through activation of the NLRP3 inflammasome. Microbes Infect. 2010;12:652–61. doi: 10.1016/j.micinf.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Sater AA, Tattoli I, Jin L, et al. Cyclic-di-GMP and cyclic-di-AMP activate the NLRP3 inflammasome. EMBO Rep. 2013;14:900–6. doi: 10.1038/embor.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Lubad M, Meyer TF, Al-Zeer MA. Chlamydia trachomatis inhibits inducible NO synthase in human mesenchymal stem cells by stimulating polyamine synthesis. J Immunol. 2014;193:2941–51. doi: 10.4049/jimmunol.1400377. [DOI] [PubMed] [Google Scholar]
- Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, et al. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–9. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- Agrawal T, Bhengraj AR, Vats V, et al. Expression of TLR 2, TLR 4 and iNOS in cervical monocytes of Chlamydia trachomatis-infected women and their role in host immune response. Am J Reprod Immunol. 2011;66:534–43. doi: 10.1111/j.1600-0897.2011.01064.x. [DOI] [PubMed] [Google Scholar]
- Aiyar A, Quayle AJ, Buckner LR, et al. Influence of the tryptophan-indole-IFNgamma axis on human genital Chlamydia trachomatis infection: role of vaginal co-infections. Front Cell Infect Microbiol. 2014;4:72. doi: 10.3389/fcimb.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zeer MA, Al-Younes HM, Braun PR, et al. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One. 2009;4:e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zeer MA, Al-Younes HM, Lauster D, et al. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9:50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, et al. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. mBio. 2013;4:e00018–3. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas S, Neff L, Vuillet M, et al. The proinflammatory cytokine response to Chlamydia trachomatis elementary bodies in human macrophages is partly mediated by a lipoprotein, the macrophage infectivity potentiator, through TLR2/TLR1/TLR6 and CD14. J Immunol. 2008;180:1158–68. doi: 10.4049/jimmunol.180.2.1158. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Byrne GI, Morrison RP. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. P Natl Acad Sci USA. 1993;90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Hunn JP, Rohde C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C, Marques-Bonet T, Alkan C, et al. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R, Ojcius DM, Byrne GI. Chlamydia. Nat Rev Microbiol. 2004;2:530–1. doi: 10.1038/nrmicro931. [DOI] [PubMed] [Google Scholar]
- Ben-Sasson SZ, Hu-Li J, Quiel J, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. P Natl Acad Sci USA. 2009;106:7119–24. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JJ, Gallo RL. Protecting the boundary: the sentinel role of host defense peptides in the skin. Cell Mol Life Sci. 2011;68:2189–99. doi: 10.1007/s00018-011-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Balsara ZR, Ulmer W, et al. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes Immun. 2006a;7:122–9. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Coers J, Balsara ZR, et al. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. P Natl Acad Sci USA. 2006b;103:14092–7. doi: 10.1073/pnas.0603338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F, Legnani D, Lombardo VM, et al. Chlamydia pneumoniae infection in acute exacerbations of COPD. Eur Respir J. 1993;6:19–22. [PubMed] [Google Scholar]
- Boehm U, Klamp T, Groot M, et al. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–95. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–78. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16:339–48. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Brade L, Zych K, Rozalski A, et al. Structural requirements of synthetic oligosaccharides to bind monoclonal antibodies against Chlamydia lipopolysaccharide. Glycobiology. 1997;7:819–27. doi: 10.1093/glycob/7.6.819. [DOI] [PubMed] [Google Scholar]
- Brankovic I, van Ess EF, Noz MP, et al. NOD1 in contrast to NOD2 functional polymorphism influence Chlamydia trachomatis infection and the risk of tubal factor infertility. Pathog Dis. 2015;73:1–9. doi: 10.1093/femspd/ftu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–91. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut Y, Faure E, Thomas L, et al. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol. 2002;168:1435–40. doi: 10.4049/jimmunol.168.3.1435. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Byrne GI, Lehmann LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect Immun. 1986;53:347–51. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest. 2003;111:1757–69. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Warner N, Inohara N, et al. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507–21. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Shivshankar P, Li Z, et al. Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun. 2008;76:515–22. doi: 10.1128/IAI.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi TY, Kim DA, Kim SK, et al. Prevalence of specific antibodies to Chlamydia pneumoniae in Korea. J Clin Microbiol. 1998;36:3426–8. doi: 10.1128/jcm.36.11.3426-3428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J. Self and non-self discrimination of intracellular membranes by the innate immune system. PLoS Pathog. 2013;9:e1003538. doi: 10.1371/journal.ppat.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Bernstein-Hanley I, Grotsky D, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol. 2008;180:6237–45. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- Coers J, Gondek DC, Olive AJ, et al. Compensatory T Cell Responses in IRG-Deficient Mice Prevent Sustained Chlamydia trachomatis Infections. PLoS Pathog. 2011;7:e1001346. doi: 10.1371/journal.ppat.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Haldar AK. Ubiquitination of pathogen-containing vacuoles promotes host defense to Chlamydia trachomatis and Toxoplasma gondii. Commun Integr Biol. 2015;8:e1115163. doi: 10.1080/19420889.2015.1115163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Starnbach MN, Howard JC. Modeling infectious disease in mice: co-adaptation and the role of host-specific IFNgamma responses. PLoS Pathog. 2009;5:e1000333. doi: 10.1371/journal.ppat.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, et al. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J Infect Dis. 2010;201(Suppl 2):S114–25. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T, O'Neill JM, Andrews CW, Jr, et al. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol. 2003;171:6187–97. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- de Vries HJ, Smelov V, Middelburg JG, et al. Delayed microbial cure of lymphogranuloma venereum proctitis with doxycycline treatment. Clin Infect Dis. 2009;48:e53–56. doi: 10.1086/597011. [DOI] [PubMed] [Google Scholar]
- Derbigny WA, Hong SC, Kerr MS, et al. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect Immun. 2007;75:1280–90. doi: 10.1128/IAI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbigny WA, Kerr MS, Johnson RM. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J Immunol. 2005;175:6065–75. doi: 10.4049/jimmunol.175.9.6065. [DOI] [PubMed] [Google Scholar]
- Derbigny WA, Shobe LR, Kamran JC, et al. Identifying a role for Toll-like receptor 3 in the innate immune response to Chlamydia muridarum infection in murine oviduct epithelial cells. Infect Immun. 2012;80:254–65. doi: 10.1128/IAI.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–11. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, et al. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, McCormack R, de Armas LR, et al. Perforin-2 restricts growth of Chlamydia trachomatis in macrophages. Infect Immun. 2013;81:3045–54. doi: 10.1128/IAI.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finethy R, Jorgensen I, Haldar AK, et al. Guanylate Binding Proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun. 2015;83:4740–9. doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondek DC, Olive AJ, Stary G, et al. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol. 2012;189:2441–9. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondek DC, Roan NR, Starnbach MN. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. J Immunol. 2009;183:1313–9. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–87. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner LM. Pathogenesis of fallopian tube damage caused by Chlamydia trachomatis infections. Contraception. 2015;92:108–15. doi: 10.1016/j.contraception.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Haggerty CL, Gottlieb SL, Taylor BD, et al. Risk of sequelae after Chlamydia trachomatis genital infection in women. J Infect Dis. 2010;201(Suppl 2):S134–55. doi: 10.1086/652395. [DOI] [PubMed] [Google Scholar]
- Hahn DL, Schure A, Patel K, et al. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS One. 2012;7:e35945. doi: 10.1371/journal.pone.0035945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Foltz C, Finethy R, et al. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. P Natl Acad Sci USA. 2015;112:E5628–37. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Piro AS, Pilla DM, et al. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9:e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar AK, Saka HA, Piro AS, et al. IRG and GBP host resistance factors target aberrant, “non-self” vacuoles characterized by the missing of “self” IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine H, Gronow S, Zamyatina A, et al. Investigation on the agonistic and antagonistic biological activities of synthetic Chlamydia lipid A and its use in in vitro enzymatic assays. J Endotoxin Res. 2007;13:126–32. doi: 10.1177/0968051907079122. [DOI] [PubMed] [Google Scholar]
- Hernandez-Cuellar E, Tsuchiya K, Hara H, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J Immunol. 2012;189:5113–7. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- Hooper LV. Epithelial cell contributions to intestinal immunity. Adv Immunol. 2015;126:129–72. doi: 10.1016/bs.ai.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Peng L, Kwak YT, et al. The DNA sensor AIM2 maintains intestinal homeostasis via regulation of epithelial antimicrobial host defense. Cell Reps. 2015;13:1922–36. doi: 10.1016/j.celrep.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, Heath RJ, Begun J, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–90. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunn JP, Koenen-Waisman S, Papic N, et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvid M, Baczynska A, Deleuran B, et al. Interleukin-1 is the initiator of Fallopian tube destruction during Chlamydia trachomatis infection. Cell Microbiol. 2007;9:2795–803. doi: 10.1111/j.1462-5822.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. P Natl Acad Sci USA. 2007;104:11430–5. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme JU, Uriri IM, Chow M, et al. Inhibition of intracellular multiplication of human strains of Chlamydia trachomatis by nitric oxide. Biochem Bioph Res Co. 1997;232:595–601. doi: 10.1006/bbrc.1997.6335. [DOI] [PubMed] [Google Scholar]
- Igietseme JU, Uriri IM, Hawkins R, et al. Integrin-mediated epithelial-T cell interaction enhances nitric oxide production and increased intracellular inhibition of Chlamydia. J Leukocyte Biol. 1996;59:656–62. doi: 10.1002/jlb.59.5.656. [DOI] [PubMed] [Google Scholar]
- Ingalls RR, Rice PA, Qureshi N, et al. The inflammatory cytokine response to Chlamydia trachomatis infection is endotoxin mediated. Infect Immun. 1995;63:3125–30. doi: 10.1128/iai.63.8.3125-3130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intemann CD, Thye T, Niemann S, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Johansson M, Schon K, Ward M, et al. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect Immun. 1997;65:1032–44. doi: 10.1128/iai.65.3.1032-1044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AC, Piro A, Clough B, et al. Human GBP1 does not localise to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol. 2016 doi: 10.1111/cmi.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani R, Adachi Y, Takata H, et al. Essential role of Toll-like receptor 2 in macrophage activation by glycogen. Glycobiology. 2012;22:146–59. doi: 10.1093/glycob/cwr122. [DOI] [PubMed] [Google Scholar]
- Kane CD, Vena RM, Ouellette SP, et al. Intracellular tryptophan pool sizes may account for differences in gamma interferon-mediated inhibition and persistence of chlamydial growth in polarized and nonpolarized cells. Infect Immun. 1999;67:1666–71. doi: 10.1128/iai.67.4.1666-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Nan X, Jin MS, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–84. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Kosaka A, Yan H, et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-beta. Immunity. 2013;38:1187–97. doi: 10.1016/j.immuni.2013.02.024. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Keestra-Gounder AM, Byndloss MX, Seyffert N, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532:394–7. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Crowley SM, Sham HP, et al. Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 2014;16:249–56. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kuo CC, Jackson LA, Campbell LA, et al. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–61. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Fukuda EY, Li J, et al. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J Immunol. 2005;174:7186–93. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- Lampe MF, Wilson CB, Bevan MJ, et al. Gamma interferon production by cytotoxic T lymphocytes is required for resolution of Chlamydia trachomatis infection. Infect Immun. 1998;66:5457–61. doi: 10.1128/iai.66.11.5457-5461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe TK, O'Connor PM, Jones NL, et al. Interleukin-1 receptor phosphorylation activates Rho kinase to disrupt human gastric tight junctional claudin-4 during Helicobacter pylori infection. Cell Microbiol. 2010;12:692–703. doi: 10.1111/j.1462-5822.2010.01429.x. [DOI] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Schripsema JH, Sigar IM, et al. A link between neutrophils and chronic disease manifestations of Chlamydia muridarum urogenital infection of mice. FEMS Immunol Med Micr. 2010;59:108–16. doi: 10.1111/j.1574-695X.2010.00668.x. [DOI] [PubMed] [Google Scholar]
- Lee Y, Sasai M, Ma JS, et al. p62 plays a specific role in interferon-gamma-induced presentation of a toxoplasma vacuolar antigen. Cell Rep. 2015;13:223–33. doi: 10.1016/j.celrep.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Leonhardt RM, Lee SJ, Kavathas PB, et al. Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of Chlamydia trachomatis. Infect Immun. 2007;75:5105–17. doi: 10.1128/IAI.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–72. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Li LX, McSorley SJ. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog. 2013;9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LX, McSorley SJ. A re-evaluation of the role of B cells in protective immunity to Chlamydia infection. Immunol Lett. 2015;164:88–93. doi: 10.1016/j.imlet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti GW, Kuru E, Hall E, et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–10. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien E, Sellati TJ, Yoshimura A, et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–25. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- Lu H, Shen C, Brunham RC. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J Immunol. 2000;165:1463–9. doi: 10.4049/jimmunol.165.3.1463. [DOI] [PubMed] [Google Scholar]
- Mabey D, Peeling RW. Lymphogranuloma venereum. Sex Transm Infect. 2002;78:90–92. doi: 10.1136/sti.78.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack R, de Armas LR, Shiratsuchi M, et al. Inhibition of intracellular bacterial replication in fibroblasts is dependent on the perforin-like protein (perforin-2) encoded by macrophage-expressed gene 1. J Innate Immun. 2013;5:185–94. doi: 10.1159/000345249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack RM, de Armas LR, Shiratsuchi M, et al. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife. 2015;4:e06508. doi: 10.7554/eLife.06508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie CR, Heseler K, Muller A, et al. Role of indoleamine 2,3-dioxygenase in antimicrobial defence and immuno-regulation: tryptophan depletion versus production of toxic kynurenines. Curr Drug Metab. 2007;8:237–44. doi: 10.2174/138920007780362518. [DOI] [PubMed] [Google Scholar]
- Maltez VI, Tubbs AL, Cook KD, et al. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity. 2015;43:987–97. doi: 10.1016/j.immuni.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16:467–75. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–6. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari P, Toussi DN, Tifrea DF, et al. Toll-like receptor 2-dependent activity of native major outer membrane protein proteosomes of Chlamydia trachomatis. Infect Immun. 2013;81:303–10. doi: 10.1128/IAI.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Izutsu H, Miyashita N, et al. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol. 1998;36:3013–9. doi: 10.1128/jcm.36.10.3013-3019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Dick MS, Dreier RF, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–70. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16:476–84. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Li G, Zhang X, et al. A TRP channel senses lysosomen by pathogens to trigger their expulsion. Cell. 2015;161:1306–19. doi: 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BB, Rathinam VA, Martens GW, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Bryant CE. Caspase-8 functions as a key mediator of inflammation and pro-IL-1beta processing via both canonical and non-canonical pathways. Immunol Rev. 2015;265:181–93. doi: 10.1111/imr.12284. [DOI] [PubMed] [Google Scholar]
- Morre SA, Murillo LS, Bruggeman CA, et al. The role that the functional Asp299Gly polymorphism in the toll-like receptor-4 gene plays in susceptibility to Chlamydia trachomatis-associated tubal infertility. J Infect Dis. 2003;187:341–2. doi: 10.1086/346044. author reply 342–343. [DOI] [PubMed] [Google Scholar]
- Morrison RP, Feilzer K, Tumas DB. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun. 1995;63:4661–8. doi: 10.1128/iai.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SG, Su H, Caldwell HD, et al. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun. 2000;68:6979–87. doi: 10.1128/iai.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlradt PF, Kiess M, Meyer H, et al. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–8. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy AK, Li W, Chaganty BK, et al. Tumor necrosis factor alpha production from CD8+ T cells mediates oviduct pathological sequelae following primary genital Chlamydia muridarum infection. Infect Immun. 2011;79:2928–35. doi: 10.1128/IAI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas I. Female genital Chlamydia trachomatis infection: where are we heading? Arch Gynecol Obstet 20122851271–85. [DOI] [PubMed] [Google Scholar]
- Nagarajan UM, Prantner D, Sikes JD, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect Immun. 2008;76:4642–8. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan UM, Sikes JD, Yeruva L, et al. Significant role of IL-1 signaling, but limited role of inflammasome activation, in oviduct pathology during Chlamydia muridarum genital infection. J Immunol. 2012;188:2866–75. doi: 10.4049/jimmunol.1103461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Virok DP, Wood H, et al. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. P Natl Acad Sci USA. 2005;102:10658–63. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Simon A, van de Veerdonk F, et al. IL-1beta processing in host defense: beyond the inflammasomes. PLoS Pathog. 2010;6:e1000661. doi: 10.1371/journal.ppat.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Cunningham D, Liang X, et al. Lipooligosaccharide is required for the generation of infectious elementary bodies in Chlamydia trachomatis. P Natl Acad Sci USA. 2011;108:10284–9. doi: 10.1073/pnas.1107478108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira CV, Zhang X, Giovannone N, et al. Protective immunity against Chlamydia trachomatis can engage both CD4+ and CD8+ T cells and bridge the respiratory and genital mucosae. J Immunol. 2015;194:2319–29. doi: 10.4049/jimmunol.1402675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CM, AbdelRahman YM, Green E, et al. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun. 2011;79:1044–56. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CM, Ingalls RR, Andrews CW, Jr, et al. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol. 2007;179:4027–34. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- O'Connell CM, Ionova IA, Quayle AJ, et al. Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J Biol Chem. 2006;281:1652–9. doi: 10.1074/jbc.M510182200. [DOI] [PubMed] [Google Scholar]
- O'Connell CM, Nicks KM. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology. 2006;152:1601–7. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- Onderdonk AB, Delaney ML, Fichorova RN. The human microbiome during bacterialvVaginosis. Clin Microbiol Rev. 2016;29:223–38. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouburg S, Spaargaren J, den Hartog JE, et al. The CD14 functional gene polymorphism −260 C >T is not involved in either the susceptibility to Chlamydia trachomatis infection or the development of tubal pathology. BMC Infect Dis. 2005;5:114. doi: 10.1186/1471-2334-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–86. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packiam M, Weinrick B, Jacobs WR, Jr, et al. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. P Natl Acad Sci USA. 2015;112:11660–5. doi: 10.1073/pnas.1514026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–61. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Anderson E, Salva PS, et al. The prevalence and identity of Chlamydia-specific IgE in children with asthma and other chronic respiratory symptoms. Respir Res. 2012;13:32. doi: 10.1186/1465-9921-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry LL, Su H, Feilzer K, et al. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-gamma-mediated inhibition. J Immunol. 1999;162:3541–8. [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. P Natl Acad Sci USA. 2014;111:6046–51. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla-Moffett D, Barber MF, Taylor GA, et al. Interferon-inducible GTPases in host resistance, inflammation and disease. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli PA, Amiel E, Schaefer TM, et al. Differential expression of Toll-like receptors 2 and 4 in tissues of the human female reproductive tract. Infect Immun. 2004;72:5799–806. doi: 10.1128/IAI.72.10.5799-5806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184:2551–60. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Darville T, Sikes JD, et al. Critical role for interleukin-1beta (IL-1beta) during Chlamydia muridarum genital infection and bacterial replication-independent secretion of IL-1beta in mouse macrophages. Infect Immun. 2009;77:5334–46. doi: 10.1128/IAI.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Sikes JD, Hennings L, et al. Interferon regulatory transcription factor 3 protects mice from uterine horn pathology during Chlamydia muridarum genital infection. Infect Immun. 2011;79:3922–33. doi: 10.1128/IAI.00140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prebeck S, Brade H, Kirschning CJ, et al. The Gram-negative bacterium Chlamydia trachomatis L2 stimulates tumor necrosis factor secretion by innate immune cells independently of its endotoxin. Microbes Infect. 2003;5:463–70. doi: 10.1016/s1286-4579(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Qu Y, Frazer LC, O'Connell CM, et al. Comparable genital tract infection, pathology, and immunity in rhesus macaques inoculated with wild-type or plasmid-deficient Chlamydia trachomatis serovar D. Infect Immun. 2015;83:4056–67. doi: 10.1128/IAI.00841-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram K, Giebel AM, Toh E, et al. Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun. 2015;83:2870–81. doi: 10.1128/IAI.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram K, Nelson DE. Chlamydia muridarum infection of macrophages elicits bactericidal nitric oxide production via reactive oxygen species and cathepsin B. Infect Immun. 2015;83:3164–75. doi: 10.1128/IAI.00382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Miranpuri GS, Sigar IM, et al. Chlamydia trachomatis persistence in the female mouse genital tract: inducible nitric oxide synthase and infection outcome. Infect Immun. 2001a;69:5131–7. doi: 10.1128/IAI.69.8.5131-5137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Sigar IM, Rana SV, et al. Role for inducible nitric oxide synthase in protection from chronic Chlamydia trachomatis urogenital disease in mice and its regulation by oxygen free radicals. Infect Immun. 2001b;69:7374–9. doi: 10.1128/IAI.69.12.7374-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–6. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DA, Tracey KJ, Pober JS. IL-1alpha and IL-1beta are endogenous mediators linking cell injury to the adaptive alloimmune response. J Immunol. 2007;179:6536–46. doi: 10.4049/jimmunol.179.10.6536. [DOI] [PubMed] [Google Scholar]
- Roan NR, Gierahn TM, Higgins DE, et al. Monitoring the T cell response to genital tract infection. P Natl Acad Sci USA. 2006;103:12069–74. doi: 10.1073/pnas.0603866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Roshick C, Wood H, Caldwell HD, et al. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect Immun. 2006;74:225–38. doi: 10.1128/IAI.74.1.225-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund S, Lindner B, Brade H, et al. Structural analysis of the lipopolysaccharide from Chlamydia trachomatis serotype L2. J Biol Chem. 1999;274:16819–24. doi: 10.1074/jbc.274.24.16819. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Tolker-Nielsen T, Dow JM. When the PilZ don't work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 2012;20:235–42. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sano H, Iwaki D, et al. Direct binding of Toll-like receptor 2 to zymosan, and zymosan-induced NF-kappa B activation and TNF-alpha secretion are down-regulated by lung collectin surfactant protein A. J Immunol. 2003;171:417–25. doi: 10.4049/jimmunol.171.1.417. [DOI] [PubMed] [Google Scholar]
- Schauvliege R, Vanrobaeys J, Schotte P, et al. Caspase-11 gene expression in response to lipopolysaccharide and interferon-gamma requires nuclear factor-kappa B and signal transducer and activator of transcription (STAT) 1. J Biol Chem. 2002;277:41624–30. doi: 10.1074/jbc.M207852200. [DOI] [PubMed] [Google Scholar]
- Scior T, Alexander C, Zaehringer U. Reviewing and identifying amino acids of human, murine, canine and equine TLR4/MD-2 receptor complexes conferring endotoxic innate immunity activation by LPS/lipid A, or antagonistic effects by Eritoran, in contrast to species-dependent modulation by lipid IVa. Comput Struct Biotechnol J. 2013;5:e201302012. doi: 10.5936/csbj.201302012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurlock AM, Frazer LC, Andrews CW, Jr, et al. Interleukin-17 contributes to generation of Th1 immunity and neutrophil recruitment during Chlamydia muridarum genital tract infection but is not required for macrophage influx or normal resolution of infection. Infect Immun. 2011;79:1349–62. doi: 10.1128/IAI.00984-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedimbi SK, Hagglof T, Karlsson MC. IL-18 in inflammatory and autoimmune disease. Cell Mol Life Sci. 2013;70:4795–808. doi: 10.1007/s00018-013-1425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck EM, Orchard RC, Lassen KG, et al. A noncanonical autophagy pathway restricts toxoplasma gondii growth in a strain-specific manner in IFN-gamma-activated human cells. mBio. 2015;6:e01157–15. doi: 10.1128/mBio.01157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy AR, Wellington DA, Kumar P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–5. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–92. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Arditi M. Innate immune responses to Chlamydia pneumoniae infection: role of TLRs, NLRs, and the inflammasome. Microbes Infect. 2012;14:1301–7. doi: 10.1016/j.micinf.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Singh SB, Ornatowski W, Vergne I, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–65. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Carlson JH, Whitmire WM, et al. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun. 2013;81:636–44. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnbach MN, Bevan MJ, Lampe MF. Protective cytotoxic T lymphocytes are induced during murine infection with Chlamydia trachomatis. J Immunol. 1994;153:5183–9. [PubMed] [Google Scholar]
- Starnbach MN, Bevan MJ, Lampe MF. Murine cytotoxic T-lymphocytes induced following Chlamydia trachomatis intraperitoneal or genital tract infection respond to cells infected with multiple serovars. Infect Immun. 1995;63:3527–30. doi: 10.1128/iai.63.9.3527-3530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary G, Olive A, Radovic-Moreno AF, et al. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science. 2015;348:aaa8205. doi: 10.1126/science.aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Myers G, Eppinger M, et al. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Micr. 2009;55:115–9. doi: 10.1111/j.1574-695X.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BD, Darville T, Ferrell RE, et al. Variants in toll-like receptor 1 and 4 genes are associated with Chlamydia trachomatis among women with pelvic inflammatory disease. J Infect Dis. 2012;205:603–9. doi: 10.1093/infdis/jir822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HR, Burton MJ, Haddad D, et al. Trachoma. Lancet. 2014;384:2142–52. doi: 10.1016/S0140-6736(13)62182-0. [DOI] [PubMed] [Google Scholar]
- Tietzel I, El-Haibi C, Carabeo RA. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One. 2009;4:e6499. doi: 10.1371/journal.pone.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver MK, Henry SC, Cantillana V, et al. Immunity-related gtpase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem. 2011;286:30471–80. doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Ahmad-Nejad P, Costa C, et al. Endocytosed HSP60s use toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. J Biol Chem. 2001;276:31332–9. doi: 10.1074/jbc.M103217200. [DOI] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, et al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165:1106–19. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij SP, Karimi O, Pleijster J, et al. TLR2, TLR4 and TLR9 genotypes and haplotypes in the susceptibility to and clinical course of Chlamydia trachomatis infections in Dutch women. Pathog Dis. 2016;74 doi: 10.1093/femspd/ftv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tang J, Geisler WM, et al. Human leukocyte antigen and cytokine gene variants as predictors of recurrent Chlamydia trachomatis infection in high-risk adolescents. J Infect Dis. 2005;191:1084–92. doi: 10.1086/428592. [DOI] [PubMed] [Google Scholar]
- Welter-Stahl L, Ojcius DM, Viala J, et al. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell Microbiol. 2006;8:1047–57. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- Wiesenfeld HC, Hillier SL, Krohn MA, et al. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis. 2003;36:663–8. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- Williams DM, Bonewald LF, Roodman GD, et al. Tumor necrosis factor alpha is a cytotoxin induced by murine Chlamydia trachomatis infection. Infect Immun. 1989;57:1351–5. doi: 10.1128/iai.57.5.1351-1355.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick PB. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis. 2010;201(Suppl 2):S88–95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Bumgarner RE, Lampe MF, et al. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J Infect Dis. 2003;187:424–34. doi: 10.1086/367962. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Okuyama M, Ma JS, et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against toxoplasma gondii. Immunity. 2012;37:302–13. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Yang D, Chen J, Shi C, et al. Autophagy gene polymorphism is associated with susceptibility to leprosy by affecting inflammatory cytokines. Inflammation. 2014;37:593–8. doi: 10.1007/s10753-013-9773-1. [DOI] [PubMed] [Google Scholar]
- Yi G, Brendel VP, Shu C, et al. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLoS One. 2013;8:e77846. doi: 10.1371/journal.pone.0077846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–45. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Yoshimura M, Kobayashi T, et al. Can bacterial vaginosis help to find sexually transmitted diseases, especially chlamydial cervicitis? Int J STD AIDS. 2009;20:108–11. doi: 10.1258/ijsa.2008.008249. [DOI] [PubMed] [Google Scholar]
- Zahringer U, Lindner B, Inamura S, et al. TLR2 - promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology. 2008;213:205–24. doi: 10.1016/j.imbio.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yeruva L, Marinov A, et al. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol. 2014;193:2394–404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]