Abstract
The Biological General Repository for Interaction Datasets (BioGRID; www.thebiogrid.org) is a freely available public database that provides the biological and biomedical research communities with curated protein and genetic interaction data. Structured experimental evidence codes, an intuitive search interface and visualization tools enable the discovery of individual gene, protein or biological network function. BioGRID houses interaction data for the major model organism species - including yeast, nematode, fly, zebrafish, mouse and human - with particular emphasis on the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe as pioneer eukaryotic models for network biology. BioGRID has achieved comprehensive curation coverage of the entire literature for these two major yeast models, which is actively maintained through monthly curation updates. As of December 2014, BioGRID houses approximately 349,000 biological interactions for budding yeast, and 69,700 interactions for fission yeast. BioGRID also supports an integrated post-translational modification (PTM) viewer that incorporates over 20,100 yeast phosphorylation sites curated through its sister database, the PhosphoGRID (www.phosphogrid.org). This protocol describes how to use the BioGRID website to query genetic or protein interactions for any gene of interest, how to visualize the associated interactions using an embedded interactive network viewer, and how to download data files for either selected interactions or the entire BioGRID interaction data set.
INTRODUCTION
The Biological General Repository for Interaction Datasets (BioGRID; www.thebiogrid.org) is an open source database that curates and disseminates collections of protein and genetic interactions from major model organism species from yeast to human (Stark et al. 2006; Chatr-Aryamontri et al. 2013). The BioGRID was originally developed as a budding yeast-specific database to house and visualize protein interaction data from high-throughput (HTP) proteomic studies (Ho et al. 2002; Breitkreutz et al. 2003a; Stark et al. 2006). Subsequently, comprehensive curation of protein and genetic interactions from the entire budding yeast literature was undertaken in order to compare emerging high-throughput interaction data to the extensive body of interaction data reported in thousands of focused studies (Reguly et al. 2006). Importantly, the evidence for each interaction in BioGRID is recorded as a structured evidence code derived from the primary experimental data. These evidence codes are concordant and interoperable with high-level stratification of the detailed PSI-MI interaction ontology (Hermjakob et al. 2004a; Kerrien et al. 2007). All curated data within BioGRID is fully archived as monthly releases and all records are date-stamped and mapped to individual curators to ensure data integrity. Curation efforts at BioGRID have since been expanded to capture biological interaction data from each of the major model organism species. These datasets serve as a readily accessible resource for interrogation of biological interactions, discovery of gene function, and computational analysis of interaction networks (Dolinski et al. 2013). The December 2014 release of BioGRID (version 3.2.120) contains over 770,000 interactions curated from both high-throughput datasets and low-throughput focused studies found in the literature. These interactions have been distilled from more than 40,000 publications covering 29 different organisms, including the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, the yeast Candida albicans SC5314, the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the mouse Mus musculus, the plant Arabidopsis thaliana and Homo sapiens (Stark et al. 2011; Chatr-Aryamontri et al. 2013). BioGRID interaction datasets are shared with the respective model organism databases (Cherry et al. 2012; Inglis et al. 2012; Lamesch et al. 2012; Wood et al. 2012; Yook et al. 2012; Marygold et al. 2013), with other interaction databases (Luc and Tempst 2004; Razick et al. 2008; Chautard et al. 2009; Matthews et al. 2009; Cerami et al. 2011; Franceschini et al. 2013) and with meta-databases (Benson et al. 2004; Matthews et al. 2009). Complete coverage of the entire literature for S. cerevisiae and S. pombe, as well as for the model plant A. thaliana, is maintained through continuous monthly updates. As of the latest BioGRID release, approximately 349,000 (230,000 unique) interactions have been curated for S. cerevisiae genes/proteins from over 12,000 publications, and approximately 69,700 (57,000 unique) interactions have been curated for S. pombe genes from nearly 2,100 publications (Table 1). Of these interactions, 60% of budding yeast and 83% of fission yeast interactions derive from genetic experiments, and for both organisms, some 80% of interactions are derived from high-throughput data sets. Recently, over 400 physical interactions have also been curated from nearly 40 papers for the pathogenic yeast model species, Candida albicans. All yeast genetic interactions include associated phenotypes curated using the structured Ascomycete Phenotype Ontology (APO) developed by SGD, the Saccharomyces Genome Database (Engel et al. 2010). In addition, over 20,100 phosphorylation sites mapped onto nearly 3,200 budding yeast proteins are documented in a sister database called PhosphoGRID (Sadowski et al. 2013), and are available through a new post-translational modification (PTM) viewer integrated within BioGRID.
Table 1.
BioGRID yeast curation statistics as of December, 2014 (BioGRID version 3.2.120). To date, over 419,000 total interactions have been curated from more than 14,000 publications. These cover 6655 S. cerevisiae proteins, 4,142 S. pombe proteins, and 379 C. albicans SC5314 proteins. The number of unique interactions is given in parentheses, and the number of interactions derived from high-throughput or low-throughput studies is given for each category. HTP, high-throughput; LTP, low-throughput. The number of unique phenotypes refers to the number of non-redundant phenotypes curated for genetic interactions using the Ascomycete Phenotype Ontology (APO).
| Total Interactions | Curated Publications | Protein Interactions | Genetic Interactions | Unique Phenotypes | |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae | 349,461 (230,302) | 12,644 | 141,153 (86,634) | 208,308 (150,413) | 600 |
| HTP | 275,014 (202,774) | 345 | 97,497 (72,393) | 177,517 (133,008) | 57 |
| LTP | 79,695 (43,449) | 12,534 | 44,115 (21,295) | 35,580 (25,480) | 598 |
| Schizosaccharomyces pombe | 69,703 (57,297) | 2,088 | 11,931 (9,108) | 57,772 (48,947) | 320 |
| HTP | 57,444 (50,264) | 48 | 5,696 (5,508) | 51,748 (44,791) | 13 |
| LTP | 12,294 (7,665) | 2,078 | 6,267 (3,864) | 6,027 (4,415) | 319 |
| Candida albicans | 417 (378) | 42 | 144 (111) | 273 (268) | 11 |
| HTP | 258 (258) | 2 | 0 (0) | 258 (258) | 1 |
| LTP | 159 (120) | 40 | 144 (111) | 15 (10) | 10 |
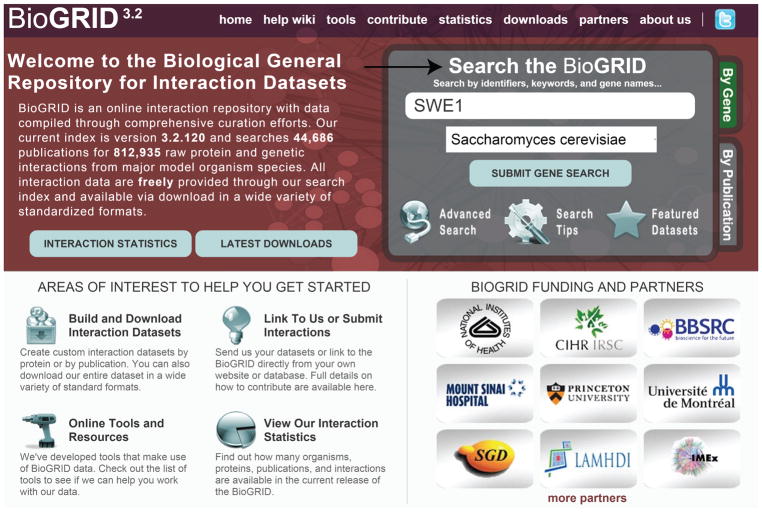
The research community can access these extensive interaction datasets using the BioGRID web interface (Fig. 1), which provides users with a tabular interaction summary for each query gene or protein, as well as a link to the abstract for each curated publication and associated PubMed identifier. Details including interaction type, evidence code and data source are provided in condensed format on each summary page. Interaction data may also be viewed using an interactive network visualization tool embedded within BioGRID, downloaded in bulk for local analysis or captured through stand-alone visualization applications, such as Osprey and Cytoscape (Breitkreutz et al. 2003b; Shannon et al. 2003; Cline et al. 2007).
Figure 1.
Search in BioGRID for interactions of a gene or protein of interest. The BioGRID home page is shown with available search options in the upper right corner (arrow). In the top menu, links are also provided to the help document, online tools, BioGRID statistics and download options. In this gene search example, “SWE1” is entered as the search term and “Saccharomyces cerevisiae” is selected as the organism.
Acknowledgments
The authors thank Chris Grove and Paul Sternberg at WormBase for ongoing collaborative development of the Genetic Interaction Ontology. This work was supported by NIH grants R01OD010929 and R24OD011194 to M.T. and K.D., the Biotechnology and Biological Sciences Research Council (grant number BB/F010486/1 to M.T.), the Canadian Institutes of Health Research (grant number FRN 82940 to M.T.), and a Genome Québec International Recruitment Award and a Canada Research Chair in Systems and Synthetic Biology to M.T.
References
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic acids research. 2004;32:D23–26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. The GRID: the General Repository for Interaction Datasets. Genome biology. 2003a;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome biology. 2003b;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic acids research. 2011;39:D685–690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic acids research. 2013;41:D816–823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K, Chatr-Aryamontri A, Tyers M. Systematic curation of protein and genetic interaction data for computable biology. BMC biology. 2013;11:43. doi: 10.1186/1741-7007-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic acids research. 2010;38:D433–436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI’s molecular interaction format--a community standard for the representation of protein interaction data. Nature biotechnology. 2004a;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic acids research. 2012;40:D667–674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC biology. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic acids research. 2012;40:D1202–1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luc PV, Tempst P. PINdb: a database of nuclear protein complexes from human and yeast. Bioinformatics. 2004;20:1413–1415. doi: 10.1093/bioinformatics/bth114. [DOI] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ, FlyBase c. FlyBase: improvements to the bibliography. Nucleic acids research. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic acids research. 2009;37:D619–622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. Journal of biology. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Breitkreutz BJ, Stark C, Su TC, Dahabieh M, Raithatha S, Bernhard W, Oughtred R, Dolinski K, Barreto K, et al. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database: the journal of biological databases and curation. 2013;2013:bat026. doi: 10.1093/database/bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic acids research. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic acids research. 2012;40:D695–699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, de la Cruz N, Duong A, Fang R, et al. WormBase 2012: more genomes, more data, new website. Nucleic acids research. 2012;40:D735–741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]