Abstract
Background
We examined the usefulness of erythrocyte-bound C4d (EC4d) to monitor disease activity in SLE.
Methods
Data and blood samples were collected from three different studies, each of which included longitudinal evaluations using the Physicians Global Assessment (PGA) of disease activity and the Safety of Estrogens in Lupus Erythematosus National Assessment (SELENA) SLE Disease Activity Index (SLEDAI), which was assessed without anti-double-stranded DNA (dsDNA) and low complement C3/C4 (clinical SELENA-SLEDAI). EC4d levels were determined using flow cytometry; other laboratory measures included antibodies to dsDNA, C3 and C4 proteins. Relationships between clinical SELENA-SLEDAI, PGA and the laboratory measures were analysed using linear mixed effect models.
Results
The three studies combined enrolled 124 patients with SLE (mean age 42 years, 97% women, 31% Caucasians and 34% African-Americans) followed for an average of 5 consecutive visits (range 2–13 visits). EC4d levels and low C3/C4 status were significantly associated the clinical SELENA-SLEDAI or PGA in each of the three study groups (p<0.05). Multivariate analysis revealed that EC4d levels (estimate=0.94±0.28) and low complement C3/C4 (estimate=1.24±0.43) were both independently and significantly associated with the clinical SELENA-SLEDAI (p<0.01) and PGA. EC4d levels were also associated with the clinical SELENA-SLEDAI (estimate: 1.20±0.29) and PGA (estimate=0.19±0.04) among patients with chronically low or normal C3/C4 (p<0.01). Anti-dsDNA titres were generally associated with disease activity.
Conclusion
These data support the association of EC4d with disease activity regardless of complement C3/C4 status and its usefulness in monitoring SLE disease. Additional studies will be required to support these validation data.
Keywords: complement, disease activity, systemic lupus erythematosus
Introduction
SLE is an autoimmune disease characterised by autoantibodies to self-antigens (such as double-stranded DNA (dsDNA)) and formation of immune complexes that activate the complement system to generate inflammatory anaphylatoxins resulting in organ damage.1
Anti-dsDNA antibodies and low complement C3 or C4 (C3/C4) have established clinical utility in the routine monitoring of SLE, and these laboratory measures have been incorporated in clinical research instruments such as the SLE Disease Activity Index (SLEDAI) for over two decades2 and in SLE classification criteria.3 However, C3 and C4 are acute phase reactant proteins, and using their levels to monitor SLE can be unreliable because their hyperconsumption during the active phase of disease can be offset by inflammation and compensatory hepatic synthesis.4 Moreover, C4 levels are dependent on copy number variations in C4A and C4B and thus can be constitutively low, irrespective of disease status.4 Anti-dsDNA is a very useful marker but is limited by its low sensitivity for SLE and performance as a stand-alone marker.5 It follows that additional laboratory measures of disease activity could be of value, not only to improve disease monitoring and treatment optimisation but also to improve outcome measures for therapeutic trial interventions.
In the past decades, complement pathway activation has been assessed using soluble split fragments,6 7 anaphylatoxins8 9 or stable C4d-bound complement activation products on erythrocytes (EC4d),10–13 and these laboratory measures represent additional approaches to assessing disease in SLE. We recently reported that EC4d was linked to clinical improvement in patients selected for active disease and complement activation. We also established the role of anti-C1q antibody titres in the monitoring of SLE renal disease.11 In this study, we compare the performance of EC4d, low complement C3/C4, and autoantibodies to dsDNA and C1q in tracking disease activity. For this, we combined data and laboratory findings from our original study11 with two additional cohorts with the objective of evaluating the generalisability of our previous findings.
Methods
All patients provided informed consent and participated in institutional-approved protocols that allowed the use of their samples and clinical data. The objective of each of the three studies was to evaluate the relationships between laboratory and disease activity measures. All patients fulfilled the 1982 American College of Rheumatology (ACR) criteria revised in 1997. The first study group (study group 1) has been reported11 and enrolled 37 patients at four sites, with all patients with SLE selected for active disease and requiring complement activation as defined by abnormal EC4d or B-lymphocyte C4d levels (>99th percentile of healthy controls). The two additional study groups we report (study group 2 and 3) enrolled 64 and 23 consecutive SLE, respectively, all coming from two academic lupus centres in the USA (Oklahoma Medical Research Foundation and Johns Hopkins University School of Medicine, respectively). Study group 2 included patients with SLE with a range of activity and treatment. All patients from the third study group were treated with methotrexate and hydroxychloroquine, and none presented with renal disease (>0.5 g urine protein).
In each study group, the relationships between the laboratory measures and disease activity were assessed longitudinally by the Physician’s Global Assessment (PGA) on a 0–3-point Visual Analogue Scale as well as by the clinical Safety of Estrogens in Lupus Erythematosus National Assessment (SELENA) SLE Disease Activity Index SELENA-SLEDAI (2), which was scored without the anti-dsDNA and complement components. An additional modification (known as the hybrid SELENA-SLEDAI) was that proteinuria was scored as positive in the presence of >0.5 g/24 hours urine protein, if attributed to SLE renal disease, but irrespective of a change from a previous visit (SLEDAI 2K definition for proteinuria).14
At each study visit, whole blood was collected in EDTA tubes and routed to the reference clinical laboratory at Exagen Diagnostics within 48 hours. EC4d levels were measured on receipt using quantitative flow cytometry as described previously15 and expressed as net mean fluorescence intensity (MFI). Antibody titres to dsDNA (Quanta Flash) and C1q (INOVA Diagnostics, San Diego, California, USA) were determined using immunoassays. The low complement (C3 or C4) status was determined using serum C3 and C4 levels measured at each of the study sites. Site investigators were blinded to EC4d, anti-dsDNA titres (Quanta Flash) and anti-C1q assay results during each study. Low complement C3/C4 and anti-dsDNA status (as determined at each of the study sites) were available to all evaluators.
The three study groups were analysed separately and in combination. Relationships between SLE disease activity and laboratory measures were analysed using linear mixed effect models (random intercept and fixed slope) with the clinical SELENA-SLEDAI and PGA as dependent variables and complement (low C3/C4, EC4d) and antibodies (anti-dsDNA (Quanta Flash) or anti-C1q titres) measures as independent predictors. Autoantibody and EC4d levels were log normalised for the analysis. Marginal R2 was calculated for each marker to evaluate the proportion (%) of variance explained by the independent predictors, and multivariate analyses were conducted. Mann-Whitney test was used to compare groups.
Results
Baseline characteristics of the population are presented in table 1 and in online supplementary tables I and II). Six hundred and twenty-four study visits were completed among the 124 enrolled patients. All patients had at least one follow-up visit after a baseline assessment, with a mean of 5 visits per patient. At baseline, patients enrolled in study group 1 (the active disease study group) had higher clinical SELENA-SLEDAI (median eight points) than those enrolled in study groups 2 and 3 (median 6 and 2 points, respectively) (p<0.01). The combined dataset (study groups 1–3; n=124 patients) provided a wider range of patients from well controlled to clinically active disease. Patients with abnormal complement and antibody titres had higher clinical disease activity than those with normal values (figure 1).
Table 1.
Patient characteristics at baseline
| Study group 1 | Study group 2 | Study group 3 | Study groups 1–3 | |
| Number of patients | 37 | 64 | 23 | 124 |
| Age | 34±2 | 42±1 | 56±3 | 42±1 |
| Gender (% of women) | 95 | 97 | 100 | 97 |
| Study visit per subject, average± SEM, median (range) | 10.5±0.6, 387 (2–3) |
2.0±0, 129 (2–3) |
4.7±0.2, 108 (3–7) |
5.0±0.4, 624 (2–13) |
| Ethnicities | ||||
| Caucasians (%) | 14 | 34 | 52 | 31 |
| African-Americans (%) | 24 | 36 | 43 | 34 |
| Hispanics (%) | 35 | 13 | 0 | 17 |
| Asians (%) | 22 | 8 | 0 | 10 |
| Others (%) | 5 | 9 | 4 | 7 |
| PGA (0–3 cm) | ||||
| Average±SEM | 1.6±0.1 | 1.4±0.1 | 0.6±0.1 | 1.3±0.1 |
| Median (range) | 1.6 (0.2–2.8) | 1.2 (0.6–2.6) | 0.5 (0.0– 2.0) | 1.3 (0.0– 2.8) |
| Clinical SELENA-SLEDAI | ||||
| Average±SEM | 8.1 ± 0.8 | 6.3±0.1 | 1.4±0.1 | 6.0±0.4 |
| Median, range | 8.0 (2–25) | 6.0 (2-17) | 2.0 (0–4) | 5.0 (0–25) |
| ANA titres (≥1:80) | 96% | 80% | 76% | 86% |
| Antibody measures | ||||
| Anti-dsDNA (units; %>35 units) | 615±180; 91 | 74±18; 38 | 14±1; 4 | 211±55;46 |
| Anti-C1q (units; %>20 units) | 58±11; 62 | 14±2; 20 | 8±2; 13 | 26%±4; 31 |
| Complement measures | ||||
| EC4d (net MFI; %>14 net MFI) | 51±19; 86 | 20±3; 39 | 14±6; 14 | 28±6; 49 |
| Low C3 or C4 (%) | 73 | 39 | 22 | 46 |
| Treatment information | ||||
| Prednisone (%; dose (mg/day)) | 47; 22.8±4.8 | 9; 16.0±2.9 | 26; 5.0±0.0 | 24; 17.8±3.2 |
| Hydroxychloroquine (%) | 58 | 75 | 100 | 75 |
| Azathioprine (%) | 11 | 0 | 0 | 5 |
| Methotrexate (%) | 8 | 3 | 100 | 24 |
| Mycophenolate (%) | 33 | 23 | 0 | 21 |
| Belimumab (%) | 8 | 2 | 0 | 3 |
Results are expressed as average, SEM and median (range) as appropriate.
dsDNA, double-stranded DNA; EC4d, erythrocyte-bound C4d; MFI, mean fluorescence intensity; PGA, Physicians Global Assessment; SELENA-SLEDAI, Safety of Estrogens in Lupus Erythematosus National Assessment—SLE Disease Activity Index.
Figure 1.
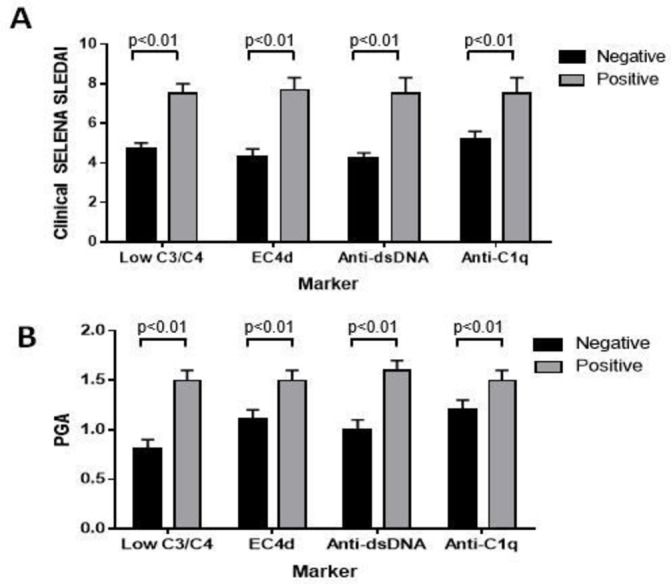
Complement and antibody abnormalities at baseline and relation to disease activity clinical SELENA-SLEDAI (A) and PGA scores (B) in relation to the presence or absence of low complement (C3/C4), abnormal EC4d (>14 net MFI), anti-dsDNA (>35 units) and anti-C1q (>20 units) levels. Results are presented as average (SEM). The presence of these abnormalities was associated with higher clinical SELENA-SLEDAI and PGA (p<0.01). dsDNA, double-stranded DNA; EC4d, erythrocyte-bound C4d; MFI, mean fluorescence intensity; PGA, Physicians Global Assessment; SELENA-SLEDAI, Safety of Estrogens in Lupus Erythematosus National Assessment—SLE Disease Activity Index.
lupus-2018-000263supp001.docx (29.3KB, docx)
The linear mixed model estimating the relationships between disease activity (by clinical SELENA-SLEDAI or PGA) and laboratory measures are presented in table 2. EC4d levels were significantly associated with the clinical SELENA-SLEDAI in all three study groups (p<0.042, marginal R2 range 2.0% to 10.7%), while low C3/C4 status only reached significance with this outcome measure in study group 1, which had been selected for complement activation (p=0.005, marginal R2=2.7%). EC4d levels were also significantly associated with the PGA in study groups 1 and 2 (p<0.05, marginal R2=2.2% and 8.7%, respectively) but was not significant in study group 3 (p=0.363, marginal R2=1.4%), which had much lower disease activity overall at baseline. Low C3/C4 status correlated significantly with PGA in all three study groups (p<0.01, marginal R2 range 4.0% to 9.1%). Altogether, EC4d and low C3/C4 status were both associated with at least one of the two disease activity measures in all study groups. Anti-dsDNA by Quanta Flash and anti-C1q titres was also associated with disease activity (table 2). However, in contrast to anti-dsDNA, anti-C1q was not associated with disease activity in the third study group of patients with SLE, all with non-renal disease and treated with methotrexate and hydroxychloroquine.
Table 2.
Linear mixed model estimates for the clinical SELENA-SLEDAI and PGA in relation to laboratory measures
| Marker | Outcome | Study group | Subject/visit, n | Intercept (SE) | Slope estimate (SE) | P values | Marginal R2 (%) |
| EC4d (log net MFI) |
Clinical SELENA-SLEDAI | 1 | 37/384 | 2.52±1.55 | 0.94±0.46 | 0.042 | 2.0 |
| 2 | 64/126 | 1.81±1.05 | 1.36±0.40 | 0.001 | 10.3 | ||
| 3 | 23/102 | 0.09±0.57 | 0.74±0.25 | 0.005 | 10.7 | ||
| 1–3 | 124/612 | 1.49±0.76 | 1.17±0.27* | <0.001 | 5.7 | ||
| PGA | 1 | 37/385 | 0.77±0.20 | 0.12±0.06 | 0.047 | 2.2 | |
| 2 | 64/128 | 0.75±0.15 | 0.18±0.06 | 0.002 | 8.7 | ||
| 3 | 23/106 | 0.50±0.24 | 0.10±0.11 | 0.363 | 1.4 | ||
| 1–3 | 124/619 | 0.64±0.11 | 0.17±0.04 | <0.001 | 5.6 | ||
| Low complement C3/C4 | Clinical SELENA-SLEDAI | 1 | 37/383 | 4.32±0.73 | 1.72±0.61 | 0.005 | 2.7 |
| 2 | 64/127 | 4.65±0.46 | 1.46±0.74 | 0.052 | 3.6 | ||
| 3 | 23/103 | 1.47±0.28 | 0.71±0.48 | 0.148 | 2.8 | ||
| 1–3 | 124/613 | 3.80±0.35 | 1.68±0.42† | <0.001 | 3.7 | ||
| PGA | 1 | 37/384 | 0.99±0.08 | 0.26±0.08 | 0.002 | 4.0 | |
| 2 | 64/127 | 1.11±0.06 | 0.27±0.10 | 0.010 | 5.7 | ||
| 3 | 23/107 | 0.58±0.11 | 0.47±0.17 | 0.007 | 9.1 | ||
| 1–3 | 124/618 | 0.95±0.05 | 0.31±0.06 | <0.001 | 5.9 | ||
| Anti-dsDNA (log net MFI) |
Clinical SELENA-SLEDAI | 1 | 37/379 | −1.11±1.43 | 1.27±0.26 | <0.001 | 17.4 |
| 2 | 64/126 | 2.03±1.03 | 0.97±0.30 | 0.002 | 9.7 | ||
| 3 | 23/102 | −0.94±1.44 | 1.01±0.55 | 0.070 | 4.6 | ||
| 1–3 | 124/607 | 0.44±0.65 | 1.08±0.15 | <0.001 | 18.2 | ||
| PGA | 1 | 37/380 | 0.69±0.17 | 0.09±0.03 | 0.005 | 5.9 | |
| 2 | 64/128 | 0.83±0.15 | 0.11±0.04 | 0.009 | 6.3 | ||
| 3 | 23/106 | −0.52±0.52 | 0.47±0.20 | 0.019 | 7.6 | ||
| 1–3 | 124/614 | 0.68±0.09 | 0.10±0.02 | <0.001 | 8.3 | ||
| Anti-C1q (log net MFI) |
Clinical SELENA-SLEDAI | 1 | 37/383 | 1.50±1.18 | 1.27±0.32 | <0.001 | 6.6 |
| 2 | 64/126 | 2.38±0.98 | 1.28±0.42 | 0.003 | 8.3 | ||
| 3 | 23/102 | 1.08±0.57 | 0.34±0.30 | 0.265 | 2.1 | ||
| 1–3 | 124/611 | 1.55±0.60 | 1.24±0.22 | <0.001 | 9.9 | ||
| PGA | 1 | 37/384 | 0.62±0.14 | 0.17±0.04 | <0.001 | 8.5 | |
| 2 | 64/128 | 0.91±0.14 | 0.13±0.06 | 0.029 | 4.3 | ||
| 3 | 23/106 | 0.76±0.24 | −0.04±0.12 | 0.765 | 0.2 | ||
| 1–3 | 124/618 | 0.70±0.09 | 0.15±0.03 | <0.001 | 7.5 |
Intercept, slope estimates, p values and marginal R2 are given.
*One log EC4d levels were associated with a 1.2-point in clinical SELENA-SLEDAI.
†Low C3/C4 status was associated with a 1.7-point clinical SELENA-SLEDAI.
dsDNA, double-stranded DNA; EC4d, erythrocyte-bound C4d; MFI, mean fluorescence intensity; PGA, Physicians Global Assessment; SELENA-SLEDAI, Safety of Estrogens in Lupus Erythematosus National Assessment—SLE Disease Activity Index.
When the three study groups were combined, univariate analysis revealed that EC4d, low C3/C4 and anti-dsDNA levels were associated with disease activity by PGA and clinical SELENA-SLEDAI (table 2). Abnormal EC4d (>14 net MFI) was associated with clinical SELENA-SLEDAI (estimate=1.29±0.39; R2=2.2%; p<0.01) and PGA (estimate=0.11±0.06; R2=0.8%; p=0.05), and a total of 37 SLE (31%) changed their EC4d status from baseline during at least one follow-up visits (of those, 8 subjects changed their status from normal EC4d at baseline to abnormal EC4d at follow-up). Multivariate analysis also revealed that EC4d levels and low complement C3/C4 were both independently and significantly (p<0.01) associated with the clinical SELENA-SLEDAI and PGA score (table 3), and adding EC4d to low complement C3/C4 resulted in a higher proportion of variance explained (R2=7.7% for clinical SELENA-SLEDAI) by comparison to low C3/C4 alone (3.7%, table 2), thus suggesting the additional information gained from EC4d.
Table 3.
Linear mixed model estimates for the clinical SELENA-SLEDAI and PGA in relation to laboratory measures by multivariate analysis
| Model | Outcome variable | Predictors | Slope estimate; p values | Marginal R2 (%) |
| EC4d + low C3/C4 | Clinical SELENA-SLEDAI |
Intercept | 1.53±0.75; | 7.7 |
| EC4d (log net MFI)+ | 0.94±0.28; p<0.01* | |||
| Low complement C3/C4 | 1.24±0.43; p<0.01 | |||
| PGA | Intercept | 0.64±0.10; | 9.1 | |
| EC4d (log net MFI)+ | 0.12±0.04; p< 0.01 | |||
| Low complement C3/C4 | 0.24±0.06; p<0.01 | |||
| EC4d + low C3/C4 +anti-dsDNA |
Clinical SELENA-SLEDAI | Intercept | −0.53±0.12; | 19.6 |
| EC4d (log net MFI)+ | 0.49±0.28; p=0.07 | |||
| Low complement C3/C4+ | 0.79±0.43; p=0.07 | |||
| Anti-dsDNA (log net MFI) | 0.90±0.17; p=0.01 | |||
| PGA | Intercept | 0.52±0.12; | 12.3 | |
| EC4d (log net MFI)+ | 0.09±0.04; p = 0.03 | |||
| Low complement C3/C4+ | 0.20±0.06; p< 0.01 | |||
| Anti-dsDNA (log net MFI) | 0.06±0.02; p = 0.01 |
Intercept, slope estimates, p values and marginal R2 are given.
*One log EC4d levels and low complement C3/C4 were associated with a 0.9-point and 1.2-point in clinical SELENA-SLEDAI, respectively; presence of both one log EC4d levels and low complement C3/C4 was associated with 2.1-point clinical SELENA-SLEDAI.
dsDNA, double-stranded DNA; EC4d, erythrocyte- bound C4d; MFI, mean fluorescence intensity; PGA, Physicians Global Assessment; SELENA-SLEDAI, Safety of Estrogens in Lupus Erythematosus National Assessment— SLE Disease Activity Index.
Of the 124 patients studied, 97 presented with either chronically low C3/C4 (n=40, followed for 9.2±0.3 visits) or normal C3/C4 at all visits (n=57, followed for 5.4±0.3 visits). In this subset (including 401 study visits), a one log EC4d level was associated with a 1.2-point increase in the SELENA-SLEDAI and a 0.19-point increase in PGA, supporting the value of EC4d when there are invariant levels of classical complement proteins (table 4).
Table 4.
Linear mixed effect models for the clinical SELENA-SLEDAI and PGA in the presence of chronically low or normal C3/C4 or fluctuating C3/C4
| Outcome variable | C3/C4 status |
n/study visits | Intercept (SE) |
Slope (SE) |
P value | Marginal R2 (%) | |
| EC4d (log net MFI) |
Clinical SELENA-SLEDAI |
Low | 40/208 | 3.48±1.50 | 0.77±0.46 | 0.097 | 2.6 |
| Normal | 57/193 | 0.47±0.97 | 1.55±0.44 | <0.001 | 9.0 | ||
| Low or normal | 97/401 | 1.60±0.79 | 1.20±0.29 | <0.001 | 7.9 | ||
| Fluctuating | 27/211 | 0.17±1.98 | 1.38±0.60 | 0.022 | 3.8 | ||
| PGA | Low | 40/210 | 0.79±0.20 | 0.15±0.06 | 0.018 | 5.3 | |
| Normal | 57/197 | 0.50±0.18 | 0.22±0.08 | 0.009 | 5.2 | ||
| Low or normal | 97/407 | 0.59±0.12 | 0.19±0.04 | <0.001 | 8.7 | ||
| Fluctuating | 27/212 | 0.70±0.27 | 0.13±0.08 | 0.128 | 1.9 | ||
| Anti-dsDNA (log titres) |
Clinical SELENA-SLEDAI |
Low | 40/207 | 0.32±1.28 | 1.16±0.25 | <0.001 | 20.0 |
| Normal | 57/190 | 2.55±1.00 | 0.40±0.33 | 0.228 | 2.0 | ||
| Low or normal | 97/397 | 0.90±0.66 | 1.00±0.016 | <0.001 | 20.8 | ||
| Fluctuating | 27/210 | −1.78±1.78 | 1.47±0.039 | <0.001 | 18.9 | ||
| PGA | Low | 40/209 | 0.76±0.18 | 0.10±0.03 | 0.004 | 8.4 | |
| Normal | 57/194 | 0.59±0.17 | 0.12±0.06 | 0.032 | 5.5 | ||
| Low or normal | 97/403 | 0.66±0.10 | 0.11±0.02 | <0.001 | 12.3 | ||
| Fluctuating | 27/212 | 0.79±0.22 | 1.07±0.05 | 0.152 | 2.8 | ||
| Anti-C1q (log titres) |
Clinical SELENA-SLEDAI |
Low | 40/207 | 2.57±1.34 | 1.02±0.38 | 0.008 | 5.4 |
| Normal | 57/192 | 1.89±0.79 | 1.02±0.40 | 0.013 | 7.1 | ||
| Low or normal | 97/399 | 1.93±0.64 | 1.12±0.24 | <0.001 | 10.8 | ||
| Fluctuating | 27/212 | 0.41±1.38 | 1.56±0.45 | <0.001 | 8.7 | ||
| PGA | Low | 40/209 | 0.67±0.18 | 0.18±0.05 | <0.001 | 9.5 | |
| Normal | 57/196 | 0.77±0.14 | 0.10±0.07 | 0.180 | 2.1 | ||
| Low or normal | 97/405 | 0.67±0.10 | 0.17±0.03 | <0.001 | 10.8 | ||
| Fluctuating | 27/213 | 0.78±0.18 | 0.12±0.06 | 0.055 | 3.1 |
Intercept, slope estimates (SE) and p value with marginal R2 are given. Of the 124 patients studied, 97 presented with either chronically low C3/C4 (n=40, followed for 9.2±0.3 visits) or normal C3/C4 at all visits (n=57, followed for 5.4±0.3 visits) and 27 presented with fluctuating C3/C4 (followed for 10.4±0.3 visits).
dsDNA, double-stranded DNA; EC4d, erythrocyte-bound C4d; MFI, mean fluorescence intensity; PGA, Physicians Global Assessment; SELENA-SLEDAI, Safety of Estrogens in Lupus Erythematosus National Assessment— SLE Disease Activity Index.
Discussion
In this report, we have analysed the relationships between laboratory measures and disease activity in three study groups. The findings from the first study group have been reported previously, and all patients with SLE from this group were selected for active disease in the presence of abnormal complement activation.11 The two additional studies reported here, all designed to evaluate complement components in relation to SLE disease activity, add to the growing body of evidence that quantitative flow cytometry determination of C4d split products on erythrocytes may be useful in the monitoring of SLE.
We analysed the data separately by study groups and in combination. While EC4d levels were independently associated with clinical SELENA-SLEDAI in all three study groups, the association with disease measured by PGA did not reach significance in the third study group, which had the lowest disease activity. Low complement C3/C4 was associated with disease measured by PGA in all study groups but did not reach significance with the clinical SELENA-SLEDAI in the third study group. The precise reasons for the inconsistencies between laboratory and clinical activity measures are unclear; nevertheless, both complement components were significantly associated with at least one of the two clinical measures in each of the study groups, thus suggesting that both EC4d and low complement C3/C4 are useful laboratory measures.
As expected, when we combined the three study groups, univariate analysis indicated that EC4d and low complement remained associated with disease activity irrespective of clinical activity measure. Multivariate analysis also revealed that adding EC4d to low complement C3/C4 resulted in a higher proportion of variance explained (R2=8% for clinical SELENA-SLEDAI) by comparison to low C3/C4 alone (4%), thus suggesting the additional information gained from EC4d.
There are potential advantages to the use of the combined measures of complement. First, the presence of normal complement component measures in the absence of the complement activation product could provide greater confidence that disease control has been achieved. To the contrary, higher clinical disease activity with low complement C3/C4 in the presence of complement activation products may reflect more active disease, suggesting that therapeutic intervention may be necessary. Our analysis also indicated that in the large subset of patients with chronically low or normal complement C3/C4 levels, EC4d levels remained associated with disease activity, thus supporting the potential of the laboratory measure when C3/C4 status is not a reliable marker of disease activity.
Abnormalities in anti-dsDNA and anti-C1q titres were also correlates of disease activity, although anti-C1q did not correlate with disease activity in the group of patients with low disease activity. This most likely is due to the lack of renal involvement in that group of patients who, for the most part, were receiving methotrexate and hydroxychloroquine to control rash and arthritis. In contrast, as seen above with EC4d, anti-dsDNA was associated with disease activity in all three study groups, and thus may have value in the monitoring of SLE in a broader group of patients with SLE, irrespective of clinical patterns and organ involvement.
In conclusion, our data expand initial work associating EC4d with SLE disease activity,12 and further support the usefulness of EC4d in the monitoring of SLE. Moving forward, it will be important to further evaluate these laboratory measures as objective endpoints in interventional clinical trials and as aids to guide optimal treatment decisions when clinical disease activity assessment is not easily discerned.
Acknowledgments
We thank Dr Arthur Weinstein for critical review of the manuscript and Dr Larry Magder for advice in the statistical analysis. We also thank Claudia Ibarra for the management of our clinical laboratory and Tyler O’Malley for technical assistance.
Footnotes
Contributors: All authors participated in the collection of data and data interpretation. TD wrote the first draft of the manuscript. All authors contributed to and approved the final manuscript.
Funding: This study was funded by Exagen Diagnostics. The Hopkins Lupus cohort is supported by National Institutes of Health grants R01 AR43727 and AR69572.
Competing interests: JTM, MAP, JB, RR-G, KK, RAF and CP have received research grants from Exagen. TD and JC are employed by Exagen Diagnostics.
Patient consent: Obtained.
Ethics approval: Internal review boards approved the study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: None.
References
- 1.Wener MH. Immune complexes in systemic lupus erythematosus In: Lahita R, Toskos G, Buyon J, Koike T, eds Systemic lupus erythematosus. 5th edn., 2010:321–38. [Google Scholar]
- 2.Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med 2005;353:2550–8. doi:10.1056/NEJMoa051135 [DOI] [PubMed] [Google Scholar]
- 3.Petri M, Orbai AM, Alarcón GS, et al. Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677–86. doi:10.1002/art.34473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson JP, Yu Y. Genetic susceptibility and class III complement genes In: Lahita R, Toskos G, Buyon J, Koike T, eds Systemic lupus erythematosus. 5th edn 2010:21–46. [Google Scholar]
- 5.Putterman C, Furie R, Ramsey-Goldman R, et al. Cell-bound complement activation products in systemic lupus erythematosus: comparison with anti-double-stranded DNA and standard complement measurements. Lupus Sci Med 2014;1:e000056 doi:10.1136/lupus-2014-000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senaldi G, Makinde VA, Vergani D, et al. Correlation of the activation of the fourth component of complement (C4) with disease activity in systemic lupus erythematosus. Ann Rheum Dis 1988;47:913–7. doi:10.1136/ard.47.11.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schramm EC, Staten NR, Zhang Z, et al. A quantitative lateral flow assay to detect complement activation in blood. Anal Biochem 2015;477:78–85. doi:10.1016/j.ab.2015.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buyon JP, Tamerius J, Belmont HM, et al. Assessment of disease activity and impending flare in patients with systemic lupus erythematosus. Comparison of the use of complement split products and conventional measurements of complement. Arthritis Rheum 1992;35:1028–37. doi:10.1002/art.1780350907 [DOI] [PubMed] [Google Scholar]
- 9.Belmont HM, Hopkins P, Edelson HS, et al. Complement activation during systemic lupus erythematosus. C3a and C5a anaphylatoxins circulate during exacerbations of disease. Arthritis Rheum 1986;29:1085–9. doi:10.1002/art.1780290905 [DOI] [PubMed] [Google Scholar]
- 10.Manzi S, Navratil JS, Ruffing MJ, et al. Measurement of erythrocyte C4d and complement receptor 1 in systemic lupus erythematosus. Arthritis Rheum 2004;50:3596–604. doi:10.1002/art.20561 [DOI] [PubMed] [Google Scholar]
- 11.Buyon J, Furie R, Putterman C, et al. Reduction in erythrocyte-bound complement activation products and titres of anti-C1q antibodies associate with clinical improvement in systemic lupus erythematosus. Lupus Sci Med 2016;3:e000165 doi:10.1136/lupus-2016-000165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao AH, Navratil JS, Ruffing MJ, et al. Erythrocyte C3d and C4d for monitoring disease activity in systemic lupus erythematosus. Arthritis Rheum 2010;62:837–44. doi:10.1002/art.27267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang DH, Chang DM, Lai JH, et al. Usefulness of erythrocyte-bound C4d as a biomarker to predict disease activity in patients with systemic lupus erythematosus. Rheumatology 2009;48:1083–7. doi:10.1093/rheumatology/kep161 [DOI] [PubMed] [Google Scholar]
- 14.Gladman DD, Ibañez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol 2002;29:288–91. [PubMed] [Google Scholar]
- 15.Dervieux T, Conklin J, Ligayon JA, et al. Validation of a multi-analyte panel with cell-bound complement activation products for systemic lupus erythematosus. J Immunol Methods 2017;446:54–9. doi:10.1016/j.jim.2017.04.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lupus-2018-000263supp001.docx (29.3KB, docx)


