Rotation of a helical cell body and mobility of cell-surface adhesins result in surface motility of the spirochete Leptospira.
Abstract
Many species of bacteria are motile, but their migration mechanisms are considerably diverse. Whatever mechanism is used, being motile allows bacteria to search for more optimal environments for growth, and motility is a crucial virulence factor for pathogenic species. The spirochete Leptospira, having two flagella in the periplasmic space, swims in liquid but has also been previously shown to crawl over solid surfaces. The present motility assays show that the spirochete movements both in liquid and on surfaces involve a rotation of the helical cell body. Direct observations of cell-surface movement with amino-specific fluorescent dye and antibody-coated microbeads suggest that the spirochete attaches to the surface via mobile, adhesive outer membrane components, and the cell body rotation propels the cell relative to the anchoring points. Our results provide models of how the spirochete switches its motility mode from swimming to crawling.
INTRODUCTION
Bacterial motility is considerably diverse: Escherichia coli and Salmonella spp. swim by rotating their flagella, which are a major motility machinery composed of a basal motor and helical filament (1, 2); Pseudomonas aeruginosa and Neisseria gonorrhoeae exhibit a twitching motility using type IV pili (3); and gliding bacteria such as Mycoplasma mobile (4) and Myxococcus xanthus (5) require a direct interaction between external complexes and surfaces. In the zoonotic spirochete Leptospira, swimming motility is well known as their major method of migration (6, 7), but an early study by Cox and Twigg (8) showed that spirochetes had a “crawling” movement on solid surfaces. The morphology and cell structure of Leptospira are unique (Fig. 1A). The outer membrane wraps around the right-handed helical protoplasmic cylinder (PC), and the cell configuration is a right-handed helix due to PC shape. Two flagella reside between the outer membrane and the peptidoglycan layer, known as periplasmic flagella (PFs) (9). P. aeruginosa uses the flagellum and pilus for swimming and twitching, respectively (10). However, PFs are the sole motility machinery of Leptospira, and none specified for motility on surfaces have been identified. How does Leptospira realize two-phase motility? To address this question, we analyzed the cell motilities and cell-surface movement of the nonpathogenic Leptospira biflexa in liquid and on surfaces.
Fig. 1. Cell structure and swimming motility of Leptospira.
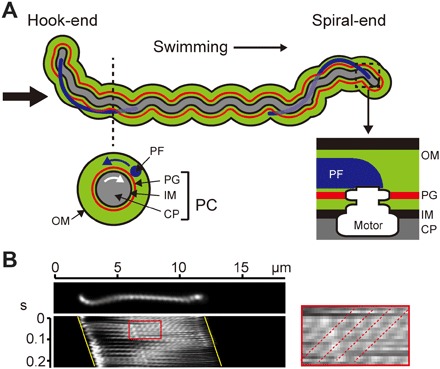
(A) Schematic diagram of Leptospira cell structure. The thin black arrow indicates the swimming direction. A cross section of the cell body is depicted below the dashed line: outer membrane (OM), PF, peptidoglycan layer (PG), inner membrane (IM), and cytoplasm (CP). The rotational direction is viewed from the Hook-end to the Spiral-end, as indicated by the thick black arrow. Blue and white arrows indicate the rotational directions of PF and the PC, respectively. At the ends of the cell body surrounded by a dashed square, the flagellar motor of each PF is embedded into PG and IM. (B) Kymograph of a cell swimming in motility medium. Yellow lines indicate cell movement. The area surrounded by a red square is enlarged on the right, and dotted lines indicate the apparent movements of the PC helix.
RESULTS
Analysis of swimming motility
When Leptospira swims, PF rotations transform the cell ends into a left-handed spiral-shape (Spiral-end) or half-circle hook-shape (Hook-end) and gyrate them counterclockwise (CCW; defined by viewing a swimming cell from the anterior side to the posterior side) (movie S1). Meanwhile, PC rotates clockwise (CW), and because PFs are attached to PC via basal rotary motors (flagellar motors), PC is believed to be rotated by counter-torques of PF rotations. Both ends of the Leptospira cell body frequently change their shape between spiral and hook shapes with a switching of rotational direction, and cell configuration is associated with the motility form; when displaying the spiral shape at one end and the hook shape at the other end, the cell swims in the direction of the Spiral-end, and when displaying symmetric configurations (for example, both cell ends exhibit the spiral shape), the cell rotates without net displacement (6, 11, 12). Figure 1B shows a kymograph of a cell swimming in a motility medium. The PC helix is observed as a series of bright spots by a dark-field illumination, and the spots move backward with PC rotation (Fig. 1B, right) (7). The backward speed of the bright spots (fpc′) is a net value resulting from the backward movement by the actual PC rotation (fpc) and the forward movement of the helical cell body by swimming (vpc) (13–15). The value of vpc is determined from the swimming speed (v) and PC pitch length by vpc = v/ppc; therefore, fpc = fpc′ + v/ppc, where the backward movement is defined as positive. The ratio of v to fpc (that is, v/fpc) indicates the distance that the cell migrates in one revolution. Moreover, the ratio of v/fpc to ppc (that is, v/fpc/ppc) indicates how much the cell slips during swimming, which can be interpreted as swimming efficiency. The average values of v and fpc were 8.3 ± 1.9 μm/s and 59 ± 12 Hz, respectively (n = 21 cells), and ppc was 0.60 ± 0.08 μm (n = 64 helices on 10 cells). Therefore, the swimming efficiency of Leptospira was 0.23, which can be compared with data from other bacteria measured in a water-based medium without any polymers; it is about twofold higher compared to Salmonella enterica (0.11) (14) and threefold for Vibrio alginolyticus (0.07) (13). Results for simultaneous measurements of swimming speeds, Spiral-end gyration rates, and PC rotation rates are shown in fig. S1.
Analysis of crawling motility
To assess Leptospira crawling, we demonstrated its movement on a glass surface (movie S2). A kymograph (Fig. 2A) shows that the apparent PC helix movement is not observed during crawling, indicating that Leptospira crawls without slip; when fpc′ = 0, v/ppc = fpc; therefore, (v/fpc)/ppc = 1 (further examples are shown in fig. S2). Although Leptospira crawling motility was observed without modification of the glass, the crawling speeds were significantly increased by coating the glass with an anti-lipopolysaccharide rabbit antibody (Ab-LPS) (Fig. 2B and movie S3). Bacterial adhesion is mediated by LPS and other cell surface components (16, 17). Crawling motility requires these adhesive molecules not only to attach to but also to detach from solid surfaces as the cell progresses (18), and a high affinity to the surface will retard crawling. Leptospira has abundant LPSs and proteins that protrude outside the cell (19, 20). Although the affinity of Leptospira adhesins to surfaces was not fully elucidated, Ab-LPS (~10 nm) could inhibit the attachment of adhesins with a higher affinity than LPS (schematically explained in Fig. 2B), thereby promoting a crawling motility.
Fig. 2. Crawling motility of Leptospira.
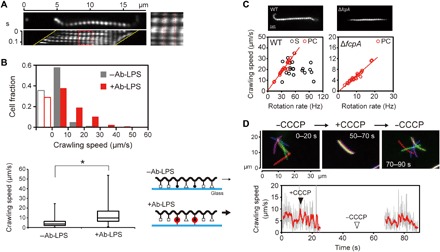
(A) Kymograph of a cell crawling on a glass surface. The PC appears to be fixed (red dotted lines), indicating movement without slip. (B) Effect of an Ab-LPS on crawling motility; 216 and 214 cells were measured on noncoated (−Ab-LPS) or Ab-LPS–coated (+Ab-LPS) glasses, respectively; open bars indicate cells adhering to glass surfaces without translation. The box-and-whisker plot shows the 25th (the bottom line of the box), 50th (middle), and 75th (bottom) percentiles and the minimum and maximum values (whiskers) of crawling speeds obtained from individual cells; statistical analysis was performed by Mann-Whitney U test (*P < 0.01). The schematics represent a hypothetical explanation of Ab-LPS effects. Various external molecules are shown by different symbols; black dots with a bar indicate LPS; red circles with a bar indicate Ab-LPS; and thin and thick arrows indicate slow and fast crawling, respectively (the details are described in the main text). (C) Wild-type (WT) and ΔfcpA cells observed by dark-field microscopy (top) and pairwise plots of crawling speeds and cell rotation rates (bottom). Red lines in the pairwise plots are regression lines fitted to PC data points. Correlation coefficients (R) for PC of WT and ΔfcpA are 0.98 and 0.97, respectively. R between Spiral-end speed and crawling speed in WT is 0.22 (no regression line shown). (D) Effects of CCCP on the cells attached to a glass surface (movie S5). Top: Results of single-cell tracking. Microscopic images captured at 0.1-s intervals were decimated to 5-s intervals and then integrated. Colors indicate time courses in the order of red, green, blue, purple, and yellow. The cell stopped crawling with the addition of 5 μM CCCP (middle), and therefore, all of the colored cell images were superposed. Bottom: Crawling speed of the cell shown in the top panel. Crawling speeds determined at 0.1-s intervals are shown in gray, and data of a 10–data-point moving average are shown in red.
As observed in Fig. 2A, Leptospira cells bend their ends into either a spiral or hook shape during crawling in the same way as during swimming. Berg et al. (12) suggested that most of the thrust for swimming in Newtonian fluid, typically water, was generated by a gyration of the Spiral-end. However, both bent ends of crawling cells seemed to just beat glass surfaces (movie S2). The pairwise plot of crawling speeds and cell body rotation rates show that crawling speeds depend on the rotation rate of PC but not on the gyration rate of the Spiral-end (Fig. 2C, left). We also measured the crawling motility of a mutant strain that lacks the flagellar coiling protein A (FcpA), which determines the coiled shape of Leptospira PF (Fig. 2C, top right, and movie S4) (21, 22). The mutant strain of L. biflexa was obtained by random insertion mutagenesis using Himar1 transposon (22). The ΔfcpA mutant remains a helix of PC, but it lacks the Spiral-end and Hook-end due to the PF shape anomaly (fig. S3). The crawling speed of the ΔfcpA mutant was strongly correlated with PC rotation (Fig. 2C). The mutant cells showed slower PC rotation rates and crawling speeds than the wild-type (WT) ones, but they also crawled without slip (fig. S4). Thus, only PC rotation propelled the Leptospira cell on the surface. Leptospira PF rotations are inhibited by protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (23). The crawling motility was inhibited by the addition of CCCP (Fig. 2D and movie S5), suggesting that the movement is caused by PF rotation within the cell body. The decrease in cell body rotation rates and crawling speeds in the ΔfcpA strain supports the mechanism that Leptospira crawling is based on flagellum-dependent motility.
Direct observation of outer membrane dynamics in crawling cells
To understand the mechanism of crawling, we first observed cell body rotation during crawling by labeling the outer membranes of cells with amino-specific Cy3–N-hydroxysuccinimide (NHS). As a result, we observed that the outer membranes of crawling cells rotated at the same speed as PC (Fig. 3 and movie S6). This result indicates that the helical cell body rotates freely on surfaces, although the cell body is somehow anchored to the surface. Concerning the mechanism by which the outer membrane rotates with PC despite the separation of these two structures (24, 25), theoretical studies on the swimming mechanism of the Lyme disease spirochete Borrelia burgdorferi predicted interactions between PF and cell membranes via viscous fluid filling the periplasmic space (26). Therefore, the outer membrane could be rotated by a hydrodynamic interaction with PC mediated by viscous fluid within the periplasmic space; namely, PC rotation drags the outer membrane.
Fig. 3. Fluorescent observation of outer membrane dynamics.
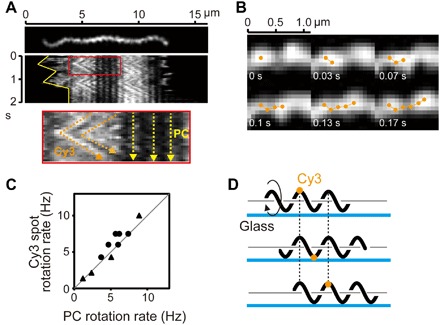
(A) Crawling cell labeled with Cy3-NHS. In a kymograph, the yellow solid line indicates cell migration (top). In an enlarged kymograph (bottom), orange and yellow dashed lines indicate translational movements of fluorescent spots and slip-less crawling of the cell, respectively. (B) A montage shows an example trajectory of a fluorescent spot. (C) The rotation rates of the PC and Cy3 fluorescent spots determined from speed versus time traces for individual cells are shown. The rotations of PC and Cy3 fluorescent spots were simultaneously analyzed at several different time periods for each trace, and data obtained from two different cells are shown by black dots and triangles (total of 11 data points). A gray line with a slope of 1 is shown. (D) Schematic explanation of fluorescent dye movement attached to a cell crawling on the surface with rotation of a helical cell body.
Movement of beads attached to the cell body via an anti-LPS antibody
What is the mechanism by which Leptospira cell body can rotate while being anchored to a surface? For surface movement, M. mobile uses abundant “leg”-like machineries on the cell surface, successively catching and releasing sialylated oligosaccharide-modified surfaces of animal tissues to propel the cell (4). M. xanthus has a gliding machinery that consists of an external complex (Agl-Glt) and intracellular motor unit (5). Flavobacterium johnsoniae glides by using the adhesive extracellular protein SprB moving along a closed helical path structure that is believed to be on the cell surface (18). Charon et al. (27) showed movements of microbeads attached to the outer membrane of Leptospira via an anti–whole-cell antibody. They carefully verified what moved the beads and reported that bead movement was caused by a viscous drag force that acted on the beads when the cell translates (the beads were dragged in the opposite direction to the cell movement). Although antigens targeted by the antibody were unspecified, they showed that the antigens residing on the cell surface are mobile, which raises the possibility that these mobile, adhesive molecules are somehow involved in crawling. Because Ab-LPS affected crawling motility (Fig. 2B), we labeled LPS with Ab-LPS–coated polystyrene beads. In free-swimming cells, wavy trajectories of the beads were observed (Fig. 4A, middle), and then, we revealed that the bead rotated in a CW direction around the cell body (Fig. 4, B to D, and movies S7 and S8; example data are also shown in figs. S5 and S6). In the cell shown in Fig. 4A, the rotation rate of the bead was about 3 Hz (Fig. 4A, bottom), whereas the Spiral-end (End2), the Hook-end (End1), and PC rotated at 16, 33, and 40 Hz, respectively (Fig. 4E). When a large aggregate of Ab-LPS beads were attached to a cell, the aggregate was almost fixed on the video screen without rotation. Nevertheless, the cell rotated and moved relative to the aggregate without slip (Fig. 4F and movie S9), as previously observed (27). These results indicate that rotations of LPS loaded with beads were delayed from the cell body rotation; LPS rotation does not synchronize with that of the cell body. Beads without an Ab-LPS coating nonspecifically bound to the cell but did not translate along the cell body (fig. S7), suggesting that the phospholipid layer of the outer membrane or adhesins with a lower mobility than LPS embedded in the outer membrane might be targets of nonspecific binding.
Fig. 4. Observation of beads attached to the cell body via an antibody.
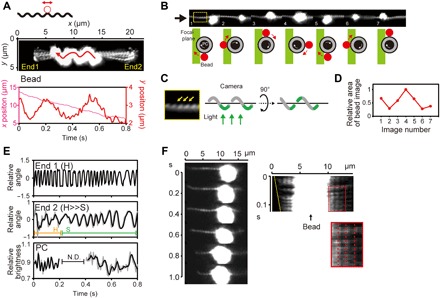
(A) The upper schematic represents the bead assay. A polystyrene bead with a diameter of 200 nm coated with Ab-LPS (red circle) was attached to the cell (black wavy line). Middle: Superposition of sequential video images. The original movie recorded at 4-ms intervals (movie S7) was decimated to 80-ms intervals, and 10 sequential images were then superposed. The red arrow indicates the trajectory and direction of bead movement. The cell ends were arbitrarily designated as End1 and End2. Bottom: Time traces of x and y positions of the bead. (B) Rotation of the bead based on the focal plane determined as shown in (C) and a change in the size of the bead image shown in (D). Bead rotation is depicted by sequential diagrams below the montage, which are observed from the direction indicated by the black arrow. (C) Enlarged image of the part indicated by the yellow square in (B). The focal plane of observation can be deduced from the visualized helix angle of PC (yellow arrows), as schematically explained. (D) Change in the area of the bead image attributed to halation caused by a z axis displacement of the bead. The x axis indicates image numbers shown in (B). (E) Time traces of End1, End2, and PC rotations in the cell shown in (A). Raw data (gray) were smoothed by moving the average (black). End1 displayed the Hook-shape (H) during recording, whereas End2 changed the shape from the Hook- to Spiral-shape (S) at around 0.2 s. PC rotation was not measured from 0.2 to 0.4 s (indicated by N.D.) due to defocusing. (F) Attachment of an aggregated bead to the cell surface. A kymograph (right) shows an apparent PC stillness (red dashed lines), that is, movement without slip.
DISCUSSION
We characterized movements of the spirochete Leptospira in liquid and over surfaces. Although swimming involves Spiral-end gyration (12), quantification of crawling using the ΔfcpA mutant showed that Leptospira only exploits PC to move on surfaces. We revealed that the outer membrane rotates with PC while being attached to surfaces and then showed the possibility that LPS could be a mobile adhesin anchoring the cell to the surface. On the basis of these results, we depicted plausible models of how Leptospira switches its motility mode from swimming to crawling (Fig. 5). In swimming (Fig. 5A), a CCW gyration of the Spiral-end and CW rotation of PC propel the cell. The outer membrane rotates with PC, which produces a resistive torque by the interaction between the cell surface and external fluid, as predicted previously (15), but it would not produce thrust. In crawling, the cell attaches to the surface via mobile, adhesive outer membrane components (for example, LPS and proteins), but PC and the outer membrane keep rotating CW (Fig. 5B), propelling the cell relative to the position where adhesive molecules attach. Because a large variety of adhesive molecules can exist on the Leptospira cell surface (19, 20), the crawling speed could be determined by a molecule with the smallest dissociation constant value; that is, the detachment of adhesive molecules from the surface is the rate-limiting process of crawling; attachments of immobile adhesive molecules to the surface would inhibit crawling. This model predicts that PC contributes to swimming as a screw propeller, whereas PC would play a role of a helical path for adhesion on surfaces. The current study did not elucidate the presence of the helical path along PC. Since the shape determination of PC involves penicillin-binding proteins and the actin homolog MreB (28), such an intracellular molecular system might synthesize a periodic structure beneath the outer membrane. Another unanswered question is what moved the beads that were attached to swimming cells in the direction of translation (Fig. 4A and fig. S8). A theoretical study predicted that in a peritrichous bacterium, cell body and flagellar bundle rotations generate flow near the cell (29). Perhaps, PC rotation might generate a directional flow in the immediate vicinity of the cell body, thereby driving bead translation.
Fig. 5. Model of the motility form transition in Leptospira.
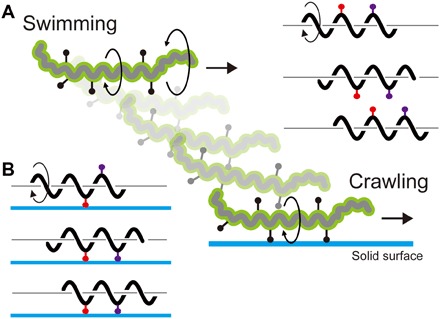
(A) Swimming is caused by CCW gyration of the Spiral-end and CW rotation of the PC, and adhesive cell-surface molecules (black dots with a bar) rotate with the cell body. Rotations of adhesive molecules are shown by red and purple symbols on the right. (B) When attaching to the surface via mobile adhesins, the cell moves relative to the anchoring points with PC rotation. In the left cartoon, first, the red adhesin attaches to the surface, and then, the purple one participates in the anchoring.
Here, we present the results for a nonpathogenic strain of Leptospira, but the pathogenic species L. interrogans also crawls on surfaces (fig. S9). Pathogenic Leptospira percutaneously invades animals through a wound site on the skin or membrane mucosa, and it then migrates to tissues through the bloodstream and penetrates and breaks the intercellular junction of the hepatocyte layer (20, 30, 31). In the infection process, swimming and crawling motilities are important for migration through the mucosa and on tissue surfaces, respectively. The WT Leptospira cells can move over surfaces faster than in liquid (fig. S1 and Fig. 2C); while the ΔfcpA mutant cells could hardly swim (21, 22), their crawling speed reached 10 μm/s (Fig. 2C). This fast translation on surfaces can be ascribed to an improvement in translation efficiency, that is, slip suppression (Fig. 2A and figs. S2 and S4). Highly efficient crawling might benefit Leptospira infection processes. Our results showed that LPS is an adhesive molecule candidate for crawling. Since pathogenic Leptospira spp. are classified into more than 250 serovars based on LPS structure, LPS heterogeneity could affect their crawling motility, as shown in Fig. 2B. Moreover, some membrane proteins are known as virulence factors (20, 30). Therefore, unveiling the diversity of external adhesive molecules in Leptospira serovars and surface properties of host tissues associated with crawling motility will help us to gain insight into the mechanism of leptospiral host specificity and pathogenesis.
MATERIALS AND METHODS
Bacteria and media
A saprophyte L. biflexa strain Patoc I and pathogenic L. interrogans serovar Manilae strain UP-MMC-NIID were used. The ΔfcpA mutant was derived from the L. biflexa strain Patoc I by random insertion mutagenesis using a Himar1 transposon (22). The cells were grown in Ellinghausen-McCullough-Johnson-Harris liquid medium at 30°C for 4 days until the stationary phase. A total of 20 mM potassium phosphate buffer (pH 7.4) was used as a motility medium. Ficoll (Sigma-Aldrich) was added to the motility medium, as necessary.
PF isolation
PF were isolated from cells and purified by the method described by Wunder et al. (21).
Electron microscopy and cryo-EM
Isolated PFs were applied onto the continuous carbon-coated electron microscopy (EM) grids and negatively stained with 2% (w/v) uranyl acetate solution. Negative-stained EM images were observed with a JEM-1011 transmission electron microscope (JEOL) operating at 100 kV using a TVIPS TemCam-F415MP charge-coupled device (CCD) camera (TVIPS).
Quantifoil grids (Quantifoil Micro Tools) were glow-discharged in a weak vacuum for 20 s immediately before use. Sample solutions of WT and ΔfcpA mutant were applied to the grid, blotted briefly with filter paper, and rapidly plunged into liquid ethane using Vitrobot Mark II (FEI Company). Cryo-EM images were collected at a liquid-nitrogen temperature using a Titan Krios electron microscope (FEI Company) equipped with a field-emission gun and a Falcon direct electron detector (FEI Company). The microscope was operated at 300 kV and a nominal magnification of 29,000× with a calibrated pixel size of 5.71 Å.
Motility assay
Swimming and crawling were analyzed by one-sided dark-field microscopy, as described previously, with some modifications (11). Cells were infused into a flow chamber made by sticking a glass slide (bottom side; Matsunami Glass Ind. Ltd.) and coverslip (upper side; Matsunami Glass Ind. Ltd.) with double-sided tape, and their movements were observed through a 100× oil immersion objective lens (UPlanFLN, Olympus) and a 5× relay lens. The microscopic images were recorded at a frame rate of 250 Hz with a high-speed complementary metal-oxide semiconductor video camera (IDP-Express R2000, Photron), and the movie was analyzed with a Visual Basic for Applications macro originally developed in Microsoft Excel.
Labeling of the outer membrane with a fluorescent dye
A 1-μl aliquot of Cy3-NHS ester (Lumiprobe) dissolved in dimethyl sulfoxide (5 μg/ml) was mixed with 100 μl of the L. biflexa culture at room temperature. Excess dyes free from cells were removed by centrifugation at 1000g for 4 min and then suspended into the motility buffer. The cells labeled with the dyes were observed with a fluorescent microscope (BX53, UPlan-FLN 100×, U-FGW, Olympus), and their fluorescent images were acquired with a CCD camera (WAT-910HX/RC, Watec) at a frame rate of 30 Hz.
Labeling of the outer membrane with microbeads
Polystyrene beads were conjugated with an anti–L. biflexa LPS antibody by the following procedure: 3 μl of carboxylated bead suspension (0.2 μm in diameter; Thermo Fisher Scientific) was diluted into 300 μl of 50 mM MES buffer (pH 5.2) and centrifuged at 17,000g for 15 min at 23°C; the pellet was suspended in 200 μl of MES buffer and mixed with 20 μl of Ab-LPS; 10 mg of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDAC) (Sigma-Aldrich) was dissolved in 1 ml of MES buffer; and 20 μl of the EDAC solution was added to the bead suspension and incubated for 30 min at 23°C. Free antibodies and EDAC were removed by centrifugation, and the pellet was suspended into 10 mM tris-HCl buffer (pH 8.0). A total of 300 μl of Leptospira cells was centrifuged at 1000g for 10 min and suspended into 500 μl of motility medium. Five microliters of the cell suspension was mixed with 15 μl of the anti-LPS–coated bead, and the mixture containing the cells and beads was infused into a flow chamber and observed using the dark-field microscope. Videos were recorded as described in the Motility assay section and analyzed by using ImageJ software (National Institutes of Health).
Supplementary Material
Acknowledgments
We thank T. Masuzawa for providing L. biflexa strain Patoc I and E. Isogai for providing anti-LPS rabbit antibody. We also thank J. Xu, K. Namba, A. Taoka, D. Nakane, and S. Kudo for the technical support and the helpful comments. Funding: This work was supported by the Grant-in-Aid for Scientific Research on Innovative Areas “Harmonized Supramolecular Motility Machinery and Its Diversity” to S.N. (15H01307). Author contributions: H.T., N.K., and S.N. designed the research. H.T., N.K., K.K., A.K., Y.S., K.T., and S.N. performed the research. H.T., N.K., K.K., A.K., Y.S., K.T., and S.N. analyzed the data. S.N., N.K., and A.K. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar7975/DC1
fig. S1. Swimming motility of Leptospira.
fig. S2. Example kymographs of Leptospira cells crawling without slip.
fig. S3. WT and “unbent” mutant (ΔfcpA) strains of Leptospira.
fig. S4. Example kymographs of ΔfcpA mutant cells crawling without slip.
fig. S5. Movement of a microbead attached to the cell surface.
fig. S6. Movement of a microbead attached to the cell surface.
fig. S7. Kymographs of Leptospira cells labeled with Ab-LPS–coated beads or noncoated beads.
fig. S8. Relationship between swimming speed and bead movement.
fig. S9. Crawling motility of the pathogenic Leptospira.
movie S1. A L. biflexa cell swimming in a 10% Ficoll solution.
movie S2. WT L. biflexa cells crawling on a glass surface.
movie S3. Effect of an anti-LPS antibody on crawling.
movie S4. ΔfcpA mutant cells crawling on a glass surface.
movie S5. Effect of CCCP on Leptospira crawling.
movie S6. Fluorescent observation of the Leptospira outer membrane using Cy3-NHS.
movie S7. Movement of a small bead aggregate on the Leptospira cell body.
movie S8. Movement of a single 200-nm bead on the Leptospira cell body.
movie S9. Movement of a large bead aggregate on the Leptospira cell body.
REFERENCES AND NOTES
- 1.Berg H. C., The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Sowa Y., Berry R. M., Bacterial flagellar motor. Q. Rev. Biophys. 41, 103–132 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Wall D., Kaiser D., Type IV pili and cell motility. Mol. Microbiol. 32, 1–10 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Miyata M., Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64, 519–537 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Faure L. M., Fiche J.-B., Espinosa L., Ducret A., Anantharaman V., Luciano J., Lhospice S., Islam S. T., Tréguier J., Sotes M., Kuru E., Van Nieuwenhze M. S., Brun Y. V., Théodoly O., Aravind L., Nollmann M., Mignot T., The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539, 530–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein S. F., Charon N. W., Multiple-exposure photographic analysis of a motile spirochete. Proc. Natl. Acad. Sci. U.S.A. 87, 4895–4899 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura S., Leshansky A., Magariyama Y., Namba K., Kudo S., Direct measurement of helical cell motion of the spirochete Leptospira. Biophys. J. 106, 47–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox P. J., Twigg G. I., Leptospiral motility. Nature 250, 260–261 (1974). [DOI] [PubMed] [Google Scholar]
- 9.Charon N. W., Cockburn A., Li C., Liu J., Miller K. A., Miller M. R., Motaleb Md. A., Wolgemuth C. W., The unique paradigm of spirochete motility and chemotaxis. Annu. Rev. Microbiol. 66, 349–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarrell K. F., McBride M. J., The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6, 466–476 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Takabe K., Tahara H., Islam Md. S., Affroze S., Kudo S., Nakamura S., Viscosity-dependent variations in the cell shape and swimming manner of Leptospira. Microbiology 163, 153–160 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Berg H. C., Bromley D. B., Charon N. W., Leptospiral motility. Symp. Soc. Gen. Microbiol. 28, 285–294 (1978). [Google Scholar]
- 13.Magariyama Y., Sugiyama S., Muramoto K., Kawagishi I., Imae Y., Kudo S., Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys. J. 69, 2154–2162 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magariyama Y., Sugiyama S., Kudo S., Bacterial swimming speed and rotation rate of bundled flagella. FEMS Microbiol. Lett. 199, 125–129 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S., Adachi Y., Goto T., Magariyama Y., Improvement in motion efficiency of the spirochete Brachyspira pilosicoli in viscous environments. Biophys. J. 90, 3019–3026 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hori K., Matsumoto S., Bacterial adhesion: From mechanism to control. Biochem. Eng. J. 48, 424–434 (2010). [Google Scholar]
- 17.Lu Q., Wang J., Faghihnejad A., Zeng H., Liu Y., Understanding the molecular interactions of lipopolysaccharides during E. coli initial adhesion with a surface forces apparatus. Soft Matter 7, 9366–9379 (2011). [Google Scholar]
- 18.Nakane D., Sato K., Wada H., McBride M. J., Nakayama K., Helical flow of surface protein required for bacterial gliding motility. Proc. Natl. Acad. Sci. U.S.A. 110, 11145–11150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vieira M. L., Fernandes L. G., Domingos R. F., Oliveira R., Siqueira G. H., Souza N. M., Teixeira A. R. F., Atzingen M. V., Nascimento A. L. T. O., Leptospiral extracellular matrix adhesins as mediators of pathogen–host interactions. FEMS Microbiol. Lett. 352, 129–139 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Picardeau M., Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 15, 297–307 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Wunder E. A. Jr., Figueira C. P., Benaroudj N., Hu B., Tong B. A., Trajtenberg F., Liu J., Reis M. G., Charon N. W., Buschiazzo A., Picardeau M., Ko A. I., A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol. Microbiol. 101, 457–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki Y., Kawamoto A., Tahara H., Kasuga K., Sato R., Ohnishi M., Nakamura S., Koizumi N., Leptospiral flagellar sheath protein FcpA interacts with FlaA2 and FlaB1 in Leptospira biflexa. PLOS ONE 13, e0194923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Islam Md. S., Morimoto Y. V., Kudo S., Nakamura S., H+ and Na+ are involved in flagellar rotation of the spirochete Leptospira. Biochem. Biophys. Res. Commun. 466, 196–200 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Raddi G., Morado D. R., Yan J., Haake D. A., Yang X. F., Liu J., Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J. Bacteriol. 194, 1299–1306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takabe K., Kawamoto A., Tahara H, Kudo S., Nakamura S., Implications of coordinated cell-body rotations for Leptospira motility. Biochem. Biophys. Res. Commun. 491, 1040–1046 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Yang J., Huber G., Wolgemuth C. W., Forces and torques on rotating spirochete flagella. Phys. Rev. Lett. 107, 268101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charon N. W., Lawrence C. W., O’Brien S., Movement of antibody-coated latex beads attached to the spirochete Leptospira interrogans. Proc. Natl. Acad. Sci. U.S.A. 78, 7166–7170 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slamti L., de Pedro M. A., Guichet E., Picardeau M., Deciphering morphological determinants of the helix-shaped Leptospira. J. Bacteriol. 193, 6266–6275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watari N., Larson R. G., The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys. J. 98, 12–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler B., Pathogenesis of leptospirosis: Cellular and molecular aspects. Vet. Microbiol. 172, 353–358 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Miyahara S., Saito M., Kanemaru T., Villanueva S. Y. A. M., Gloriani N. G., Yoshida S., Destruction of the hepatocyte junction by intercellular invasion of Leptospira causes jaundice in a hamster model of Weil’s disease. Int. J. Exp. Pathol. 95, 271–281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar7975/DC1
fig. S1. Swimming motility of Leptospira.
fig. S2. Example kymographs of Leptospira cells crawling without slip.
fig. S3. WT and “unbent” mutant (ΔfcpA) strains of Leptospira.
fig. S4. Example kymographs of ΔfcpA mutant cells crawling without slip.
fig. S5. Movement of a microbead attached to the cell surface.
fig. S6. Movement of a microbead attached to the cell surface.
fig. S7. Kymographs of Leptospira cells labeled with Ab-LPS–coated beads or noncoated beads.
fig. S8. Relationship between swimming speed and bead movement.
fig. S9. Crawling motility of the pathogenic Leptospira.
movie S1. A L. biflexa cell swimming in a 10% Ficoll solution.
movie S2. WT L. biflexa cells crawling on a glass surface.
movie S3. Effect of an anti-LPS antibody on crawling.
movie S4. ΔfcpA mutant cells crawling on a glass surface.
movie S5. Effect of CCCP on Leptospira crawling.
movie S6. Fluorescent observation of the Leptospira outer membrane using Cy3-NHS.
movie S7. Movement of a small bead aggregate on the Leptospira cell body.
movie S8. Movement of a single 200-nm bead on the Leptospira cell body.
movie S9. Movement of a large bead aggregate on the Leptospira cell body.


