Abstract
Exploring the origin and extent of reproductive isolation within the same species is valuable to capture early events to the onset of speciation. In multiple genetic models, reproductive isolation was recently observed at the intraspecific scale, indicating that the raw potential for speciation segregates readily within populations, which could be a rule rather than an exception in a broad context. We briefly recapitulate the molecular evidence of intrinsic post-zygotic isolation in major model organisms including Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster and their close relatives. We then focus on recent advances in yeast and review the genetic basis of post-zygotic isolation within and between multiple members of the Saccharomyces genus, especially in Saccharomyces cerevisiae. We discuss the role of various mechanisms involved in the onset of reproductive isolation including DNA sequence divergence, chromosomal rearrangement, cytonuclear as well as nuclear–nuclear genetic incompatibilities and provide a comparative view along a continuum of genetic differentiation, which encompasses intraspecific populations, recent delineating nascent species as well as closely related sister species in the same subphylum.
Keywords: speciation, reproductive isolation, intraspecific
Recent systematic explorations of large natural yeast populations have been useful for dissecting the mechanistic complexity and multiplicity of their reproductive isolation.
INTRODUCTION
Speciation, the evolutionary process by which new species emerge, lies at the heart of the observed biodiversity. Under the biological species concept, the formation of new species requires the establishment of reproductive barriers that limit the gene flow among populations (Coyne and Orr 2004). In other words, new species form when individuals from diverging populations become reproductively isolated and unable to produce viable or fertile offspring, eventually allowing nascent species to be genetically and phenotypically distinct. While this concept is widely applied for sexually reproducing organisms, it is not until the past two decades that precise molecular characterizations of the genetic basis of reproductive isolation have become possible (Presgraves 2010).
Reproductive isolation can act prior to mating (pre-zygotic), which prevent the formation of a zygote or soon after mating (post-zygotic) leading to reduced offspring viability or fertility (Coyne and Orr 2004). While many external factors, such as differences in life history and temporal patterns may cause pre-zygotic isolation (Coyne and Orr 2004), genetic analyses mainly focused on intrinsic post-zygotic isolation (Presgraves 2010; Maheshwari and Barbash 2011). During the past years, much progress has been made on the subject, leading to the identification of multiple mechanisms between closely related species in various taxa (Bomblies and Weigel 2007a,b; Greig 2009; Chou and Leu 2010; Presgraves 2010; Maheshwari and Barbash 2011; Ouyang and Zhang 2013; Chen et al. 2014; Zanders et al. 2014). However, contrasting to the perception of its mechanistic multiplicity, only a few examples have been characterized to the molecular level, and the tempo and mode of reproductive isolation were still poorly understood. Are the identified mechanisms the original cause of reproductive isolation, or just a consequence of subsequent divergence within nascent species? Which types of genetic changes are of particular interest in the onset of reproductive isolation? What is the relative role of selection versus drift through initial stage to the completion reproductive isolation?
To address these questions, it is essential to systematically explore the onset and accumulation of reproductive isolation at various evolutionary scales over a broad taxonomic range. Within the past few years, such efforts have started to be undertaken (Table 1). In fact, with the increased availability of large collections of isolates from various species, cases of partial reproductive isolation at the intraspecific scale were recently observed in model systems such as Drosophila (Brideau et al. 2006; Corbett-Detig et al. 2013; Phadnis et al. 2015), Arabidopsis (Leppala, Bokma and Savolainen 2013; Alcazar et al. 2014; Chae et al. 2014), Caenorhabditis (Ross et al. 2011; Snoek et al. 2014; Chang, Rodriguez and Ross 2015) and Saccharomyces (Almeida et al. 2014; Charron, Leducq and Landry 2014; Hou et al. 2014; Paliwal, Fiumera and Fiumera 2014; Bui et al. 2015; Hou et al. 2015). While the number of cases identified is still low, it has been clear that the raw potential for speciation segregates readily within populations, which seems to be a rule rather than an exception in a broad context.
Table 1.
Evidences of reproductive isolation within and between species in model organisms.
| Species pair | Evidence | Genes | Chromosome | Phenotype | References | |
|---|---|---|---|---|---|---|
| Arabidopsis | A. thaliana × A. thaliana | TRD | HPA1,HPA2 | Autosome | Viability | Demogines et al. (2008) |
| A. thaliana × A. thaliana | Diallele cross | DM1-9,SRF3 | Autosome | Fitness | Hittinger (2013); Brion et al. (2015) | |
| A. lyrata × A. lyrata | TRD | - | Autosome | Viability | Chae et al. (2014) | |
| Caenorhabditis | C. elegans × C. elegans | TRD | PEEL1, ZEEL1 | Autosome | Viability | Hittinger et al. (2010) |
| C. elegans × C. elegans | TRD | - | Autosome | Viability | Charron, Leducq and Landry (2014) | |
| C. briggsae × C. briggsae | Cybrid | - | Cytonuclear | Fitness | Chang et al. (2015) | |
| C. briggsae × C. briggsae | TRD | - | Autosome | Viability | Chen et al. (2014) | |
| C. briggsae × C. nigoni | Introgression | - | X-linked | Viability | Friedrich et al. (2015) | |
| Drosophila | D. melanogaster × D. melanogaster | TRD | - | Autosome | Viability | Bomblies and Weigel (2007b) |
| D. melanogaster × D. simulans | Suppressor | Lhr, Hmr, gzgf | X-linked | Viability | Brideau et al. (2006); Bomblies and Madlung (2014) |
TRD: transmission ration distortion.
Here, we briefly recapitulate the genetic origins of intrinsic post-zygotic reproductive isolation in major model organisms including Arabidopsis thaliana (Bikard et al. 2009; Chae et al. 2014), Caenorhabditis elegans (Seidel et al. 2011; Chang, Rodriguez and Ross 2015), Drosophila melanogaster (Corbett-Detig et al. 2013) and their close relatives (Brideau et al. 2006; Kozlowska et al. 2012; Matute, Gavin-Smyth and Liu 2014; Bi et al. 2015; Phadnis et al. 2015). We then concentrate on yeasts and conduct a more comprehensive review on the current state of the genetic basis of post-zygotic reproductive isolation in the Saccharomyces genus and recent advances at the intraspecific scales within multiple species of this group (Almeida et al. 2014; Charron, Leducq and Landry 2014; Leducq et al. 2016), especially in Saccharomyces cerevisiae (Hou et al. 2014, 2015; Paliwal, Fiumera and Fiumera 2014; Bui et al. 2015). We focus on the mechanistic diversity as well as their underlying evolutionary origins that act intraspecifically, and try to provide a comparative view on the onset of reproductive isolation along a continuum of genetic differentiation, which encompasses intraspecific populations, recent delineating nascent species as well as closely related sister species of the same subphylum.
Brief overview of reproductive isolation in different model organisms
On the conceptual ground, the most prominent genetic explanation of intrinsic post-zygotic reproductive isolation is the presence of genetic incompatibilities, popularized in the 1940s by Theodosius Dobzhansky and Hermann Müller (Dobzhansky 1937; Muller 1942). The hitherto known as the Dobzhansky–Müller model posits that populations could evolve independently and accumulate different mutations that are well adapted in their original genetic backgrounds but do not function properly together in hybrids. The loss of viability or fertility in the offspring may simply be caused by the accumulation of such incompatible mutations, which arose as a by-product of genomic differentiation (Seehausen et al. 2014). Not only that this model offers an elegant solution on how genetic basis for reproductive isolation could originate from an intermating population, but also integrates the notion that incompatible alleles may accumulate with increased genomic divergence (Nosil, Harmon and Seehausen 2009; Seehausen et al. 2014). Examples of such incompatible gene pairs have been identified between closely related species in various taxa (Bomblies and Weigel 2007a,b; Greig 2009; Chou and Leu 2010; Presgraves 2010; Maheshwari and Barbash 2011; Ouyang and Zhang 2013; Chen et al. 2014; Zanders et al. 2014), and more recently among populations of the same species in several major genetic models such as D. melanogaster (Corbett-Detig et al. 2013; Phadnis et al. 2015), A. thaliana (Bikard et al. 2009; Chae et al. 2014), C. elegans (Seidel et al. 2011; Chang, Rodriguez and Ross 2015) (Table 1).
Different evolutionary forces could putatively explain the observed incompatibilities. Adaptive processes such as niche specialization to pathogens were of particular importance in the evolution of plant immune systems, where defense-related genes acquired in different populations could cause hybrid necrosis through autoimmune responses (Bomblies and Weigel 2007a,b; Alcazar et al. 2009, 2014; Chae et al. 2014). Many other cases were related to neutral processes such as genetic drift or the propagation of selfish genetic elements (Seidel, Rockman and Kruglyak 2008; Bikard et al. 2009). For example, reciprocal inactivation of a duplicated essential gene pair HPA1/HPA2 in natural accessions of A. thaliana could lead to seed abortion in the F2 offspring when none of the functional copies were present (Bikard et al. 2009). Differences in recombination rates or mutation loads could also put emphasizes on certain types of mechanisms, for example cytonuclear incompatibilities involving interacting genes located on mitochondrial and nuclear genomes (Rand, Clark and Kann 2001; Lee et al. 2008; Chou and Leu 2010; Chou et al. 2010; Paliwal, Fiumera and Fiumera 2014; Chang, Rodriguez and Ross 2015). In fact, due to the dynamic structure and high mutation rate, mitochondrial genomes accumulated mutations more rapidly compared to the nuclear genome, in turn causing incompatibilities. Such cytonuclear incompatibilities were found to cause F2 sterility in interspecific yeast hybrids (Lee et al. 2008; Chou et al. 2010), and could be a common cause of hybrid weakness in Drosophila (Rand, Clark and Kann 2001).
Besides incompatibilities at the gene level, large genomic changes could also lead to post-zygotic reproductive isolation (Coyne and Orr 2004). For example, differences of the ploidy level or chromosome numbers among parental species were common in causing reproductive isolation in plants (Bomblies and Madlung 2014) and animals (Coyne and Orr 2004), where unbalanced gene dosages in the offspring could lead to inviability or sterility. Between A. thaliana and its sister species A. lyrata, differences in chromosome numbers (five for A. thaliana and eight for A. lyrata) were accounted for the observed reproductive isolation, where F1 hybrids were viable but sterile (Bomblies and Weigel 2007a,b). Other localized chromosomal rearrangements, for example, translocations and inversions, have also drawn much interest, as parental species that differ by such structural variation would most likely produce offspring with unequal distribution of essential genes upon meiosis (Coyne and Orr 2004). The role of chromosomal rearrangements is indeed well established leading to post-zygotic reproductive isolation in various taxa, especially in plants (Hoffmann and Rieseberg 2008) and Drosophila (Kulathinal, Stevison and Noor 2009).
However, while extremely insightful, reproductive isolation studies in complex model systems suffered from several drawbacks. Studies using such models often restricted to a low number of parental combinations due to experimental workloads, therefore large-scale analysis has rarely been undertaken (Chae et al. 2014). Moreover, due to considerable genome size and complexity, precise identifications of the molecular mechanisms involved are challenging especially for structural variations such as inversions and translocations. As a result, only a low number of cases have been fully characterized to date, and an overview of the relative importance of different mechanisms to the onset and propagation of reproductive isolation across the observed natural diversity in these species is far from being reached.
Yeasts, ideal model to explore inter- and intraspecies reproductive isolation
An emerging model system, which allows the integration of the genetic and genomic diversity within and between closely related species, is the budding yeast S. cerevisiae and its close relatives. Compared to other complex models, yeasts present numerous advantages due to their short generation time, small and compact genomes and laboratory amenable sexual reproductions. Rather than relying on a low number of crosses and transmission ratio distortion in the offspring as it is the case for complex organisms, yeasts offer the possibility to systematically examine a large number of crosses and use pooled mapping strategies that require much less sequencing efforts. Natural populations of multiple yeasts species can be isolated from various biotopes and geographical locations (Liti et al. 2009; Schacherer et al. 2009). The vast natural distribution with the ever-growing availability of whole genome sequencing data makes yeasts an ideal model system to obtain a comprehensive view on how reproductive isolation emerges at different evolutionary scales by taking into account the roles of ecology, domestication and other selective or neutral processes.
Saccharomyces genus and reproductive isolation
The Saccharomyces sensu stricto complex comprises seven known species (S. cerevisiae, S. paradoxus, S. arboricola, S. kudiavzevii, S. mikatae, S. eubayanus and S. uvarum) to date, all of which could cross to form viable hybrids under laboratory conditions, with limited mating preferences (Murphy et al. 2006; Greig 2009; Hittinger 2013) (Fig. 1). Although outcrossing events are rare (Knop 2006; Hittinger 2013), natural hybrids between closely related Saccharomyces species are readily observed (Gonzalez et al. 2006; Belloch et al. 2009; Baker et al. 2015). Many of the hybrids were found to be involved in industrial-related processes, such as beer (Baker et al. 2015) and wine making (Gonzalez et al. 2006). Among Saccharomyces species, introgressions are frequently reported, for example, between S. cerevisiae and S. paradoxus (Zhang et al. 2010; Barbosa et al. 2016). Recent evidence indicates that such events could occur across multiple populations within a species (Almeida et al. 2014). In fact, genome-wide screen in a large number of S. uvarum isolates identified multiple chromosomal regions ascribed to different Saccharomyces species, such as S. kudriavzevii, S. cerevisiae and S. eubayanus. It is worth noting that a large number of introgressions observed were found in isolates from anthropic niches, including various alcoholic fermentation processes (Almeida et al. 2014).
Figure 1.
Intraspecific divergence and evidences of post-zygotic reproductive isolation within species of the Saccharomyces genus. References [1–9]: Greig et al. (2003); Heck et al. (2006); Liti et al. (2006); Demogines et al. (2008); Cubillos et al. (2011); Wang et al. (2012); Hou et al. (2014, 2015); Paliwal et al. (2014). [10 and 11]: Charron et al. (2014); Leducq et al. (2016). [12 and 13]: Hittinger et al. (2004, 2010). [14]: Almeida et al. (2014).
Strong post-zygotic reproductive isolations are observed between species within this complex. Interspecific hybrids typically yield less than 1% of viable meiotic offspring in most parental combinations (Greig 2009), and ∼7% for the least diverged species pair S. eubayanus and S. uvarum (Libkind et al. 2011; Hittinger 2013). High levels of DNA sequence divergence are considered as the main cause of loss of hybrid offspring viability, which impairs proper chromosomal segregation through mechanism of anti-recombination by the mismatch repair system (MMR) (Greig et al. 2003; Liti et al. 2006). In fact, viable hybrid offspring often show high numbers of aneuploidies and reduced recombination rate (Hunter et al. 1996; Delneri et al. 2003), the effect of which could be rescued by deleting components of the MMR (Hunter et al. 1996; Greig et al. 2003).
It is widely admitted that the degree of post-zygotic isolation is correlated with the level of divergence between the parental pair, as the effect of anti-recombination progresses with increased DNA sequence divergence (Liti, Barton and Louis 2006; Greig 2009; Hittinger 2013). However, this overly simplified generalization might be due to sampling bias of the parental isolate pairs. As a matter of fact, it is increasingly evident that multiple mechanisms operate concurrently at both intra- and interspecific levels, leading to partial or complete offspring loss depending on the parental populations involved. Therefore, the correlation between sequence divergence and reproductive isolation might be plausible when other mechanisms were absent, and likely to play a relatively minor role to the initial stage of reproductive isolation.
For example, besides sequence divergence, chromosomal rearrangements (Fischer et al. 2000; Delneri et al. 2003) and cytonuclear incompatibilities (Lee et al. 2008; Chou et al. 2010) also contribute to the observed post-zygotic reproductive isolation in the Saccharomyces genus. Classic Dobzhansky–Müller incompatibilities could also play a role, although no clear examples have been found so far (Greig et al. 2002; Greig 2007; Kao, Schwartz and Sherlock 2010). In fact, allotetraploid hybrids between S. cerevisiae and S. paradoxus (with two copies of each parental genome) showed high offspring viability contrasting to diploid hybrids, strongly suggesting that dominant Dobzhansky–Müller incompatibilities were not present between this species pair (Greig et al. 2002). Nevertheless, while chromosome replacements in S. cerevisiae with its S. paradoxus homologs in haploids were mostly viable (Greig 2007; Kao, Schwartz and Sherlock 2010), the existence of recessive epistatic interactions impacting offspring fertility and fitness cannot be ruled out (Li, Wang and Zhang 2013).
Nevertheless, as interspecific reproductive isolation is nearly complete in Saccharomyces, it has been difficult to disentangle the effect of simple divergence from functional genetic differentiation. As a result, the role of mechanisms identified using interspecific approaches remains largely indecisive at the incipient stage of reproductive isolation (Fischer et al. 2000; Delneri et al. 2003), and recent works have turned their focus on intraspecific studies using natural populations within the same species of Saccharomyces yeasts.
Evidence of intraspecific reproductive isolation within yeast natural populations
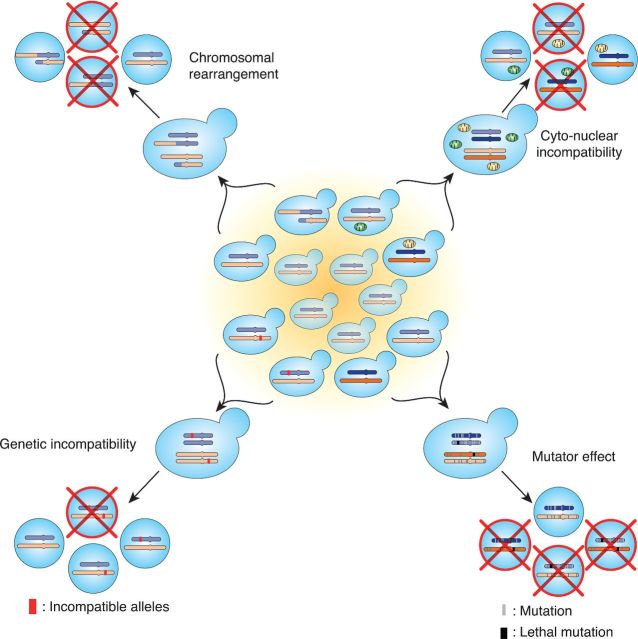
With the increasing availability of whole genome sequencing data, multiple yeast species have become the workhorses for functional and evolutionary genomic studies (Almeida et al. 2014; Brion et al. 2015; Friedrich et al. 2015; Jeffares et al. 2015; Strope et al. 2015). Evidence of intraspecific reproductive isolation leading to offspring loss upon crosses has been quite frequently observed within collection of isolates of various yeast species (Hittinger, Rokas and Carroll 2004; Liti, Barton and Louis 2006; Hittinger et al. 2010; Cubillos et al. 2011; Wang et al. 2012; Almeida et al. 2014). However, such cases were often found when generating recombined offspring for linkage mapping and were usually dismissed as ‘annoying crosses’ without further dissection of the underlying causes. Again, these observations indicate that mechanisms leading to reproductive isolation segregate readily at the intraspecific scale. Using various approaches, many recent studies addressed specifically how such mechanisms could emerge and lead to intraspecific reproductive isolation, which will be discussed individually in the following section (Fig. 2).
Figure 2.
Molecular mechanisms leading to reproductive isolation within natural populations of S. cerevisiae.
The role of chromosomal rearrangements in intrinsic post-zygotic isolation
Although it is well accepted that large chromosomal rearrangements such as translocations and inversions could contribute at least partly to the observed offspring loss in Saccharomyces hybrids (Fischer et al. 2000; Delneri et al. 2003), their role at the incipient stage of speciation has received much debate (Delneri et al. 2003). For one reason is that the overall distribution of chromosomal rearrangements usually does not correlate with the level of reproductive isolation or with the scales of genetic divergence observed (Fischer et al. 2000; Delneri et al. 2003). In fact, many species pairs within the Saccharomyces sensu stricto complex harbor individuals with collinear genomes but are completely reproductively isolated (Greig 2009). Moreover, artificially generated collinear parental pairs by reverting the observed translocation between the S. cerevisiae and S. mikatae species clearly showed that translocation events only have a marginal effect on the loss of offspring viability (Delneri et al. 2003).
Paradoxically, it appears that while genomic configurations were sometimes conserved between species, individuals from the same species could be surprisingly diverse in terms of chromosomal profiles (Charron, Leducq and Landry 2014; Hou et al. 2014). When studying a large collection of natural isolates in S. cerevisiae, three different types of translocation were identified in 10 out of 60 isolates, which explains the total effect of reproductive isolation observed (Hou et al. 2014). Similar observations have been made in S. paradoxus populations, where the level of chromosomal rearrangements was partly but significantly correlated with the level of reproductive isolation across 25 isolates (Charron, Leducq and Landry 2014). Most identified rearrangements arise through neutral events such as ectopic recombination between repetitive sequences such Ty elements, with few exceptions that were adaptive in specific environmental contexts by altering expression patterns of genes present at the junction of the rearranged regions (Perez-Ortin et al. 2002).
While chromosomal rearrangements cannot be the only explanation of reproductive isolation observed in many Saccharomyces species pairs, there has been a recent example illustrating that such rearrangements could be directly involved during incipient speciation in allopatric populations of North American S. paradoxus (Leducq et al. 2016). In this case, two allopatric populations separated by glaciation differing by a translocation and an inversion in their genomes gave rise to a hybrid population upon secondary contact. These rearrangements were then fixed in the hybrids and introgressed by repeated backcrossing events with one of the parental population that does not have these rearrangements. Eventually, the hybrid population became reproductively isolated with both parental species through time, illustrating that speciation through chromosomal rearrangement could indeed be possible in yeast (Leducq et al. 2016).
Cytonuclear incompatibility and offspring fitness
Compelling evidence suggests that cytonuclear incompatibilities could play a major role during speciation. Incompatible combinations between mitochondrial and nuclear genomes were found to lead to hybrid problems in a wide range of species (Rand, Clark and Kann 2001; Chang, Rodriguez and Ross 2015), including yeast (Lee et al. 2008; Chou et al. 2010). In fact, as mitochondrial genomes were more prone to mutations, nuclear genomes have to evolve accordingly because proper interactions between the two were often essential for survival or fitness (Chou and Leu 2010). Because of such constant arms race between mitochondrial and nuclear genomes, cytonuclear incompatibilities were more likely to evolve as diverging population could take different trajectories of cytonuclear coevolution. Among the Saccharomyces genus, examples of cytonuclear incompatibilities were found between S. cerevisiae and S. paradoxus as well as between S. cerevisiae and S. uvarum (formerly S. bayanus), each with independent origins (Chou et al. 2010). For example, the incompatibility between S. cerevisiae and S. paradoxus was due to inefficient splicing of the COX1 intron in S. paradoxus by the S. cerevisiae version of the MRS1 gene, which arose with the loss of corresponding intron in S. cerevisiae. In all cases, the observed cytonuclear incompatibility dampens offspring respiratory capacities, leading to partial sterility (Chou et al. 2010).
At the intraspecific level, no evident cases of specific cytonuclear gene pairs leading to reproductive isolation have been found so far. However, global epistasis between nuclear and mitochondrial genomes was commonly observed leading to increased phenotypic variance within S. cerevisiae (Paliwal, Fiumera and Fiumera 2014). By testing pairwise combinations of mitochondrial and nuclear genomes in 10 divergent isolates, it was shown that novel combinations often lead to reduced fitness, and the effect of which was not correlated with the level of genetic divergence between the tested pairs. These observations suggest coevolutions between mitochondrial and nuclear genomes were already significant at the intraspecific scale; however, whether such observed fitness variation could eventually lead to reproductive isolation remains unclear.
Antagonistic effects of mutator phenotype related to mismatch repair
In addition to mitochondrial and nuclear genome pairs, genes in the nuclear genome could also coevolve and lead to hybrid problems. One classic example was illustrated by the MMR in S. cerevisiae. Using allelic survey across a number of natural isolates, it has been shown that specific combinations of the PMS1 and MLH1 genes, essential players of MMR system, were conserved across the species and possibly maintained by balancing selection (Heck et al. 2006). When the original combinations are disrupted, interaction between incompatible allelic pairs could result in a mutator phenotype due to malfunctioning in the MMR. Accumulation of undesired mutations could then lead to sporadic offspring loss, the effect of which could depend on specific backgrounds (Demogines et al. 2008).
Other than offspring loss related to the accumulation of deleterious mutations, the mutator phenotype could also lead to accelerated adaptation in stress conditions due to increased mutation rates (Bui et al. 2015). In fact, it was shown that strains with incompatible combination of PMS1 and MLH1 thrives more rapidly on high osmotic stress condition by acquiring advantageous mutations in the PMR1 gene earlier than strains with compatible combinations.
In principle, such mutator phenotype could be considered as a special form of Dobzhansky–Müller incompatibility. However, the effect of incompatible allelic combination results in increased mutation rates and only act indirectly on offspring viability. The most curious feature of this type of interaction is that the incompatibility could lead to opposite effects on offspring viability depending on different environmental contexts. The interplay between genetic interactions and environmental selections could therefore be important in the onset of reproductive isolation, at least in this specific configuration.
Condition-specific genetic incompatibilities and the role of selection
Similarly, different environmental conditions could also have an impact on the effect of classic Dobzhansky–Müller incompatibilities in yeast. Genetic incompatibilities related to negative epistatic interactions were mostly invisible on permissive laboratory conditions that optimize growth in S. cerevisiae (Hou et al. 2014). However, by taking into account of different environmental factors, such interactions were much more common than previously thought, leading to condition specific reproductive isolations. In fact, systematic survey across 25 crosses on 20 conditions revealed over 24% of the cases showing offspring loss with various severities, the effect of which were specific to independent crosses and conditions (Hou et al. 2015). Using segregation analysis followed by pooled sequencing strategy, the first example of two loci Dobzhansky–Müller incompatibilities within a yeast species was identified related to respiratory conditions. In this case, the incompatibility was due to a nonsense mutation in the COX15 gene and a tRNA suppressor SUP7, leading to the loss of 25% of the offspring in conditions that require respiration (Hou et al. 2015). Both mutations were extremely rare across natural populations in this species, although there were some evidence suggesting that the specific derived combination was maintained by positive selection (Hou et al. 2015).
Interestingly, most identified cases of negative epistasis appeared not to be related to two loci interactions but instead showing a higher genetic complexity even at the intraspecific scale (Hou et al. 2015; Hou and Schacherer 2016). Moreover, despite the relatively high frequency of occurrence, most incompatible cases were not shared across different isolates, indicating unique genetic origins (Hou and Schacherer 2016). These observations highlight again the role of environmental selection to the onset of reproductive isolation in yeast. It is worth noting that natural populations within the same species govern raw speciation potential through condition-specific epistatic interactions. Nevertheless, precise molecular dissection of more such incompatibility cases is still required to get a global view of types and distributions of genes involved.
CONCLUSION AND PERSPECTIVES
Yeasts along with other major genetic model organisms provide unique comparative insights into the genetic basis of post-zygotic reproductive isolation across a broad evolutionary scale. In particular, systematic exploration by looking at large natural yeast populations across multiple environmental conditions was particular useful and fruitful to dissect the mechanistic complexity of reproductive isolation. Nevertheless, despite significant advances, intraspecific reproductive isolation is still underexplored. In particular, most molecular exploration of reproductive isolation cases were restricted the Saccharomyces cerevisiae, and even so many evident cases in S. cerevisiae still have to be characterized. Would the patterns be different in other yeast species where the level of intraspecific genetic diversity is usually higher? How relevant are the mechanisms found in yeast to the onset of post-zygotic reproductive isolation in other species in general? What are the roles of the genetic bases of reproductive isolation in shaping other phenotypic traits? Further explorations of the natural population diversity across a broad taxonomic range will be promising to provide some answers to these questions, and may impart deeper understandings regarding the patterns and constraints of genetic differentiation as well as their role in speciation and biodiversity.
FUNDING
The authors thank the (NIH Grant ) and the (ANR Grant ) for financial support. TF is supported by a grant from the and JH by a fellowship from the medical association .
Conflict of interest. None declared.
REFERENCES
- Alcazar R, Garcia AV, Parker JE, et al. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. P Natl Acad Sci USA. 2009;106:334–9. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar R, von Reth M, Bautor J, et al. Analysis of a plant complex resistance gene locus underlying immune-related hybrid incompatibility and its occurrence in nature. PLos Genet. 2014;10:e1004848. doi: 10.1371/journal.pgen.1004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P, Goncalves C, Teixeira S, et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat Commun. 2014;5:4044. doi: 10.1038/ncomms5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E, Wang B, Bellora N, et al. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Mol Biol Evol. 2015;32:2818–31. doi: 10.1093/molbev/msv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa R, Almeida P, Safar SV, et al. Evidence of Natural Hybridization in Brazilian Wild Lineages of Saccharomyces cerevisiae. Genome Biol Evol. 2016;8:317–29. doi: 10.1093/gbe/evv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloch C, Perez-Torrado R, Gonzalez SS, et al. Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl Environ Microb. 2009;75:2534–44. doi: 10.1128/AEM.02282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Ren X, Yan C, et al. A Genome-wide hybrid incompatibility landscape between Caenorhabditis briggsae and C. nigoni. PLos Genet. 2015;11:e1004993. doi: 10.1371/journal.pgen.1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Patel D, Le Mette C, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–6. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Madlung A. Polyploidy in the Arabidopsis genus. Chromosome Res. 2014;22:117–34. doi: 10.1007/s10577-014-9416-x. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. Arabidopsis: a model genus for speciation. Curr Opin Genet Dev. 2007a;17:500–4. doi: 10.1016/j.gde.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Weigel D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet. 2007b;8:382–93. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–5. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Brion C, Pflieger D, Friedrich A, et al. Evolution of intraspecific transcriptomic landscapes in yeasts. Nucleic Acids Res. 2015;43:4558–68. doi: 10.1093/nar/gkv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui DT, Dine E, Anderson JB, et al. A genetic incompatibility accelerates adaptation in yeast. PLos Genet. 2015;11:e1005407. doi: 10.1371/journal.pgen.1005407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, et al. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159:1341–51. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Rodriguez J, Ross J. Mitochondrial-nuclear epistasis impacts fitness and mitochondrial physiology of interpopulation Caenorhabditis briggsae hybrids. G3. 2015;6:209–19. doi: 10.1534/g3.115.022970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Leducq JB, Landry CR. Chromosomal variation segregates within incipient species and correlates with reproductive isolation. Mol Ecol. 2014;23:4362–72. doi: 10.1111/mec.12864. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Lin YS, et al. A two-locus interaction causes interspecific hybrid weakness in rice. Nat Commun. 2014;5:3357. doi: 10.1038/ncomms4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JY, Hung YS, Lin KH, et al. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JY, Leu JY. Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. BioEssays. 2010;32:401–11. doi: 10.1002/bies.200900162. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Zhou J, Clark AG, et al. Genetic incompatibilities are widespread within species. Nature. 2013;504:135–7. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Cubillos FA, Billi E, Zorgo E, et al. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol. 2011;20:1401–13. doi: 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, et al. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Demogines A, Wong A, Aquadro C, et al. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLos Genet. 2008;4:e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- Fischer G, James SA, Roberts IN, et al. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–4. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- Friedrich A, Jung P, Reisser C, et al. Population genomics reveals chromosome-scale heterogeneous evolution in a protoploid yeast. Mol Biol Evol. 2015;32:184–92. doi: 10.1093/molbev/msu295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez SS, Barrio E, Gafner J, et al. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006;6:1221–34. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- Greig D. A screen for recessive speciation genes expressed in the gametes of F1 hybrid yeast. PLos Genet. 2007;3:e21. doi: 10.1371/journal.pgen.0030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D. Reproductive isolation in Saccharomyces. Heredity. 2009;102:39–44. doi: 10.1038/hdy.2008.73. [DOI] [PubMed] [Google Scholar]
- Greig D, Borts RH, Louis EJ, et al. Epistasis and hybrid sterility in Saccharomyces. Proc Biol Sci/Roy Soc. 2002;269:1167–71. doi: 10.1098/rspb.2002.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D, Travisano M, Louis EJ, et al. A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol. 2003;16:429–37. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- Heck JA, Argueso JL, Gemici Z, et al. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. P Natl Acad Sci USA. 2006;103:3256–61. doi: 10.1073/pnas.0510998103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013;29:309–17. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Goncalves P, Sampaio JP, et al. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–8. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Rokas A, Carroll SB. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. P Natl Acad Sci USA. 2004;101:14144–9. doi: 10.1073/pnas.0404319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol S. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Friedrich A, de Montigny J, et al. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr Biol. 2014;24:1153–9. doi: 10.1016/j.cub.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Friedrich A, Gounot JS, et al. Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nat Commun. 2015;6:7214. doi: 10.1038/ncomms8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Schacherer J. Negative epistasis: a route to intraspecific reproductive isolation in yeast? Curr Genet. 2016;62:25–9. doi: 10.1007/s00294-015-0505-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, et al. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–33. [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Rallis C, Rieux A, et al. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat Genet. 2015;47:235–41. doi: 10.1038/ng.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Schwartz K, Sherlock G. A genome-wide analysis reveals no nuclear dobzhansky-muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLos Genet. 2010;6:e1001038. doi: 10.1371/journal.pgen.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. BioEssays. 2006;28:696–708. doi: 10.1002/bies.20435. [DOI] [PubMed] [Google Scholar]
- Kozlowska JL, Ahmad AR, Jahesh E, et al. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evolution. 2012;66:1180–95. doi: 10.1111/j.1558-5646.2011.01514.x. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MA. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq J-B, Nielly-Thibault L, Charron G, et al. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat Microbiol. 2016;1:15003. doi: 10.1038/nmicrobiol.2015.3. [DOI] [PubMed] [Google Scholar]
- Lee HY, Chou JY, Cheong L, et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–73. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Leppala J, Bokma F, Savolainen O. Investigating incipient speciation in Arabidopsis lyrata from patterns of transmission ratio distortion. Genetics. 2013;194:697–708. doi: 10.1534/genetics.113.152561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Z, Zhang J. Toward genome-wide identification of bateson-dobzhansky-muller incompatibilities in yeast: a simulation study. Genome Biol Evol. 2013;5:1261–72. doi: 10.1093/gbe/evt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valerio E, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. P Natl Acad Sci USA. 2011;108:14539–44. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–50. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Annu Rev Genet. 2011;45:331–55. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- Matute DR, Gavin-Smyth J, Liu G. Variable post-zygotic isolation in Drosophila melanogaster/D. simulans hybrids. J Evol Biol. 2014;27:1691–705. doi: 10.1111/jeb.12422. [DOI] [PubMed] [Google Scholar]
- Muller H. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Murphy HA, Kuehne HA, Francis CA, et al. Mate choice assays and mating propensity differences in natural yeast populations. Biol Lett. 2006;2:553–6. doi: 10.1098/rsbl.2006.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24:145–56. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Zhang Q. Understanding reproductive isolation based on the rice model. Annu Rev Plant Biol. 2013;64:111–35. doi: 10.1146/annurev-arplant-050312-120205. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Fiumera AC, Fiumera HL. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics. 2014;198:1251–65. doi: 10.1534/genetics.114.168575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortin JE, Querol A, Puig S, et al. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12:1533–9. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Baker EP, Cooper JC, et al. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science. 2015;350:1552–5. doi: 10.1126/science.aac7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–80. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–87. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Koboldt DC, Staisch JE, et al. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLos Genet. 2011;7:e1002174. doi: 10.1371/journal.pgen.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, et al. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–5. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–92. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- Seidel HS, Ailion M, Li J, et al. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–94. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Orbidans HE, Stastna JJ, et al. Widespread genomic incompatibilities in Caenorhabditis elegans. G3. 2014;4:1813–23. doi: 10.1534/g3.114.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, et al. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015;25:762–74. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Liu WQ, Liti G, et al. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Zanders SE, Eickbush MT, Yu JS, et al. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife. 2014;3:e02630. doi: 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Skelton A, Gardner RC, et al. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010;10:941–7. doi: 10.1111/j.1567-1364.2010.00681.x. [DOI] [PubMed] [Google Scholar]