Abstract
In order to elucidate the distribution of Cryptococcus neoformans and C. gattii in the Mediterranean basin, an extensive environmental survey was carried out during 2012–2015. A total of 302 sites located in 12 countries were sampled, 6436 samples from 3765 trees were collected and 5% of trees were found to be colonized by cryptococcal yeasts. Cryptococcus neoformans was isolated from 177 trees and C. gattii from 13. Cryptococcus neoformans colonized 27% of Ceratonia, 10% of Olea, Platanus and Prunus trees and a lower percentage of other tree genera. The 13 C. gattii isolates were collected from five Eucalyptus, four Ceratonia, two Pinus and two Olea trees. Cryptococcus neoformans was distributed all around the Mediterranean basin, whereas C. gattii was isolated in Greece, Southern Italy and Spain, in agreement with previous findings from both clinical and environmental sources. Among C. neoformans isolates, VNI was the prevalent molecular type but VNII, VNIV and VNIII hybrid strains were also isolated. With the exception of a single VGIV isolate, all C. gattii isolates were VGI. The results confirmed the presence of both Cryptococcus species in the Mediterranean environment, and showed that both carob and olive trees represent an important niche for these yeasts.
Keywords: Cryptococcus, C. neoformans, C. gattii, environment, Europe, epidemiology, molecular typing
The present survey established a wide network that, for the first time, collected abundant information concerning the environmental distribution and ecology of Cryptococcus neoformans/C. gattii species complex in the Mediterranean area.
INTRODUCTION
Cryptococcosis is a life-threatening fungal infection caused by the basidiomycetous yeasts in the Cryptococcus neoformans and C. gattii species complex. The infection is likely acquired from the environment by inhalation of spores or dehydrated yeast cells that are able to penetrate the pulmonary alveoli and then disseminate through the bloodstream causing soft tissue infections, pneumonia and most often meningoencephalitis (Kwon-Chung et al. 2014).
Cryptococcosis caused by C. neoformans is a major cause of mortality in patients with AIDS. An estimated one million cases of cryptococcal meningitis occur annually among people with HIV infection worldwide, resulting in nearly 625 000 deaths (Park et al. 2009). Since the introduction of antiretroviral therapy, the cases of cryptococcosis and the number of deaths in people with advanced HIV infection have decreased substantially in developed countries. While cryptococcosis cases in HIV-infected patients have been decreasing, an increase in the number of cases has been reported in non-HIV patients due to the rising number of susceptible patients such as patients with hematological malignancies, organ transplant recipients and patients affected by autoimmune diseases, but also in patients without any other risk factor except that they were exposed to the pathogen (Bratton et al. 2012; Henao-Martínez and Beckham 2015).
In Europe, the epidemiology of cryptococcosis is difficult to establish for two reasons: there are only a few outdated reports on epidemiology of cryptococcosis from a limited number of countries and the lack of coordination to collect epidemiological data among scientists from EU countries. The epidemiological data thus far available on cryptococcosis are restricted to Croatia, France, Germany, Italy, Serbia, Spain, Portugal, the Netherlands and the United Kingdom (Baró et al. 1999; FIMUA Network 2002; Dromer et al. 2004; Mlinaric-Missoni et al. 2011; Hagen et al. 2012b; Patel et al. 2013; Arsic Arsenijevic et al. 2014; Bitar et al. 2014; Sanchini et al. 2014; Maduro et al. 2015). Data from the rest of the EU are either scarce or completely lacking, especially from Central and Eastern European countries where a higher incidence of cryptococcosis is expected due to a heavier burden of HIV infection compared to Western Europe (de Colombani et al. 2004).
A unique attempt for a prospective European survey was performed during a survey from 1997 to 1999 in which 655 cases from 17 countries were reported and 311 cryptococcal isolates were collected for molecular typing (Viviani et al. 2006). Although the survey represented a milestone in the elucidation of the European epidemiology of cryptococcosis, the results underestimated the burden of the disease since many countries did not participate in the study. At the national level, a recent study carried out in France reported 1850 cases of cryptococcosis from 2001 to 2010 and an incidence of 0.3 per 100 000 population/year with a fatality rate of 15% (Bitar et al. 2014), while 129 cases were recorded in a study carried out in Germany from 2004 to 2010 (Sanchini et al. 2014).
Because of its geographical location, Europe is also subjected to extensive immigration of people from both Asia and Africa where cryptococcosis represents the third highest cause of death among HIV-infected patients (Park et al. 2009; Assogba et al. 2015). This inevitably favors the spread of new genotypes in Europe through the introduction of the pathogen via vehicles, clothing and goods potentially contaminated as also been shown in the Vancouver outbreak caused by C. gattii (Kidd et al. 2007). Furthermore, the high flow of people to and from Europe for business and tourism allows the emergence of cryptococcosis cases acquired in endemic areas (Dromer, Ronin and Dupont 1992; Hagen et al. 2012a). The recent cryptococcosis outbreaks occurring on Vancouver Island (Canada), and the Pacific Northwest of North America showed how this fungal threat could spread rapidly in the environment once it has found a favorable niche (Byrnes and Marr 2011; Bartlett et al. 2012; Hagen et al. 2012a, 2013). The Centers for Disease Control and Prevention in the USA worked with local public health authorities to implement a plan to monitor the epidemiology of C. gattii in the states of Washington and Oregon where the reporting of this fungal disease is now mandatory. The coordination of such actions is a lengthy process in Europe and needs to be improved for early documentation of outbreaks as have been reported due to C. gattii.
Few studies to assess the occurrence of the C. neoformans/C. gattii species complex in the environment have been performed in Europe. Cryptococcus neoformans was mainly reported to be associated with bird excreta (Colom Valiente et al. 1997; Garcia-Hermoso et al. 1997; Pernice et al. 1998; Boekhout et al. 2001; Montagna et al. 2003; Lagrou et al. 2005; Cafarchia et al. 2006), and only few isolates were recovered from arboreal sources (Criseo et al. 1995; Criseo and Gallo 1997; Lo Passo et al. 1997; Campisi et al. 2003; Bauwens et al. 2004, Chowdhary et al. 2012, Colom et al. 2012). Cryptococcus gattii was recovered for the first time from the European environment in Southern Italy (Montagna et al. 1997; Romeo, Scordino and Criseo 2011), and then also in the Netherlands (Chowdhary et al. 2012) and Spain (Colom et al. 2012). However, these surveys were limited to a restricted territory and carried out at different periods, as such the results are geographically and temporally fragmented.
This study represents the first collaborative effort aimed to understand the environmental distribution of C. neoformans and C. gattii on trees around the Mediterranean basin and in continental Europe. In addition, isolates and associated metadata collected during the survey represent an important source for the comparison and correlation of European and global clinical data.
MATERIALS AND METHODS
Network and study design
The ISHAM Working Group for Genotyping of C. neoformans and C. gattii (http://www.isham.org/WorkingGroups/Genotyping_neoformans_gattii) established a network to survey the distribution of C. neoformans and C. gattii in the environment focusing on sampling around the Mediterranean basin. Thirty-two centers from nine European countries and three non-European countries (Israel, Libya and Turkey) participated in the study. Each participating center was required to collect samples from trees and soil especially in urban area where the finding of the pathogens could represent a menace for humans, and to record a defined set of metadata, including sampling site and date, type of sample, tree species, daily mean temperature, number of collected samples and number of positive samples. Sample collection and cultivation were performed in each participating center according to predefined methods described below. Both the metadata and the isolates were sent to the coordinating center of the study at the Medical Mycology Laboratory, Università degli Studi di Milano (Italy). All isolates were coded and stocked, and then processed for molecular analyses. Three additional non-European centers, one from Australia and two from the USA, joined the study and participated in the molecular strain typing effort.
Environmental samples
Samples were collected in the geographical areas where the different participating centers were located mainly from the widest public gardens in urban areas but also from some rural areas. In each site, a minimum of 10 and a maximum of 100 samples were collected depending of the extension of the sampled area and the density of the trees using a non-random sampling methodology.
The sources of samples were hollows and fissures of trees, flowers, leaves, bark, fruits, decaying wood, soil underneath trees and bird excreta on or near the tree. All the sampled trees were identified as far as possible to the species level.
Hollows and fissures on the tree trunk
Samples were collected by rubbing the inner of the hollows or fissures of the trees with a sterile cotton-tipped swab moistened in a solution of sterile distilled water supplemented with chloramphenicol (10 mg L−1). The swab was placed into a tube with 3 mL of the solution and the tube was shaken for 5 min without removing the swab. The swab was removed and the suspension was left to sediment at least for 10 min. A total of 100 μl of the supernatant and 100 μl of the diluted supernatant (1:10 in sterile distilled water) were inoculated onto two different Niger seed agar plates (Kwon-Chung and Bennett 1992). The plates were incubated at 37°C for at least 10 days. All brown colored colonies grown on the plates were isolated for further species identification.
Flowers and leaves
About 10–20 g of the sample were collected and sealed in zip-lock bags. A portion of the sample (5 g) was transferred in a sterile mortar and fragmented with a pestle. The fragments were suspended in 50 mL sterile distilled water and vortexed for about 2 min at maximum speed. Sediment was allowed to settle for 15–20 min. A total of 2 mL of the supernatant was mixed with 8 mL of sterile distilled water containing chloramphenicol (10 mg L−1). A total of 100 μl of the supernatant and 100 μl of the diluted supernatant were inoculated in two different Niger seed agar plates. The plates were incubated at 37°C for at least 10 days. All brown colonies grown on the plates were collected for identification.
Bark and decaying wood
Samples were obtained by scraping the surface of the wood with a scalpel. The obtained shavings were sealed in a zip-lock bag. Following vigorous grinding with a mortar, 1 g of the sample was suspended in 50 mL of sterile distilled water containing chloramphenicol at 10 mg L−1, shaken for 2 min and allowed to settle for 30 min. A total of 100 μl of the supernatant and 100 μl of the diluted supernatant (1:10 in sterile distilled water) were inoculated onto two different Niger seed agar plates. The plates were incubated at 37°C for at least 10 days. All brown colonies grown on the plates were collected for identification.
Soil
Approximately 10–20 g of soil was collected and sealed in a zip-lock bag. Part of the soil (5 g) was suspended in 50 mL sterile distilled water and mixed by vortexing for about 2 min at maximum speed. Sediment was allowed to settle for at least 15 min. A total of 2 mL of supernatant was mixed with 8 mL of sterile distilled water containing chloramphenicol (10 mg L−1). A total of 100 μl of the supernatant and 100 μl of the diluted supernatant were inoculated on two different Niger seed agar plates. The plates were incubated at 37°C for at least 10 days. All brown colonies grown on the plates were collected for identification.
Isolation, species identification, coding and storage
Brown colonies were streaked for isolation on a fresh Niger seed agar plate in order to collect pure single colonies. Isolates were then examined by microscopy for assessing the yeast morphology and capsule presence, tested for urease activity and the ability to grow at 37°C and assimilate myo-inositol as a carbon source.
The cryptococcal species was identified by inoculating a fresh colony onto canavanine-glycine-bromothymol blue agar differential medium (Kwon-Chung and Bennett 1992).
A code identifying the country, the place of origin, the type of sample and the tree code number was assigned to each isolate, which were then suspended and stored in a vial containing 3 mL of sterile distilled water at room temperature.
Molecular analyses
Genomic DNA was extracted as previously reported (Viviani et al. 1997). Molecular type and mating type of all isolates was determined by four multiplex PCRs specific for both C. neoformans and C. gattii as described elsewhere (Cogliati et al. 2000; Esposto et al. 2004; Feng et al. 2013; Cogliati, D'Amicis and Tortorano 2015). Molecular types were assigned according to the standard nomenclature of the ISHAM working group for genotyping of C. neoformans and C. gattii (Meyer et al. 2009). Strains H99 (VNI-αA), JEC20 (VNIV-aD), JEC21 (VNIV-αD), IUM 96–2828 (VNII-aA), WM 626 (VNII-αA), WM779 (VGIV-αC), NIH312 (VGIII-αB), NIH191 (VGIII-aC), WM201 (VGI-αB) and IUM 00–5363 (VGII-aB) were used as reference strains.
Mating assay
A mating assay was performed to test the fertility of selected environmental isolates from the survey. The isolates were streaked onto a 90-mm plate containing 20 ml Murashige-Skoog agar medium (0.44% Murashige-Skoog basal medium; Sigma-Aldrich, St. Louis, MO, USA; and 4% agar in distilled water) and then mixed with a tester strain of the opposite mating type. Co-cultures were incubated at 25°C in the dark for at least 3–4 weeks and checked periodically for the formation of hyphae and basidiospores. Basidiospores were collected by cutting a square of the agar on which hyphae were produced (avoiding to touch the yeast colony edge), and transferring it to a tube containing 2 mL sterile distilled water. After gently stirring the tube, the supernatant was transferred to a new tube and checked microscopically for the presence of basidiospores and the absence of yeast cells and hyphae. A 100 μl volume of the spore suspensions were plated on Sabouraud dextrose agar and incubated at 37°C for 48 h. Ten single colonies grown on the plate were collected and processed for molecular typing. A nearly 1:1 ratio of MATa and MATα spores confirmed the successful mating between the two tested strains. The strain pairs JEC20 (VNIV-aD) and JEC21 (VNIV-αD), and H99 (VNI-αA) and IUM 96–2828 (VNII-aA) were used as tester strains.
Taxonomy
Although a new taxonomic classification of C. neoformans/C. gattii species complex has recently been proposed (Hagen et al. 2015), it is still under discussion. Therefore, in this study we continue to adopt the classical taxonomy which classifies the agents of cryptococcosis into two species, C. neoformans and C. gattii, and C. neoformans into two varieties, C. neoformans var. grubii and C. neoformans var. neoformans.
Statistics
Differences between the percentages of colonized trees observed in different tree populations, months of isolation and mean daily temperature ranges were statistically analyzed by χ2 test using the online statistical calculator at www.vassarstats.net. The χ2 test was also applied to compare the results obtained in different sampling areas in order to assess whether the results obtained in a specific area could have influenced the overall results.
RESULTS
Sampling distribution
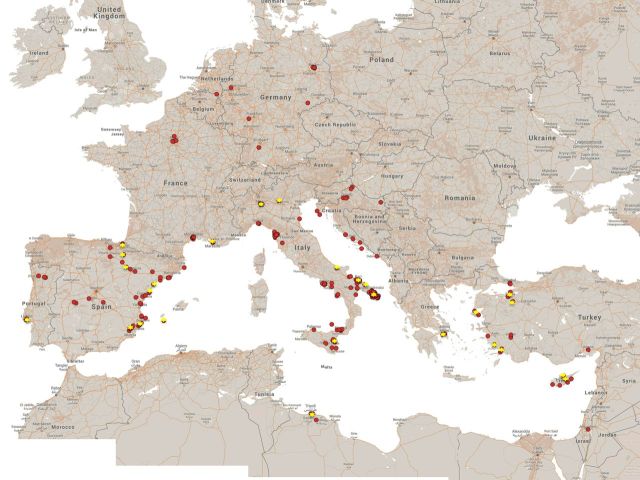
The survey was performed during 2012–2015. A total of 302 sites in 12 countries were sampled, primarily in Italy (n = 152), followed by Spain (n = 47), France (n = 27), Turkey (n = 19), Croatia (n = 18), Portugal (n = 11), Germany (n = 10), Greece (n = 7), Cyprus (n = 6), Libya (n = 3) and Israel plus the Netherlands with one site each (Fig. 1).
Figure 1.
A map of the Mediterranean basin showing the sampling sites (red dots). Stars indicate the positive sites. The coordinates for each site were recorded and then plotted on the map using GoogleMaps (www.google.com).
Trees
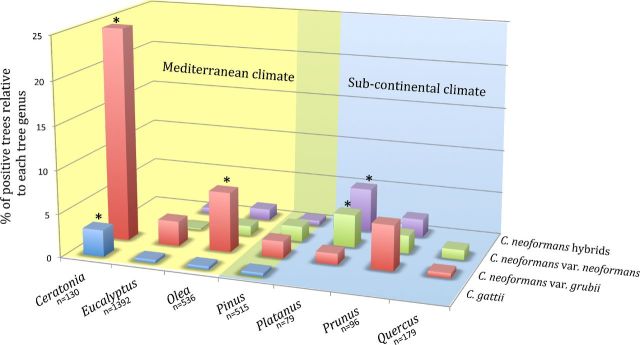
Samples were collected from 3765 trees representing more than 100 different genera (Table S1, Supporting Information). Most of the trees were Eucalyptus (37%), Olea (olive tree, 14%), Pinus (13%), Quercus (oak, 5%), Ceratonia (carob tree, 3%), Prunus (2%) and Platanus (plane tree, 2%). The highest percentage of sampled trees was in Italy (43%), followed by Turkey (12%), Cyprus (10%), Libya (9%), Spain (6.5%) and Greece (6%). A total of 188 trees (5%) were colonized by C. neoformans or C. gattii. Colonized trees were found in Cyprus, France, Greece, Italy, Libya, Portugal, Spain and Turkey, with the highest percentage of positive trees recorded in Spain and Greece (16.7% and 16.8%, respectively). The percentage of colonized trees relative to each tree genus was the following: Ceratonia (n = 130, 27.7%), Platanus (n = 79, 10.1%), Prunus (n = 96, 9.4%), Olea (n = 536, 9.3%), Pinus (n = 515, 4.7%), Eucalyptus (n = 1392, 3.7%) and Quercus (n = 179, 1.1%). Sporadic positive samples were also found on Aesculus hippocastanum, Carpinus betulus, Juglans nigra, Juniperus spp., Gleditsia triacanthos and Pyrus communis.
Samples
A total of 6436 samples were collected from trunk hollows (62%), bark (11.5%), leaves (8%), flowers (1.3%), soil under trees (16%) and other samples (1.2%) that included fruits, decaying wood, debris near trees or bird excreta on trees. The majority of the samples, 44.4% and 9.6%, were collected in Italy and Greece, respectively. A total of 220 samples (3.4%) were positive. Colony-forming units per sample were variable and ranged from 1 to 100. Cryptococcus neoformans or C. gattii was recovered from cultures of trunk hollow swabs, bark scrapings, soil and decaying wood, whereas no isolates were recovered from leaves and flowers. Trunk hollows had a percentage positivity of 4.1% (n = 3986), bark of 3% (n = 738) and soil of 2.4% (n = 1039). In addition, nine samples from debris of decaying wood were positive.
Distribution of C. neoformans and C. gattii isolates
During the survey, 512 isolates from 188 trees were recovered representing 474 C. neoformans and 38 C. gattii isolates. The percentage of colonized trees relative to the two species was 4.6% for C. neoformans and 0.4% for C. gattii with a ratio of 13.5:1, respectively (n = 3765). Cryptococcus neoformans was isolated from 177 trees belonging mainly to the genera Eucalyptus, Olea (olive trees), Ceratonia (carob trees) and Pinus, but also from Aesculus, Carpinus, Juglans, Juniperus, Gleditsia, Platanus, Prunus, Pyrus and Quercus (Fig. 2). The trees colonized by C. neoformans isolates were found in Cyprus, France, Greece, Italy, Libya, Portugal, Spain and Turkey. Cryptococcus gattii was isolated from 13 trees belonging to four different genera: five trees of Eucalyptus, four trees of Ceratonia (carob trees), two trees of Olea (olive trees) and two trees of Pinus pinea (Fig. 2). The trees were distributed in Spain (Alicante, Tarragona and Mendivil in Navarra), Italy (Bari and Catania) and Greece (Athens and Salamina Island). One carob tree in Spain and one olive tree in Italy were colonized by both C. neoformans and C. gattii.
Figure 2.
Percentage of colonized trees per tree genera. In the histogram, only the tree genera that are represented at least by 2% of the total sampled trees (n = 1765) have been included. The trees of genus Pinus can adapt to different climate and in the graphic are located in an overlapping zone. Asterisk indicates that the value is statistically higher (P < 0.05) compared to the values observed in the other tree populations. n = Number of sampled trees for each tree genus.
A comparison of the percentage of positive trees for each tree genus in the different sampling areas was also performed in order to exclude the influence of the results obtained in a specific area on the overall analysis. No statistical difference was observed (P > 0.05) between the percentage of positive trees in the different sampling areas confirming that the differences observed are likely associated with the tree species considered.
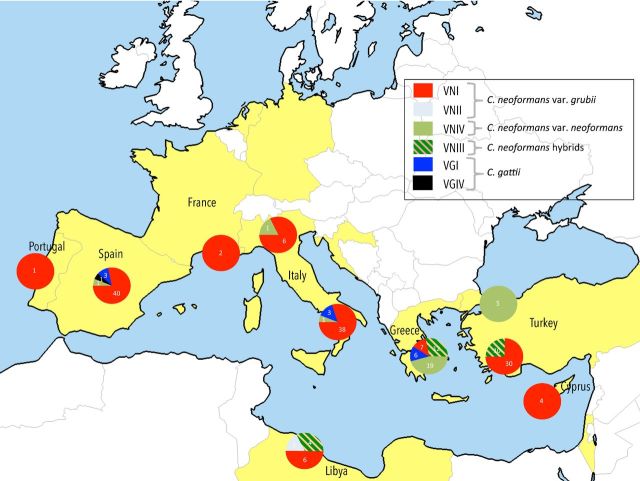
Prevalence and distribution of molecular types
The map in Fig. 3 shows the distribution of molecular types found around the Mediterranean basin. The prevalent molecular type was VNI with 330 isolates from 129 trees distributed in all seven countries that yielded positive samples. Molecular type VNII was only isolated from two trees in Tripoli, Libya, whereas 107 VNIV isolates were recovered from 27 trees located in Greece, Italy, Spain and Turkey. In addition, 35 AD-hybrids (molecular type VNIII) were isolated from 26 trees in Greece, Libya and Turkey. VGI was the prevalent molecular type for C. gattii with 37 isolates obtained from 12 trees. One isolate from a Spanish Ceratonia siliqua tree was identified as VGIV. Trees colonized by two molecular types, VNI and VGI (two trees), or VNI and VNIV (four trees), or VNI and VNIII (three trees), were also found.
Figure 3.
Distribution and prevalence of C. neoformans and C. gattii molecular types around the Mediterranean basin. Numbers inside the circles indicate the numbers of positive trees. Participating countries are presented in yellow.
Prevalence and distribution of mating types
Both mating type a and mating type α were found among C. neoformans var. grubii, C. neoformans var. neoformans and C. gattii isolates collected during the survey. The mating type allelic pattern αA was the prevalent mating type (327 isolates) among C. neoformans var. grubii, whereas the mating type aA was identified in seven isolates, six from Spain and one from Italy. Cryptococcus neoformans var. neoformans αD mating type was recovered in Italy (71 isolates), Spain (one isolate) and Turkey (6 isolates), whereas mating type aD was found only in Greece (29 isolates). The 35 AD-hybrid isolates presented two different mating-type allelic patterns, three were heterozygous αADa and 32 were homozygous αAAα. One olive tree in Italy and four carob trees in Spain were colonized by both αA and aA isolates, whereas two Eucalyptus trees in Greece were colonized by both αA and aD isolates. Coexistence of αA and αD as well as αA and AD-hybrids was observed in Italy (two trees) and Turkey (three trees), respectively.
Regarding C. gattii, ten trees were colonized by mating type αB strains, two trees in Italy by mating type aB and one in Spain by mating type αC. One of the two Italian aB isolates shared the same olive tree with C. neoformans var. grubiiaA and αA strains. Similarly, in Spain one C. gattii αB strain coexisted with one C. neoformans var. grubii αA strain in a carob tree.
Results of mating assays
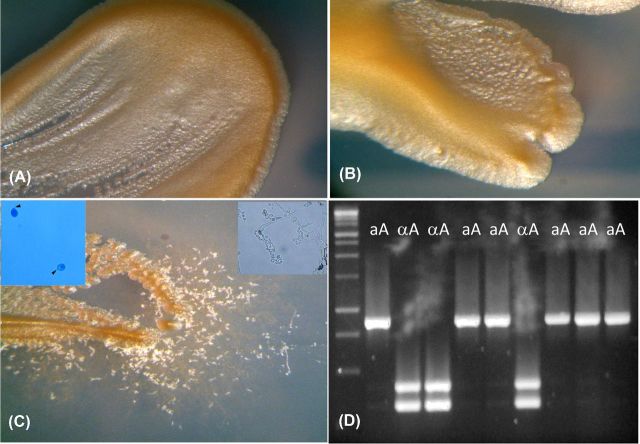
Seven mating assays were performed to test the fertility of the isolates that originated from the trees where two opposite mating type isolates shared the same niche. Five assays were intra-variety mating assays between aA and αA isolates, whereas the other two were inter-variety assays between αA and aD isolates. All intra-variety assays produced filaments and basidiospores. In addition, molecular analysis showed that the progeny included both aA and αA mating types in a Mendelian ratio (Fig. 4). In contrast, none of the inter-varietal matings was fertile after 4 weeks of observation.
Figure 4.
Results of one representative mating assay performed on Murashige Skoog agar medium. One VNI-αA isolate and one VNI-aA isolate which shared the same niche in the same tree was used for crossing. (A) VNI-αA culture. (B) VNI-aA culture. (C) Filament production in the mixed culture (VNI-αA × VNI-aA). The upper-right inset shows a basidium with long chains of basidiospores. The upper-left inset shows basidiospores collected in water suspension. Arrows indicate the stalk-like ends of the basidiospores. (D) Multiplex PCR to determine the mating type allelic pattern of the progeny. Each lane represents DNA from a single basidiospore culture. Molecular Ladder: 100 bp DNA ladder (Promega Italia, Milano, Italy).
Seasonality and daily mean temperature
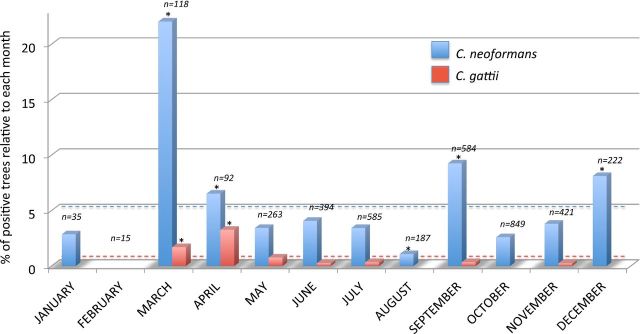
Data of seasonality are presented in Fig. 5. Samples were collected monthly, year round, by each of the collaborating groups. The highest number of samples was recorded in July (19.9%) followed by September (19.4%) and October (14.4%). The lowest number of samples was recorded in February (0.8%) and January (1.6%). The percentage of trees colonized by C. neoformans showed a significant increase (P < 0.05) in March (22%), April (6.5%), September (9.2%) and December (8.1%) when compared to the overall percentage of colonization (4.6%) observed for this species. In the other months, the value was lower with a minimum recorded in February (no positive trees) and August (1.1%, P < 0.05). Cryptococcus gattii colonization of trees was higher in March (1.7%, P < 0.05), April (3.3%, P < 0.05) and May (0.8%, P > 0.05) than that observed in overall tree population (0.4%). No C. gattii positive trees were found in January, February, August, October and December.
Figure 5.
Percentage of trees colonized by C. neoformans and C. gattii recorded in each month of the year. The blue and red dashed lines represent the overall percentage of colonization observed for C. neoformans and C. gattii, respectively. Asterisk indicates that the value is statistically different from the mean (P < 0.05) compared to the values observed in the other months. n = Number of sampled trees for each month.
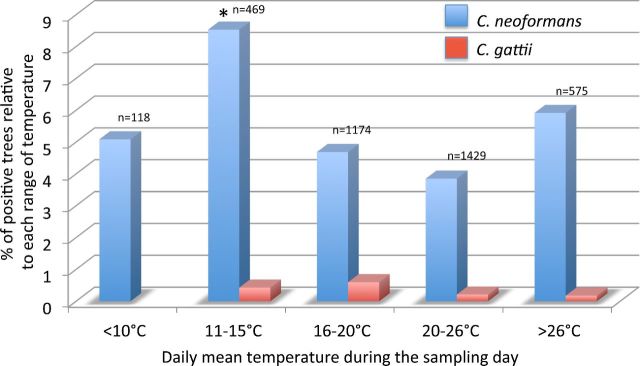
Samples were collected within a range of daily mean temperatures of 7°C–32°C. Depending on the mean temperature of the sampling day, the data relative to each sampling were placed in one of five temperature ranges: <10°C, 11°C–15°C, 16°C–20°C, 21°C–26°C and >26°C. The percentage of colonized trees relative to each temperature range was calculated for both the Cryptococcus species and displayed in a histogram (Fig. 6). The results showed that, although C. neoformans was recovered in all temperature ranges with no statistically difference (P > 0.05), C. gattii was recovered only when the mean daily temperature was above 10°C.
Figure 6.
Percentage of trees colonized by C. neoformans and C. gattii in relation to the daily average temperature during the sampling days. Asterisk indicates that the value is statistically higher (P < 0.05) compared to the values observed in the other groups. n = Number of sampled trees for each range of temperatures.
The analyses were repeated for the different sampling areas and, after comparison, the results were not significantly different (P > 0.05).
DISCUSSION
This study is the first European collaborative prospective environmental survey investigating the distribution of the C. neoformans/C. gattii species complex. Although the survey was not exhaustive, it covered a wide territory of Europe including a large part of the Mediterranean coast as well as some continental areas. This first environmental survey generated an immense quantity of data, which are now available for detailed follow-up analysis in future studies.
The results showed that both C. neoformans and C. gattii are present in the Mediterranean environment in association with trees. Cryptococcus neoformans is more prevalent than C. gattii with a 13-fold higher percentage of colonized trees. Our findings suggest that trees could represent an important environmental niche and a stable reservoir for both species, and that bird excreta could represent a secondary and temporary niche especially for C. neoformans. The association of trees with Cryptococcus is also implicated by the findings of the yeasts as infectious or colonizing agents in animals reported from other parts of the world, such as koalas, squirrels, monkeys and parrots whose lives revolve around trees. Goats have the habit of eating tree barks and they also develop cryptococcosis (Roussilhon, Postal and Ravisse 1987; López-Martínez and Castañón-Olivares 1995; Torres-Rodríguez et al. 1999; Krockenberger, Canfield and Malik 2003; Stilwell and Pissarra 2014; Iatta et al. 2015; Maestrale et al. 2015).
This study reveals a difference in the association of the two Cryptococcus species with specific trees. In particular, Ceratonia (carob tree) and Olea (olive tree), two tree genera typical for the Mediterranean region, together with Eucalyptus trees produced the highest number of positive samples. However, when the values were normalized as a percentage of the positive tree genus and stratified for each of the Cryptococcus species and varieties, the results showed that Ceratonia is an important niche for both C. gattii and C. neoformans var. grubii, but not for C. neoformans var. neoformans and AD-hybrids, since the latter were not recovered from these trees. In addition, C. gattii was recovered only from trees typical for the Mediterranean climate (Ceratonia, Olea, Eucalyptus and Pinus pinea), whereas C. neoformans var. grubii colonized 12 different tree genera confirming the ability of this pathogen to adapt to different environments and, hence, contributing to its global distribution. The importance of Ceratonia siliqua as a niche for C. gattii, shown in the present survey, is in agreement with the data reported by previous environmental studies carried out in Spain (Colom et al. 2012; Linares et al. 2015). In contrast, C. neoformans var. neoformans and AD-hybrids showed a preference to colonize trees typical of the subcontinental climate, such as Platanus, Prunus and Quercus. This could reflect the ability of C. neoformans var. neoformans to tolerate lower temperature better than C. neoformans var. grubii and C. gattii as previously shown by other authors (Martinez, Garcia-Rivera and Casadevall 2001). However, the survey has the limit that the samples were not collected with a rigorous randomized method and therefore the conclusions here reported must be considered as an attempt to describe the observed data and a starting point for future more extensive studies.
Although the two C. neoformans varieties and C. gattii have differences in tree preference, the climatic zones do not have sharp boundaries and they overlap along the entire Mediterranean basin. Therefore, in the Mediterranean environment, the different C. neoformans and C. gattii populations are continuously in contact with each other. This is confirmed by the finding that isolates belonging to different species or varieties shared a niche of the same tree, as well as by the presence of hybrids in the same area.
Hybridization between C. neoformans var. grubii and C. neoformans var. neoformans is well documented in Europe by the identification of numerous AD-hybrids in clinical isolates which have a prevalence of about 30% (Viviani et al. 2006). In contrast, only a few hybrids have been isolated from pigeon droppings (Baró et al. 1999; Ferreira et al. 2014) due to the paucity of environmental studies carried out in Europe. This study reports for the first time the association of AD-hybrids with trees and their presence in the same area where the putative parental C. neoformans var. grubii and C. neoformans var. neoformans isolates may coexist. This finding suggests that hybridization between the two C. neoformans varieties is occurring in the European environment and this may play an important role as a mechanism of evolution of this species (Cogliati, Lin and Viviani 2009; Li et al. 2012; Desnos-Ollivier et al. 2015). Although the inter-varietal mating assays carried out in this study did not succeed, this does not mean that the isolates studied are not compatible but probably means that the in vitro assay conditions here adopted were not optimal for hybridization. The high variability of conditions and substrates encountered in the environment may better favor this process.
The presence of C. gattii in Greece, Southern Italy and Spain confirms the previous results obtained in these geographical areas where the pathogen was isolated from both clinical and environmental sources (Montagna et al. 1997; Torres-Rodríguez et al. 1999; Velegraki et al. 2001; Colom et al. 2005; Viviani et al. 2006; Solla et al. 2008; Ropstad et al. 2011; Colom et al. 2012; Iatta et al. 2012; Romeo et al. 2012; Hagen et al. 2012a). The survey also confirmed that VGI is the prevalent C. gattii molecular type, which is in agreement with a previous analysis carried out by Hagen et al. (2012a) that identified an endemic VGI cluster in Mediterranean Europe. However, one isolate was identified as VGIV-αC (Linares et al. 2015) suggesting that other molecular types are also able to colonize the Mediterranean basin. Further molecular analysis of the isolates collected during this survey by MLST will elucidate the relationships and the clusters present in the European environment.
Cryptococcus neoformans VNI was the prevalent molecular type distributed all around the Mediterranean basin from Portugal to Libya confirming the ubiquitous presence of this pathogen in the region (Cogliati 2013). In this study, a high prevalence of the VNIV molecular type was found in Greece, where it represented the most common molecular type, and Northern Turkey, where it was the only molecular type present, whereas it occurred less frequently in Italy and Spain. On the basis of these results, it could be speculated that this molecular type is spreading from subcontinental areas of the South-Eastern Mediterranean towards the Western part of Europe. Further sampling and an accurate niche modeling analysis is in progress to corroborate the above hypothesis.
Our results showed that mating type a and α are present in the Mediterranean environment for both C. neoformans varieties and C. gattii. Interestingly, we found that the occurrence of two strains with different mating types in the same tree is not rare, as already observed for C. gattii in Australia (Halliday et al. 1999); therefore, trees are possible niches to complete the sexual cycle. This hypothesis is supported by the observation that most of the isolates with different mating types sharing the same niche were able to produce filaments and recombinant basidiospores in the mating assays. In addition, other authors showed that C. neoformans and C. gattii are able to proliferate and mate in vitro in a Cryptococcus-Arabidopsis system (Xue et al. 2007).
These findings suggest that the current view of a clonal population structure observed for both C. neoformans and C. gattii could be due to the genotyping results being mainly obtained from clinical isolates and the relatively low number of available environmental isolates investigated (Cogliati 2013). The analysis of a larger number of environmental isolates might show a more relevant involvement of sexual reproduction in the evolution and propagation of the C. neoformans/C. gattii species complex. Our data are corroborated by previous studies reporting recombination among C. neoformans var. grubii (VNI and VNB) and C. gattii (VGI and VGII) populations isolated from the environment (Litvintseva et al. 2003; Saul, Krockenberger and Carter 2008; Carriconde et al. 2011).
A different trend can be observed with respect to seasonality depending on the Cryptococcus species considered. Sampling during different seasons did not greatly influence the recovery of C. neoformans although a peak of positive trees was observed during spring. Similarly, C. gattii recovery was more likely during spring and early summer, but was absent during the colder seasons. Both species were isolated less in August, which is the hottest and driest month in the Mediterranean area, suggesting the difficulty to cultivate these yeasts during such climatic conditions. A recent study carried out in Colombia reported a similar observation with a low probability to recover Cryptococcus from trees during the seasons with reduced rainfall (Noguera, Escandón and Castañeda 2015). When daily mean temperatures were considered, the results confirmed that C. neoformans can be recovered during a wide range of temperatures, whereas C. gattii seems to be absent or difficult to cultivate at temperatures below 10°C.
In conclusion, the present survey established a wide laboratory network that, for the first time, collected extensive information concerning the environmental distribution and ecology of the C. neoformans/C. gattii species complex in Europe and the Mediterranean area. The results represent the basis for future studies on environmental niches of Cryptococcus in Europe and an important step towards the comparison of clinical and environmental isolates.
Supplementary Material
SUPPLEMENTARY DATA
FUNDING
The work by K.J. Kwon-Chung and A. Varma was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, NIH. The work by A. Sampaio and C. Pereira was in part supported by CITAB under the project UID/AGR/04033/2013. W. Meyer was supported by funding from the NH and MRC, Australia, grant # APP1031943. K. Ferreira-Paim was supported by a CAPES Science without Borders visiting fellow (N◦ 9313133) from Brazil.
DISCLOSURES
The findings and conclusions of this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Conflict of interest. None declared.
REFERENCES
- Assogba K, Belo M, Wateba MI, et al. Neuromeningeal cryptococcosis in sub-Saharan Africa: Killer disease with sparse data. J Neurosci Rural Pract. 2015;6:221–4. doi: 10.4103/0976-3147.153231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsic Arsenijevic V, Pekmezovic MG, Meis JF, et al. Molecular epidemiology and antifungal susceptibility of Serbian Cryptococcus neoformans isolates. Mycoses. 2014;57:380–7. doi: 10.1111/myc.12171. [DOI] [PubMed] [Google Scholar]
- Baró T, Torres-Rodríguez JM, Morera Y, et al. Serotyping of Cryptococcus neoformans isolates from clinical and environmental sources in Spain. J Clin Microbiol. 1999;37:1170–2. doi: 10.1128/jcm.37.4.1170-1172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett KH, Cheng PY, Duncan C, et al. A decade of experience: Cryptococcus gattii in British Columbia. Mycopathologia. 2012;173:311–9. doi: 10.1007/s11046-011-9475-x. [DOI] [PubMed] [Google Scholar]
- Bauwens L, Vercammen F, Wuytack C, et al. Isolation of Cryptococcus neoformans in Antwerp Zoo's nocturnal house. Mycoses. 2004;47:292–6. doi: 10.1111/j.1439-0507.2004.00991.x. [DOI] [PubMed] [Google Scholar]
- Bitar D, Lortholary O, Le Strat Y, et al. Population-based analysis of invasive fungal infections, France, 2001–2010. Emerg Infect Dis. 2014;20:1149–55. doi: 10.3201/eid2007.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, Theelen B, Diaz M, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- Bratton EW, El Husseini N, Chastain CA, et al. Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS One. 2012;7:e43582. doi: 10.1371/journal.pone.0043582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ, 3rd, Marr KA. The Outbreak of Cryptococcus gattii in Western North America: Epidemiology and Clinical Issues. Curr Infect Dis Rep. 2011;13:256–61. doi: 10.1007/s11908-011-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafarchia C, Romito D, Iatta R, et al. Role of birds of prey as carriers and spreaders of Cryptococcus neoformans and other zoonotic yeasts. Med Mycol. 2006;44:485–92. doi: 10.1080/13693780600735452. [DOI] [PubMed] [Google Scholar]
- Campisi E, Mancianti F, Pini G, et al. Investigation in Central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis. Eur J Epidemiol. 2003;18:357–62. doi: 10.1023/a:1023652920595. [DOI] [PubMed] [Google Scholar]
- Carriconde F, Gilgado F, Arthur I, et al. Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population - an emerging outbreak in Australia. PLoS One. 2011;6:e16936. doi: 10.1371/journal.pone.0016936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Randhawa HS, Boekhout T, et al. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerg Infect Dis. 2012;18:172–4. doi: 10.3201/eid1801.111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Scientifica. 2013:675213. doi: 10.1155/2013/675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, Allaria M, Tortorano AM, et al. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med Mycol. 2000;38:97–103. doi: 10.1080/mmy.38.2.97.103. [DOI] [PubMed] [Google Scholar]
- Cogliati M, D'Amicis R, Tortorano AM. Cryptococcus gattii sero-mating type allelic pattern determined by multiplex PCR. Clin Microbiol Infect. 2015;21:190.e1–4. doi: 10.1016/j.cmi.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Cogliati M, Lin X, Viviani MA. Heitman J, Kozel TR, Kwon-Chung KJ, et al. Cryptococcus: from Pathogen to Model Yeast. Washington, USA: ASM Press; 2009. Hybridization and its importance in the Cryptococcus species complex; pp. 359–70. [Google Scholar]
- Colom MF, Frasés S, Ferrer C, et al. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J Clin Microbiol. 2005;43:3548–50. doi: 10.1128/JCM.43.7.3548-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colom MF, Hagen F, Gonzalez A, et al. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Med Mycol. 2012;50:67–73. doi: 10.3109/13693786.2011.574239. [DOI] [PubMed] [Google Scholar]
- Colom Valiente MF, Alberdi M, Meseguer I, et al. Isolation of Cryptococcus neoformans from environmental samples in Alicante. Rev Iberoam Micol. 1997;14:63–4. [PubMed] [Google Scholar]
- Criseo G, Bolignano MS, De Leo F, et al. Evidence of canary droppings as an important reservoir of Cryptococcus neoformans. Zentralbl Bakteriol. 1995;282:244–54. [PubMed] [Google Scholar]
- Criseo G, Gallo M. Serotyping of Cryptococcus neoformans isolates from environmental and clinical sources in extreme southern Italy (Calabria and Sicily, central Mediterranean area) Mycoses. 1997;40:95–100. doi: 10.1111/j.1439-0507.1997.tb00194.x. [DOI] [PubMed] [Google Scholar]
- de Colombani P, Banatvala N, Zaleskis R, et al. European framework to decrease the burden of TB/HIV. Eur Respir J. 2004;24:493–501. doi: 10.1183/09031936.04.00064504. [DOI] [PubMed] [Google Scholar]
- Desnos-Ollivier M, Patel S, Raoux-Barbot D, et al. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. MBio. 2015;6:e00311. doi: 10.1128/mBio.00311-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromer F, Mathoulin-Pélissier S, Fontanet A, et al. Epidemiology of HIV-associated cryptococcosis in France (1985–2001): comparison of the pre- and post-HAART eras. AIDS. 2004;18:555–62. doi: 10.1097/00002030-200402200-00024. [DOI] [PubMed] [Google Scholar]
- Dromer F, Ronin O, Dupont B. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J Med Vet Mycol. 1992;30:395–7. [PubMed] [Google Scholar]
- Esposto MC, Cogliati M, Tortorano AM, et al. Determination of Cryptococcus neoformans var. neoformans mating type by multiplex PCR. Clin Microbiol Infect. 2004;10:1092–4. doi: 10.1111/j.1469-0691.2004.00972.x. [DOI] [PubMed] [Google Scholar]
- Feng X, Fu X, Ling B, et al. Rapid differentiation of cryptic species within Cryptococcus gattii by a duplex PCR assay. J Clin Microbiol. 2013;51:3110–2. doi: 10.1128/JCM.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AS, Sampaio A, Maduro AP, et al. Genotypic diversity of environmental Cryptococcus neoformans isolates from Northern Portugal. Mycoses. 2014;57:98–104. doi: 10.1111/myc.12106. [DOI] [PubMed] [Google Scholar]
- FIMUA Cryptococcosis Network European Confederation of Medical Mycology (ECMM) prospective survey of cryptococcosis: report from Italy. Med Mycol. 2002;40:507–17. [PubMed] [Google Scholar]
- Garcia-Hermoso D, Mathoulin-Pélissier S, Couprie B, et al. DNA typing suggests pigeon droppings as a source of pathogenic Cryptococcus neoformans serotype D. J Clin Microbiol. 1997;35:2683–5. doi: 10.1128/jcm.35.10.2683-2685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Ceresini PC, Polacheck I, et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One. 2013;8:e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Colom MF, Swinne D, et al. Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerg Infect Dis. 2012a;18:1618–24. doi: 10.3201/eid1810.120068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Illnait-Zaragozí MT, Meis JF, et al. Extensive genetic diversity within the Dutch clinical Cryptococcus neoformans population. J Clin Microbiol. 2012b;50:1918–26. doi: 10.1128/JCM.06750-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F, Khayhan K, Theelen B, et al. Recognition of seven species in the Cryptococcus neoformans/C. gattii species complex. Fungal Genet Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Halliday CL, Bui T, Krockenberger M, et al. Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol. 1999;37:2920–6. doi: 10.1128/jcm.37.9.2920-2926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Martínez AF, Beckham JD. Cryptococcosis in solid organ transplant recipients. Curr Opin Infect Dis. 2015;28:300–7. doi: 10.1097/QCO.0000000000000171. [DOI] [PubMed] [Google Scholar]
- Iatta R, Hagen F, Fico C, et al. Cryptococcus gattii infection in an immunocompetent patient from Southern Italy. Mycopathologia. 2012;174:87–92. doi: 10.1007/s11046-011-9493-8. [DOI] [PubMed] [Google Scholar]
- Iatta R, Immediato D, Puttilli MR, et al. Cryptococcus neoformans in the respiratory tract of squirrels, Callosciurus finlaysonii (Rodentia, Sciuridae) Med Mycol. 2015;53:666–73. doi: 10.1093/mmy/myv045. [DOI] [PubMed] [Google Scholar]
- Kidd SE, Bach PJ, Hingston AO, et al. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg Infect Dis. 2007;13:51–7. doi: 10.3201/eid1301.060823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krockenberger MB, Canfield PJ, Malik R. Cryptococcus neoformans var. gattii in the koala (Phascolarctos cinereus): a review of 43 cases of cryptococcosis. Med Mycol. 2003;41:225–34. doi: 10.1080/369378031000137242. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, editors. Medical Mycology. Philadelphia, USA: Lea & Fabiger; 1992. [Google Scholar]
- Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4:a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrou K, Van Eldere J, Keuleers S, et al. Zoonotic transmission of Cryptococcus neoformans from a magpie to an immunocompetent patient. J Intern Med. 2005;257:385–8. doi: 10.1111/j.1365-2796.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Li W, Averette AF, Desnos-Ollivier M, et al. Genetic diversity and genomic plasticity of Cryptococcus neoformans AD Hybrid Strains. G3. 2012;2:83–97. doi: 10.1534/g3.111.001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares C, Colom MF, Torreblanca M, et al. Environmental sampling of Ceratonia siliqua (carob) trees in Spain reveals the presence of the rare Cryptococcus gattii genotype AFLP7/VGIV. Rev Iberoam Micol. 2015;32:269–72. doi: 10.1016/j.riam.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, et al. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell. 2003;2:1162–8. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Passo C, Pernice I, Gallo M, et al. Genetic relatedness and diversity of Cryptococcus neoformans strains in the Maltese Islands. J Clin Microbiol. 1997;35:751–5. doi: 10.1128/jcm.35.3.751-755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Martínez R, Castañón-Olivares LR. Isolation of Cryptococcus neoformans var. neoformans from bird droppings, fruits and vegetables in Mexico City. Mycopathologia. 1995;129:25–8. doi: 10.1007/BF01139333. [DOI] [PubMed] [Google Scholar]
- Maduro AP, Gonçalves L, Inácio J, et al. HIV/AIDS-associated cryptococcosis in Portugal spanning the pre- to post-HAART era: a retrospective assessment at the genotypic level based on URA5-RFLP. Curr Microbiol. 2015;71:449–57. doi: 10.1007/s00284-015-0873-z. [DOI] [PubMed] [Google Scholar]
- Maestrale C, Masia M, Pintus D, et al. Genetic and pathological characteristics of Cryptococcus gattii and Cryptococcus neoformans var. neoformans from meningoencephalitis in autochthonous goats and mouflons, Sardinia, Italy. Vet Microbiol. 2015;177:409–13. doi: 10.1016/j.vetmic.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Martinez LR, Garcia-Rivera J, Casadevall A. Cryptococcus neoformans var. neoformans (serotype D) strains are more susceptible to heat than C. neoformans var. grubii (serotype A) strains. J Clin Microbiol. 2001;39:3365–7. doi: 10.1128/JCM.39.9.3365-3367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, Aanensen DM, Boekhout T, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:561–70. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlinaric-Missoni E, Hagen F, Chew WH, et al. In vitro antifungal susceptibilities and molecular typing of sequentially isolated clinical Cryptococcus neoformans strains from Croatia. J Med Microbiol. 2011;60:1487–95. doi: 10.1099/jmm.0.031344-0. [DOI] [PubMed] [Google Scholar]
- Montagna MT, Santacroce MP, Caggiano G, et al. Cavernicolous habitats harbouring Cryptococcus neoformans: results of a speleological survey in Apulia, Italy, 1999–2000. Med Mycol. 2003;41:451–5. doi: 10.1080/13693780310001602785. [DOI] [PubMed] [Google Scholar]
- Montagna MT, Viviani MA, Pulito A, et al. Cryptococcus neoformans var. gattii in Italy - Note 2 - Environment investigation related to an autochtonous clinical case in Apulia. J Mycol Méd. 1997;7:93–96. [Google Scholar]
- Noguera MC, Escandón P, Castañeda E. Cryptococcosis in Atlántico, Colombia: an approximation of the prevalence of this mycosis and the distribution of the etiological agent in the environment. Rev Soc Bras Med Tro. 2015;48:580–6. doi: 10.1590/0037-8682-0178-2015. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–30. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Patel S, Shin GY, Wijewardana I, et al. The prevalence of cryptococcal antigenemia in newly diagnosed HIV patients in a Southwest London cohort. J Infect. 2013;66:75–9. doi: 10.1016/j.jinf.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernice I, Lo Passo C, Criseo G, et al. Molecular subtyping of clinical and environmental strains of Cryptococcus neoformans variety neoformans serotype A isolated from southern Italy. Mycoses. 1998;41:117–24. doi: 10.1111/j.1439-0507.1998.tb00312.x. [DOI] [PubMed] [Google Scholar]
- Romeo O, Scordino F, Chillemi V, et al. Cryptococcus neoformans/Cryptococcus gattii species complex in southern Italy: an overview on the environmental diffusion of serotypes, genotypes and mating-types. Mycopathologia. 2012;174:283–91. doi: 10.1007/s11046-012-9547-6. [DOI] [PubMed] [Google Scholar]
- Romeo O, Scordino F, Criseo G. Environmental isolation of Cryptococcus gattii serotype B, VGI/MATα strains in southern Italy. Mycopathologia. 2011;171:423–30. doi: 10.1007/s11046-010-9389-z. [DOI] [PubMed] [Google Scholar]
- Ropstad EO, Leiva M, Peña T, et al. Cryptococcus gattii chorioretinitis in a ferret. Vet Ophthalmol. 2011;14:262–6. doi: 10.1111/j.1463-5224.2011.00885.x. [DOI] [PubMed] [Google Scholar]
- Roussilhon C, Postal JM, Ravisse P. Spontaneous cryptococcosis of a squirrel monkey (Saimiri sciureus) in French Guyana. J Med Primatol. 1987;16:39–47. [PubMed] [Google Scholar]
- Sanchini A, Smith IM, Sedlacek L, et al. Molecular typing of clinical Cryptococcus neoformans isolates collected in Germany from 2004 to 2010. Med Microbiol Immunol. 2014;203:333–40. doi: 10.1007/s00430-014-0341-6. [DOI] [PubMed] [Google Scholar]
- Saul N, Krockenberger M, Carter D. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot Cell. 2008;7:727–34. doi: 10.1128/EC.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solla I, Morano LE, Vasallo F, et al. Cryptococcus gattii meningitis: observation in a Spanish patient. Enferm Infec Micr Cl. 2008;26:395–6. doi: 10.1157/13123846. [DOI] [PubMed] [Google Scholar]
- Stilwell G, Pissarra H. Cryptococcal meningitis in a goat—a case report. BMC Vet Res. 2014;10:84. doi: 10.1186/1746-6148-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rodríguez JM, Baró T, Morera Y, et al. Molecular characterization of Cryptococcus neoformans var. gattii causing epidemic outbreaks of cryptococcosis in goats. Rev Iberoam Micol. 1999;16:164–5. [PubMed] [Google Scholar]
- Velegraki A, Kiosses VG, Pitsouni H, et al. First report of Cryptococcus neoformans var. gattii serotype B from Greece. Med Mycol. 2001;39:419–22. doi: 10.1080/mmy.39.5.419.422. [DOI] [PubMed] [Google Scholar]
- Viviani MA, Cogliati M, Esposto MC, et al. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–9. doi: 10.1111/j.1567-1364.2006.00081.x. [DOI] [PubMed] [Google Scholar]
- Viviani MA, Wen H, Roverselli A, et al. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J Med Vet Mycol. 1997;35:355–60. [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, et al. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–73. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.