INTRODUCTION
The BioGRID database (www.thebiogrid.org) is an extensive repository of curated genetic and protein interactions for the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, and the yeast Candida albicans SC5314, as well as for several other model organisms and human. This protocol describes how to use the BioGRID website to query genetic or protein interactions for any gene of interest, how to visualize the associated interactions using an embedded interactive network viewer, and how to download data files for either selected interactions or the entire BioGRID interaction data set.
RELATED INFORMATION
BioGRID (Chatr-Aryamontri et al. 2013) is a member of the International Molecular Exchange (IMEX) consortium (http://www.imexconsortium.org/) (Orchard et al. 2012), which is a network of public databases that provide curated protein interaction data in a standardized Proteomics Standards Initiative Molecular Interaction (PSI-MI) format (http://www.psidev.info/). The BioGRID Post-Translational Modification (PTM) viewer displays phosphorylation sites housed in PhosphoGRID (www.phosphogrid.org), a database of experimentally verified phosphosites curated from the S. cerevisiae primary literature (Sadowski et al. 2013). BioGRID is also a source of protein and genetic interaction data for the following model organism databases: Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org) (Cherry et al. 2012), FlyBase (http://flybase.org/) (Marygold et al. 2013), PomBase (http://www.pombase.org/) (Wood et al. 2012), TAIR (http://www.arabidopsis.org/) (Lamesch et al. 2012), and WormBase (http://www.wormbase.org) (Yook et al. 2012). Visualization tools compatible with BioGRID formats include Osprey (http://biodata.mshri.on.ca/osprey/servlet/Index) (Breitkreutz et al. 2003b), Cytoscape (http://www.cytoscape.org/) (Shannon et al. 2003; Cline et al. 2007) using the BioGRIDPlugin2 (Winter et al. 2011), and GeneMania (http://www.genemania.org/) (Montojo et al. 2010; Zuberi et al. 2013).
MATERIALS
Equipment
Computer (internet-connected with web browser)
BioGRID is compatible with all modern web-browsers including Firefox, Chrome, Safari and Internet Explorer, but is not compatible with Internet Explorer 6 and earlier versions.
METHOD
Search interactions for a gene or protein of interest
This procedure describes how to search BioGRID for interactions that involve a gene or protein of interest, view additional details about interactions and associated post-translational modifications, visualize the local network of interactions in the BioGRID viewer or Cytoscape, and download selected interactions or the entire collection of curated interaction data.
-
1
Use any modern web browser, except Internet Explorer 6, to open the following URL: http://thebiogrid.org/
-
2
Locate the BioGRID search form in the upper right corner.
BioGRID may be searched “By gene” or “By publication”. These options are available as vertical tabs on the right hand side of the search form. When searching by publication, it is possible to search for a PubMed Identifier or by keywords found in the title or abstract. When searching for a specific protein or gene, the search can be restricted to an organism of interest, or performed on all 29 organisms simultaneously. Additional instructions are provided from the “Advanced Search Options” and “Search Tips” links. Any of the following common types of search terms will be recognized by the gene search field:
Gene names (e.g., SWE1), gene accession numbers from NCBI (e.g., 853252) or RefSeq (e.g., NM_001181620)
Protein identifiers from UniProtKB (e.g., P32944) or NCBI RefSeq (e.g., NP_012348)
Systematic identifiers from the corresponding model organism database, such as the Saccharoymyces Genome Database (SGD) (e.g., YJL187C).
A complete list of supported search terms is provided under the “Search Tips” link.
-
3
Enter a search term, e.g. SWE1, and select Saccharomyces cerevisiae from the organism pull down menu, then press the “SUBMIT GENE SEARCH” button.
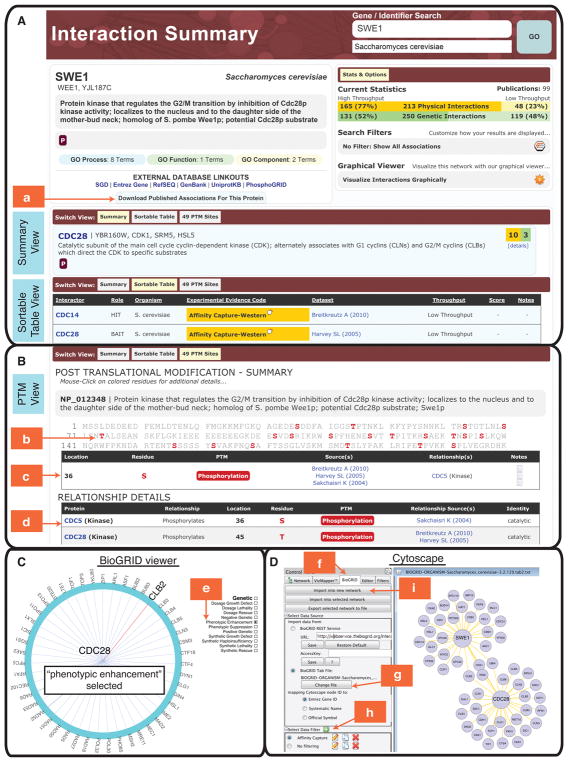
This action will return an Interaction Summary page (Fig. 1A), which provides a summary of curated interactions for the gene(s) or protein(s) of interest.
At the top left of the page, a brief description of the gene/protein is given with links to relevant external resources, such as SGD, GenBank and UniProtKB, as well as an option to download associated interactions.
In the upper right “Stats & Options” panel, the curated interaction statistics are provided for the gene or protein of interest. The total number and kind of interactions are listed and are color-coded throughout the page in green for genetic interactions and in yellow for protein interactions. The interactions may also be filtered via the “Search Filters” option according to whether they are derived from low versus high throughput experimental studies. A link to a graphical viewer which allows for visualization of the gene- and/or protein-specific interaction network, is also available, and is described in more detail in step 7.
A full summary table displays the list of unique interactors, with the total number of independent evidence codes for genetic and protein interactions shown on the far right with further information provided in the “details” link underneath (Fig. 1A Summary View).
-
4
Click on the “details” link on the right side of the table to obtain information about each of the listed interaction partners.
This action reveals a detailed list of the different experimental evidence codes associated with each interaction, including the context of the interactor as either a bait or hit, and whether the experimental evidence is from a low or high throughput study. The different types of interaction data curated by BioGRID are described in the “Experimental Evidence Codes” help document (http://wiki.thebiogrid.org/doku.php/experimental_systems). Pointing to specific icons in the right-hand “Notes” column reveals further applicable details under different icons:
Plus Icon: phenotypes curated by BioGRID using the Ascomycete Phenotype Ontology (APO) developed by SGD (Engel et al. 2010)
Globe icon: post-translational modifications associated with the in vitro “Biochemical Activity” experiment type, most commonly phosphorylation or ubiquitination.
Histogram icon: quantitative scores published in the original low throughput study or high-throughput data set, when available.
Tablet icon: additional notes provided by BioGRID curators to explain experimental details or significance scores.
A “Switch View” bar at the top of the table may be used to navigate from the “Summary” view to a “Sortable Table”, as shown for SWE1 (Fig. 1A). By clicking on the column headers, this view can be sorted by interactor, role (bait/hit), organism, experimental evidence code, dataset publication, throughput (high/low), or interaction scores provided in the original publications. The “Switch View” bar may also be used to access the “PTM Sites” page corresponding to the Post-Translational Modification (PTM) Viewer for the query gene of interest.
-
5
Click on the “PTM Sites” button to access the Post-Translational Modification (PTM) viewer (Fig. 1B).
The BioGRID PTM viewer integrates over 20,100 phosphorylation sites on approximately 3,200 yeast proteins, curated by PhosphoGRID (version 2.0) from both low and high throughput studies (Sadowski et al. 2013). More than 2,500 of these phosphosites are associated with specific kinases and/or phosphatases, and over 3,200 of the sites occur on 1,100 proteins under specific environmental conditions. The PTM viewer will be expanded to include sites of ubiquitin modification and other modifications in the future.
Phosphorylation sites in the protein sequence are shown in red (Fig. 1B, arrow b). Click on a modified residue in the protein sequence to display links to publications with supporting evidence.
Refer to the table below the protein sequence that lists the individual residues and their phosphorylation sites (Fig. 1B, arrow c), along with the interacting proteins and their relationship, such as kinase or phosphatase, as applicable. Additional notes associated with each phosphosite are provided in the right hand “Notes” column under a tablet icon.
Scroll down to a separate “Relationship Details” table below (Fig. 1B, arrow d) which provides information about the enzymes involved in the PTMs, identifying whether each protein listed in the left column phosphorylates or dephosphorylates the query protein of interest.
-
6
Navigate to another gene of interest in BioGRID by clicking on any gene name link listed in the different table views.
For example, in the “PTM sites” view, click on any interacting gene listed on the left hand side of the “Relationship Details” table, e.g. CDC28 (Fig. 1B, arrow d). This takes the user to the “Interaction Summary” page for that particular gene.
Figure 1.
(A) Interaction summary results page that displays information for the query gene or protein. Results for the query gene SWE1 are shown. The Summary View displays unique interactors for the query gene or protein, sorted from highest to lowest number of curated interactions. The Sortable Table View allows results to be ordered based on interactor, role (bait/hit), organism, experimental evidence code, dataset publication, throughput (high/low), or interaction scores given in the original publications. Genetic and protein interactions are color-coded in green and yellow, respectively, throughout the display. Filtering options in the top right “Stats and Options” box may be used to sort the table based on genetic or protein interactions and/or high or low-throughput studies. Genetic and protein interactions for the query gene can be downloaded in all supported BioGRID download formats by clicking “Download Published Associations for this Protein” (arrow a).
(B) Integrated post-translational modification (PTM) viewer within BioGRID. The Swe1 protein sequence is displayed with documented phosphorylation sites shown in red (arrow b). The table below the sequence shows additional details for each phosphosite, including the corresponding enzyme and its relationship to the query protein (arrow c). The relationship table provides further details, such as whether the interactor in the left column phosphorylates or dephosphorylates the main query protein (arow d). The PTM viewer currently incorporates phosphosites curated by PhosphoGRID, but will be expanded to include other types of PTMs, such as ubiquitination sites. also as curated by BioGRID.
(C) Interaction network viewer within BioGRID. A filtered view is shown for interactions of CDC28, which is located at the center of the viewer. Filters are applied by choosing options to the left and right side of the graphic. In this instance, interactions have been filtered to show low throughput interactions only and further restricted to genetic interactions associated with the “Phenotypic Enhancement” experimental evidence code (arrow e). The network can be expanded to show secondary interactor interactions, which reveals local network density and potential protein or genetic interaction sub-networks. Mouse-overs of each node in the network can be used to highlight all connections for that node.
(D) Use of Cytoscape with the BiogridPlugin2.2. After copying the BiogridPlugin2.2.jar into the plugins directory of Cytoscape, select the “BioGRID” tab in the control panel (arrow f). In the “Import data from” window select the “BioGRID Tab File” and choose the tab2 file of interest (arrow g). In the “Select Data Filter” window, click on the green plus symbol (arrow h). After setting up the filter described in step 11 in the text, click the “Import in new network” button to view the interaction network (arrow i).
Interaction network visualization
An interactive network viewer is embedded within the BioGRID interface to allow users to immediately visualize interactions of interest. The viewer shows the interaction network of the gene of interest based on its direct genetic and/or protein interaction partners.
-
7
Click on the “Graphical Viewer” link in the top right “Stats & Options” panel of any gene page (e.g. CDC28).
This action opens up a separate tab or window that graphically displays the interaction network around the query gene in a circular layout. The query gene is represented by a node at the center of the circle, and the interacting genes are displayed as nodes around the outside of the circle; each line (edge) between a pair of genes represents an individual interaction (Fig. 1C).
View basic information regarding the gene of interest and the total number of interactions in the left corner of the display. Statistics are recalculated according to any filtering options selected in the viewer.
Filter the network display via options provided in the top left corner, such as low or high experimental throughput, or whether to include secondary interactions, i.e. interspecies, self or interactor interactions. Note that the latter option can significantly increase the number of interactions displayed.
Select physical or genetic interactions in the upper right panel to limit the number of displayed interactions based on interaction type, or further reduce the numbers by selecting specific experimental evidence codes.
Only those genes or proteins that pass the user defined filters will be displayed in the network viewer.
In the example given in Fig. 1C, the displayed interactions for CDC28 have been restricted to low-throughput genetic interactions annotated as “phenotypic enhancement” (arrow e).
Mouse over any gene (e.g. CLB2) to highlight the edge representing the interaction in red, and double-click on any gene in the network to show the same type of filtered interactions for that particular gene, if applicable. The newly selected gene will be displayed in the center of the circle.
In addition to BioGRID’s integrated network viewer linked from each gene page, selected interaction data may be downloaded in formats compatible with either the Cytoscape (http://www.cytoscape.org/) (Shannon et al. 2003; Cline et al. 2007)) or Osprey (Breitkreutz et al. 2003b; Stark et al. 2006) network visualization systems. These formats include the BioGRID TAB 2.0 delimited file format for Cytoscape, and an Osprey Custom Network File format for the Osprey visualization system. A BioGRID Cytoscape plugin 2.0 (http://wiki.thebiogrid.org/doku.php/biogridplugin2) is available to facilitate the import of BioGRID interaction data into Cytoscape in order to visualize interaction networks and integrate interactions with gene expression profiles (Winter et al. 2011).
-
8
To visualize a network in Cytoscape, first download BioGRID interaction data in tab2 format. Visit the BioGRID Download page (http://thebiogrid.org/download.php) and click on “Current Release”, then click on the “BIOGRID-ORGANISM-(VERSION NUMBER).tab2.zip” to begin downloading the available interaction data which will be broken into distinct files by organism. Open the zip file and locate “BIOGRID-ORGANISM-Saccharomyces_cerevisiae-(VERSION NUMBER).tab2.txt”. This file contains all of the budding yeast interactions from BioGRID and will be used in this example.
-
9
Download a copy of Cytoscape version 2.8.2 from the following url: http://www.cytoscape.org/download.html. Note that the BioGRID Cytoscape plugin is not currently compatible with version 3.x of Cytoscape but will be updated in the future.
-
10
After installing Cytoscape, download the BiogridPlugin version 2.2 located in the folder “BiogridPlugin2” within “Cytoscape Plugins” on the BioGRID download page (http://thebiogrid.org/download.php).
Copy the downloaded file, “BiogridPlugin2.2.jar” into the plugins directory of Cytoscape, located in the folder in which Cytoscape is installed. Typically this will be located in “C:\Program Files\Cytoscape_v2.8.2\plugins” on Windows-based computers, or “/Applications/Cytoscape_v.2.8.2/plugins” on a Mac.
Launch Cytoscape and select the “BioGRID” tab in the control panel (Fig. 1D, arrow f) for a list of input variables for the BioGRID plugin.
In the “Import data from:” window (Fig. 1D, arrow g) select the “BioGRID Tab File:” option box and change the file to the one that was downloaded in step 8.
Interaction data can be filtered using this plugin. As an example, set up a filter to show only Affinity Capture-MS interactions for the budding yeast proteins CDC28 and SWE1.
-
11
Click on the green and white plus symbol just to the right of the “Select Data Filter” window (Fig. 1D, arrow h). This will bring up the filter window.
Assign the filter a name: in this example use “Affinity Capture-MS Filter”. In the “Filter by gene” window, add the protein names CDC28 and SWE1 and make sure the drop down is set to “Include only” and check the box for “and their primary interactors”.
In the “Filter by evidence code (include checked)” window, select the experimental system “Affinity Capture-MS”(Fig. 1D, arrow h). Remove any genetic evidence codes. Click “OK” to save this filter.
Once the filter is set up, click the “Import in new network” button (Fig. 1D, arrow i), which will produce an interaction network containing the query genes of interest and their interactors.
To better visualize the network, select one of the built-in layouts in Cytoscape. Navigate to the “Layout” menu at the top and select “Cytoscape Layouts->Forced-Directed Layout->(unweighted)”. Cytoscape will generate a network display with each query gene at the center of its set of interactors (Fig. 1D).
Download options and formats
Interaction data in BioGRID are updated on a monthly basis and may be downloaded for each query gene or protein, or by publication, in various formats.
-
12
To obtain interactions for a single query gene, on a BioGRID gene page click on the “Download Published Associations for this Protein” button on any gene page above the “switch view” option in the upper left (Fig. 1A, arrow a). This opens up a “BioGRID Downloads” page.
Select the desired file format (documentation available at http://wiki.thebiogrid.org/doku.php/downloads). The BioGRID Tab 2.0 format is the most commonly downloaded tab-delimited text format and is provided as the default option. Other formats include the IMEX-compatible PSI-MI XML or MITAB formats.
Click the “Download Interactions” button to obtain the requested file.
The data may also be downloaded based on publication. This requires searching BioGRID using the “By Publication” option on the home page in the upper right search form. Entering any PubMed ID returns the publication of interest. The “Download Interactions for the Publication” option is available below the abstract. The entire curated interaction data set may also be freely downloaded, and source code for BioGRID is available without any restrictions (http://wiki.thebiogrid.org/doku.php/development_overview).
-
13
To obtain the complete collection of curated interactions, click on the “Downloads” menu option at the top right of any BioGRID page to go to the main download page (http://thebiogrid.org/download.php).
Select the “Current Release” folder to expand a list of downloadable file formats. A description of the different formats is available in the right hand panel of the page.
The interaction data may be downloaded based on organism type or experimental system. In addition, the entire collection is available in various formats, including the BioGRID Tab 2.0 format, and the IMEX-compatible PSI-MI XML or MITAB formats.
-
14
To build a specific data set based on a subset of genes, proteins or publications, click on the “Custom download generator” link towards the bottom of the main download page. This opens up a BioGRID Custom Interaction Dataset Creator page (http://thebiogrid.org/downloadgenerator.php).
Select the PublicationID(s) or Gene Identifier(s) option; the former is for PubMed IDs, and the latter is for official gene symbols, systematic names, and external database IDs. Identifiers that may be used are listed at http://wiki.thebiogrid.org/doku.php/identifiers
Up to five PubMed IDs or ten Gene IDs may be entered per search.
The search may be further refined by selecting an organism of interest, and then choosing an output file format.
Click the “Build Custom Dataset” button to obtain the specific interaction data set.
Interaction data in BioGRID can also be retrieved automatically via the BioGRID REST service. Details regarding the REST service are documented on a wiki page (http://wiki.thebiogrid.org/doku.php/biogridrest) and are also published (Winter et al. 2011).
The documentation includes information on how to obtain a unique access key, which is required for all queries to the web service, and how to fetch and filter interactions for a single query gene. Several examples are given along with a table listing the complete set of query parameters that may be used. A formal description of the REST service is also provided in Web Application Description Language (WADL) at http://webservice.thebiogrid.org/application.wadl. BioGRID is also part of the PSICQUIC project (https://code.google.com/p/psicquic/) (del-Toro et al. 2013), which is part of the HUPO Proteomics Standard Initiative (HUPO-PSI) to standardize automated access to molecular interaction databases by providing a standard web service and common Molecular Interactions Query Language (MIQL).
Community and user feedback
The BioGRID values feedback from the yeast research community and other users to correct curation errors, extend interaction coverage, request new features, or support and/or contribute published or pre-publication interaction data.
Users may provide direct feedback to biogrid.admin@gmail.org, which is monitored daily by the BioGRID team. The BioGRID programming team will assist with extraction of data features, resolution of data discrepancies, and links to BioGRID pages or the REST service. The BioGRID curation team will correct any reported errors in the subsequent monthly release and assist with confidential upload of new datasets to coincide with publication date. Finally, BioGRID welcomes collaborators with an interest in contributing domain expertise or curation capacity. Further details can be found at http://wiki.thebiogrid.org/doku.php/contribute.
DISCUSSION
The BioGRID will continue to expand the curation of protein and genetic interactions from the biomedical literature, as well as associated attributes such as PTMs, protein variants, phenotypes, and chemical or drug interactions. In addition to BioGRID, the following IMEX partners also actively curate and freely disseminate yeast protein interaction data:
MINT (Molecular INTeraction Database: http://mint.bio.uniroma2.it/) (Chatr-aryamontri et al. 2007; Licata et al. 2012)
DIP (Database of Interacting Proteins: http://dip.doe-mbi.ucla.edu/) (Xenarios et al. 2002; Salwinski et al. 2004)
IntAct (IntAct Molecular Interaction Database: http://www.ebi.ac.uk/intact/) (Hermjakob et al. 2004; Kerrien et al. 2012)
The MPact yeast protein interaction database also supports the PSI-MI standard (Guldener et al. 2006) (http://mips.helmholtz-muenchen.de/genre/proj/mpact), and provides protein interaction data contained in the MIPS Comprehensive Yeast Genome Database (CYGD) (Guldener et al. 2005).
BioGRID is currently unique amongst interaction databases in that it is the only open access resource that curates both genetic and protein interactions for different yeast species from published high- and low-throughput studies. In particular, BioGRID provides comprehensive interaction curation coverage for the budding and fission yeast literature, which is updated via continuous monthly increments that are fully archived (Reguly et al. 2006). BioGRID is also the only current resource that fully captures phenotypes using the Ascomycete Phenotype Ontology (APO) for all curated yeast genetic interactions.
The ongoing development of the PhosphoGRID database allows full integration of documented yeast phosphorylation sites into the BioGRID PTM viewer, and thereby provides an integrated display of all curated phosphosites and their corresponding interactions in a single resource. In conjunction with Wormbase, BioGRID has also developed a new Genetic Interactions (GI) ontology that will allow consistent curation of genetic interactions across various model organisms and interaction databases, enabling better comparisons of phenotypic information across different species (Grove et al. 2015). The GI ontology has recently been incorporated into the Proteomics Standards Initiative – Molecular Interaction (PSI-MI) ontology, with the intention that the GI ontology will be adopted as a community standard.
The BioGRID will continue to facilitate the use of yeast model systems for understanding the role of biological interaction networks in human biology and disease. Protein and genetic interactions, and entire interaction networks, are often physically and functionally conserved (Bandyopadhyay et al. 2006), such that the detailed interrogation of interactions in genetically tractable systems can prove extremely informative in biomedical contexts. To this end, efforts are now underway at BioGRID for parallel curation of yeast and human interactions that are implicated in human disease. These focused curation drives may be either biological process-centric, such as curation of the ubiquitin-proteasome system (UPS) that controls the stability, localization and activity of most of the proteome, or disease-centric, such as curation of interaction networks implicated in HIV or other infectious diseases, neurobiological disorders and metabolic enzymes, all of which are currently in progress. New features planned for BioGRID include the incorporation of ubiquitination sites, which are comprehensively curated as part of the UPS project, into the integrated PTM display. Many expansive interaction networks implicated in prevalent human diseases will be the focus of future curation drives across multiple model organism species, from yeast to human. The integration of these network datasets with other data types, including expression data, quantitative phenotype data, and high resolution sequence data should help to enable predictive medicine and future drug discovery efforts.
Acknowledgments
The authors thank Chris Grove and Paul Sternberg at WormBase for ongoing collaborative development of the Genetic Interaction Ontology. This work was supported by NIH grants R01OD010929 and R24OD011194 to M.T. and K.D., the Biotechnology and Biological Sciences Research Council (grant number BB/F010486/1 to M.T.), the Canadian Institutes of Health Research (grant number FRN 82940 to M.T.), and a Genome Québec International Recruitment Award and a Canada Research Chair in Systems and Synthetic Biology to M.T.
References
- Bandyopadhyay S, Sharan R, Ideker T. Systematic identification of functional orthologs based on protein network comparison. Genome research. 2006;16:428–435. doi: 10.1101/gr.4526006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome biology. 2003b;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2013 update. Nucleic acids research. 2013;41:D816–823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic acids research. 2007;35:D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del-Toro N, Dumousseau M, Orchard S, Jimenez RC, Galeota E, Launay G, Goll J, Breuer K, Ono K, Salwinski L, et al. A new reference implementation of the PSICQUIC web service. Nucleic acids research. 2013;41:W601–606. doi: 10.1093/nar/gkt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic acids research. 2010;38:D433–436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, Oughtred R, Winter A, Baryshnikova A, Dolinski K, Tyers M, Sternberg PW. An Ontology of Genetic Interactions. 2015 (manuscript in preparation) [Google Scholar]
- Guldener U, Munsterkotter M, Kastenmuller G, Strack N, van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic acids research. 2005;33:D364–368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic acids research. 2006;34:D436–441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A, et al. IntAct: an open source molecular interaction database. Nucleic acids research. 2004;32:D452–455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic acids research. 2012;40:D841–846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic acids research. 2012;40:D1202–1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic acids research. 2012;40:D857–861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ, FlyBase c. FlyBase: improvements to the bibliography. Nucleic acids research. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nature methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. Journal of biology. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Breitkreutz BJ, Stark C, Su TC, Dahabieh M, Raithatha S, Bernhard W, Oughtred R, Dolinski K, Barreto K, et al. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database: the journal of biological databases and curation. 2013;2013:bat026. doi: 10.1093/database/bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic acids research. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AG, Wildenhain J, Tyers M. BioGRID REST Service, BiogridPlugin2 and BioGRID WebGraph: new tools for access to interaction data at BioGRID. Bioinformatics. 2011;27:1043–1044. doi: 10.1093/bioinformatics/btr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic acids research. 2012;40:D695–699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic acids research. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, de la Cruz N, Duong A, Fang R, et al. WormBase 2012: more genomes, more data, new website. Nucleic acids research. 2012;40:D735–741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic acids research. 2013;41:W115–122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]