Abstract
The three-dimensional organization of chromosomes within the nucleus and its dynamics during differentiation are largely unknown. To visualize this process in molecular detail, we generated high-resolution maps of genome – nuclear lamina interactions during subsequent differentiation of mouse embryonic stem cells via lineage-committed neural precursor cells into terminally differentiated astrocytes. This reveals that a basal chromosome architecture present in embryonic stem cells is cumulatively altered at hundreds of sites during lineage commitment and subsequent terminal differentiation. This remodeling involves both individual transcription units and multi-gene regions, and affects many genes that determine cellular identity. Often, genes that move away from the lamina are concomitantly activated; many others however remain inactive yet become unlocked for activation in a next differentiation step. These results suggest that lamina-genome interactions are widely involved in the control of gene expression programs during lineage commitment and terminal differentiation.
Introduction
Embryonic stem cells (ESCs) can give rise to all somatic cell types through the process of differentiation (Hochedlinger and Plath, 2009; Welstead et al., 2008). Each cell type has its own unique gene expression program, and the transition from one cell type to another typically involves the up- and downregulation of hundreds of genes. It is still largely unclear how a repertoire of active and potentially active genes is maintained in a given cell type, and how it is reprogrammed during the transition from one cell type to another.
Control of transcription programs is mediated by three major mechanisms that act in concert. One mechanism is based on transcription factors that bind to specific sequence motifs and thereby regulate particular sets of genes. Several transcription factors have been identified that keep ESCs in their undifferentiated state, and when artificially introduced in certain combinations they can reprogram differentiated cells back to pluripotent cells, albeit with low efficiency (Hochedlinger and Plath, 2009; Welstead et al., 2008). The second regulatory mechanism involves methylation of DNA and posttranslational modifications of histones, which together may provide an epigenetic memory that helps to stabilize the differentiation state of a cell and its progeny (Mohn and Schubeler, 2009; Probst et al., 2009). Indeed, inhibition of DNA methyltransferase or histone deacetylases facilitates the reprogramming of differentiated cells into ESCs (Huangfu et al., 2008; Mikkelsen et al., 2008; Shi et al., 2008). The third major mechanism of gene regulation involves higher-order chromatin organization. Large-scale folding of chromatin may affect gene expression by locating genes to specific nuclear subcompartments that are either stimulatory or inhibitory to transcription (Dekker, 2008; Fraser and Bickmore, 2007). While transcription factors and epigenetic modifications have been extensively studied in relation to the differentiation of ESCs, almost nothing is known about the role of higher-order chromatin organization.
A particularly interesting aspect of chromatin organization is the role of the nuclear lamina (NL), which is a fibrous multi-protein network that lines the nucleoplasmic surface of the inner nuclear membrane in most metazoan cells. Lamins are the major components of the NL. Lamins and a variety of other NL proteins can interact with DNA, histones, transcription factors and chromatin proteins (Prokocimer et al., 2009; Taddei et al., 2004). Close contacts between the NL and chromatin have been observed by electron microscopy (Belmont et al., 1993). Recent genome-wide molecular mapping studies in Drosophila and human cells have identified hundreds of genomic regions that interact with the NL (Guelen et al., 2008; Pickersgill et al., 2006). In human fibroblasts the genome is organized into about 1,300 discrete lamina-associated domains (LADs) that range in size from ~80kb-30Mb and together contain thousands of genes (Guelen et al., 2008). Thus, the NL represents a major structural element for the organization of the genome inside the nucleus.
Evidence is accumulating that the NL is not only an anchoring point for chromatin, but also directly involved in gene repression. In Drosophila and human cells, the vast majority of NL-associated genes are transcriptionally inactive and enriched in repressive histone marks such as H3K27me3 and H3K9me2 (Guelen et al., 2008; Pickersgill et al., 2006; Wen et al., 2009). Depletion of B-type lamin in Drosophila leads to derepression of lamina-associated genes (Shevelyov et al., 2009), and misregulation of gene expression has also been observed in mammalian cells that lack lamins or other NL proteins (Frock et al., 2006; Vergnes et al., 2004). Conversely, elegant experiments in mouse and human cells have shown that artificial tethering of genes to the lamina can cause the downregulation of some (but not all) reporter and endogenous genes (Finlan et al., 2008; Kumaran and Spector, 2008; Reddy et al., 2008). Taken together, these data point to a direct role of the nuclear lamina in gene repression.
These observations raise the question whether and how the contacts between the genome and the NL are remodeled during differentiation. Fluorescence in situ hybridization (FISH) microscopy studies have suggested that certain genomic loci can move to or from the NL depending on the cell type (Kosak et al., 2002; Williams et al., 2006; Zink et al., 2004). However, with its limited spatial resolution (~0.2µm) FISH cannot discriminate molecular contacts of genes with the NL from mere spatial proximity. Moreover, the poor sequence resolution (~50-100kb) of FISH and the inability to visualize more than a few loci in parallel have prevented the construction of detailed models of chromosome reshaping during differentiation. A previous molecular study in Drosophila cells (Pickersgill et al., 2006) also pointed to changes in genome – NL interactions during differentiation, but lacked the resolution required to identify general principles that govern these changes.
Here we report a highly detailed analysis of the changes in the spatial organization of the entire genome during the process of lineage commitment and subsequent terminal differentiation of mouse cells. Specifically, we constructed high-resolution maps of genome-NL contacts in nuclei of mouse ESCs, as well as multipotent Neural Precursor Cells (NPCs) and terminally differentiated astrocytes (ACs) that were sequentially derived from these ESCs. For further comparison, we also analyzed embryonic fibroblasts, which represent a distinct differentiation lineage. The results reveal a highly orchestrated stepwise reorganization of NL-genome interactions upon progressive differentiation, and suggest an important role of the NL in the establishment and switching of gene expression programs during differentiation.
Results
Molecular maps of genome-NL interactions in ESCs and differentiated cells
We constructed genome-wide maps of NL interactions using DamID of Lamin B1. This approach was previously demonstrated to reliably identify NL-associated sequences (Guelen et al., 2008; Pickersgill et al., 2006). Briefly, a fusion protein consisting of mouse LaminB1 and E. coli DNA adenine methyltransferase (Dam) is expressed in cultured mouse cell lines. We confirmed by immunofluorescence confocal microscopy that this fusion protein is correctly integrated into the NL, similar to endogenous Lamin B1 (Figures S1A, S1B). The NL-tethered Dam will adenine-methylate DNA sequences that are in close contact with the NL. Next, genomic DNA is purified, and adenine-methylated fragments are specifically amplified (Vogel et al., 2007) and hybridized to custom-designed oligonucleotide arrays (Gräf et al., 2007) with 2.1 million probes that query the entire mouse genome at median intervals of ~1.2kb. In these hybridizations methylated DNA fragments from cells expressing unfused Dam (which freely diffuses throughout the nucleus) are used as a reference to correct for effects of local DNA accessibility (Greil et al., 2006; Vogel et al., 2007). The resulting Dam-LaminB1 over Dam methylation ratios are taken as a measure for the relative contact frequency of each probed sequence with the NL.
We conducted these mapping experiments in mouse ESCs, as well as NPCs and postmitotic ACs that were sequentially derived from the ESCs by standard protocols (Ying and Smith, 2003). The NPCs were derived from the same population of ESCs on which DamID was performed, and the ACs were subsequently derived twice independently from these NPCs. Thus, the 3 celltypes are isogenic. Each celltype was however separately transduced with Dam and Dam-LaminB1 vectors, to prevent carry-over of adenine-methylation patterns from one differentation stage to the next. In order to assure that these cell types were essentially homogeneous populations of a single differentiation state, we monitored the expression of the NPC marker protein Nestin and the AC marker protein GFAP. This confirmed that the NPC population was 92% Nestin-positive and the AC population was 92% GFAP-positive (Figures S1C, S1D). As a fourth cell type we chose mouse 3T3 embryonic fibroblasts (MEFs), representing a non-neural lineage.
For each cell type full-genome LaminB1 interaction maps were constructed by combining two independent DamID experiments, which were highly correlated (Pearson correlations between replicates ranging from 0.77 to 0.92, Figures S1E, S1F). All single-channel data were quantile-normalized to facilitate comparisons between the cell types. Figure 1A shows a representative chromosomal map for each cell type. Similar to what has been reported for human cells (Guelen et al., 2008), we find that Lamin B1 interactions of the genome occur through hundreds of large domains, ranging in size from 40kb to 15Mb. Detailed analysis of these data will be described below.
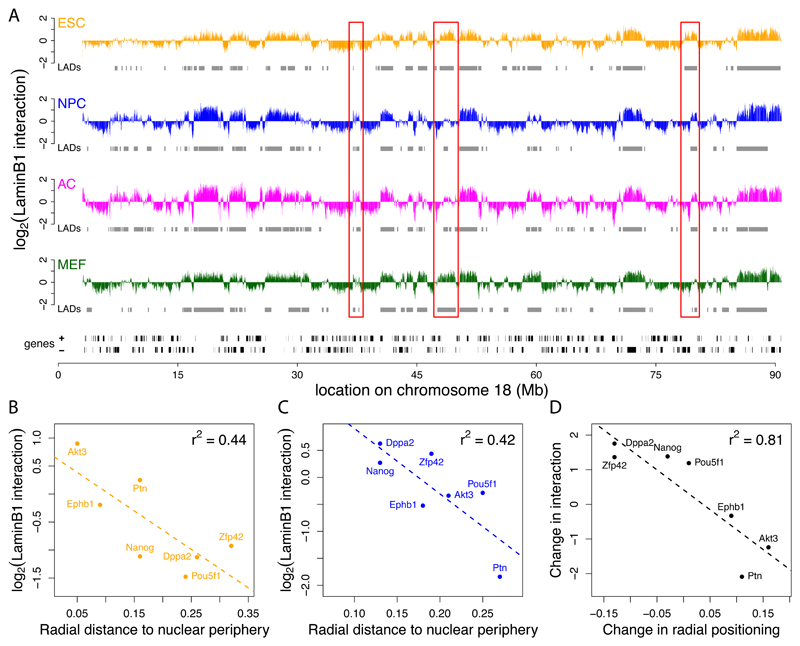
Figure 1. Chromosomal maps of NL interactions.
(A) LaminB1 binding profiles for ESCs (orange), NPCs (blue), ACs (magenta), and MEFs (green) along mouse chromosome 18. Y-axis depicts the log2 transformed Dam-LmnB1 over Dam methylation ratio. Grey rectangles below each track represent LADs for each cell type, black rectangles at the bottom represent genes. Red boxes mark some regions of reorganization during differentiation. (B-D) FISH microscopy data validates NL interaction maps. Scatter plots showing correlation between the mean LaminB1 binding score per gene and the radial distance to the nuclear periphery as determined by FISH (Hiratani et al., 2008) in ESCs (B) and NPCs (C) for 7 loci. (D) Concordance between changes in NL interactions and changes in radial position upon ESC to NPC differentiation (p = 0.005621).
DamID maps are validated by microscopy
Although previous reports have extensively validated DamID mapping with B-type lamins as a means to identify NL interactions (Guelen et al., 2008; Pickersgill et al., 2006), we conducted additional analyses to confirm this. First, we compared our data to a published Fluorescence In Situ Hybridization (FISH) nuclear localization study of seven loci in ESCs and NPCs (Hiratani et al., 2008). This demonstrated a high correlation between the nuclear radial position of these loci and the Lamin B1 DamID score in both cell types (Figures 1B, 1C). Importantly, several loci that shifted radial positions between the two cell types as reported by FISH show a proportional change in DamID log-ratio (Figure 1D). Second, we performed two-color FISH in MEFs using as probes the DamID samples from MEFs that we used for microarray hybridizations. Confocal microscopy sections show clear enrichment at the nuclear periphery when adenine-methylated DNA from cells expressing Dam-LaminB1 is used for hybridization, unlike adenine-methylated DNA from cells expressing unfused Dam (Figures S1H-J). Although it cannot be ruled out that a small number of regions with high DamID signals interact with Lamin B1 that is located in the nuclear interior, these results further validate the use of Lamin B1 DamID for mapping of genome-NL interactions.
Different cell types have similar, but not identical, chromosome architecture
The Lamin B1 binding maps show that in each cell type many large contiguous stretches of DNA interact with the NL (Figure 1A and Figure S1K). A domain detection algorithm (Guelen et al., 2008) identified approximately 1,100-1,400 LADs in each of the four cell types, ranging in size from about 40kb to 15Mb and covering ~40% of the genome (Table S1 and S2). Below, we will use these discrete LAD definitions for further analyses, but we emphasize that this binary classification into LADs and inter-LADs is an oversimplification; we believe that it is more accurate to assume that each genomic locus has a specific probability to contact the NL, and that this probability is reflected by the Lamin B1 DamID log-ratios. Nevertheless, the numbers and size distributions of LADs in the four mouse cell types are remarkably similar to those previously found in human fibroblasts, in which ~1,300 LADs of sizes ~80kb-30Mb were identified that also cover ~40% of the human genome.
Among the four mouse cell types, the genomic patterns of NL interactions are strongly congruent. This is illustrated by the genome-wide correlations in DamID log ratios between cell types, which range from 0.64 (ESCs to ACs) to 0.9 (NPCs to ACs) (Figure S1G). Furthermore, LADs overlap by 73-87% between cell types, quantified as the percentage of LAD-included basepairs in one cell type that remain in LADs in another cell type. We note that the DamID data from ESCs had a slightly lower dynamic range (~0.6-1.1 log2 unit) compared to the data from the other three cell types. This may indicate that NL interactions in ESCs are somewhat less robust, or more variable in time or between individual cells. However, we cannot rule out technical reasons, such as a somewhat different expression level of the Dam proteins in ESCs, or effects of the more flattened morphology of nuclei in the differentiated cell types compared to ESCs in culture. Taken together, the overall architecture of mouse chromosomes within the interphase nucleus is, with respect to NL interactions, remarkably constant over a variety of cell types, from pluripotent stem cells to terminally differentiated cells. Because ESCs and NPCs proliferate while ACs do not, these data also imply that mitosis does not have a strong impact on the overall NL interaction pattern as detected in unsynchronized cell populations.
Despite these global similarities, there are also localized differences in NL interactions between cell types at many sites in the genome. Some of these differences are highlighted by red boxes in Figure 1A. In the two sequential differentiation steps of the ESC→NPC→AC lineage, we observe many loci that increase or decrease their interaction with the NL. Thus, on top of an unaltered "backbone" chromosome architecture, there are many local alterations in NL contacts that are cell type specific. These changes will be discussed in detail below.
LADs are repressive domains in all four cell types
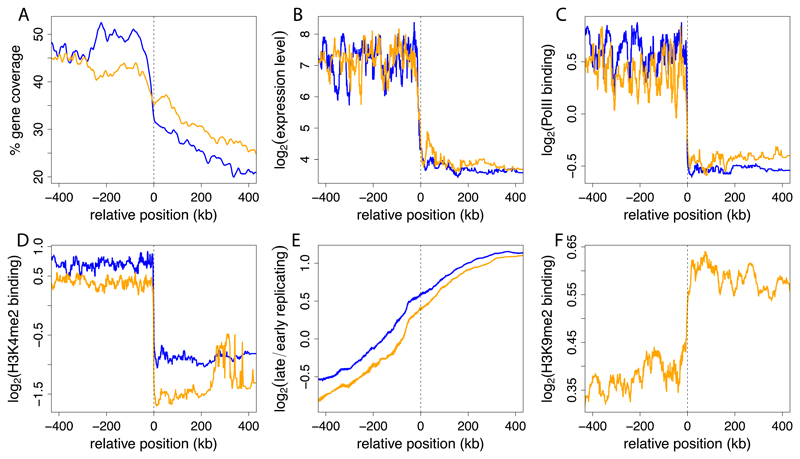
LADs in all four cell types have lower gene density than inter-LAD regions (Figure 2A). Despite a low overall gene density, LADs in each cell type contain between 13-18% of all genes in the genome. Analysis of microarray mRNA expression profiles (Mikkelsen et al., 2007) revealed that the vast majority of these genes are expressed at very low levels. The median expression level of genes inside LADs is about 5-10 fold lower than outside LADs (Figures 2B and S2). Consistent with this, promoters in LADs mostly lack RNA polymerase II and the histone mark H3K4me2 that is predominantly present on active genes (Berger, 2007; Martin and Zhang, 2005) (Figures 2C, 2D). In contrast, late-replicating DNA and the histone modification H3K9me2, both typical for repressed chromatin (Hiratani et al., 2008; Wen et al., 2009) are enriched in LADs (Figures 2E, 2F). We note that replication timing, although globally correlating with LAD organization, does on average not follow the sharp transitions across LAD borders as seen for transcription or histone modifications (Figure 2E). This implies that in the vicinity of LAD borders, replication timing is not a reliable predictor of NL interactions. Taken together, these results indicate that LADs represent a strongly repressive chromatin environment in all tested cell types. This is in agreement with earlier studies of NL-associated parts of the genome in Drosophila and human cells (Guelen et al., 2008; Pickersgill et al., 2006), suggesting an evolutionarily conserved function of the NL.
Figure 2. LADs are repressive chromatin regions in all four cell types.
Profiles across aligned LAD border regions (all borders; left and mirrored right border regions combined). To align data to LAD borders, genome-wide positions of all analysed features were converted to coordinates relative to the nearest border. Positive coordinates, inside LADs; negative coordinates, outside LADs. Colored lines show moving-window (A) mean averages with window sizes of 10 kb and (B-F) median averages with window sizes of 2% of all data within the plotting range. Data are shown for ESCs (orange) and NPCs (blue). (A) local gene density; (B) gene expression levels as determined by microarray profiling (Mikkelsen et al., 2007); (C) RNA Polymerase II and (D) H3K4me2 levels on promoters (Mohn et al., 2008); (E) replication timing (Hiratani et al., 2008); and (F) H3K9me2 levels (Wen et al., 2009). H3K9me2 data were not available for NPCs.
Reorganization occurs in individual transcription units and multi-gene regions
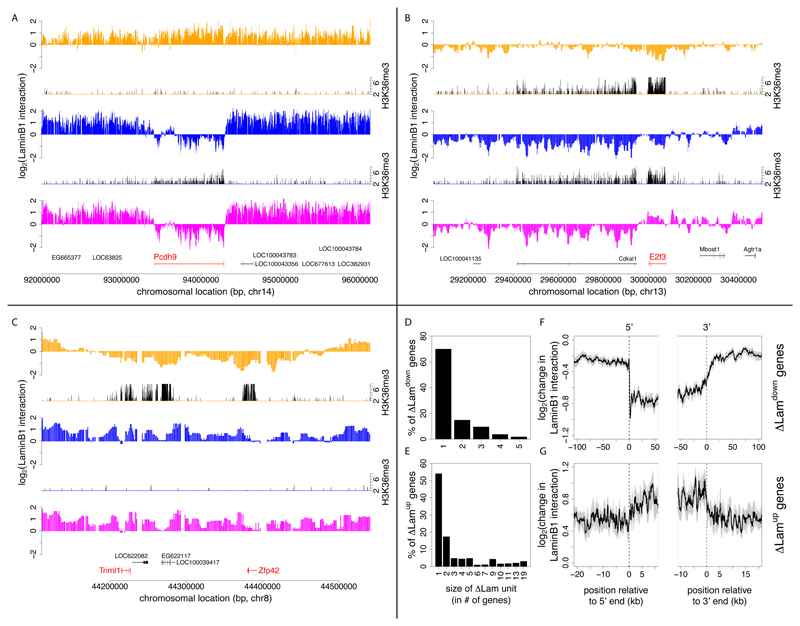
Next, we focused on genomic regions with cell type specific NL interactions. We will refer to loci with significantly altered NL interactions according to our DamID data as “relocating” loci. Visual inspection suggested that these relocating regions often overlap precisely with a single transcription unit. For example, the brain-specific gene Pcdh9 is released from the NL upon differentiation of ESCs into NPCs (Figure 3A). The detachment of Pcdh9 involves the entire transcription unit, but the flanking non-transcribed regions remain mostly unaffected. Similarly, the cell-cycle regulating gene E2f3 becomes specifically associated with the NL upon differentiation of NPCs into ACs, and this relocation occurs mostly independently from directly surrounding sequences (Figure 3B). These changes in lamina interaction of isolated transcription units are commonly observed, but we also found several instances where two or more neighboring genes together with the intergenic DNA show a consistent change in NL interaction. An example of this is a region of ~150kb that includes the ESC-specific genes Triml1 and Zfp42. This region shows increased NL interaction over its entire length after differentiation of ESCs into NPCs (Figure 3C).
Figure 3. Changes in NL interactions involve single transcription units and multi-gene regions.
(A-C) Examples of genes that change NL interaction levels during differentiation. The log2 transformed Dam-LmnB1 over Dam methylation ratios are plotted for selected loci in ESCs (orange), NPCs (blue) and ACs (magenta). Corresponding H3K36me3 maps are shown for ESCs and NPCs (plotted in black, on orange respectively blue baselines) to indicate local transcription activity. (A) Neuron/glia-specific gene Pcdh9; (B) cell-cycle regulating gene E2f3; (C) stem cell marker genes Triml1 and Zfp42. H3K36me3 data was taken from (Mikkelsen et al., 2007). (D-E) Size distribution of relocating units during ESC→NPC transition, calculated as the number of neighboring genes with concordant significant decreases (D) or increases (E) in NL interaction levels. (F, G) Average profiles of the change in LaminB1-interaction along singleton ΔLamdown and ΔLamup genes. (F, G) Grey areas mark estimated 95% confidence intervals, the cut-offs on the x-axes are based on half the median gene size of the genes in each group.
To extend these observations of individual genes to a global analysis we devised a statistical test to identify all genes in the genome that show significant changes in NL binding during the ESC→NPC or NPC→AC differentiation steps. For the ESC→NPC transition this yielded 847 genes with significantly reduced and 633 genes with significantly increased NL interaction levels. For the NPC→AC transition these numbers are 239 and 390, respectively. We will refer to genes with significantly increased NL interaction as ΔLamup, and genes with significantly decreased NL interaction as ΔLamdown (Table S3). We emphasize that our statistical test evaluates quantitative changes in the DamID signals for each gene, rather than the binary LAD versus inter-LAD status, which is defined on a coarser scale. As a consequence, some genes may show significant changes in DamID signals but not a change in their LAD status.
We wondered to what extent the genes that exhibit changes in NL interactions are clustered along the genome. Here, we define a cluster as a set of neighboring genes for which each gene as well as each intergenic region that separates the genes shows a significant change in NL interacts (and all in the same direction). A systematic survey revealed that the majority (~54-82%) of ΔLamdown and ΔLamup genes undergo changes in NL interactions as singletons, while the remainder occur in clusters of typically 2-5 genes (Figures 3D, 3E and S3A, S3B). In total, we identified 101 clusters of ΔLamdown genes and 87 clusters of ΔLamup genes for the ESC→NPC transition, and 19 respectively 45 for the NPC→AC transition. Random permutation analysis indicates that this degree of clustering is more than may be expected by chance (p < 10-4 for all cases).
In order to study the general relocation behavior of singleton ΔLamup and ΔLamdown genes in relation to their flanking sequences, we calculated average profiles of the change in Lamin-B1-interaction log-ratios inside and around these genes. Strikingly, reduction of Lamin B1 interaction occurs primarily within the transcription units, while flanking sequences are much less affected (Figure 3F). Thus, for these genes the changes in NL interactions are highly localized. Similar, albeit less pronounced patterns are observed for ΔLamup genes (Figure 3G). These general patterns are also observed for the NPC→AC transitions (Figures S3C, S3D).
Taken together, these results reveal two different modes by which NL interactions are modified during differentiation. The first and most prevalent mode involves individual genes and is confined to the transcription unit; the second mode involves larger regions that include multiple genes together with the intergenic DNA that separates these genes.
Reorganization in sequential differentiation steps is progressive
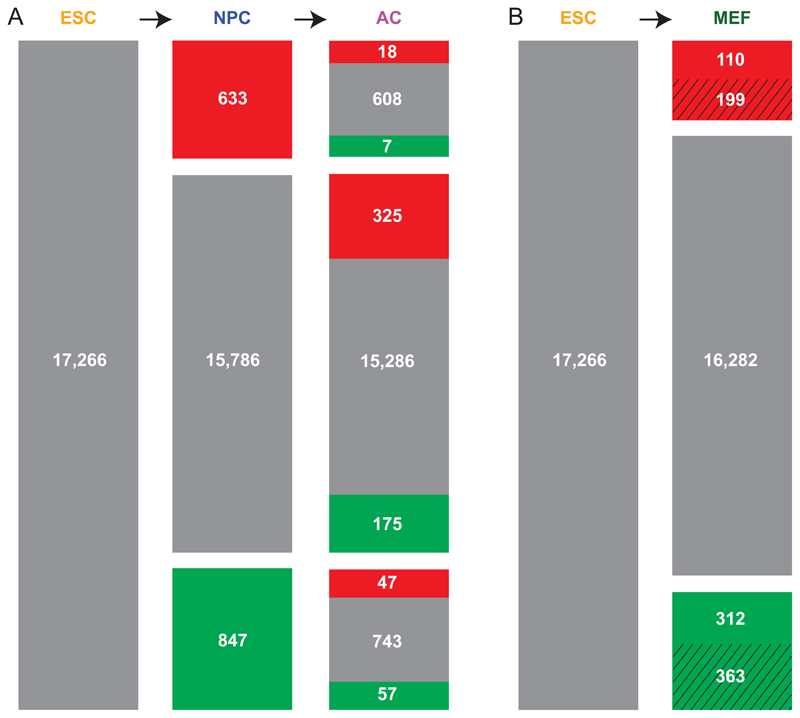
Because ESCs give rise to all somatic cell types, one may regard the chromosomal organization in ESCs as the “basal” architecture that is modified during differentiation. Interestingly, during the two sequential differentiation steps in the ESC→NPC→AC lineage there is a progressive departure from this basal state (Figure 4A). Hundreds of genes relocate during the ESC→NPC transition, and most of these genes are not significantly changed in the NPC→AC transition. Rather, a new set of genes is relocated in the second differentiation step. Thus, the reorganization of genome-NL interactions in these two sequential differentiation steps is mostly cumulative, i.e., the chromosome organization of ACs differs more from the basal architecture than the organization in NPCs. When comparing MEFs to ESCs, the remodeling of NL interactions involves genes that are substantially different from those that relocate in the ESC→NPC→AC lineage (Figure 4B). This is further illustrated by hierarchical clustering of the lamina interaction profiles, showing that MEFs and the NPC→AC lineage diverge into two separate branches (Figure S1G). Thus, modifications of the basal NL interactions upon departure of the ESC state are cumulative and reflect the cell type lineage.
Figure 4. Cumulative reorganization of NL-gene interactions in subsequent differentiation steps.
Out of a curated set of 17,266 genes, the numbers of genes are indicated that exhibit statistically significant increased (red) or decreased (green) levels of NL interactions compared to the preceding differentiation state. (A) ESC→NPC→AC lineage; (B) ESC→MEF lineage, which is a virtual lineage because the MEFs were not directly derived from the ESC culture. Striped areas represent genes that are also modified in one or both steps of the ESC→NPC→AC lineage.
Relocating genes are important for cellular identity
Next, we investigated whether the genes with changes in NL interaction are important for the physiology of the respective cell types. We first turned our attention to genes that encode factors that are important for pluripotency in ESCs. Strikingly, several of these "stemness" genes exhibit significantly increased interactions with the NL in NPCs compared to ESCs (Figure 5A). These genes include Nanog, Klf4, and Oct4 (Pou5f1), which are all downregulated during the ESC→NPC transition and can promote the reprogramming of differentiated cells into pluripotent cells when overexpressed (Hochedlinger and Plath, 2009; Welstead et al., 2008). Similar increases in NL interactions are seen for several other genes that are specifically expressed in stem cells (Takahashi and Yamanaka, 2006). For most of these “stemness” genes, the increase in NL interaction is concomitant with a loss of transcription activity. Myc is an exception, as its expression increases during the ESC→NPC transition and its interaction with the NL slightly decreases. In ACs and MEFs most "stemness" genes are also downregulated and exhibit increased NL interaction compared to ESCs (Figure S4), indicating a tight link between repression and NL interaction of these genes in various cell types that are not ESCs.
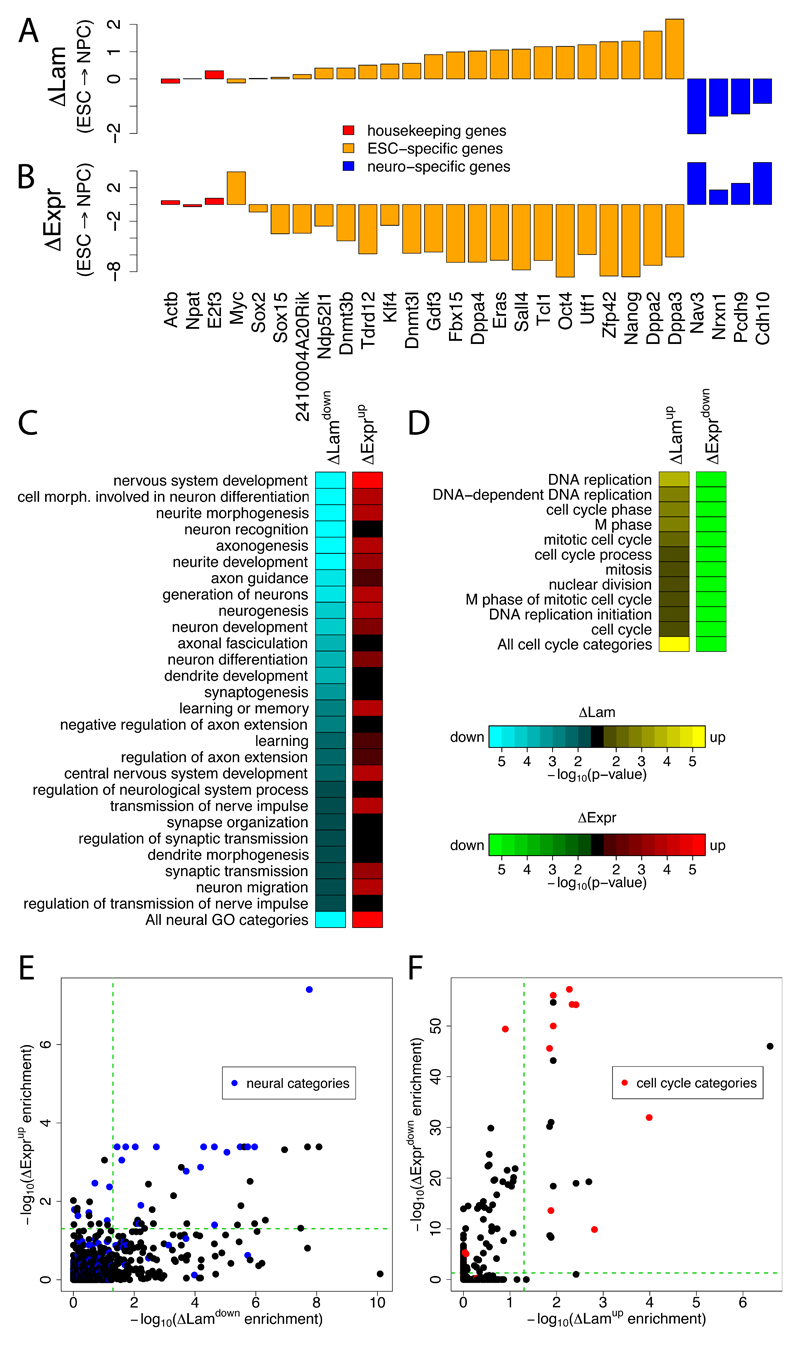
Figure 5. Genes with altered NL interaction have cell type specific functions and expression levels.
(A-B) Log2 changes in LaminB1 interaction (ΔLam) (A) and gene expression levels (ΔExpr) (B) upon ESC→NPC differentiation, for ESC-specific (orange) (Takahashi and Yamanaka, 2006), neuron-specific (blue) and housekeeping (red) genes. (C,D) GO enrichment analysis. Green-to-red and cyan-to-yellow color scales reflect statistical signifance of changes in gene expression and NL interactions, respectively. (C) GO categories representing neural functions that show significant decreases in NL interactions during the ESC→NPC transition. (D) GO categories linked to cell cycle functions that show significant increases in NL interactions during the NPC→AC transition. (E-F) Scatter plots comparing -log10 corrected p-values for changes in NL interaction and changes in gene expression levels during the ESC→NPC (E) and NPC→AC (F) transitions. Blue dots, GO categories related to neural functions; red dots, GO categories related to cell cycle. Green dashed line marks cutoff for statistical significance (p<0.05).
Many other genes exhibit changes in NL interaction during the ESC→NPC and NPC→AC transitions. We conducted a Gene Ontology (GO) analysis (2008) to systematically identify functional categories of genes that are enriched for either decreased or increased NL interactions (Figures 5C-F; Table S4). For the ESC→NPC transition this yielded 184 GO categories with significantly decreased NL interactions. Of these, 27 categories relate to aspects of neural physiology, indicating a genome-wide tendency for genes with neural functions to detach from the NL upon ESC→NPC differentiation (Figures 5C, 5E). To confirm this, we constructed a non-redundant GO meta-category that combines all GO categories related to neuronal processes (Supplementary Methods). Out of 499 GO-annotated genes with decreased NL interactions during the ESC→NPC transition, 98 (19.6%) are in this neuronal meta-category. This is a highly significant enrichment compared to the genome-wide frequency (11.3%; P=1.3 x 10-8, Fisher’s Exact test).
Interestingly, during NPC→AC differentiation, significantly increased NL interactions are observed for 11 GO categories that relate to various cell cycle functions such as DNA replication and mitosis (Figures 5D, 5F, see also Figure 3B). A non-redundant GO category containing all cell cycle related genes shows significant enrichment for increased NL interactions. This raises the possibility that the proliferative arrest of ACs may be in part due to the compartmentalization of a substantial set of cell cycle genes to the repressive environment at the NL.
For many of the significant GO categories, increased or decreased NL interactions correspond to reduced respectively elevated gene expression of the genes in the category (Figures 5C-F), consistent with the repressive nature of NL interacting chromatin. However, we note that this relationship is only partial, in particular for the ESC→NPC transition, where a substantial number of GO categories exhibit significant detachment from the NL without significant upregulation of expression (Figure 5E lower right quadrant). Below we will address this issue in more detail. Taken together, this analysis shows that the changes in NL-chromatin interactions reflect the changes in cellular identity.
Silent genes that detach from the NL are unlocked for activation
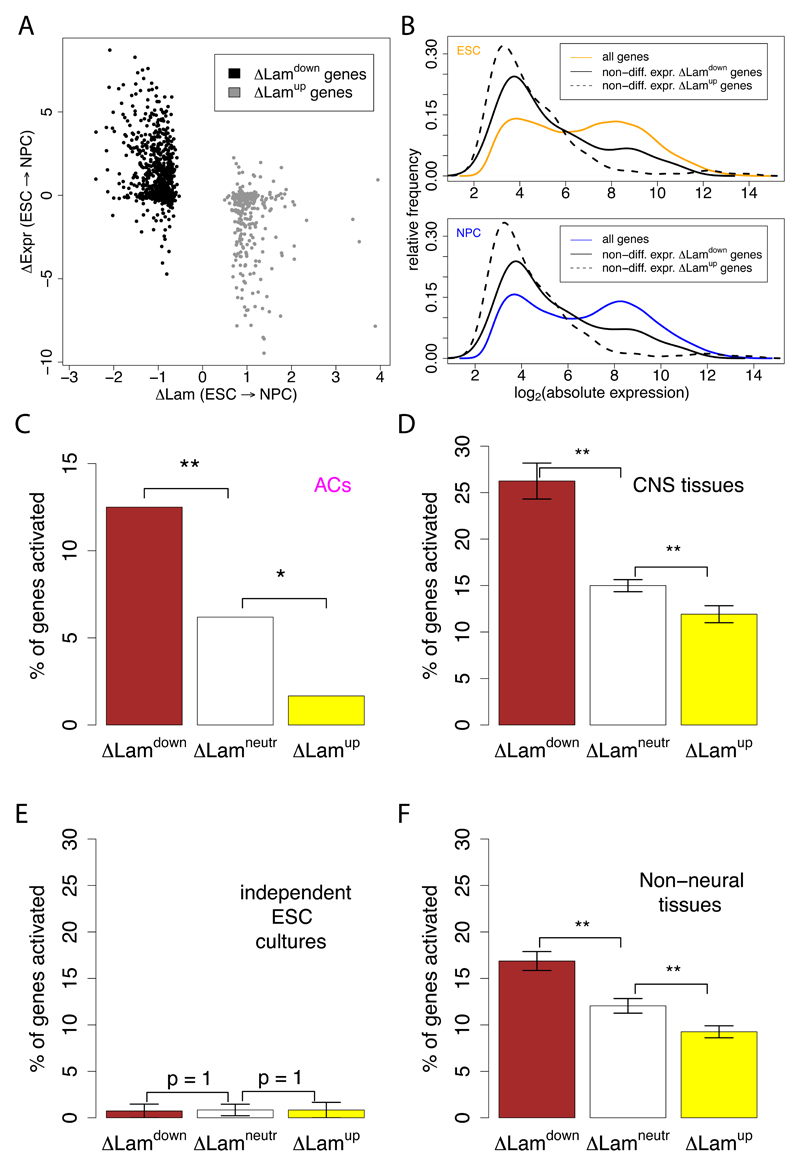
We investigated the link between gene regulation and NL interactions in detail for the ESC→NPC differentiation step. Many ΔLamdown genes display increased expression, and conversely many ΔLamup genes display decreased expression levels (Figure 6A). However, this is not a strict relationship. First, analysis of microarray expression data (Mikkelsen et al., 2007) indicates that 4,052 out of 13,798 tested genes are significantly downregulated in NPCs. Of these, only 215 exhibit a significant increase in NL interactions. Thus, increased association with the NL is not a general consequence of the loss of transcriptional activity.
Figure 6. Silent genes are preferentially activated after detachment from the NL.
(A) Scatterplot of the change in NL interaction (ΔLam) versus the change in gene expression (ΔExpr) during the ESC→NPC transition. Only genes with significantly increased (ΔLamup) and decreased (ΔLamdown) NL interactions are shown. (B) Distribution of log2 absolute expression levels of non-differentially expressed ΔLamup and ΔLamdown genes in ESCs (top) and NPCs (bottom), compared to all genes. (C-F) Differentiation-related activation of ΔLamdown, ΔLamneutral, and ΔLamup genes, as defined for the ESC→NPC transition. that are silent in both ESCs and NPCs. Bars show the percentage of genes that is activated in ACs (C); 10 central nervous system tissues (D); two independent ESC lines (E) and 77 non-neural tissues (F). In (D-F), each tissue or cell line was analyzed separately, and the results were averaged; error bars represent the standard error of the mean; ** indicates p<0.01 and * indicates p<0.05. Expression data for D, E, and F were taken from (Lattin et al., 2008). Note that values on the y-axes in D, E and F and not directly comparable to those in C, due to slightly different selection criteria for activated genes (see Methods).
Second, we observed that about one third of genes that exhibit significantly increased or decreased NL interactions do not have detectable changes in expression. We noticed that these relocating genes with unaltered expression typically exhibit very low activity in both ESCs and NPCs (Figure 6B, see also Figures S5A, S5B). An interesting possibility is that genes that detach from the NL without concomitant activation may become available for expression at a later stage, e.g. upon further differentiation. Conversely, silent genes that are moved to the NL may become more stably inactivated than silent genes that stay in the nuclear interior. We tested this model by studying the expression fate of these gene groups upon further differentiation to ACs. For this purpose, we first defined the set of genes that are expressed at very low levels in both ESCs and NPCs, using stringent criteria based on the combination of mRNA expression data and H3K36me3 levels (Figures S5C, S5D). For simplicity we will refer to these genes as "silent". We then investigated whether the probability of these genes to become upregulated during the transition to ACs is linked to their previous repositioning during ESC→NPC transition (Figure 6C). Strikingly, silent ΔLamdown genes are indeed more likely to become activated in ACs in comparison to silent genes with unaltered NL interactions (ΔLamneutr genes). In contrast, silent ΔLamup genes have a reduced probability to become active in ACs. In other words, silent genes that move away from the NL tend to become "unlocked" for expression at a next differentiation step, and conversely silent genes with increased NL interactions tend to become "locked" in their repressed state.
Because NPCs can give rise to a variety of neuronal and glial cell types besides ACs, we repeated this analysis using gene expression data for 10 different central nervous system tissues (Lattin et al., 2008). This yielded the same pattern as observed for expression in ACs: silent genes are more likely to become active in neural tissues if they were relocated away from the NL in the preceding ESC→NPC differentiation step (Figure 6D). A control analysis of two expression datasets from independent ESC cultures showed no activation, as expected (Figure 6E). Interestingly, distinct (often partially overlapping) subsets of the unlocked genes are activated in different neural tissues (data not shown), suggesting that each neural or glial cell type may deploy a specific subset of the unlocked genes.
We considered two interpretations of these results. First, the unlocking of the set of ΔLamdown genes could be important for the process of commitment of NPCs to the neural/glial lineage. In this scenario the unlocked genes would have functions that are specific for neurons or glia cells, and hence the activation of unlocked genes should occur predominantly in neural tissues. Alternatively, the unlocked genes may serve in a broader set of cell types, but may have been locked in ESCs because their expression would be somehow detrimental to ESCs. To discriminate between these two models, we studied the expression status of these genes in 77 non-neural tissues (Figure 6F). While non-neural tissues still exhibit a preference to activate ΔLamdown genes compared to ΔLamneutr genes, this preference is significanty less pronounced than in neural tissues (p = 3 x 10-4, Wilcoxon test). Most unlocked genes are expressed in a minority of tissues (Figure S5E), indicating that they tend to have specialized functions. Taken together, these results suggest an unlocking mechanism, involving dissociation of silent genes from the NL upon ESC→NPC differentiation, which primes these genes for activation later in development. This unlocking appears to be partially linked to the commitment of NPCs to the neural/glial lineage, and partially to the departure from ESC identity. The unlocking mechanism is distinct from “polymerase poising” (Core et al., 2008; Muse et al., 2007; Zeitlinger et al., 2007), because the silent genes that become detached from the NL in NPCs lack detectable amounts of RNA polymerase II (Pol II) at their promoters (Figures S5A, S5B).
Discussion
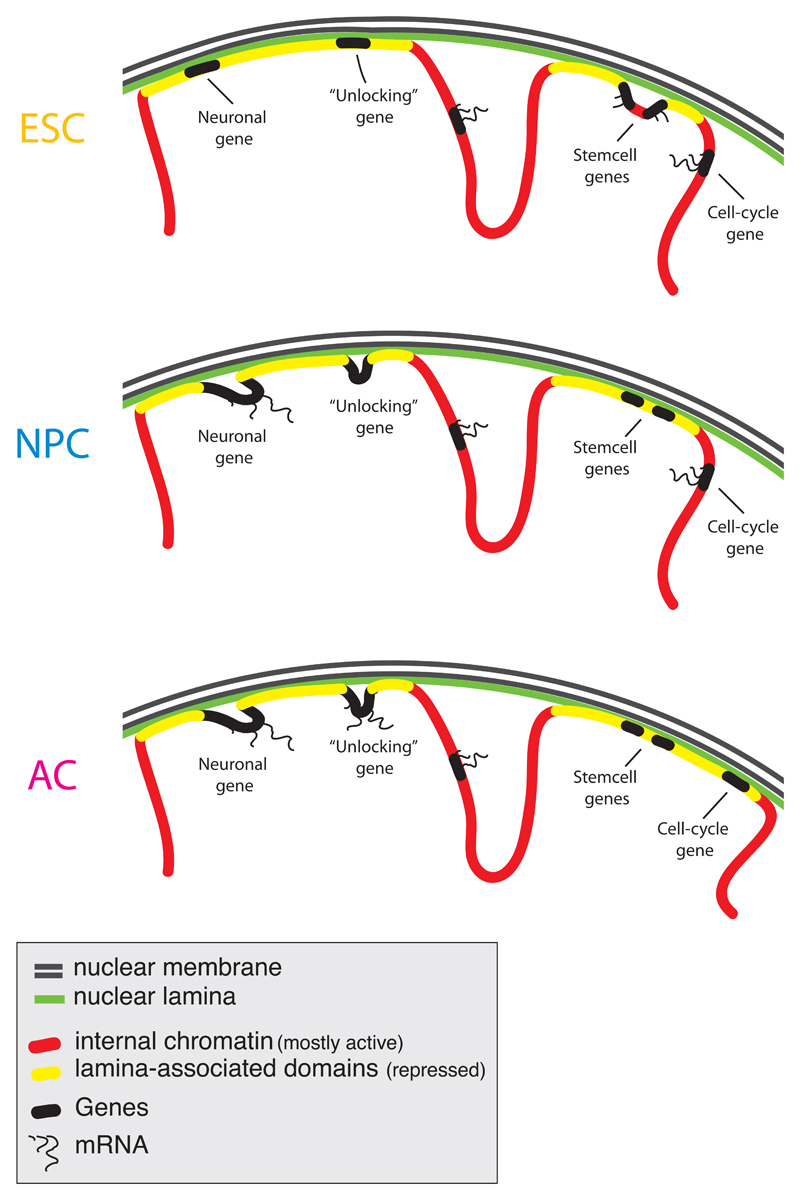
The high-resolution Lamin B1 interaction maps presented here reveal that pluripotent ESCs, multipotent precursor cells and terminally differentiated cells share a common global architecture of their chromosomes, characterized by substantially overlapping interactions with the NL through more than 1,000 large genomic domains. At a finer level, each differentiation step involves the highly orchestrated reorganization of NL-chromatin interactions of hundreds of genes. This reorganization is cumulative over sequential differentiation steps, and involves single transcription units as well as extended DNA regions that encompass multiple genes (Figure 7). Furthermore, NL interactions are tightly linked to gene repression, and the reorganization of these interactions during differentiation involves many genes that are important for celllular identity. Finally, we demonstrate that a substantial number of genes are not immediately activated upon detachment from the NL, but rather become unlocked for activation at a later stage (Figure 7).
Figure 7. Model of dynamic reshaping of NL-genome interactions during differentiation.
Overview of the changes in NL interactions for major gene classes during ESC→NPC and NPC→AC differentiaton steps.
Cell identity and gene repression at the NL
As a rule, NL-associated genes in all four mouse cell types have low transcriptional activity, similar to what has been observed in human and Drosophila cells (Guelen et al., 2008; Pickersgill et al., 2006). Recent evidence indicates that the NL can play a causal role in gene repression. Tethering of genes to the NL can, at least in certain genomic contexts, lead to reduced gene expression (Finlan et al., 2008; Kumaran and Spector, 2008; Reddy et al., 2008), and depletion of Lamin B in Drosophila causes activation of a gene cluster that is normally silent and located at the NL (Shevelyov et al., 2009).
While the NL may contribute to the repressed state of interacting genes, it cannot be ruled out that the NL interactions of some genomic regions are altered as a consequence rather than as a cause of changes in transcriptional activity. In fact, both directions of causality may be true: the NL may enhance the repression of genes, while lack of transcriptional activity in turn may strengthen NL interactions. Such a positive feedback loop may help to stably repress specific genes, thereby securing the cellular transcription program. In this context it is interesting to note that many 'stemness' genes interact more strongly with the NL in non-ESC cell types. This could help to lock these genes in a permanently repressed state once ESCs differentiate.
We provide evidence that silent genes that detach from the NL are more likely to become active in a subsequent differentiation step than genes with unaltered NL interactions. This observation of "unlocking" underscores the notion that the NL may help to secure the repression of specific genes. NL interactions may thus help to constrain the repertoire of genes that can be activated, and thereby restrict the possibilities of further differentiation.
Relocation of singleton and clustered genes
During differentiation, NL interactions are frequently modulated at the level of individual genes. We identified many single transcription units that detach from the NL without substantial impact on the flanking intergenic DNA or neighboring genes. We also identified dozens of clusters of genes that relocate as a unit during differentiation. Earlier observations have also indicated that both individual genes and clusters of genes can relocate with respect to the nuclear periphery (Pickersgill et al., 2006; Williams et al., 2006; Zink et al., 2004).
Robustness of NL interactions in ES cells
A number of reports have suggested that chromatin in ESCs may be globally in a more plastic state than in differentiated cells. For example, in undifferentiated ESCs some structural chromatin proteins are less stably bound (Meshorer et al., 2006), intergenic transcription has been reported to be more widespread (Efroni et al., 2008), and replication timing domains appear somewhat more fragmented compared to differentiated cells (Hiratani et al., 2008). Large H3K9me2 domains have been claimed to be prevalent in chromatin of differentiated cells and nearly absent in ESCs (Wen et al., 2009), but this conclusion has been questioned (Filion and van Steensel, 2010). We observed a clearly defined "basal state" pattern of genome-NL interactions in ESCs, indicating that despite the plasticity observed in other parameters, chromosomes are organized in a reproducible manner in ESCs. However, we did notice a slightly lower dynamic range in Lamin B1 DamID signals in ESCs compared to the other cell types. This could reflect somewhat less robust NL interactions in ESCs, but technical explanations may also account for this minor difference. Future studies may address the dynamics of the genome at the NL in more detail, and should provide insight into the molecular mechanisms that control genome – NL interactions.
Methods
DamID
DamID was performed as described (Vogel et al., 2007). Two independent biological replicates were performed for each cell type on separate days (Supplemental Experimental Procedures). Data are available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), accession GSE17051.
Expression data
Affymetrix array expression data for ESC, NPC, MEF (Mikkelsen et al., 2007) and AC (Meissner et al., 2008) were obtained from GEO (accession numbers GSE8024 and GSE11483, respectively). Tissue gene expression data (Lattin et al., 2008) were obtained from GEO (accession number GSE10246). H3K36me3 data were obtained from (Mikkelsen et al., 2007).
Analysis of singleton vs. clustered genes
We stringently defined a ΔLam gene cluster as the genes in a contiguous stretch of DNA in which all genes, as well as all intergenic regions, show significant and concordant changes in DamID signal. The intergenic region is disregarded if it is too small to test for significant changes in lamina interaction, i.e., when it has fewer than five array probes. Singleton ΔLam genes are defined as genes which exhibit significantly altered NL interactions without significant concordant changes in NL interactions of the neighboring genes, irrespective of the behaviour of intergenic regions.
Analysis of gene (un)locking
To analyze the potential unlocking and locking of genes, a set of genes that are consistently silent in both ESCs and NPCs was defined based on the bimodal distributions of both Affymetrix microarray expression and H3K36me3 ChIP-seq data. We estimated the point that best separates the two modes into a ‘low’ and ‘high’ region (Figures S5C, S5D, blue lines). Silent genes are defined as genes that consistently lie in the ‘low’ region for both gene expression and H3K36me3, in both ESCs and NPCs. Additionally, they are required to show no statistically significant change in expression between both cell types. Activated genes in ACs are defined as genes in the ‘high’ part of the bimodal AC gene expression distribution, and with statistically significantly increased expression levels compared to NPCs. For the tissue gene expression data, which constitute a separate dataset, we defined active genes simply as being in the ‘high’ part of the bimodal distribution. Statistical significance of differences in gene activation between ΔLamdown, ΔLamneutr and ΔLamup silent gene groups was estimated by Fisher’s Exact test (for ACs) or a paired Wilcoxon test (for multiple tissues).
Supplementary Material
Acknowledgements
We thank Guillaume Filion for advice on statistics; Koen Braat and Anke Sparmann for help with cell culture and characterization; Lenny Brocks for help with confocal microscopy; Boris Joffe for help with image analysis; Maarten Fornerod and members of the BvS lab for helpful discussions. Supported by the Netherlands Bioinformatics Centre, the Netherlands Genomics Initiative and a European Young Investigator Award to BvS.
References
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat Genet. 2010;42:4. doi: 10.1038/ng0110-4. author reply 5-6. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006;20:486–500. doi: 10.1101/gad.1364906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene_Ontology_Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf S, Nielsen FG, Kurtz S, Huynen MA, Birney E, Stunnenberg H, Flicek P. Optimized design and assessment of whole genome tiling arrays. Bioinformatics. 2007;23:i195–204. doi: 10.1093/bioinformatics/btm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, Moorman C, van Steensel B. DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol. 2006;410:342–359. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282–3287. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci U S A. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Welstead GG, Schorderet P, Boyer LA. The reprogramming language of pluripotency. Curr Opin Genet Dev. 2008;18:123–129. doi: 10.1016/j.gde.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.