Abstract
Antagonism of protein-protein interactions (PPIs) with small molecules is becoming more feasible as a therapeutic approach. However, successful PPI inhibitors tend to target proteins containing deep peptide-binding grooves or pockets as opposed to the much more common large, flat protein interaction surfaces. Here we review one of the most abundant PPI domains in the human proteome, the WD40 repeat domain (WDR), which has a central peptide-binding pocket. Recently, two WDR proteins, WDR5 and EED, have been successfully targeted by potent, specific, cell-active, drug-like chemical probes. Could WDRs represent a novel target class for drug discovery? While clinical validation remains to be seen, a cautious optimism is justified, considering the ubiquitous involvement of WDR proteins across multiple disease-associated pathways. The druggability and structural diversity of WDR binding pockets suggest that this prevalent domain class could open-up areas of biology that have so far resisted drug discovery efforts.
Introduction
Protein-protein interactions (PPIs) are essential mediators of both physiologic and pathologic biology, yet, until recently have been considered very challenging to target therapeutically with small molecules. Recent approval of protein-protein interaction inhibitory drugs such as venetoclax (a BCL2-BAX antagonist)1,2 and continuing clinical progression of others such as MDM2-TP53 antagonists3,4 and BET bromodomain antagonists5–7 demonstrate that certain types of protein-protein interactions can be effectively targeted with small molecules. A common feature of these druggable protein-protein interactions is the presence of a reasonably sized ‘pocket’ or groove on the surface of the targeted protein that binds to a short peptide sequence of its respective interacting partner protein. Because these protein pockets have the appropriate size, shape and physicochemical features to bind well to drug-like small molecules, the latter can effectively compete for binding with the physiological peptide regions of the target protein, thereby disrupting the protein-protein interaction.
Our greater appreciation of the structural and chemical features of targetable PPIs together with increasing knowledge of functional protein interaction networks argues strongly that there are likely to be many more therapeutic opportunities embedded in the human protein interactome. The likelihood of exploiting these opportunities is enhanced by recent improvements in high throughput methods for screening of small molecules that bind to proteins, such as thermal stabilization8, mass spectrometry detected affinity selection9, DNA encoded libraries10 and high throughput fragment screening11. Methods for directly screening for disruption of a PPI have also advanced significantly over the past decade, including the use of fluorescence polarization12, and Alpha (Perkin Elmer) and NanoLuc (Promega) technologies.
Among the most abundant protein interaction domains in the human proteome is the WD40 repeat (WDR) domain, with over 360 domains currently annotated. The WDR domain is a typically seven bladed β-propeller domain with an overall donut shape. Significantly, the ‘donut hole’ or central pore of the WDR domain frequently mediates interactions with peptide regions of key interaction partners, and often has appropriate size and physicochemical features for high affinity binding to drug-like small molecules. WDR domains are often essential subunits of multiprotein complexes involved in a wide range of signaling pathways including DNA damage sensing and repair, ubiquitin signaling and protein degradation, cell cycle, epigenetic regulation of gene expression and chromatin organization, and immune related pathways. Here, we review recent progress and future opportunities for therapeutic targeting of human WDR proteins.
WDR domains are ubiquitous and disease-associated
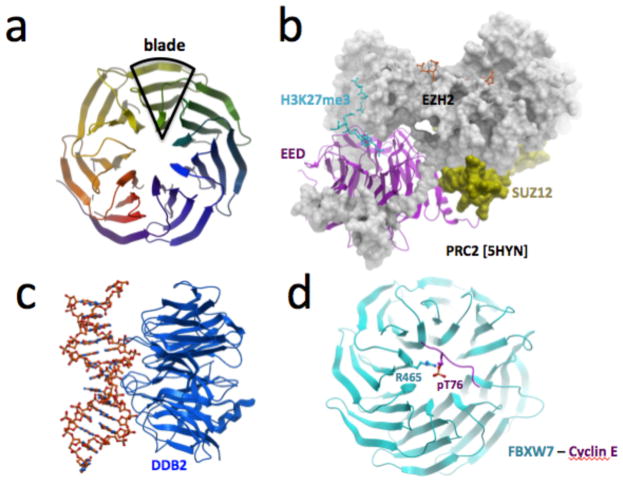
The blades of a WDR domain each contain a conserved glycine-histidine and tryptophan-aspartate (WD) motif. Structural adaptability allows WDRs to retain their β-propeller fold upon deletion or insertion of WD repeats, the number of which can vary from five to eight (Fig. 1a)13,14. WDR domains typically act as scaffolds, often within large multiprotein complexes15. The top, bottom, and side surfaces of the donut can simultaneously act as interaction sites for diverse binding partners, including proteins, peptides, RNA and DNA, suggesting that multiple surfaces can potentially be targeted by chemical inhibitors. For instance, EZH2, SUZ12, and an activating histone peptide all exploit distinct surfaces of EED within the PRC2 complex (Fig. 1b), while the WDR domain of DDB2 binds damaged DNA (Fig. 1c). WDRs can also specifically recognize post-translational modifications on proteins: EED binds tri-methylated lysines16, WDR5 binds methylated arginines17, while yeast Cdc4 and human FBXW7 bind phosphothreonine/phosphoserine degron motifs via their central cavity18,19 (Fig. 1d).
Figure 1. WDR domains are β-propeller interaction hubs.
(a) WDRs form a β-propeller structure, generally composed of seven blades. (b) The WDR protein EED uses top, bottom, and side surfaces to interact with other components of the PRC2 complex [PDB 5HYN]. (c,d) WDR domains can also interact with DNA (DDB2 bound to damaged DNA [PDB 3EIU1]), and peptides (FBXW7 bound to a phosphorylated Cyclin-E peptide [PDB 2OVR]).
Probably due to their versatility as protein interaction scaffolds, WDRs are the fourth most abundant domain in the human proteome15. A systematic search identifies 361 WDR-containing proteins in human (Supplementary Table 1), but this number is probably a conservative estimate, as WDR sequences are poorly conserved outside of their defining “WD” di-peptide motifs, and are poorly annotated in public databases. An algorithm that is comparatively sensitive to WD repeats was recently developed to annotate WDR proteins20,21. The number of β-propeller domains is even larger when the structurally related Kelch domains and other domains are taken into consideration22. Additionally, in yeast, where the interactome is best characterized, WDR domains are engaged in more protein-protein interactions than any other domain15, emphasizing the ubiquitous role of these domains in connecting the global protein interaction network.
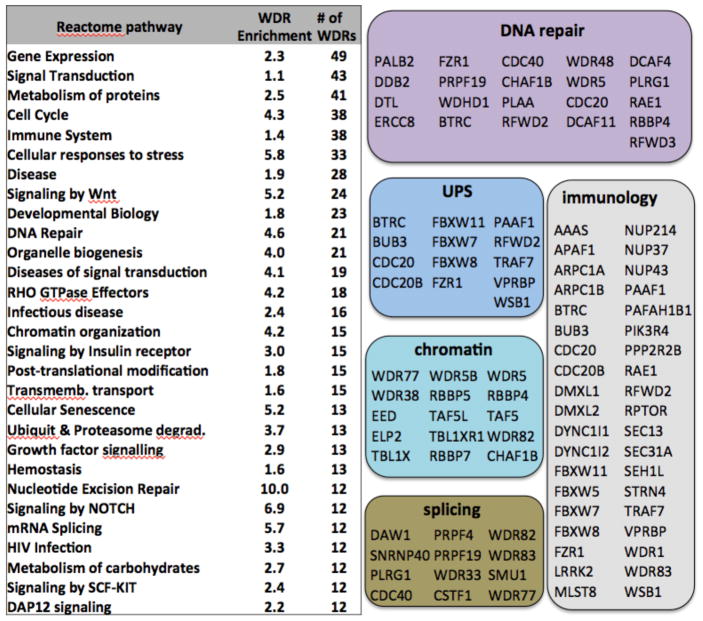
As a consequence of their prevalence in the human proteome, WDR domains are involved in a wide spectrum of cellular networks, many of which are perturbed in human diseases. Cellular pathways from the Reactome database23 were ranked based on the number of WDR proteins involved (Fig. 2, Supplementary Table 2). At the top of the list, at least 49 WDR proteins are known to participate in the regulation of gene expression. Among these, 15 are components of chromatin complexes, including EED and WDR5, for which potent inhibitors have been reported24–26. WDRs are also found in pathways that are targeted by existing drugs, such as DNA repair or the immune system, in emerging areas of drug discovery, such as chromatin mediated signaling or splicing, and in cellular networks that have so far proven largely undruggable, such as the extensive ubiquitin-proteasome system (UPS) that exerts post-translational control over most of the proteome. In some cases, disease association is direct and causative. For instance, recognition of a Cyclin E phosphothreonine degron motif by the central pocket of the WDR domain of FBXW7 leads to Cyclin E degradation, which restrains DNA replication and thereby helps ensure genome stability. Loss-of-function mutations in FBXW7, including a mutation hot-spot at R465, the side-chain that recognizes the phosphothreonine, lead to accumulation of Cyclin-E, and are recurrent driver events in endometrial, colorectal and other cancers27 (Fig. 1d). In another example, mapping missense variations from clinical and population genetics onto the 3D structure of the WDR protein TBL1XR1 clearly separates pathogenic mutations that are responsible for severe neurological disorders in children from benign polymorphisms28.
Figure 2. WDR proteins perform in diverse cellular functions.
The number of WDR proteins associated with Reactome pathways and corresponding enrichment compared with the whole proteome are indicated. Specific disease-associated examples are shown in colored boxes.
WDR proteins are not only associated with disease gene networks, but many are target candidates for therapy in cancer, metabolic disorders, neurological diseases and regenerative medicine (Table 1).
Table 1. WDR proteins that may represent targets of interest.
References are provided in the text.
| WDR | Function | Disease association |
|---|---|---|
| CDC20 | Required for full ubiquitin ligase activity of the anaphase promoting complex/cyclosome (APC/C) | CDC20 knockdown inhibited - and CDC20 overexpression increased - the ability of human glioblastoma stem cells to generate brain tumors in an orthotopic xenograft model in vivo |
| CHAF1B | Histone chaperone | Suppression of CHAF1B enhances conversion of B cells into macrophages and fibroblasts into neurons |
| EED | Component of the PRC2 complex | Some lymphomas and SWI/WNF mutated tumors are sensitive to compounds targeting PRC2 |
| GNB2L1 | Binds to and stabilizes activated protein kinase C | Local administration of GNB2L1 siRNA into mice calvariae reduced the numbers of osteoclasts and bone loss |
| GNB3 | Guanine nucleotide-binding protein | Mouse model implicates GNB3 duplication in a childhood obesity syndrome |
| LRRK2 | Positively regulates autophagy through activation of the CaMKK/AMPK signaling pathway | Gain of function mutations in kinase domain cause Parkinson’s disease |
| PAFAH1B1 | Required for activation of Rho GTPases and actin polymerization in cerebellar and hippocampal neurons | Increase in PAFAH1B1 dosage in the developing brain results in brain abnormalities in mice and humans |
| PALB2 | Recruit BRCA2 and RAD51 to DNA breaks | PALB2 mutation predicts exceptional in vivo response to PARP inhibition |
| PPP2R2A | Regulatory subunit of serine/threonine-protein phosphatase 2A | Loss of PPP2R2A inhibits homologous recombination DNA repair and predicts tumor sensitivity to PARP inhibition |
| RBBP4,7 | Chromatin signalling (PRC2, HAT, HDAC NuRD, CAF-1, complexes) | Knockdown sensitizes glioblastoma to Temozolomide |
| RBBP5 | Component of the MLL complex | MLL-translocated and C/EBPa leukemias, prostate cancer and p53 gain-of-function mutant tumors are sensitive to compounds targeting the MLL complex |
| SEC31A | Formation of transport vesicles from the endoplasmic reticulum (ER) | Oncogenic fusion of WDR with ALK or JAK2 in lymphoma |
| VPRBP | Substrate recognition component of E3 ubiquitin-protein ligase complexes and atypical serine/threonine-protein kinase | Knockdown and chemical inhibition reactivate growth regulatory genes and impede tumor growrth. Recruited by HIV-2 to counteract the effects of host cellular defense. |
| WDR5 | Component of the MLL complex | MLL-translocated and C/EBPa leukemias, prostate cancer and p53 gain-of-function mutant tumors are sensitive to compounds targeting the MLL complex |
| WDR77 | Component of the PRMT5 methyltransferase complex | PRMT5 inhibitors have antitumor activity in lymphoma. MTAP-deleted cancers may be sensitive to PRMT5 inhibition. |
Epigenetic targets in oncology
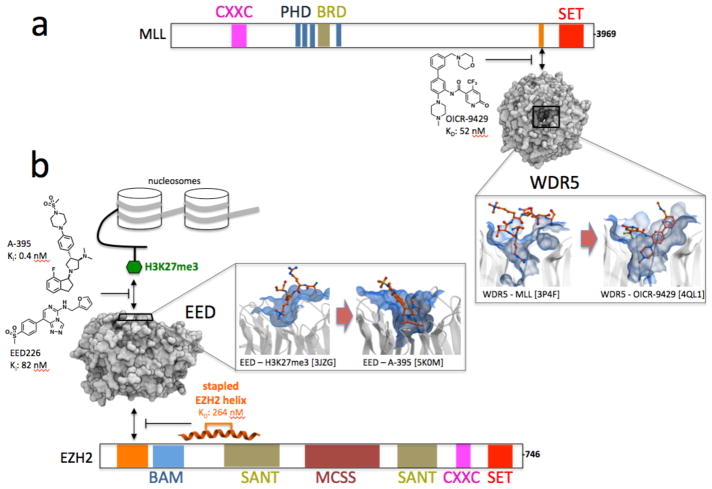
Epigenetic mechanisms are now recognized as being central to the etiology of many cancers29,30 and proteins involved in chromatin mediated signaling are a promising target class for cancer therapy31,32. At least 15 WDR proteins are components of chromatin complexes (Fig. 2), and some may be valid drug targets. For example, pharmacological inhibition of WDR5 has recently emerged as a promising strategy in oncology. WDR5 is a binding partner of the histone methyltransferases MLL1-4 and SET1A/B. WDR5 recruits the MLL1 complex at leukemic loci in over 5% of acute myeloid leukemias through p30, an oncogenic variant of the transcription factor C/EBPα. MLL1 in turn methylates lysine 4 of histone 3 (H3K4), an epigenetic signal associated with active gene transcription, resulting in a p30-dependent inhibition of myeloid differentiation. The drug-like chemical probe OICR-9429 occupies the MLL1 binding pocket of WDR5 (Fig. 3a), disrupts the MLL1 complex in cells with sub-micromolar IC50, reduces MLL1 activity at p30-occupied loci, and selectively kills patient-derived acute myeloid leukemia expressing p3024.
Figure 3. Pioneer epigenetic chemical probes against WDR proteins.
(a) OICR9429 is a potent and selective WDR5 antagonist that competes with MLL and disrupts the transcriptionally activating MLL complex, which includes MLL, WDR5, RBBP5 and ASH2L, in cells24. (b) Amino pyrrolidine A-395 and triazolopyrimidine EED226 antagonize binding of the histone 3 peptide trimethylated at lysine 27 (H3K27me3) to the WDR protein EED, which in turn inhibits the function of the transcriptionally repressive complex PRC2, composed of EZH2, EED and other proteins25,26. A synthetically stapled helical peptide binding at the opposite surface of EED inhibits EZH2 binding, and PRC2 function37,38.
Another study with the WDR5 inhibitor exploited the observation that prevalent cancer-promoting gain-of-function mutations of TP53 lead to up-regulation of MLL1 and MLL2, a global increase of histone methylation, and proliferation of cancer cells. Disrupting the MLL1 complex through pharmacological targeting of WDR5 with OICR-9429 selectively lowered the proliferation of such TP53 gain-of-function mutant cancer cells33. A related study was based on the observation that leukemogenesis induced by the MLL-AF9 fusion protein requires co-expression of the wild-type MLL1 allele34. MM-401, a macrocyclic inhibitor of WDR5, inhibited wild-type MLL1 and induced myeloid differentiation of mixed lineage leukemia cells, with no apparent toxicity on normal bone marrow cells35. Notably, another WDR protein, RBBP5 is also component of the MLL1, MLL2 and MLL3 complexes. While WDR5 is essential only to the catalytic activity of MLL135, RBBP5 is an obligatory structural component of all MLL1 proteins36, and chemical antagonists of RBBP5 could have a more profound effect than WDR5-targeting agents.
More recently, the WDR protein EED was uncovered as a promising therapeutic target. EED is a component of the Polycomb repressive complex 2 (PRC2) and is necessary for the methyltransferase activity of the PRC2 catalytic subunit EZH2, a histone methyltransferase that epigenetically silences gene expression and is the target of inhibitors currently in Phase I/II clinical trials against diffuse large B-cell lymphomas39–42. Research groups from Novartis, and Abbvie in collaboration with the Structural Genomics Consortium have independently shown that drug-like inhibitors targeting the WDR domain of EED antagonize binding of an activating peptide that is necessary for the proper propagation of the methyl mark deposited by PRC2 (Fig. 3b). The compounds inhibit PRC2 in cells with low nanomolar IC50s, and remain active against PRC2-mutated cell lines that are resistant to catalytic inhibitors currently in clinical trial in oncology25,26. EED also interacts with EZH2, and an EZH2-derived stapled helix targeting EED disrupts PRC2 in cells37,38. Though encouraging, how these results will translate in pre-clinical and clinical development is still an open question.
The WDR protein RBBP4 is another component of PRC2 and other chromatin complexes, including the nucleosome remodeling and deacetylase (NuRD) complex, and the chromatin assembly complex (CAF-1)43. The structural role of RBBP4 in different complexes and its function in transcriptional reprogramming of cancer cells is not clear, but probably varies with cancer types and grades. Recent data reveal that RBBP4 regulates chromatin assembly during repair of DNA damage and genetic disruption of RBBP4 sensitizes glioblastoma cells to DNA lesions following treatment with the standard of care temozolomide44.
WDR77, another WDR protein involved in epigenetic control of gene expression, directly binds to the methyltransferase PRMT5 to promote symmetrical dimethylation of arginine side-chains on histone and non-histone substrates. PRMT5 is upregulated in lymphomas and some solid tumors, preclinical PRMT5 inhibitors show dose-dependent antitumor activity in animal model of mantle cell lymphoma45, and a dose escalation study in patients with solid tumors and non-Hodgkin’s lymphoma was recently initiated for PRMT5 inhibitors (clinicaltrials.gov identifier NCT02783300). The PRMT5-WDR77 complex was also shown to remodel gene expression to promote epithelial-to-mesenchymal transition and cancer cell invasion46. Resistance mechanisms may arise for catalytic PRMT5 inhibitors, and targeting WDR77 to inhibit PRMT5 function may represent a strategy for the development of second-generation compounds against tumors that are dependent on PRMT5 activity.
Other oncology targets
Many WDR proteins are implicated in cancer-associated cellular pathways beyond epigenetic mechanisms. Thirteen or more WDR proteins play a role in the ubiquitin proteasome system. One of these, Cell division cycle protein 20 homolog (CDC20), is a co-activator of the anaphase promoting complex (APC) E3 ubiquitin ligase that recruits substrates for subsequent ubiquitination and drives mitosis47. CDC20 is upregulated in numerous cancer types, is repressed by ectopic introduction of the tumor suppressor p53, and silencing of CDC20 induces G2/M arrest and suppresses cancer cell growth48. CDC20 is one of two genes that are systematically upregulated in glioblastoma versus low-grade gliomas, drives the invasiveness and self-renewal of glioblastoma stem-like cells, and CDC20 knockdown induces cell cycle arrest and apoptosis in glioblastoma tumor initiating cells49–51. These results suggest that small molecules disrupting the interaction between CDC20 and APC, or antagonizing CDC20-mediated recruitment of APC substrates could have therapeutic value against glioblastoma, and potentially other cancer types.
Genetic aberrations recurrently found in cancer often drive tumor initiation or tumor growth. For instance, chromosomal translocations recurrently fuse the WDR domain of SEC31 homolog A (SEC31A), a protein involved in the formation of transport vesicles from the endoplasmic reticulum, with the kinase domain of Janus kinase 2 (JAK2) in Hodgkin lymphoma52, and with the kinase domain of ALK in large B-cell lymphomas53,54. In both cases, chemical inhibitors targeting the kinase domain of the oncogenic fusion protein have anti-proliferative effect52,54, and pharmacological targeting of the WDR domain may be an alternative strategy against tumors with SEC31A translocations.
Partner and localizer of BRCA2 (PALB2) is another WDR protein that is recurrently mutated in cancer. Homozygous mutations of PALB2 cause Fanconi anemia and predispose to childhood cancer55,56, while heterozygous mutations resulting in truncation of the WDR domain are found in 1% familial breast cancer57. PALB2 interacts with BRCA2, a breast cancer susceptibility gene, and is required for BRCA2 mediated homologous recombination, double-strand break repair, and tumor suppression58. Poly(ADP-ribose) polymerase 1 (PARP1) facilitates DNA repair, and PARP1 inhibitors are in advanced clinical trials in BRCA2 mutated breast cancer 59. Cancer-associated PALB2 mutants lose their ability to bind BRCA2, and PALB2 mutation resulting in loss of the WDR domain predicts exceptional in vivo response to PARP1 inhibitors60,61. These data suggest that compounds targeting the WDR domain of PALB2 could be of value in tumors where genetic lesions affect other components of the DNA-repair machinery.
Regenerative medicine
WDR proteins are promising candidate targets in the emerging field of regenerative medicine. Chromatin assembly factor 1 subunit B (CHAF1B) mediates chromatin assembly in DNA replication62, and an RNAi screen identified CHAF1A and CHAF1B as the top targets whose loss induces the reprogramming of mouse fibroblasts into pluripotent stem cells63. Suppression of CHAF1B also enhanced the conversion of B cells into macrophages and fibroblasts into neurons63. CHAF1B knockdown increased chromatin accessibility at enhancer domains associated with activation of pluripotency genes, confirming an epigenetic function of CHAF1B in maintaining somatic cell identity. CHAF1B is a subunit of the Chromatin assembly factor 1 (CAF-1) complex, and it is likely that the WDR domain of CHAF1B plays a structural role in stabilizing CAF-1. This suggests that compounds targeting the WDR of CHAF1B could inhibit CAF-1 function and predispose to a pluripotent chromatin landscape.
Interestingly, WDR5 has also recently been associated with pluripotency. Disruption of the MLL complex with the WDR5 antagonist MM-401 leads to global redistribution of mono-methylated H3K4 at enhancer elements, represses lineage determinant genes, and reprograms mouse epiblast stem cells to naive pluripotency, such that over 50% of treated epiblasts exhibit naive embryonic stem cell features after 3 days of treatment64.
Neurological disorders
The WDR protein platelet activating factor acetylhydrolase 1b regulatory subunit 1 (PAFAH1B1, also know as LIS1) is required for actin polymerization in cerebellar and hippocampal neurons, and functions with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration65. Deletion and point mutations of PAFAH1B1 cause neuronal migration defects, and can lead to neurological disorders and brain malformation66,67. Conversely, overexpression of PAFAH1B1 resulting from genomic amplification is associated with cerebellar atrophy in human, and transgenic mice overexpressing PAFAH1B1 have impaired neural migration and smaller brain68. Mapping of point mutations associated with neural migration defects indicate that the WDR domain of PAFAHB1 plays a central role in neural migration68, and a compound that targets the WDR domain of PAFAH1B1 may reduce the pathogenic effect of PAFAH1B1 amplification.
Mutation of Gly2019 to serine in leucine rich repeat kinase 2 (LRRK2) is responsible for 1% of sporadic and 4% of hereditary Parkinson’s disease69. LRRK2 contains multiple domains, including a kinase, a GTPase and a WDR domain. Mutations driving Parkinson’s disease augment the kinase activity of LRRK2, and the neuronal toxicity of mutant LRRK2 can be reduced by alterations that decrease its kinase activity70,71, placing the catalytic activity at root of the pathology. Chemical inhibitors targeting the kinase or GTP binding domains of LRRK2 protect against models of Parkinson’s disease, further supporting mutated LRRK2 as a target for therapy72–74. An LRKK2 construct that lacks the WDR domain is unable to auto-phosphorylate, and loss of the WDR domain blocks the neurotoxicity of multiple LRRK2 mutations75,76. Pharmacological targeting of the WDR domain could therefore represent an alternative strategy to ameliorate LRRK2 defects that cause Parkinson’s disease.
Other disease areas
The WDR protein G protein subunit beta 3 (GNB3) is a Gβ subunit of a G protein complex that transduces GPCR signals to intracellular signaling events and is highly expressed in the brain. The C825T synonymous polymorphism results in a splice variant with increased G protein activation and is associated with hypertension, diabetes and obesity77,78. Duplication of a genomic region encompassing GNB3 is recurrently observed in children with a syndrome associated with obesity, intellectual disability, and seizures, and transgenic mice carrying an extra copy of GNB3 have an increased body mass index79. These results connect GNB3 gene dosage to obesity. Like all other Gβ subunits, GNB3 is composed entirely of a seven bladed WDR domain, and chemical probes that inhibit GNB3 interactions would be useful to investigate the merit of targeting this WDR protein in syndromes associated with GNB3-duplication. More generally, similar arguments would apply to the other Gβ subunits that mediate signaling responses to the ~800 known GPCRs in humans.
Receptor for activated C kinase 1 (RACK1) is a ubiquitous WDR protein that acts as a binding scaffold with a diverse array of interactors, including protein kinases, membrane receptors and 40S ribosomal subunits, and is associated with multiple, cell type–specific functions80,81. Receptor activator of nuclear factor κB (NF-κB) ligand (RANKL) is a cytokine that elicits differentiation of bone marrow cells into osteoclasts and activation of bone resorption, leading to bone loss, via a nuclear factor κB (NF-κB) and mitogen-activated protein kinases (MAPKs) signaling pathway involving dual specificity mitogen-activated protein kinase kinase 6 (MAP2K6)82. RACK1 acts as a scaffolding protein that recruits MAP2K6 to the MAPK signaling cascade triggered by RANKL, and silencing RACK1 reduces RANKL-induced bone loss in mice83. Small molecules disrupting the scaffolding function of RACK1, a protein exclusively composed of a WDR domain, may phenocopy the anti-osteoclastic effect of siRNAs. RACK1 also facilitates translation by internal ribosome entry site (IRES)-containing viruses and is an essential host factor for HCV infection, such that it may represent a broad antiviral target84. Considering the ubiquitous nature and multiple functions of RACK1, the effects of its inhibition may depend on cell-type context, although silencing of RACK1 does not seem to overly affect cell viability and proliferation84.
DDB1 and CUL4 associated factor 1 (DCAF1, also known as VPRBP) is another WDR protein and putative anti-viral host target. DCAF1 is composed of an atypical kinase domain that phosphorylates nucleosomal histone 2A85, and a WDR domain that recruits substrate proteins to the CUL4A-RBX1-DDB1-DCAF1 E3 ubiquitin ligase complex for subsequent proteasomal degradation86. In human immunodeficiency virus 1 (HIV-1), the viral protein Vpr binds DCAF1 to hijack the CUL4A-RBX1-DDB1-DCAF1 complex to anti-viral proteins such as uracil DNA glycosylase (UNG), thereby aberrantly committing protective host proteins for proteasome-mediated degradation87. Similarly, the HIV-2 protein Vpx binds DCAF1 to usurp the CUL4A-RBX1-DDB1-DCAF1 complex for degradation of the anti-viral protein SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1)88. Compounds that target the WDR domain of DCAF1 may thus antagonize the illicit degradation of host defense factors by different viruses.
Pharmacologically targeting WDR domains
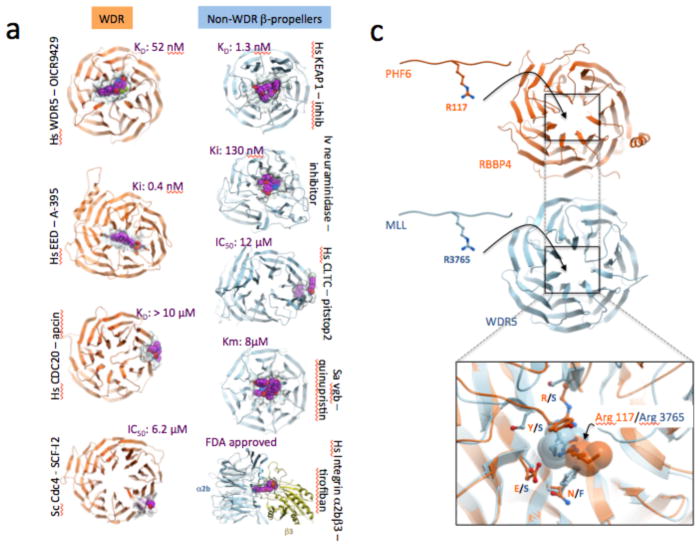
Considering that WDR domains are ubiquitous and often disease-associated, it comes as a surprise that only four WDR proteins have been targeted pharmacologically so far: human WDR5, EED, CDC20 and yeast Cdc424–26,89,90 These four published cases can be further grouped into two categories, depending on the specific location of the inhibitors. The WDR5 and EED inhibitors occupy the central pocket and bind with nanomolar potency24–26, while the other inhibitors occupy side cavities and bind in the micromolar range89,90 (Figure 4a). OICR-9429, a potent and selective chemical probe targeting WDR5, was derived from a hit from a medium-throughput screen of a diverse library of 16,000 compounds using a fluorescence polarization assay to measure the displacement of MLL1 peptide, followed by multiple rounds of structure-guided optimization24,91–93. OICR-9429 binds WDR5 with an affinity of 50 nM as measured by isothermal titration calorimetry, inhibits the co-immunoprecipitation of an MLL peptide with WDR5 with an IC50 of 223 nM, and elicits a clear phenotypic response in cellular assays at 5 μM24. A-395, a potent and novel protein-protein interaction inhibitor that binds EED to inhibit the PRC2 complex, was initially identified via a high-throughput, small molecule screen utilizing a thermal shift assay (TSA) with EED protein, and subsequently optimized by incorporating a strategic sp3-rich ring constraint and appending polar functionalities to increase potency while decreasing cLogP25,94. EED26, another EED-targeting chemical probe, was discovered via a high-throughput campaign to find compounds inhibiting the catalytic activity of the reconstituted PRC2 complex26. Elucidating the mechanism of action of the screening hit enabled structure-guided fragmentation, regrowth and optimization into a potent, selective EED inhibitor95,96. A-395 and EED226 display target-based cellular activity and significant in vivo efficacy in mouse tumor models and are valuable chemical tools to further interrogate the role of the PRC2 complex in tumor initiation and maintenance25,26.
Figure 4. Exemplar inhibitors of WDR and related β-propeller proteins.
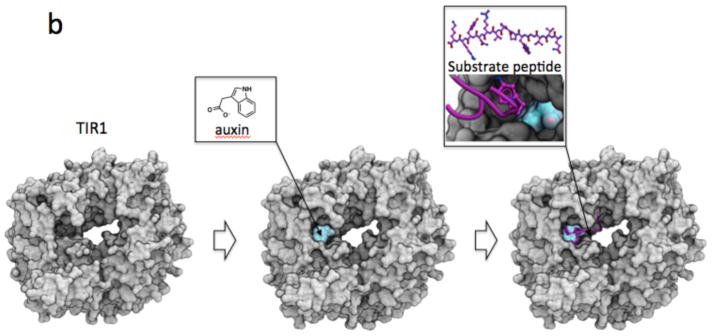
(a) To date, compounds exploiting the central pocket of WD40 and other β-propeller domains are more potent than compounds that bind side-cavities. (b) The small molecule hormone agonist auxin binds the central cavity of TIR1 to promote recruitment of a substrate peptide. (c) The central pockets of RBBP4 and WDR5 are both occupied by an arginine (from PHF6 and MLL respectively), but are not conserved.
Small molecule ligands that bind WDRs on the side of the donut naturally tend to insert between the WD40 blades and may occlude substrate interactions, as in the case of the Cdc20 inhibitor apcin, which competitively blocks interaction of the destruction box (D-box) motif in APC substrates90. The conformational flexibility of WDRs also lends itself to allosteric control of protein interactions, as illustrated in the case of Cdc4, the yeast ortholog of human FBWX7. A small molecule called SCF-I2 binds to a cryptic allosteric pocket on the side of the WD40 domain of Cdc4 and thereby triggers an elaborate series of main chain rearrangements that result in partial occlusion of the main binding pocket that recognizes Cdc4 phospho-degron (CPD) motifs in substrates89. Interestingly, NMR evidence suggests that the allosteric pocket also interacts weakly with CPD motifs, suggesting a mechanism whereby multi-CPD substrates may engender the dynamic exchange of sites in the main binding pocket in a single WD4097. Whether other WDR proteins are susceptible to allosteric inhibition remains to be seen.
Extending the analysis to β-propeller domains that are structurally related but do not belong to the WDR family, ligands that target the central cavity of the Kelch domain protein KEAP1 and viral neuraminidase are nanomolar inhibitors98–100, while pitstop2, a compound that exploits a side pocket of the clathrin terminal domain (CLTC), binds with micromolar potency101. The cyclic peptide antibiotic quinupristin, a component of the streptogramin combination therapy Synercid, targets the 50S ribosomal subunit, but also binds with micromolar affinity to the central cavity of the virginiamycin B lyase β-propeller domain, leading to linearization of the cyclic antibiotic, and drug resistance100. The anti-platelet drug Tirofiban utilizes a third mechanism of action where juxtaposed cavities from the β-propeller domain of integrin α2b and an unrelated domain of integrin β3 both contribute to the binding pocket102. In yet another structural mechanism, the small molecule hormone auxin acts as an agonist of the β-propeller protein TIR1 by occupying a small cleft in the central cavity at the interface of TIR1 and its substrate peptide IAA7, thereby stabilizing the TIR1-IAA7 interaction to enable IAA7 ubiquitination by the SCFTIR1 complex103. Similarly, jasmonate acts as a molecular glue at the interface of the β-propeller F-box protein COI1 and a JAZ1 substrate peptide104.
Importantly for drug discovery, the WDR pockets are structurally diverse, owing to the low sequence identity of WDR domains, and these pockets are intrinsically malleable due to the conformational flexibility of the β-propeller domain. This structural diversity even extends to cases when pockets share similar ligands. For instance, arginine side-chains of PHF6 and MLL serve as anchoring residues in the central pockets of RBBP4 and WDR5 respectively, but significant differences in side-chains at key positions surrounding the bound arginines are found between RBBP4 and WDR5 (Figure 4c). This suggests that achieving selectivity within this target class may not be as much of a challenge as for other protein classes that share binding pockets with similar chemical features, such as kinases or bromodomains. Indeed, the highly potent antagonists of EED do not bind to WDR5, nor does the potent WDR5 inhibitor OICR9429 bind to EED24,25.
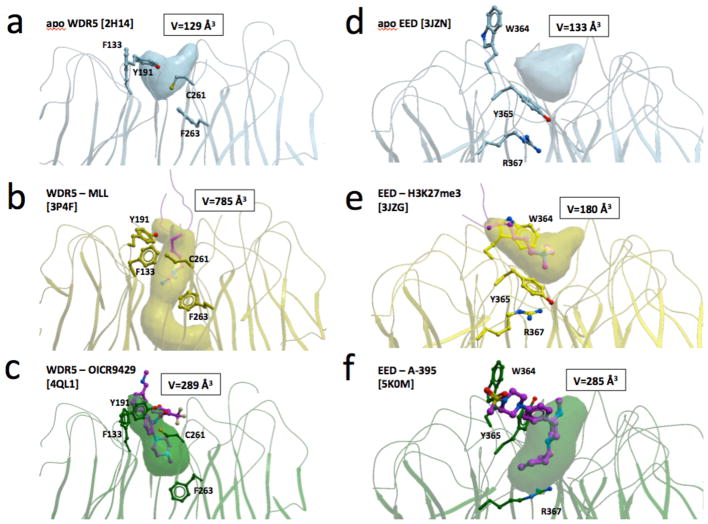
Conformational rearrangement of a limited number of side-chains can have dramatic impact on the size and geometry of the WDR central pocket. For instance, four residues (F133, Y191, C261 and F263) control the volume of the top pocket of WDR5. In particular, F263 obstructs the cavity in the apo state, is partially open in the inhibitor-bound state, and is forced by the arginine of the MLL peptide into a fully open state where the bottom of the cavity disappears and the pocket is replaced with a tunnel that connects both sides of the WDR domain (Fig 5a–c)24,105. Similarly, the geometry of the EED inhibitor-binding site is regulated by the conformation of W364, Y365 and R367. While the pocket is shallow in the apo and histone-bound states, conformational rearrangement of Y365 and adjacent residues (engaged either directly or indirectly in aromatic cage formation) produces a larger, deeper cavity that can accommodate small-molecule inhibitors (Fig 5d–f)25,26,94–96.
Figure 5. Structural dynamics of WDR binding pockets.
Conformational rearrangement of a few side-chains can induce dramatic changes in the volume and enclosure of the central pockets of WDR5 (a–c) or EED (d–f). Conformations of mobile side-chains and associated pocket volumes are shown for apo, peptide-bound and inhibitor-bound structures.
Taken collectively, all these pioneering efforts to target WDR and other β-propeller domains suggest that the central cavity of the propeller may be the most tractable site for drug discovery, albeit the possibility of targeting other WD40 domain surfaces cannot be ruled out.
The merits of targeting a protein interaction domain
Considering their ubiquitous nature, disease association, structural diversity and encouraging druggability, it is surprising that WDRs have so far been largely neglected by the drug discovery community. One explanation may be that WD40 domains are typically scaffolding rather than catalytic subunits of protein complexes. To inhibit a methyltranferase, a deubiquitinase or a histone acetyltransferase, the natural inclination is to target the catalytic domain, particularly if a well-defined druggable pocket is evident. However, targeting the catalytic site is not always readily achievable, and for many enzyme classes this is still very challenging. For instance, despite its potential as an oncology target, no catalytic inhibitor of the methyltransferases MLL1 has been reported to date. Nevertheless, the methyltransferase function of MLL1 can be antagonized in cells by the WDR5 inhibitor OICR-942924. Similarly, some of the intense but so far unsuccessful drug discovery efforts that have focused on ubiquitin ligases and deubiquitinases for the past decade might be more effective if redirected towards the numerous WDR domains implicated in the UPS (Fig. 3).
Targeting WDRs that bind to druggable enzymes can also have merit. The epigenetic regulator PRC2 is an instructive example to consider. Prolonged treatment of cancer cells with catalytic domain inhibitors of the PRC2 complex elicits a resistance mechanism triggered by mutations at the inhibitor-binding site. However, allosteric inhibitors targeting the WDR subunit, EED25,26, retain on-target inhibition in cells resistant to catalytic inhibitors. Moreover, unlike catalytic inhibitors, prolonged treatment with an EED inhibitor failed to select for resistance mutations25. In another example, synthetic lethal pairs between components of the SWI/SNF chromatin remodeling complex and EZH2 rely in some cases on a structural rather than enzymatic function of EZH2, and EED inhibitors that disrupt the PRC2 complex, such as an EZH2-stapled helix, are active when catalytic EZH2 inhibitors are not37. Thus, targeting the WDR domain interaction hub of PRC2 offers several complementary approaches compared to targeting the catalytic domain.
Outlook
Recent successful pharmacological targeting of WDR5 and EED with drug-like molecules does not imply that all WDR domains are druggable. The variability in shape and electrostatics of the WDR central pocket suggests that this protein family harbours a continuum of structures with varying degrees of chemical tractability. Considering the conformational malleability observed at the WDR5 and EED inhibitor binding sites (Fig. 5), structural studies of apo or peptide-bound structures may not always accurately predict the druggability of the central pocket. Where WDR domains behave as docking platforms for electrostatically-charged peptides, it will be challenging to find inhibitors sufficiently polar to compete with substrates, while simultaneously sufficiently hydrophobic to cross cell membranes. It is also unclear whether the weak-binding compounds targeting side-pockets of WDR domains reflect fundamentally poor druggability of these sites, or simply the paucity of drug-discovery efforts focused on these sites to date. Considering the size of the WDR family, and recent progresses in targeting protein-protein interactions, we expect that additional WDR proteins will prove druggable in the future.
An unknown variable in targeting WDR domains is the potential effects of perturbing multiple protein complexes. For instance, RBBP7 is part of the type B histone acetyltransferase complex required for chromatin assembly following DNA replication, but is also a component of the NURD histone deacetylase and nucleosome remodeling complex and the PRC2 methyltransferase complex, all three of which are associated with transcriptional repression. In another example, the WDR protein RBBP5 is a component with varying degrees of essentiality in the MLL1, MLL2, MLL3, MLL4, SET1A and SET1B complexes36. The combined disruption of multiple independent molecular machineries may result in either increased overall efficacy or unexpected phenotypic outcomes, depending on the context. This reality argues strongly for the use of chemical probes to validate WDR domain targets using appropriate disease models in order to explore potential therapeutic windows106–108.
The presence of WDR repeat proteins in diverse E3 ubiquitin ligase complexes, and the seminal discovery of natural small molecule ligands that bridge E3-substrate interactions in plants, raises the possibility that bi-dentate small molecules that simultaneously bind the WDR domain of the E3 and a therapeutic target protein of interest may enable the proteasome-mediated elimination of the target protein. Indeed, thalidomide and its derivatives have been discovered to act in exactly this fashion, by bridging an ectopic interaction between the CUL4 substrate receptor subunit cereblon (CRBN) and the transcription factors Ikaros and Aiolos, which leads to their degradation and efficacy against B-cell malignancies109,110. This molecular glue concept has been elaborated in the form of “protein-targeting chimeric molecules”, or Protacs111,112, which in some cases have yielded a phenotypic outcome that is superior to mono-functional ligands inhibiting a specific domain of the target protein113. This principle may be more general than anticipated since it was recently found that anticancer sulfonamides ectopically target the oncoprotein RBM39 to the WDR protein DCAF15 for ubiquitination by the CUL4 complex114,115. It is not clear yet whether these sulfonamide drugs bind the WDR domain or another region of DCAF15, but E3-associated WDR domains may represent interesting general alternatives to CRBN, with diverse tissue distributions and availability profiles for harnessing chemically the ubiquitin-proteasome system.
In conclusion, the ubiquitous, functionally versatile, and often disease-associated WDR domain has a structurally diverse pocketome, and is probably more druggable than meets the eye, thanks to its intrinsic conformational flexibility. Consequently WDRs offer potential avenues to target catalytic, scaffolding and substrate or activator recognition functions of diverse enzyme complexes. Recent encouraging results for WDR inhibitors in epigenetic regulation hold great potential but still need to be validated in the clinic. The chemical tractability of WDR domains as a target class should be further explored, especially in areas of disease biology that have so far proven undruggable by conventional approaches.
Supplementary Material
Acknowledgments
The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute [OGI-055], Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, Sao Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. MT is supported by grants from the Canadian Institute for Health Research (MOP 126129), the Canadian Cancer Society Research Institute (703906), Genome Canada, NIH (R01OD010929) and holds a Canada Research Chair in Systems and Synthetic Biology. The authors acknowledge Clarissa Jakob for her careful review of the manuscript.
Footnotes
Author contributions
MS, MT, MT and CHA contributed to the manuscript.
References
- 1.Deeks ED. Venetoclax: First Global Approval. Drugs. 2016;76:979–87. doi: 10.1007/s40265-016-0596-x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts AW, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2016;374:311–22. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–41. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 5.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 9.Annis DA, Nickbarg E, Yang X, Ziebell MR, Whitehurst CE. Affinity selection-mass spectrometry screening techniques for small molecule drug discovery. Curr Opin Chem Biol. 2007;11:518–26. doi: 10.1016/j.cbpa.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–54. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 11.Pearce NM, et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat Commun. 2017;8:15123. doi: 10.1038/ncomms15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lea WA, Simeonov A. Fluorescence polarization assays in small molecule screening. Expert Opin Drug Discov. 2011;6:17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juhasz T, Szeltner Z, Fulop V, Polgar L. Unclosed beta-propellers display stable structures: implications for substrate access to the active site of prolyl oligopeptidase. J Mol Biol. 2005;346:907–17. doi: 10.1016/j.jmb.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94:161–70. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 15.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–74. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Migliori V, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–44. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 18.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–43. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 19.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–56. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, et al. WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res. 2015;43:D339–44. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Jiang F, Zhuo Z, Wu XH, Wu YD. A method for WD40 repeat detection and secondary structure prediction. PLoS One. 2013;8:e65705. doi: 10.1371/journal.pone.0065705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CK, Chan NL, Wang AH. The many blades of the beta-propeller proteins: conserved but versatile. Trends Biochem Sci. 2011;36:553–61. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grebien F, et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat Chem Biol. 2015;11:571–8. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- 26.Qi W, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 27.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–64. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, et al. Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Hum Mol Genet. 2016;25:927–35. doi: 10.1093/hmg/ddv625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 30.Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 31.Shortt J, Ott CJ, Johnstone RW, Bradner JE. A chemical probe toolbox for dissecting the cancer epigenome. Nat Rev Cancer. 2017;17:160–183. doi: 10.1038/nrc.2016.148. [DOI] [PubMed] [Google Scholar]
- 32.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–11. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiel AT, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–59. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao F, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53:247–61. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, et al. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016;530:447–52. doi: 10.1038/nature16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–6. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–50. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garapaty-Rao S, et al. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem Biol. 2013;20:1329–39. doi: 10.1016/j.chembiol.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 40.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 41.Konze KD, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–34. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 43.Wolffe AP, Urnov FD, Guschin D. Co-repressor complexes and remodelling chromatin for repression. Biochem Soc Trans. 2000;28:379–86. [PubMed] [Google Scholar]
- 44.Kitange GJ, et al. Retinoblastoma Binding Protein 4 Modulates Temozolomide Sensitivity in Glioblastoma by Regulating DNA Repair Proteins. Cell Rep. 2016;14:2587–98. doi: 10.1016/j.celrep.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan-Penebre E, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–7. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 46.Chen H, Lorton B, Gupta V, Shechter D. A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2017;36:373–386. doi: 10.1038/onc.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 48.Kidokoro T, et al. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene. 2008;27:1562–71. doi: 10.1038/sj.onc.1210799. [DOI] [PubMed] [Google Scholar]
- 49.Mao DD, et al. A CDC20-APC/SOX2 Signaling Axis Regulates Human Glioblastoma Stem-like Cells. Cell Rep. 2015;11:1809–21. doi: 10.1016/j.celrep.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Q, et al. CDC20 maintains tumor initiating cells. Oncotarget. 2015;6:13241–54. doi: 10.18632/oncotarget.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marucci G, et al. Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Arch. 2008;453:599–609. doi: 10.1007/s00428-008-0685-7. [DOI] [PubMed] [Google Scholar]
- 52.Van Roosbroeck K, et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood. 2011;117:4056–64. doi: 10.1182/blood-2010-06-291310. [DOI] [PubMed] [Google Scholar]
- 53.Bedwell C, et al. Cytogenetically complex SEC31A-ALK fusions are recurrent in ALK-positive large B-cell lymphomas. Haematologica. 2011;96:343–6. doi: 10.3324/haematol.2010.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Roosbroeck K, et al. ALK-positive large B-cell lymphomas with cryptic SEC31A-ALK and NPM1-ALK fusions. Haematologica. 2010;95:509–13. doi: 10.3324/haematol.2009.014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 56.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–4. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 57.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 60.Smith MA, et al. Initial testing (stage 1) of the PARP inhibitor BMN 673 by the pediatric preclinical testing program: PALB2 mutation predicts exceptional in vivo response to BMN 673. Pediatr Blood Cancer. 2015;62:91–8. doi: 10.1002/pbc.25201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 62.Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–75. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheloufi S, et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 2015;528:218–24. doi: 10.1038/nature15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, et al. MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell. 2016;18:481–94. doi: 10.1016/j.stem.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka T, et al. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–21. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haverfield EV, Whited AJ, Petras KS, Dobyns WB, Das S. Intragenic deletions and duplications of the LIS1 and DCX genes: a major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur J Hum Genet. 2009;17:911–8. doi: 10.1038/ejhg.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 68.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–77. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Healy DG, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7:583–90. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith WW, et al. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–3. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 71.West AB, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li T, et al. Novel LRRK2 GTP-binding inhibitors reduced degeneration in Parkinson’s disease cell and mouse models. Hum Mol Genet. 2014;23:6212–22. doi: 10.1093/hmg/ddu341. [DOI] [PubMed] [Google Scholar]
- 73.Deng X, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011;7:203–5. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee BD, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kett LR, et al. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum Mol Genet. 2012;21:890–9. doi: 10.1093/hmg/ddr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jorgensen ND, et al. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One. 2009;4:e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klenke S, Kussmann M, Siffert W. The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet Genomics. 2011;21:594–606. doi: 10.1097/FPC.0b013e3283491153. [DOI] [PubMed] [Google Scholar]
- 78.Siffert W, et al. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18:45–8. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 79.Goldlust IS, et al. Mouse model implicates GNB3 duplication in a childhood obesity syndrome. Proc Natl Acad Sci U S A. 2013;110:14990–4. doi: 10.1073/pnas.1305999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–41. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–73. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 82.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–42. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 83.Lin J, Lee D, Choi Y, Lee SY. The scaffold protein RACK1 mediates the RANKL-dependent activation of p38 MAPK in osteoclast precursors. Sci Signal. 2015;8:ra54. doi: 10.1126/scisignal.2005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Majzoub K, et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159:1086–95. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim K, et al. VprBP has intrinsic kinase activity targeting histone H2A and represses gene transcription. Mol Cell. 2013;52:459–67. doi: 10.1016/j.molcel.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Angers S, et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–3. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 87.Wu Y, et al. The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat Struct Mol Biol. 2016;23:933–940. doi: 10.1038/nsmb.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schwefel D, et al. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature. 2014;505:234–8. doi: 10.1038/nature12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orlicky S, et al. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–7. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sackton KL, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–9. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolshan Y, et al. Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction. ACS Med Chem Lett. 2013;4:353–7. doi: 10.1021/ml300467n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Getlik M, et al. Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1) J Med Chem. 2016;59:2478–96. doi: 10.1021/acs.jmedchem.5b01630. [DOI] [PubMed] [Google Scholar]
- 93.Senisterra G, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449:151–9. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Curtin ML, et al. SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding. Bioorg Med Chem Lett. 2017;27:1576–1583. doi: 10.1016/j.bmcl.2017.02.030. [DOI] [PubMed] [Google Scholar]
- 95.Huang Y, et al. Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy. J Med Chem. 2017;60:2215–2226. doi: 10.1021/acs.jmedchem.6b01576. [DOI] [PubMed] [Google Scholar]
- 96.Li L, et al. Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED. PLoS One. 2017;12:e0169855. doi: 10.1371/journal.pone.0169855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Csizmok V, et al. An allosteric conduit facilitates dynamic multisite substrate recognition by the SCFCdc4 ubiquitin ligase. Nat Commun. 2017;8:13943. doi: 10.1038/ncomms13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davies TG, et al. Monoacidic Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery. J Med Chem. 2016;59:3991–4006. doi: 10.1021/acs.jmedchem.6b00228. [DOI] [PubMed] [Google Scholar]
- 99.Kerry PS, et al. Structural basis for a class of nanomolar influenza A neuraminidase inhibitors. Sci Rep. 2013;3:2871. doi: 10.1038/srep02871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Korczynska M, Mukhtar TA, Wright GD, Berghuis AM. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc Natl Acad Sci U S A. 2007;104:10388–93. doi: 10.1073/pnas.0701809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.von Kleist L, et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146:471–84. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 102.Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. J Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–5. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 104.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–5. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Avdic V, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–8. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 106.Edwards AM, et al. Preclinical target validation using patient-derived cells. Nat Rev Drug Discov. 2015;14:149–50. doi: 10.1038/nrd4565. [DOI] [PubMed] [Google Scholar]
- 107.Bunnage ME, Chekler EL, Jones LH. Target validation using chemical probes. Nat Chem Biol. 2013;9:195–9. doi: 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- 108.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 109.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–9. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–81. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2001;98:8554–9. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saenz DT, et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia. 2017 doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Uehara T, et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat Chem Biol. 2017 doi: 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- 115.Han T, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356 doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.