Abstract
One major challenge during genome duplication is the stalling of DNA replication forks by various forms of template blockages. As these barriers can lead to incomplete replication, multiple mechanisms have to act concertedly to correct and rescue stalled replication forks. Among these mechanisms, replication fork regression entails simultaneous annealing of nascent and template strands, which leads to regression of replication forks and formation of four-way DNA junctions. In principle, this process can lead to either positive outcomes, such as DNA repair and replication resumption, or less desirable outcomes, such as misalignment between nascent and template DNA and DNA cleavage. While our understanding of replication fork regression and its various possible outcomes is still at an early stage, recent studies using combinational approaches in multiple organisms have begun to identify the enzymes that catalyze this DNA transaction and how these enzymes are regulated, as well as the specific manners by which fork regression can influence replication. This review summarizes these recent progresses. In keeping with the theme of this series of reviews, we focus on studies in yeast and compare to findings in higher eukaryotes. It is anticipated that these findings will form the basis for future endeavors to further elucidate replication fork remodeling and its implications for genome maintenance.
Keywords: replication fork regression, DNA motor proteins, Smc5/6, checkpoint kinases
DNA motor protein-mediated replication fork regression can restart stalled replication but may also generate dangerous outcomes. Positive and negative regulators can influence the outcomes depending on the specific molecular settings.
INTRODUCTION
Studies in model organisms and human cells have provided abundant evidence that template barriers frequently obstruct DNA replication. Examples of these barriers include DNA secondary structures and topological stress, proteins tightly bound to DNA, collision with transcriptional machineries and different types of DNA lesions generated spontaneously or by exogenous agents (Zeman and Cimprich 2014). Consequently, cellular machineries that cope with these barriers become indispensible for faithful genome replication and transmission of genetic information. Though the full atlas of these machineries is not yet known, the factors identified thus far have been shown to have pivotal roles in preventing the deleterious genetic changes that fuel the development of cancers and other forms of human diseases (Jackson and Bartek 2009; Zeman and Cimprich 2014). Thus, the examination of various processes by which cells overcome DNA replication barriers has been an important biomedical research topic for many years.
The cellular processes dealing with replication barriers vary depending on the situation. Several commonly observed processes have been described in recent reviews, and are briefly summarized here (Lambert and Carr 2013; Berti and Vindigni 2016) (Fig. 1). First, repriming downstream of replication barriers allows replication to continue. Single-strand DNA (ssDNA) gaps formed between sites of stalled forks and repriming can be repaired by post-replicative gap filling (Fig. 1A). Second, specialized DNA polymerases, called TLS (translesion DNA synthesis) polymerases, are able to replicate past certain types of damaged nucleotides in the template DNA (Fig. 1B). Third, neighboring replication forks can complete the remaining synthesis left from stalled replication forks (Fig. 1C). Fourth, specialized DNA helicases can remove or bypass DNA structures or protein barriers, allowing resumption of synthesis (Fig. 1D). Fifth, template switching mediated by post-replicative repair and recombination proteins enables the stalled nascent strand to use its sister chromatid as a template for synthesis (Fig. 1E). Sixth, the replication fork can be remodeled in the form of replication fork regression (or fork regression). This mechanism entails newly synthesized strands annealing to each other, accompanied by parental strand re-annealing (Fig. 1F). Although this process has the potential to restart a stalled replication fork by switching the template or other mechanisms, it can also generate undesirable consequences (see below).
Figure 1.
Multiple processes cope with DNA replication barriers. A replication barrier on the template DNA is depicted as a star, and a few examples of barriers are indicated. Parental and nascent DNA strands are depicted as blue and red lines. Newly synthesized DNA in each situation is indicated by dashed lines. (A) DNA replication can continue when a repriming event occurs downstream of replication fork. Subsequently, the ssDNA gap left behind can be repaired by homologous recombination. (B) Specialized translesion DNA synthesis (TLS) polymerases can replace the replicative polymerases to synthesize a stretch of DNA using some types of damaged DNA as template. Subsequent switch from TLS polymerases to replicative polymerase resume further synthesis (not depicted here). (C) As eukaryotic cells initiate replication at multiple sites, some unreplicated regions caused by stalled replication forks can be synthesized by the converging replication forks (light blue). (D) When replication barriers are in the form of DNA secondary structures or proteins tightly bound to DNA, DNA helicases (green circle) can remove them, allowing replisome to resume DNA synthesis. (E) When replication forks encounter lesions on one template strand, recombination-based post-replicative repair enables the switch of DNA template. This entails one nascent strand invades the sister chromatid and use the other nascent strand as template for replication. After DNA synthesis, re-annealing of the invading nascent DNA strand and the original template strand leads to the bypass of the damage sites. (F) Replication fork regression (or reversal) entails the annealing of two nascent DNA strands and re-annealing of two parental DNA strands. A four-way ‘chicken foot’ like junction is formed from a three-way junction. More details of this type of replication fork remodeling are explained in Fig. 2.
Among the aforementioned processes, replication fork regression is the least understood. Fork regression has been used to explain certain phenotypes manifested by Escherichia coli and yeast mutants defective in replication (reviewed in Atkinson and McGlynn 2009), since the idea was first suggested 40 years ago (Higgins, Kato and Strauss 1976). More recently, the use of multidisciplined approaches in model organisms and human cells has begun to delineate mechanisms of fork regression, the antagonistic regulation to which it is subjected and possible biological outcomes. Several studies in yeast have integrated genetic approaches with in vitro fork regression assays as well as a physical examination of DNA structures by 2D gel electrophoresis and electron microscopy (EM). These findings have implicated several DNA helicases and their regulators in fork regression. They also pinpointed situations under which fork regression events occur with increased frequency and linked these events to recombinational repair. Regressed forks detectable by EM appear to be more prevalent in higher eukaryotic cells than in yeast (Ray Chaudhuri et al.2012). This difference is likely associated with a larger number of enzymes capable of catalyzing fork regression reactions in higher eukaryotic cells (see below). In these systems, like in yeast, the fork regression processes are subject to positive and negative regulations and are linked to multiple outcomes.
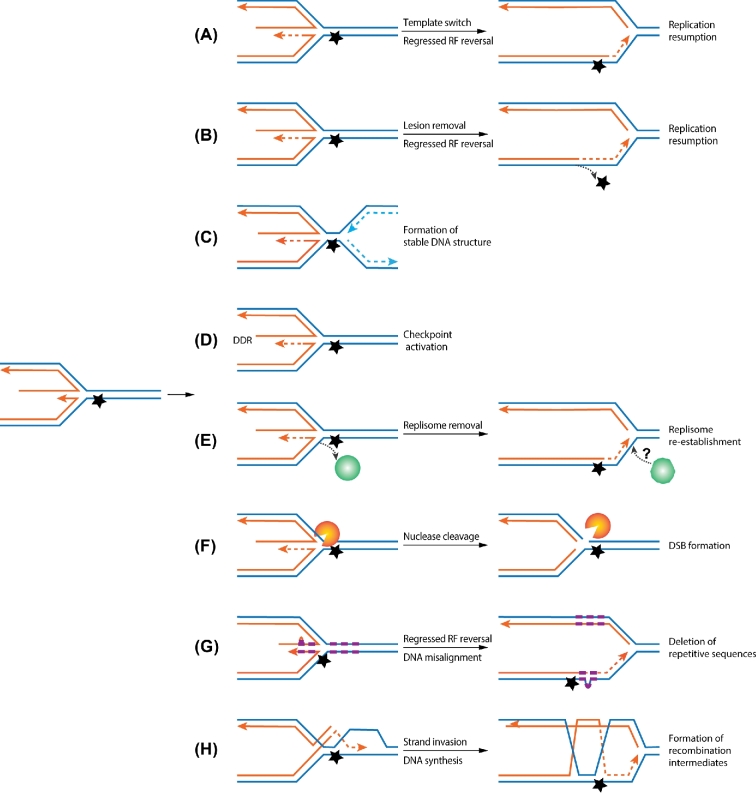
POTENTIAL OUTCOMES OF REPLICATION FORK REGRESSION
In principle, replication fork regression can generate either helpful or harmful outcomes depending on the downstream molecular events. As these outcomes have been discussed in previous reviews (Neelsen and Lopes 2015; Berti and Vindigni 2016), only an overview is given here (Fig. 2). Upon fork regression, the nascent lagging strand can serve as a template for the nascent leading strand to bypass the lesion on the parental strand (Fig. 2A). In addition, template re-annealing converts damaged regions from ssDNA to double-strand DNA (dsDNA) that can lead to DNA repair, as double-strandedness is required for several repair processes (Fig. 2B). In either situation, new DNA can be made and regressed replication forks can be reversed back to allow replication assumption (Fig. 2A and B). It is also possible that regressed replication forks can generate a more stable DNA structure, considering that stalled replication forks with large ssDNA regions can be more susceptible to breakage than regressed fork structures. This outcome is particularly beneficial if nascent strand DNA synthesis can occur once fork regression takes place. A stable DNA structure also increases the chances for neighboring replication forks to synthesize the unreplicated regions (Fig. 2C). It is also possible that dsDNA ends formed by fork regression or resected DNA ends could activate the checkpoint kinases (Fugger et al.2015). Such events could be useful to coordinate cell cycle delay and local increase of origin firing, both of which can help to complete replication (Fig. 2D). Theoretically, these outcomes could occur at the same time or independently depending on the fork stalling situation. Further studies are needed to understand these possibilities.
Figure 2.
Replication fork regression can lead to different molecular outcomes. Symbols and schematic features are the same as in Fig. 1. (A) Upon replication fork (RF) regression, the nascent leading strand can use the nascent lagging strand DNA as template to synthesize a stretch of DNA, thus bypassing the lesions on the parental strand. Subsequent reversal of the regression forks allows the replication fork to resume DNA synthesis. (B) Regression of replication forks can generate dsDNA regions on templates that include the region containing the damaged DNA. Changing from ssDNA to dsDNA strand forms allows the damaged DNA to be repaired using the other strand as template by the classical DNA repair pathways. (C) Replication fork regression may generate stable DNA structure, thus preventing DNA degradation and providing time for the adjacent forks to converge. (D) As dsDNA ends can be generated during replication fork regression, they can elicit the DNA damage checkpoint responses (DDR) that can induce beneficial consequences in coping with genome stress. (E) Replication fork regression can lead to eviction of replisome or its components from replication forks. This may generate the need to reload replication machineries for continued DNA synthesis. (F) Replication fork regression can lead to DSBs when the four-way ‘chicken foot’ DNA structure is cleaved by structure-specific DNA nucleases, such as the Mus81-Mms4 complex. (G) When fork regression occurs at repetitive sequences, misalignment could occur, and deletion or expansion of the DNA repeats can be produced. (H) The juxtaposition of homologous nascent and template strands as a consequence of replication fork regression can generate opportunity for recombination. One scenario is depicted where a nascent strand invades the parental strands to form recombination intermediates, which can be deleterious if not resolved.
Replication fork regression has also been predicted to generate harmful consequences (Fig. 2E–H). First, this process likely requires removal of the replisome from DNA, which can be difficult to reinstall (Fig. 2E). It is also possible that ‘reinstalled’ replisome may not be as proficient as the initial machinery (Miyabe et al.2015). Second, regressed forks can be subject to attack by structure-specific nucleases that recognize branched structures, which would lead to double-strand break (DSB) formation (Hanada et al.2007; Couch et al.2013; Neelsen et al.2013) (Fig. 2F). Though DSB can provide a way to restart replication through recombination-based mechanism, as the most deleterious form of DNA damage, DSB can cause cellular lethality if it is not repaired in time. Third, at repetitive sequences, fork regression can generate misalignment, leading to expansion or retraction of DNA repeats (Mirkin 2007; McMurray 2010; Follonier et al.2013) (Fig. 2G). Fourth, a ssDNA tail formed during fork regression can be used by recombination machinery to invade parental strands, generating recombination structures. This can lead to proper DNA repair and reestablishment of replication forks, but also can lead to chromosomal nondisjunction when left unresolved (Fig. 2H) (Sun et al.2008; Lambert et al.2010; Mizuno et al.2013).
Thus far, only some of the theoretical outcomes of fork regression described above have experimental support. For example, several nucleases, such as those containing MUS81 and SLX4, have been implicated in the cleavage of the four-way DNA junction formed during fork regression (Hanada et al.2007; Couch et al.2013; Neelsen et al.2013). In fission yeast, when programed replication fork stalling is adjacent to a recombination reporter, fork regression can lead to increased recombination events (Sun et al.2008; Lambert et al.2010; Mizuno et al.2013). Interestingly, these events tend to be highly mutagenic for unknown reasons. Evidence for other proposed fork regression outcomes, such as partial disassembly and reassembly of replisomes, is currently limited. Regardless, the yin-yang effects associated with fork regression suggest that this process needs to be tightly regulated in cells. Below, we describe the enzymes that possess fork regression activity based on in vitro and/or in vivo evidences and how they are regulated.
ENZYMES AND MECHANISMS INVOLVED IN REPLICATION FORK REGRESSION
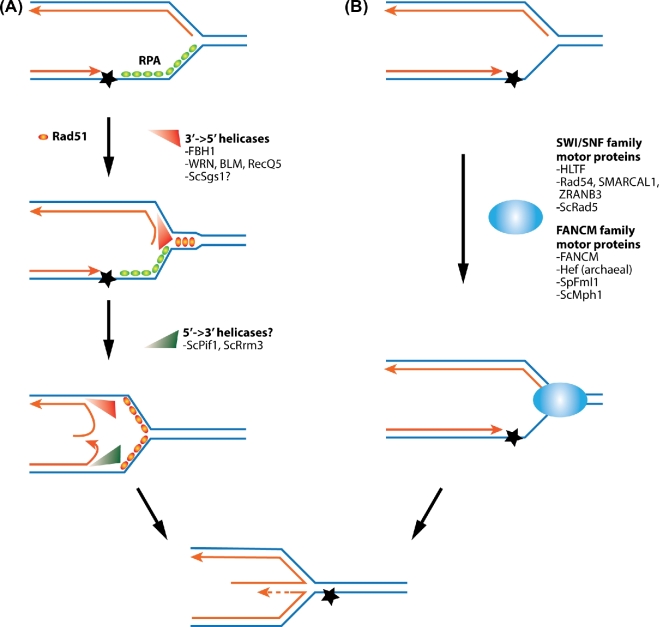
DNA helicases
DNA helicases use ATP hydrolysis to move along DNA in a defined polarity (3΄-5΄ or 5΄-3΄) and catalyze DNA strand separation (DNA unwinding), though some DNA helicases can also carry out DNA annealing (DNA rewinding). In vitro evidence suggests that DNA helicases can affect fork regression in several ways (Fig. 3). Upon leading strand blockage, the DNA polymerases that synthesize leading and lagging strands can be uncoupled, allowing further synthesis of the lagging strand (Fig. 3A). 3΄-5΄ DNA helicases can then unwind a portion of the lagging strand so that the nascent and template strands are in the form of ssDNA (Fig. 3A). DNA helicases with the opposite polarity can do the same on leading strands to produce two ssDNA strands. As the two nascent strands are complementary to each other, they can anneal; the same reaction can occur between the template sequences. Though annealing can occur spontaneously, recent evidence suggests that the human Rad51 protein binds to ssDNA, likely on the template strands, and promotes their annealing thus stimulating fork regression (Zellweger et al.2015) (Fig. 3A).
Figure 3.
Possible molecular mechanisms underlying replication fork regression. (A) Replicating fork regression can be catalyzed in part by 3΄-5΄ DNA helicases, such as FBH1 and the RecQ family proteins. They can unwind lagging strands to generate ssDNA, the substrate for annealing with nascent leading strand. It is possible that Rad51 binding to single-stranded parental DNA facilitates re-annealing of parental strands, an event that may drive nascent strand annealing. Budding yeast 5΄-3΄ DNA helicases Pif1 and Rrm3 may also play a role in unwinding leading strands. (B) Two family of DNA motor proteins, including the SWI/SNF family and FANCM family enzymes, can catalyze replication fork regression in vitro. These motor proteins use ATP hydrolysis to translocate along DNA. They appear to catalyze the annealing of nascent strands and re-annealing of parent strands at the same time.
Thus far, biochemical tests have shown that several human 3΄-5΄ DNA helicases catalyze fork regression reactions. These enzymes can effectively convert model substrates mimicking stalled replication forks, either in the form of synthetic DNA or plasmid DNA, into regressed fork structures. These enzymes include FBH1 and three RecQ family helicase members, namely WRN, BLM and RecQ5 (Kanagaraj et al.2006; Machwe et al.2006, 2007; Ralf, Hickson and Wu 2006; Popuri et al.2008; Fugger et al.2015) (Fig. 3A). Interestingly, WRN can also catalyze the reversal of regression forks in vitro, in a similar fashion as the T4 phage helicase UvsW (Manosas et al.2012; Shin et al.2016). This type of activity can be important for resetting the fork after fork regression, and is likely common among monomeric SF2 family DNA helicases (Manosas et al.2013). Another enzyme capable of driving the reversal of regressed forks is another RecQ family enzyme, RECQ1, and this activity can be inhibited by PARP (Ray Chaudhuri et al.2012; Berti et al.2013). It is important to further understand how these enzymes, alone or with other factors, can couple fork regression with the restoration of replication forks after the completion of DNA repair.
In yeast, two 5΄-3΄ DNA helicases, Pif1 and Rrm3, are thought to act concertedly to generate regressed replication forks in the cells when DNA damage checkpoint function is removed, though whether they can regress DNA fork structures in vitro remains to be determined (Rossi et al.2015) (Fig. 3A). Currently, it is unclear if the yeast RecQ protein Sgs1 is capable of catalyzing either fork regression or the reversal of this reaction.
SWI/SNF family motor proteins
Another way to generate fork regression is through DNA motor proteins that can simultaneously drive the annealing of newly synthesized strands and that of template strands (Fig. 3B). Specific members of the SWI/SNF family of DNA motor proteins, namely Rad5 in yeast and its mammalian homolog HLTF, human Rad54, SMARCAL1/HARP and ZRANB3/AH2 proteins, are primary examples in eukaryotic cells (Bugreev, Rossi and Mazin 2011; Betous et al.2012; Ciccia et al.2012) (Fig. 3B). Biochemical and structural data have shown that besides their DNA motor domains that can interact with DNA, HLTF, SMARCAL1 and ZRANB3 each contains a unique substrate recognition domain for recognition of DNA fork structures (Betous et al.2012; Yuan, Ghosal and Chen 2012; Mason et al.2014; Hishiki et al.2015; Kile et al.2015; Badu-Nkansah et al.2016). These proteins also exhibit differences in terms of how they are recruited to stalled replication forks, how they catalyze the reaction, and their co-factors and additional activities (reviewed in Neelsen and Lopes 2015). For example, Rad5 also acts as an ubiquitin E3 that adds polyubiquitin chain onto the DNA polymerase processing factor PCNA, an act critical for the DNA template switch process mentioned above (reviewed in Bonner and Zhao 2016). ZRANB3 also acts as a structure-specific ATP-dependent endonuclease during replication stress response (Weston, Peeters and Ahel 2012).
SMARCAL1 and RAD54, but not ZRANB3, Rad5 or its homologs, exhibit activity that can drive the reversal of regressed forks (Bansbach et al.2009; Bugreev, Rossi and Mazin 2011; Betous et al.2013). In the case of SMARCAL1, a key determinant of the directionality of the reactions (fork regression vs reversal of regressed fork) is how its co-factor replication protein A (RPA) is engaged with ssDNA at DNA forks. Specifically, RPA binding of leading strand template DNA directs the SMARCAL1 fork regression reaction (Betous et al. 2013; Bhat, Betous and Cortez 2015). However, RPA binding of nascent leading strand DNA favors reversal of fork regression (Betous et al.2013; Bhat, Betous and Cortez 2015). For ZRANB3 and HLTF, their involvement in fork regression is linked to p53 and the associated translesion polymerase tao, though detailed mechanisms by which these associations affect ZRANB3 and HLTF functions remain to be determined (Hampp et al.2016). Studies as exemplified by the above have begun to dissect the involvement of complex activities among DNA motor proteins and interactions with regulators during fork regression and rescue of stalled replication forks.
FANCM family motor proteins
A third group of proteins that can drive fork regression in vitro are the multifunctional FANCM family proteins (Fig. 3B) (reviewed in Xue, Sung and Zhao 2015). Human FANCM is one of the 17 proteins mutated in Fanconi anemia (FA) patients (reviewed in Wang and Smogorzewska 2015). FA is a syndrome characterized by cancer predisposition, developmental abnormalities, bone marrow failure and increased genomic instability. The most studied FANCM homologs are the budding yeast Mph1, fission yeast Fml1 and archaeabacteria Hef. Mph1 and Fml1 exhibit 3΄-5΄ DNA helicase activities, while FANCM does not. However, all three can catalyze fork regression, suggesting that they use DNA motor functions to migrate branch points of joint DNA structures during this reaction (Gari et al.2008b, Sun et al.2008; Zheng et al.2011). Consistent with this view, RPA does not prevent their functions in fork regression, while it inhibits those mediated by 3΄-5΄ helicases that initiate the process by strand unwinding (Gari et al.2008a).
Fork regression mechanisms of FANCM family proteins share similarities and exhibit differences from those of SWI/SNF family proteins described above. Both types of DNA motor proteins promote annealing between the nascent strands and between the template strands. A snapshot of how FANCM family proteins engage with branched DNA has been provided by atomic force microscopy (AFM) and EM data (Xue et al.2014). Mph1 appears to form oligomers only upon binding to the junction of modeled replication fork structures. This is analogous to how the bacterial RuvAB complex interacts with DNA junctions during fork regression and branch migration (Yamada et al.2002). This mode of action is different from that of SWI/SNF members, such as SMARCAL1. SMARCAL1 is more similar to T4 phage UvsW and bacterial RecG, as their interaction with ssDNA binding proteins can guide their activities. The HARP domain found in SMARCAL1 and UvsW can serve as the wedge in fork remodeling (Buss, Kimura and Bianco 2008; Manosas et al.2012, 2013; Betous et al.2013; Mason et al.2014).
Beside fork remodeling activities, FANCM family proteins also catalyze other DNA transactions. For example, their ability to displace D-loop structures is key to control the levels of crossover products during recombinational repair (Sun et al.2008; Prakash et al.2009; Sebesta et al.2011; Lorenz et al.2012). In addition, FANCM promotes replicative traversal across inter-strand crosslinked sites on DNA (Huang et al.2010). These proteins also have structural roles. For example, FANCM can recruit the FA core complex, and influence replication and the damage checkpoint partly through binding to the checkpoint kinase HCLK1 (Collis et al.2008; Sobeck et al.2009; Huang et al.2010; Luke-Glaser et al.2010; Schwab, Blackford and Niedzwiedz 2010; Wang et al.2013). These additional functions make it challenging to pinpoint how each one specifically contributes to the replication defects associated with their mutants. The following sections expound upon the yeast Mph1/Fml1 and Rad5 proteins and discuss their roles in fork regression and replication.
THE Mph1 DNA HELICASE AND ITS REGULATION
The budding yeast Mph1 and the homologous fission yeast Fml1 have been studied biochemically and genetically. They show potent ability to catalyze the regression of modeled fork structures and branch migration of Holliday junction (Sun et al.2008; Zheng et al.2011). Mph1 and Fml1 also efficiently dissociate D-loops in vitro, an activity that provides the basis for their roles in suppression of crossover levels in vivo (Sun et al.2008; Prakash et al.2009). Though regressed fork structures have not been directly examined in mutants of Mph1 or Fml1 by EM, these cells do exhibit increased levels of fork-associated recombination. In the case of Fml1, its ATPase activity is required for the increase of Rad51-mediated gene conversion at a programmed replication fork blockage (Sun et al.2008). Similarly, the ATPase activity of Mph1 is required for increased levels of recombination intermediates near replication forks stalled by template lesions in cells lacking certain recombination regulators (Chen et al.2009; Mankouri, Ngo and Hickson 2009; Choi et al.2010; Chavez, Agrawal and Johnson 2011). In both cases, a logical explanation is that the Fml1/Mph1 motor activity channels stalled replication forks into recombinational repair, since other observed biochemical activities of the proteins, such as D-loop dissociation, cannot easily account for the findings.
One regulator of Mph1 is the Smc5/6 complex, an octomeric chromosomal structural protein complex containing Smc5, Smc6 and six other non-Smc subunits (Zhao and Blobel 2005). Mph1 and Smc5/6 physically interact in vitro and in vivo (Chen et al.2009; Xue et al.2014). Genetically, the deficiencies of smc5/6 mutant cells, such as increased levels of recombination intermediates near impaired replication forks and replication stress sensitivity, are suppressed by mph1 ATPase mutant or deletion (Chen et al.2009; Choi et al.2010; Chavez, Agrawal and Johnson 2011). Biochemical studies further showed that Smc5/6 inhibits Mph1-mediated fork regression, but not the D-loop dissociation, by preventing Mph1 from forming oligomers at fork junctions (Xue et al.2014) (Fig. 3B). These findings are consistent with observation that Smc5/6 genetically interacts with Mph1 under fork stalling conditions but not during crossover control in yeast cells. Together, these data suggest that Smc5/6 is a regulator specific for Mph1-mediated fork regression (Xue et al.2014). Positive regulators of Mph1 and Fml1 have also been found. Both Mph1 and Fml1, like FANCM, interact with the histone-fold complex MHF (Singh et al.2010; Yan et al.2010; Bhattacharjee et al.2013; Xue et al.2015). In addition, Mph1 and MHF also associate with a protein called Mte1, which directly binds to DNA and helps MHF in promoting Mph1 functions (Silva et al. 2016; Xue et al. 2016; Yimit et al.2016). MHF and Mte1 can counteract Smc5/6's effects on Mph1, suggesting an antagonistic regulation of these motor proteins (Xue et al.2016). How exactly these different regulators guide Mph1 functions in different processes are exciting questions to be addressed in the future. As these proteins are conserved, it is possible that similar regulation may occur in other organisms.
THE Rad5 DNA HELICASE IN FORK REGRESSION AND IN OTHER PROCESSES
Rad5 possesses three major functions that pertain to DNA replication. As described above, its ubiquitin E3 function enables PCNA polyubiquitination, which initiates a recombination-based template switch (Hoege et al.2002; Stelter and Ulrich 2003; Branzei, Vanoli and Foiani 2008). In this capacity, Rad5 acts together with other members of the post-replicative repair pathway. These include the Rad6 and Rad18 pair of ubiquitin E2 and E3 enzymes that monoubiquitinate PCNA and the Ubc13/Mms2 dimeric ubiquitin E2 that collaborates with Rad5 to add ubiquitin chains onto monoubiquitinated PCNA (reviewed in Bergink and Jentsch 2009). While monoubiquitinated PCNA leads to the recruitment of translesion DNA polymerases that can bypass some template lesions, polyubiquinated PCNA binds to other proteins such as the yeast Mgs1 and human ZRANB3 (Ciccia et al.2012; Saugar et al.2012; reviewed in Ulrich 2014). These interactions contribute to the mechanisms of DNA template switching, a major means to deal with replication blockages. In addition to its E3 role, Rad5 can also directly promote TLS through an interaction with the TLS factor Rev1 (Kuang et al.2013; Xu et al.2016). Moreover, as described above, the Rad5 DNA motor activity supports regression of replication fork-like structures in vitro (Blastyak et al.2007).
While its interaction with Rev1 requires an N-terminal domain of Rad5, both its ubiquitin E3 and DNA motor functions require its C-terminal region (Blastyak et al.2007; Kuang et al.2013; Xu et al.2016). Within this region, the ubiquitin E3 domain is inserted within its DNA motor domain. It has been shown that the Walker B motif of the Rad5 helicase domain promotes PCNA interaction with Ubc13, and that such an effect likely facilitates the nearby Rad5 E3 domain in ubiquitin transfer (Ball et al.2014; Choi et al.2015). Although the Walker B motif of Rad5 plays a structural role in supporting its E3 function, the Rad5 helicase activity per se is not required for PCNA polyubiquitination, rather it has a role outside template switching that may be related to its fork regression functions (Choi et al.2015).
Whether these three functions of Rad5 coordinate at a stalled fork and how the protein can deploy a particular activity are not currently well understood. However, it is clear from several studies that these activities make separate contributions to genotoxic resistance (Minca and Kowalski 2010; Ortiz-Bazan et al.2014). Another question to be addressed in the future is whether the Rad5 helicase activity can promote other DNA transactions related to DNA replication beyond fork regression. Additionally, it is unclear what the relationship is between Rad5 and Mph1. Genetic findings suggest that their mutants are additive in causing genotoxic sensitivity (Choi et al.2010), suggesting that they have separate contributions. Since disrupting Smc5/6's inhibition of Mph1 partially suppresses the MMS sensitivity of a Rad5 helicase mutant, hyperactive Mph1 may compensate Rad5 mutant's fork regression defects (Xue et al.2014). Future work on Rad5 should lead to a better understanding of the situations in which it deploys its multiple functions and the outcomes of each reaction.
OTHER PROTEIN FACTORS INVOLVED IN REPLICATION FORK REGRESSION
DNA damage checkpoint proteins affect replication fork regression through multiple means. Mutating the budding yeast checkpoint kinase Rad53 increases the levels of regressed replication forks and ssDNA regions on templates (Sogo, Lopes and Foiani 2002). A recent study further demonstrates that Rad53 phosphorylates the Rrm3 and Pif1 helicases upon replication stress, which likely prevents their engagement in fork regression (Rossi et al.2015). In the absence of Rad53, the exonuclease Exo1 can degrade ssDNA intermediates required for fork regression (Cotta-Ramusino et al.2005). Taken together, these findings suggest that the Rad53 checkpoint pathway favors replication fork regression by limiting Exo1 functions and disfavors it by limiting Pif1 and Rrm3 functions. It is possible that these opposing methods of regulation could occur at different replication fork stalling situations as described above.
A role for the DNA damage checkpoint on fork regression is also seen in fission yeast. Phosphorylation of the fission yeast nuclease Dna2 by the DNA checkpoint kinases targets it to stalled replication forks to remove ssDNA required for fork regression (Hu et al.2012). The nuclease function of Dna2 in fork regression is also seen in human cells. In this case, Dna2, in conjunction with WRN, can degrade reversed replication forks and promote replication restart (Thangavel et al.2015).
Besides regulating DNA helicases and nucleases, DNA damage checkpoint signaling can also prevent fork regression through reducing topological stress. RNA transcripts connected to DNA templates can be tethered to nuclear pore complexes. When this occurs in front of replication forks, increased topological stress is thought to favor replication fork regression. Rad53-mediated phosphorylation of several nucleoporins disfavors this tethering, thus reducing the opportunity for topologically induced fork regression (Bermejo et al.2011).
Replisome proteins have been also shown to keep replication fork regression in check. The trimeric Ctf4 is a member of the replisome and serves as a hub to link several other proteins to the replicative DNA helicase (Simon et al.2014; Villa et al.2016). Among Ctf4-associated factors is the primase-Polα complex (Simon et al.2014). It was found that mutations of Ctf4 and primase can lead to increased levels of regressed replication forks (Fumasoni et al.2015). This could be caused by a reduction in repriming events or a stronger tendency of the replisome being dislodged from stalled forks. It is not known whether a similar scenario occurs in human cells. In summary, current findings implicate multiple players, such as Smc5/6, checkpoint kinases and replisome members, in regulating fork regression either directly or indirectly.
CONCLUDING REMARKS
Combined approaches using biochemistry, genetics and physical analysis of DNA suggest that eukaryotes contain multiple types of DNA helicases and motor proteins that affect replication fork regression. Thus far, a dozen enzymes in higher eukaryotes and a few in yeasts have been examined to some depth. It is likely that additional enzymes will continue to be added to the atlas of the enzymes that participate in fork regression and its regulation. Specific mechanisms of fork regression have been examined for only a few of these enzymes. These studies suggest that different DNA helicases and motor proteins may be most suitable to function during particular fork blocking situations. Additional biochemical and biophysical studies are needed to further delineate detailed mechanisms and compare the roles of these enzymes.
It is not surprising that fork regression is subject to both positive and negative regulation, since in principle this process can lead to fork restart but can also elicit deleterious consequences, such as DNA breaks or recombination intermediates. Tight regulation may allow other fork rescuing pathways, such as TLS or template switching, to restart replication, while maintaining fork regression as a backup pathway. Thus far, negative regulations have been found in yeast, targeting different DNA helicases by the Smc5/6 complex and DNA damage checkpoint kinases (Xue et al.2014; Rossi et al.2015). Positive regulations have been reported in human cells, wherein PARP interaction of RECQ1 prevents the latter from catalyzing the reversal of fork regression (Berti et al.2013). This regulation may provide one explanation for the higher amount of regressed forks seen by EM in human cells compared with yeast. Another possible explanation is that more proteins catalyzing this reaction are present in human cells than in yeast (see above). In addition, as yeast cells exhibit robust recombination, it is possible that regressed replication forks could be channeled into this pathway more readily than in human cells. The abundant recombination intermediates associated with fork regression activity in yeast are consistent with this idea (Chen et al.2009; Choi et al.2010; Xue et al.2014). Moreover, while human Rad51 can promote fork regression (Zellweger et al.2015), it is unclear whether this is also true in yeast. Thus, while fork regression can take place in both single-celled organisms like yeast and higher eukaryotes such as humans, the relative abundance and outcome can vary depending on the number of enzymes catalyzing these reactions, their positive and negative regulators, other DNA transaction pathways and replisome functions. Using yeast as a model system can be a powerful way to delineate aspects of the biology associated with fork regression and their effects on genome replication. Future studies will undoubtedly provide a deeper understanding into the mechanisms that dictate this process, its regulation and how it is integrated with other fork rescue mechanisms.
Acknowledgments
We thank the Zhao lab members for constructive comments.
FUNDING
This study was supported by the US National Institutes of Health grant GM080670.
Conflict of interest. None declared.
REFERENCES
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 2009;37:3475–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badu-Nkansah A, Mason AC, Eichman BF et al. . Identification of a substrate recognition domain in the replication stress response protein zinc finger Ran-binding domain-containing protein 3 (ZRANB3). J Biol Chem 2016;291:8251–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LG, Xu X, Blackwell S et al. . The Rad5 helicase activity is dispensable for error-free DNA post-replication repair. DNA Repair (Amst) 2014;16:74–83 [DOI] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA et al. . The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Gene Dev 2009;23:2405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 2009;458:461–7 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R et al. . The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 2011;146:233–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Ray Chaudhuri A, Thangavel S et al. . Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 2013;20:347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Vindigni A. Replication stress: getting back on track. Nat Struct Mol Biol 2016;23:103–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Couch FB, Mason AC et al. . Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep 2013;3:1958–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP et al. . SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Gene Dev 2012;26:151–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KP, Betous R, Cortez D. High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling. J Biol Chem 2015;290:4110–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Osman F, Feeney L et al. . MHF1-2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol 2013;3:130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blastyak A, Pinter L, Unk I et al. . Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 2007;28:167–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JN, Zhao X. Replication-associated recombinational repair: lessons from budding yeast. Genes (Basel) 2016;7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature 2008;456:915–20 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res 2011;39:2153–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res 2008;36:7029–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J Biol Chem 2011;286:5119–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B et al. . Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. P Natl Acad Sci USA 2009;106:21252–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen YH et al. . The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell 2010;21:2306–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Batke S, Szakal B et al. . Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance. Nucleic Acids Res 2015;43:2666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y et al. . Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 2012;47:396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ et al. . FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 2008;32:313–24 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C et al. . Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 2005;17:153–9 [DOI] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R et al. . ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Gene Dev 2013;27:1610–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follonier C, Oehler J, Herrador R et al. . Friedreich's ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat Struct Mol Biol 2013;20:486–94 [DOI] [PubMed] [Google Scholar]
- Fugger K, Mistrik M, Neelsen KJ et al. . FBH1 catalyzes regression of stalled replication forks. Cell Rep 2015;10:1749–57 [DOI] [PubMed] [Google Scholar]
- Fumasoni M, Zwicky K, Vanoli F et al. . Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Pol alpha/Primase/Ctf4 complex. Mol Cell 2015;57:812–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M et al. . Remodeling of DNA replication structures by the branch point translocase FANCM. P Natl Acad Sci USA 2008a;105:16107–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ et al. . The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 2008b;29:141–8 [DOI] [PubMed] [Google Scholar]
- Hampp S, Kiessling T, Buechle K et al. . DNA damage tolerance pathway involving DNA polymerase iota and the tumor suppressor p53 regulates DNA replication fork progression. P Natl Acad Sci USA 2016;113:E4311–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL et al. . The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 2007;14:1096–104 [DOI] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol 1976;101:417–25 [DOI] [PubMed] [Google Scholar]
- Hishiki A, Hara K, Ikegaya Y et al. . Structure of a novel DNA-binding domain of helicase-like transcription factor (HLTF) and its functional implication in DNA damage tolerance. J Biol Chem 2015;290:13215–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL et al. . RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002;419:135–41 [DOI] [PubMed] [Google Scholar]
- Hu JZ, Sun L, Shen FF et al. . The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell 2012;149:1221–32 [DOI] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B et al. . The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell 2010;39:259–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R, Saydam N, Garcia PL et al. . Human RECQ5 beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res 2006;34:5217–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile AC, Chavez DA, Bacal J et al. . HLTF's ancient HIRAN domain binds 3΄ DNA ends to drive replication fork reversal. Mol Cell 2015;58:1090–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang LS, Kou HP, Xie ZW et al. . A non-catalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5. DNA Repair (Amst) 2013;12:27–37 [DOI] [PubMed] [Google Scholar]
- Lambert S, Carr AM. Impediments to replication fork movement: stabilisation, reactivation and genome instability. Chromosoma 2013;122:33–45 [DOI] [PubMed] [Google Scholar]
- Lambert S, Mizuno K, Blaisonneau J et al. . Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 2010;39:346–59 [DOI] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W et al. . The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 2012;336:1585–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S et al. . FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J 2010;29:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J et al. . The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 2006;45:13939–46 [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Lloyd RG et al. . Replication fork regression in vitro by the Werner syndrome protein (WRN): Holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res 2007;35:5729–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in Saccharomyces cerevisiae. Mol Biol Cell 2009;20:1683–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Bianco PR et al. . RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun 2013;4:2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Croquette V et al. . Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 2012;338:1217–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AC, Rambo RP, Greer B et al. . A structure-specific nucleic acid-binding domain conserved among DNA repair proteins. P Natl Acad Sci USA 2014;111:7618–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 2010;11:786–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell 2010;38:649–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007;447:932–40 [DOI] [PubMed] [Google Scholar]
- Miyabe I, Mizuno K, Keszthelyi A et al. . Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat Struct Mol Biol 2015;22:932–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Miyabe I, Schalbetter SA et al. . Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 2013;493:246–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Bio 2015;16:207–20 [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IM, Herrador R et al. . Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol 2013;200:699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Bazan MA, Gallo-Fernandez M, Saugar I et al. . Rad5 plays a major role in the cellular response to DNA damage during chromosome replication. Cell Rep 2014;9:460–8 [DOI] [PubMed] [Google Scholar]
- Popuri V, Bachrati CZ, Muzzolini L et al. . The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 2008;283:17766–76 [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E et al. . Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Gene Dev 2009;23:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 2006;281:22839–46 [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R et al. . Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 2012;19:417–23 [DOI] [PubMed] [Google Scholar]
- Rossi SE, Ajazi A, Carotenuto W et al. . Rad53-mediated regulation of Rrm3 and Pif1 DNA helicases contributes to prevention of aberrant fork transitions under replication stress. Cell Rep 2015;13:80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I, Parker JL, Zhao SK et al. . The genome maintenance factor Mgs1 is targeted to sites of replication stress by ubiquitylated PCNA. Nucleic Acids Res 2012;40:245–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 2010;29:806–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Haracska L et al. . Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst) 2011;10:567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Lee J, Yoo S et al. . Active control of repetitive structural transitions between replication forks and Holliday junctions by Werner syndrome helicase. Structure 2016;24:1292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Altmannova V, Luke-Glaser S et al. . Mte1 interacts with Mph1 and promotes crossover recombination and telomere maintenance. Gene Dev 2016;30:700–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Zhou JC, Perera RL et al. . A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 2014;510:293–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM et al. . MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi Anemia pathway via FANCM. Mol Cell 2010;37:879–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Landais I et al. . The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem 2009;284:25560–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 2002;297:599–602 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003;425:188–91 [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F et al. . The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 2008;32:118–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M et al. . DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol 2015;208:545–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Two-way communications between ubiquitin-like modifiers and DNA. Nat Struct Mol Biol 2014;21:317–24 [DOI] [PubMed] [Google Scholar]
- Villa F, Simon AC, Bazan MAO et al. . Ctf4 Is a hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol Cell 2016;63:385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell 2015;160:354. [DOI] [PubMed] [Google Scholar]
- Wang Y, Leung JW, Jiang Y et al. . FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell 2013;49:997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Gene Dev 2012;26:1558–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lin AY, Zhou CY et al. . Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic Acids Res 2016;44:5231–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner J et al. . Restriction of replication fork regression activities by a conserved SMC complex. Mol Cell 2014;56:436–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner JN et al. . Selective modulation of the functions of a conserved DNA motor by a histone fold complex. Gene Dev 2015;29:1000–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Gene Dev 2015;29:1777–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue XY, Papusha A, Choi KY et al. . Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Gene Dev 2016;30:687–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Miyata T, Tsuchiya D et al. . Crystal structure of the RuvA-RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell 2002;10:671–81 [DOI] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C et al. . A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell 2010;37:865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimit A, Kim T, Anand RP et al. . MTE1 functions with MPH1 in double-strand break repair. Genetics 2016;203:147–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell 2012;47:410–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K et al. . Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 2015;208:563–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol 2014;16:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. P Natl Acad Sci USA 2005;102:4777–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XF, Prakash R, Saro D et al. . Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst) 2011;10:1034–43 [DOI] [PMC free article] [PubMed] [Google Scholar]