Abstract
Molecular chaperones are involved in the protection of cells against protein damage through their ability to hold, disaggregate, and refold damaged proteins or their ability to facilitate degradation of damaged proteins. Little is known about how these processes are spatially coordinated in cells. Using a heat-sensitive nuclear model protein luciferase fused to the traceable, heat-stable enhanced green fluorescent protein (N-luc-EGFP), we now show that heat inactivation and insolubilization of luciferase were associated with accumulation of N-luc-EGFP at multiple foci throughout the nucleus. Coexpression of Hsp70, one of the major mammalian chaperones, reduced the formation of these small foci during heat shock. Instead, the heat-unfolded N-luc-EGFP accumulated in large, insoluble foci. Immunofluorescence analysis revealed that these foci colocalized with the nucleoli. Time-lapse analysis demonstrated that protein translocation to the nucleolus, in contrast to the accumulation at small foci, was fully reversible upon return to the normal growth temperature. This reversibility was associated with an increase in the level of active and soluble luciferase. Expression of a carboxyl-terminal deletion mutant of Hsp70(1–543), which lacked chaperone activity, had no effect on the localization of N-luc-EGFP, which suggests that the Hsp70 chaperone activity is required for the translocation events. Our data show that Hsp70 not only is involved in holding and refolding of heat-unfolded nuclear proteins but also drives them to the nucleolus during stress. This might prevent random aggregation of thermolabile proteins within the nucleus, thereby allowing their refolding at the permissive conditions and preventing indirect damage to other nuclear components.
Under conditions of normal cell growth, quality control over protein structure and function is maintained by the activities of molecular chaperones and proteases. Molecular chaperones prevent aggregation and enable refolding of unfolded proteins whereas proteases eliminate irreversibly damaged proteins (1, 2). Under stress conditions, such as heat shock, the amount of unfolded proteins increases dramatically and the balance between chaperones, proteases, and unfolded proteins is disturbed, which can lead to the formation of protein aggregates. In response to this imbalance, cells rapidly induce synthesis of heat shock proteins, which function as molecular chaperones in the repair of the existing protein damage and provide a temporal resistance against the effects of subsequent stress (3, 4).
One of the major heat shock proteins involved in protection of mammalian cells against stress-induced protein damage is Hsp70. Hsp70 prevents aggregation and assists in protein refolding by repeated binding and release of exposed hydrophobic stretches of amino acids in unfolded proteins (5). These activities require cycles of ATP binding and hydrolysis, which are catalyzed by the cochaperone Hsp40 (6, 7). The biochemical activities of Hsp70 and its regulation by cochaperones have been studied extensively in vitro with purified proteins (6, 8–12) and in “black-box” cell models that looked at the solubility and activity of reporter proteins in lysed cells (13–17). Despite these studies, only little is known about where exactly in the cell unfolded proteins are processed by Hsp70. Here, we investigated the fate of nuclear proteins in heat-stressed cells. The biochemically well characterized, heat-sensitive protein model luciferase, adapted for expression in the nucleus of mammalian cells (18), was fused to the traceable protein EGFP (enhanced green fluorescent protein; N-luc-EGFP) to follow its fate during and after heat stress. We found that heat shock results in accumulation of heat-unfolded luciferase at multiple foci throughout the nucleus, which is reduced by coexpression of Hsp70. Interestingly, in the presence of Hsp70, the unfolded luciferase translocates to form large, insoluble foci during heat stress. Upon return to the physiological growth temperature, N-luc-EGFP relocates from the large foci and is reactivated in an Hsp70-dependent manner. As these large foci colocalized with the nucleoli and Hsp70, our results indicate that the translocation of unfolded proteins to this subnuclear compartment may be an additional step in the Hsp70-mediated protection of stressed cells.
Materials and Methods
Plasmids and Constructs.
pRSVnlsLL/V encodes firefly luciferase fused to a nuclear localization sequence (18). pN-luc-EGFP encodes nuclear luciferase fused to EGFP under control of a cytomegalovirus promoter. pN-luc-EGFP was constructed by using pRSVnlsLL/V as a template for PCR amplification of nuclear luciferase with the forward primer 5′-ATT AGG AAG GCA ACA GAC GGG TCT GAC ATG GAT TGG ACG-3′ and the reverse primer 5′-ATG GCG GTA CCG TCG ACT TCC TAC TTT GGA CTT TCC GCC CTT-3′. The reverse primer destroyed the stop codon and introduced a new SalI site (Eurosequence, Groningen, The Netherlands). The PCR product was digested with HindIII and SalI and ligated into pEGFP-N2 (CLONTECH). Construction of pCMV70 has been described (13). pCDNAHsp70(1–543) was created by ligation of an HindIII-EcoRI fragment from pMS(1–543) (gift from R. I. Morimoto, Northwestern University, Evanston, IL) (6) into pCDNA (Invitrogen).
Cell Culture and Transfections.
O23 hamster fibroblasts were cultured in DMEM supplemented with 10% FBS (Life Technologies, Gaithersburg, MD). Transient transfections were performed by using Lipofectamine according to the procedure of the manufacturer (GIBCO/BRL). In cotransfection experiments, the total amount of plasmid DNA was kept constant by the addition of pCDNA-3 (Invitrogen).
Luciferase Activity and Solubility Assays.
Before and during heat shock experiments, cells were grown in medium containing 20 μg/ml cycloheximide and 20 mM 4-morpholinepropanesulfonic acid (Mops; pH 7.0). For luciferase activity measurements, quadruplicate samples were taken and cells were lysed and assayed as described (18). For determination of the solubility of luciferase, cells were trypsinized and lysed in 100 μl of BLUC (25 mM Tris/H3PO4, pH 7.8/10 mM MgCl2/1% Triton X-100/15% glycerol/1 mM EDTA)/1.5 × 106 cells. Supernatant and pellet fractions were separated by centrifugation at 15,000 rpm for 15 min and analyzed by SDS/PAGE and Western blot analysis.
Western Blot Analysis and Immunofluorescence Analysis.
Cells were trypsinized and lysed by the addition of BLUC. Lysates were taken up in SDS/PAGE sample buffer and sonicated before SDS/PAGE–Western blot analysis. Luciferase-EGFP was detected by using a polyclonal antibody to luciferase (Cortex Pharmaceuticals, Irvine, CA). Hsp70 was detected by C92, a mAb specific for the heat-inducible form of Hsp70 (StressGen Biotechnologies, Victoria, Canada). Binding of anti-mouse or anti-rabbit secondary antibodies for Western analysis was made visible by enhanced chemiluminescence (ECL, Amersham Pharmacia). Indirect immunofluorescence was performed as described (18). Nucleolin was detected by using a mAb to antinucleolin (Santa Cruz Biotechnology) and a tetramethylrhodamine B isothiocyanate (TRITC)-conjugated anti-mouse secondary antibody. Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. Images of GFP, TRITC, and DAPI fluorescence were obtained by using a Leica confocal laser-scanning microscope (Leica TCS SP2). For quantification of the fluorescent staining patterns, cells were washed twice with PBS, fixed in 3.7% formaldehyde/PBS, washed three times for 10 min with PBS, and stained with DAPI for staining of nuclear DNA.
Protein Extraction.
At 48 h after transfection, individual unheated or heated cells were followed during a stepwise extraction procedure under the fluorescent microscope. They were washed twice with PBS followed by a 3-min wash with CSK buffer [10 mM Pipes/100 mM NaCl/300 mM sucrose/3 mM MgCl2/1 mM EDTA/protease inhibitor mixture (Amersham Pharmacia)/0.5% Triton X-100]. After extraction, the remaining DNA was stained by Hoechst (3.2 μg/ml) in CSK buffer.
Live Imaging and Time-Lapse Analysis.
A coverslip with transfected cells was transferred to an isolated, temperature-controlled incubator positioned under a reverted microscope and supplied with medium containing cycloheximide (20 μg/ml). The incubator was attached to a water bath to regulate the temperature. The actual temperature of the culture medium was monitored constantly. To prevent cooling by the lens and immersion oil, the temperature of the lens was controlled as well. The pH in the incubator was maintained by the presence of Mops at pH 7.0 during heat shock and by blowing CO2 (5% in air) over the culture medium during recovery. After time lapse analysis, cells were washed twice with PBS and stained with Hoechst (6.4 μg/2 ml PBS) for 5 min. For detection of expression and localization of Hsp70, subsequent immunofluorescent staining was applied under the microscope according to the standard procedure. Images were made by using a fluorescence microscope (Olympus IMT-2) attached to a sensitive, silicon-intensified tube camera (SIT-66; Dage–MTI, Michigan City, IN), connected to a digital signal, remote-control processor (DSP 100; Dage–MTI), and a CUE II image analysis system (Olympus, New Hyde Park, NY).
Results
Nuclear Luciferase-EGFP Fusion Protein Retains the Heat-Sensitive Characteristics of the Nonfused Luciferase.
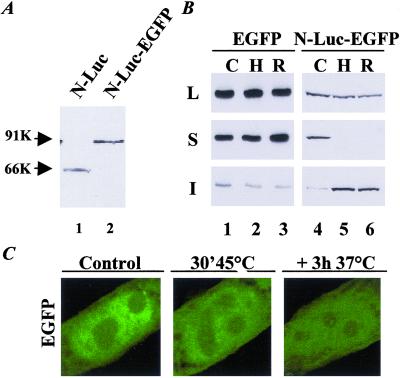
To examine where in the cell unfolded nuclear proteins are processed, we generated a fusion of the well characterized heat-sensitive model protein firefly luciferase, which contained a nuclear localization sequence (18), and the EGFP (N-luc-EGFP). The fusion protein was expressed successfully in hamster lung fibroblast (Fig. 1A). To determine whether luciferase had retained its heat-sensitive properties when fused to EGFP, cells transfected with N-luc-EGFP were lysed for analysis of detergent solubility of the fusion protein before, immediately after, and at 3 h after a heat shock at 45°C for 30 min (Fig. 1B). Immediately after heat shock, the fraction of soluble N-luc-EGFP was decreased below detection (Fig. 1B, lane 5). During the recovery period at 37°C, more than 95% of N-luc-EGFP remained insoluble (Fig. 1B, lane 6). Because heat had a similar effect on the nonfused nuclear luciferase (ref. 18; data not shown), this revealed that fusion to EGFP had not affected the heat-sensitive properties of the nuclear luciferase. The solubility, fluorescence intensity, and intranuclear distribution of nonfused EGFP were unaffected by heat (Fig. 1 B, lanes 1–3, and C), which indicated that the luciferase part of N-luc-EGFP was responsible for the thermosensitive characteristics of the fusion protein.
Figure 1.
Characterization of the nuclear expressed luciferase-EGFP fusion protein. Hamster lung fibroblasts were transfected with plasmids encoding nuclear luciferase (N-luc), EGFP, or N-luc-EGFP. Forty-eight hours after transfection, the cells were lysed before (C), directly after a heat shock of 45°C for 30 min (H), or after 3 h of recovery at 37°C (R). Cycloheximide (20 μg/ml) was added to the cells to prevent novel protein synthesis. (A) Western analysis with an antibody to luciferase of lysates of cells expressing N-luc or N-luc-EGFP. (B) Western analysis with an antibody to EGFP of total cell lysates (L) and detergent-soluble (S) and detergent-insoluble fractions (I) of EGFP- or N-luc-EGFP-transfected cells (lanes 1–3) or with an antibody to luciferase (lanes 4–6). (C) Confocal microscopy pictures of the subnuclear distribution of EGFP before (control), immediately after (30′ 45°C), and 3 h after (+3h 37°C) heat shock.
Heat-Induced Unfolding of Luciferase Changes the Subnuclear Distribution of N-luc-EGFP: Effects of Hsp70 Overexpression.
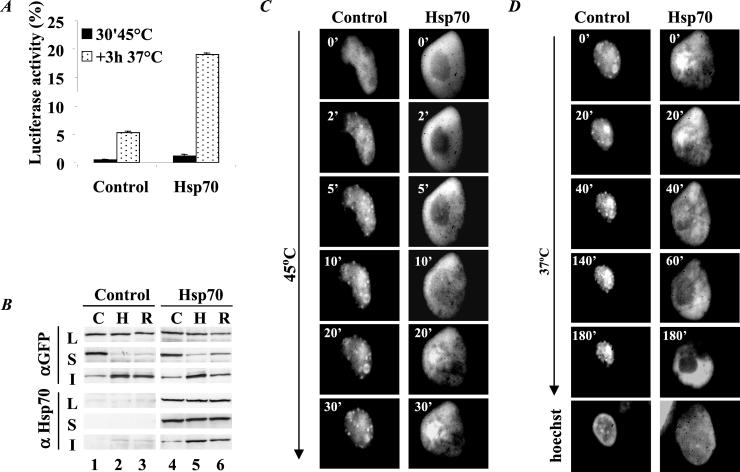
We have demonstrated that Hsp70 functions as a molecular chaperone in the resolubilization and refolding of heat-denatured proteins in the cytosol and nucleus of heat-stressed mammalian cells (13, 16, 19). Transfection-mediated coexpression of Hsp70 similarly enhanced the reactivation and resolubilization of heat-denatured N-luc-EGFP (Fig. 2 A and B, compare lanes 1–3 with 4–6). We investigated whether the reactivation and resolubilization of heat-denatured N-luc-EGFP also was reflected in the spatial distribution of the protein. Using time-lapse analysis, we found that in control cells that showed a diffuse nuclear staining before heat shock, N-luc-EGFP accumulated at multiple small foci during heat shock, which were not reversible upon recovery at 37°C up to 3 h (Fig. 2 C and D, control).
Figure 2.
Effect of Hsp70 on the activity, solubility, and distribution of N-luc-EFGP. O23 cells were transfected to express either N-luc-EGFP alone (Control) or in combination with Hsp70 (Hsp70). Before heat shock (C), after a heat shock at 45°C for 30 min (H), and after recovery at 37°C for 3 h (R), cells were processed and analyzed for luciferase activity (A) or detergent solubility (B). Cycloheximide (20 μg/ml) was present in the culture medium before, during, and after heat shock to prevent novel protein synthesis. (A) Luciferase activity in cell lysates expressed as a percentage of the activity before heat shock. Data represent the average of two independent experiments. Error bars indicate SD. (B) SDS/PAGE–Western blot analysis of total cell lysates (L) and detergent-soluble (S) and detergent-insoluble fractions (I) with an antibody to luc-EGFP (αGFP) and Hsp70 (αHsp70). (C and D) Time-lapse analysis of staining profiles during and after heat shock of N-luc-EGFP-transfected cells. At 48 h after transfection, the cells were transferred to a temperature- and pH-controlled incubator and placed under a fluorescent microscope. The cells were heated for 30 min at 45°C (C) and subsequently cooled down to 37°C for recovery (D). Pictures were taken at the indicated time points, using an Olympus Cue II image analysis system. The apparent nuclear shrinkage of the control cells with time is a result of rounding up and cell movement of the cells during the time-lapse period.
In the presence of Hsp70, N-luc-EGFP accumulated into large foci during heat shock (Fig. 2C, Hsp70). During recovery, the fluorescent protein relocated back to a diffuse staining pattern (Fig. 2D, Hsp70). Reappearance of the diffuse distribution coincided with the reappearance of soluble and active protein in the same cell population (Fig. 2 A and B). These data suggest that in the presence of Hsp70, accumulation of heat-unfolded proteins in small foci is reduced. Instead, these proteins accumulate in large inclusions, from which they can be recovered at physiological conditions.
Multiple Foci and Large Foci Are Detergent-Insoluble.
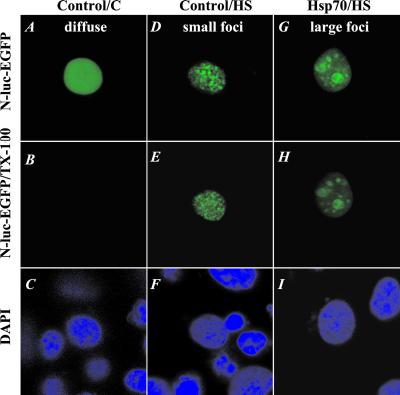
Having established a difference in the localization patterns of N-luc-EGFP and reversibility of these patterns in control vs. Hsp70-expressing cells, we wondered what could be the basis for the difference in reversibility. One explanation could be that there are differences in the biochemical constitution of the small and large foci, for example, that N-luc-EGFP is more soluble in the large foci. To investigate this, the fluorescent staining patterns of individual nuclei were followed during a stepwise protein-extraction procedure in situ (Fig. 3). Detergent extraction of diffusely stained nuclei of unheated cells removed the fluorescent signal to below detection, indicating that most of the N-luc-EGFP was detergent-soluble in these nuclei (Fig. 3 A–C). The same was observed for Hsp70-expressing cells that showed a diffuse staining pattern after recovery from heat shock (data not shown). When present in multiple foci, the N-luc-EGFP staining pattern remained unchanged by detergent extraction (Fig. 3 D–F). This revealed that the small foci were insoluble, which was consistent with the analysis of the solubility of N-luc-EGFP in cell lysates (Fig. 2B). Interestingly, in the case of the large foci, the major form in cells expressing Hsp70, the Triton X-100-extraction step did not alter the appearance of the large foci (Fig. 3 G and H). Also, subsequent extraction with DNase I digestion and high salt treatment did not resolubilize the N-luc-EGFP from either small or large foci (data not shown). These data therefore suggest that the large foci represented a detergent-insoluble state, similar to the small foci.
Figure 3.
Protein extractability of cells with different staining patterns. Images of distribution patterns of heat-unfolded N-luc-EGFP in living cells (A, D, and G), after extraction with CSK buffer containing 0.5% Triton X-100 (B, E, and H), and after subsequent staining with DAPI (C, F, and I). (A–C) Unheated control cell: diffuse staining pattern. (D–F) Cells heated at 45°C for 30 min: control cell with multiple foci. (G–I) Hsp70-expressing cell with large foci.
Small Foci Are Nonnucleolar and Large Foci Colocalize with the Nucleoli.
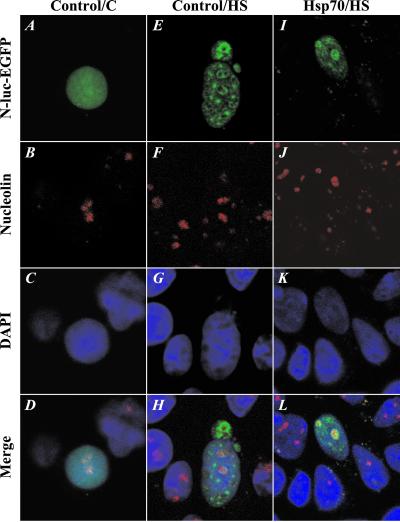
Because the small and large foci were similar with respect to their detergent and salt extractability, we next asked whether they differed with respect to underlying subnuclear compartments or structures. Light and electron microscopic analysis of immunolabeled cells showed that in heated cells with multiple foci, the staining was concentrated at multiple sites throughout the nucleus, which did not overlap with the nucleoli. In contrast, in cells in which the protein was concentrated in large foci after heat shock, the staining was highly enriched in the nucleoli (data not shown). To confirm that the large foci of heat-denatured luciferase indeed colocalized with the nucleoli, we stained cells transfected with N-luc-EGFP with an antibody to nucleolin, a protein known to localize in the nucleolus (20). Confocal microscopy revealed that in unheated control cells, N-luc-EGFP did not colocalize with nucleolin (Fig. 4 A–D). Similarly, when control cells were heated, most of the N-luc-EGFP aggregated in small foci, which did not overlap with the antinucleolin staining (Fig. 4 E–H). In heated cells that overexpressed Hsp70, the large foci of N-luc-EGFP did colocalize with the antinucleolin staining. In fact, we found N-luc-EGFP to locate at the periphery of the nucleolin-stained area (Fig. 4 I–L). From these data, we conclude that the large foci and multiple foci, although they were both detergent- and high salt-insoluble, are distinct in their subnuclear localization. The smaller foci are localized in the nonnucleolar region of the nucleus whereas the large foci colocalize with the nucleoli.
Figure 4.
Large foci colocalize with the nucleoli. Cells were fixed and stained with an antibody to nucleolin. (A–D) Unheated control cell. (E–H) Control cell heated for 30 min at 45°C. (I–L) Hsp70-expressing cell heated for 30 min at 45°C.
Translocation of N-luc-EGFP to and from the Nucleolus Requires Hsp70 Chaperone Activity.
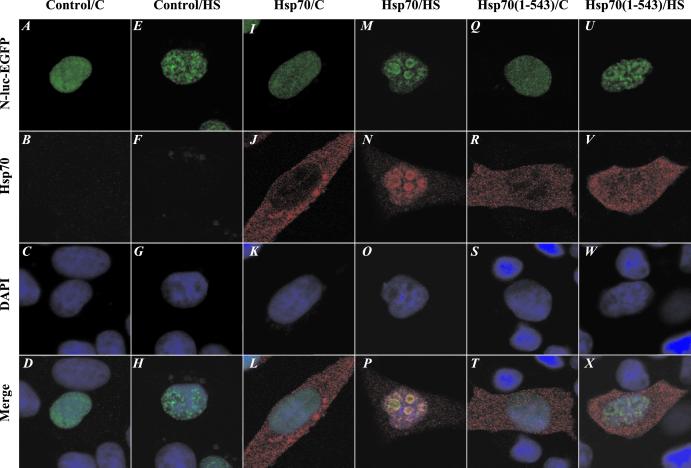
To assess whether Hsp70-dependent translocation of unfolded N-luc-EGFP to the nucleoli required Hsp70 chaperone activity, we quantified the localization pattern of N-luc-EGFP in nontransfected cells and cells transfected to express Hsp70 or an inactive mutant of Hsp70 [Hsp70(1–543)] that lacks 95 aa of its C-terminal domain that is involved in the regulation of substrate binding (6). In nontransfected cells, the number of nuclei with a diffuse distribution pattern of N-luc-EGFP (Fig. 5A) dropped from 96% under normal conditions to 6% after heat shock. In the majority of cells, N-luc-EGFP instead localized into multiple small, nonnucleolar foci (Fig. 5E). This redistribution pattern remained unchanged after 3 h of recovery at 37°C, which was in line with the observation that in this cell population only little N-luc-EGFP was reactivated and resolubilized within the same time frame (Table 1 and Fig. 2). Similarly, in Hsp70-transfected cells, the fraction of cells with a diffuse N-luc-EGFP distribution was reduced to below 5% by exposure to 45°C for 30 min. Unlike control cells, however, the major distribution pattern was a nucleolar distribution rather than localization into small, nonnucleolar foci (Table 1 and Fig. 5 I and M). Return of the cells to normal growth temperature resulted in an increase in the number of cells with a diffuse staining pattern to 36% at 3 h after heat shock, which coincided with enhanced N-luc-EFGP reactivation and resolubilization (Table 1 and Fig. 2). In cells transfected to express the carboxyl-terminal deletion mutant of Hsp70 [Hsp70(1–543)], of which the ability to interact with unfolded substrates is reduced and which lacks chaperone activity (6) (Table 1), the distribution patterns of N-luc-EGFP after heat shock and recovery were similar to the distribution patterns in control transfected cells (Fig. 5 Q and U), suggesting that Hsp70 chaperone activity is required for translocation of heat-denatured N-luc-EGFP to the nucleolus (Table 1).
Figure 5.
Nucleolar accumulation of N-luc-EGFP requires the C-terminal regulatory domain of Hsp70. N-luc-EGFP-transfected cells were treated, fixed, and stained with an antibody to Hsp70. (A–D) Unheated control cell. (E–H) Control cell heated for 30 min at 45°C. (I–L) Unheated, Hsp70-expressing cell. (M–P) Hsp70-expressing cell heated for 30 min at 45°C. (Q–T) Unheated cell expressing the chaperone-deficient Hsp70ΔC. (U–X) Hsp70ΔC-expressing cell heated for 30 min at 45°C.
Table 1.
Quantification of fluorescent staining patterns of N-luc-EGFP: Effects of Hsp70 and Hsp70(1–543) expression
| Cells | Treatment† | Immunofluorescent staining pattern, %
|
Luciferase activity‡ | |||||
|---|---|---|---|---|---|---|---|---|
| Diffuse | Only small foci | Small foci > nucleolar | Small foci-nucleolar | Small foci < nucleolar | Only nucleolar | |||
| Control | Unheated | 96 | 4 | 100% | ||||
| 30 min at 45°C | 8 | 35 | 49 | 9 | 0.7% ± 0.1 | |||
| +3 h at 37°C | 6 | 58 | 30 | 6 | 11% ± 0.7 | |||
| Hsp70* | Unheated | 98 | 2 | 100% | ||||
| 30 min at 45°C | 32 | 35 | 25 | 5 | 2% ± 0.3 | |||
| +3 h at 37°C | 38 | 4 | 18 | 20 | 4 | 20 | 25% ± 1 | |
| Hsp70(1–543)* | Unheated | 94 | 6 | 100% | ||||
| 30 min at 45°C | 33 | 49 | 16 | 0.7% ± 0.1 | ||||
| +3 h at 37°C | 6 | 25 | 47 | 17 | 5 | 9% ± 1.1 | ||
Only cells in which Hsp70 expression was detected by immunofluorescence were judged for these conditions.
For each condition of each cell population, 45–55 cells were judged by taking confocal microscopy images.
Luciferase activity was measured in the parallel experiment as described in the Materials and Methods and in Fig. 2 (mean ± SD).
Immunofluorescence analysis of the Hsp70 and Hsp70(1–543)-transfected cell population showed that both proteins were expressed predominantly in the cytosol under normal growth conditions (Fig. 5 I–L and Q–T). After heat shock, Hsp70 colocalized with N-luc-EGFP in the nucleoli (Fig. 5 M–P), whereas the localization of the inactive Hsp70(1–543) did not visibly change localization in response to heat shock, which further supported a role for Hsp70 chaperone activity in translocation heat-denatured N-luc-EGFP to the nucleoli. Altogether, these data indicate that an interaction between the unfolded substrate and Hsp70 is required for translocation of the substrate to the nucleoli.
Discussion
Using a model protein to monitor the localization, activity, and solubility of the same heat-sensitive nuclear protein during heat stress, this study shows that heat causes dynamic changes in the distribution of heat-denatured nuclear proteins, which are strongly influenced by the presence of the Hsp70 chaperone. Heat unfolding of nuclear luciferase fused to EGFP was associated with accumulation of the fusion protein at multiple foci throughout the nucleus. Overexpression of Hsp70, which enhanced resolubilization and refolding of the protein, diminished this aggregation throughout the nucleus in favor of nucleolar accumulation from which the heat-denatured proteins relocated upon return to the permissive conditions. These effects were not observed in cells expressing a chaperone defective Hsp70(1–543). Intriguingly, when the nuclear isoform of the Hsp70-inhibitory protein Bag1 was coexpressed with Hsp70, the N-luc-EGFP remained nucleolar (data not shown). This is consistent with the finding that Bag-1 inhibits the Hsp70-dependent refolding of heat-denatured luciferase in vitro (12, 21) and in vivo (15, 16). Our data therefore indicate that a functional Hsp70 chaperone is required for translocation of the substrate to the nucleoli and that this is associated with the ability of Hsp70 to refold the unfolded substrate.
Our observations are in line with what has been reported previously about subcellular distribution of Hsp70. Under normal growth conditions, Hsp70 is present mainly in the cytosol. During heat stress, Hsp70 translocates from the cytosol into the nucleus and the nucleolus, from which it retracts during continued recovery (22, 23). This movement of Hsp70 in and out the nucleolus has been proposed to be associated with repair of heat-induced nucleolar damage (24). Indeed, several observations suggest that the nucleolus is especially sensitive to heat. Upon heat shock, the nucleoli become swollen and, instead of the small granules present in normal nucleoli, large electron-dense structures become visible. Nucleolar processes, such as ribosomal RNA synthesis and assembly of ribosomal precursor particles, are inhibited. Both the morphological changes and the inhibition of nucleolar functions can be restored upon return to normal growth temperatures (23). Overexpression of Hsp70 in COS cells accelerates the restoration of the normal nucleolar morphology, rRNA transcription, and ribosome assembly (24). Together with the observation that ATP is essential for exit of Hsp70 from the nucleolus, which also is required for Hsp70-mediated refolding of unfolded proteins, this has supported the hypothesis that movement of Hsp70 in and out of the nucleolus is associated with the repair of heat-induced nucleolar protein damage. Although our observations are in agreement with these existing data, they point to a novel interpretation. We propose that the morphological changes of the nucleoli during heat shock may, at least in part, be a result of translocation of unfolded nonnucleolar proteins to the nucleoli. Recently, a similar translocation to the nucleolus under heat-stress conditions has been observed for the protein kinase CK2 involved in cell proliferation and development, which also relocates from the nucleolus after a return to normal growth conditions (25). As we present here that overexpression of Hsp70 stimulates translocation of unfolded proteins to the nucleolus, Hsp70 may play an active role in this process. In addition, we found that in the presence of Hsp70, N-luc-EGFP relocated from the nucleoli during recovery, which was associated with resolubilization and reactivation of the unfolded proteins. This indicates that the translocation of Hsp70 to and from the nucleolus may be associated with storage of nonnucleolar unfolded proteins and, as was reported previously, with subsequent repair. Consistent with our observations suggesting that denatured luciferase may be stored in the nucleolus for later processing, a variety of other proteins have been reported to be sequestered actively in the nucleolus (26).
Storage of unfolded proteins in the nucleolus during ongoing stress could have many advantages. Clearly, concentration of unfolded proteins at one locus in the nucleus will reduce damage because it will prevent their random aggregation with other nuclear proteins and macromolecular structures. Storage of unfolded proteins in the nucleolus will interfere with the assembly of ribosomes essential for protein synthesis. Indeed, inhibition of ribosome assembly has been reported as one of the consequences of exposure stress (24). However, during ongoing stress, this may be beneficial because it will inhibit protein synthesis at its earliest step and, hence, limit the number of thermolabile nascent chains that could damage the cell further.
Changes in the subcellular distribution of damaged proteins are not restricted to the nuclear compartment. In the cytosol, misfolded or heat-unfolded proteins are transported to the proximity of centrosome, which has been associated with degradation of these proteins by the proteasome (27–30). The translocation of heat-unfolded proteins to the nucleolus described here similarly suggests an additional step in the processing of damaged proteins. Therefore, changes in the spatial distribution and concentration at specific sites within the cell of unfolded or misfolded proteins may be a common step in the control over the quality of protein structure and function in cells.
Acknowledgments
We thank Willy Lemstra for assistance with quantification of the distribution patterns; Geert Mesander of the Flow Cytometry Unit, University Hospital Groningen, for assistance with the quantimet 600 software; Jan Nieborg of the instrument mechanical shop for technical contributions to the live imaging set-up; Guus de Wit and Geert Kors for preparing the electron microscopy specimen; and Bert Hellinga for photography. This study was supported by a grant from the Dutch Cancer Society (NKB Grant 95-1082) to H.H.K.
Abbreviations
- EGFP
enhanced green fluorescent protein
- DAPI
4′,6-diamidino-2phenylindole
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 2.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 3.Morimoto R I. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 4.Parsell D A, Lindquist S. In: The Biology of Heat Proteins and Molecular Chaperones. Morimoto R I, Tissieres A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 457–494. [Google Scholar]
- 5.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 6.Freeman B C, Myers M P, Schumacher R, Morimoto R I. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman B C, Morimoto R I. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 9.Hohfeld J, Minami Y, Hartl F U. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 10.Hohfeld J, Jentsch S. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minami Y, Hohfeld J, Ohtsuka K, Hartl F U. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 12.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aimesempe C, Xie Z H, Morimoto R I, Reed J C. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michels A A, Kanon B, Konings A W T, Ohtsuka K, Bensaude O, Kampinga H H. J Biol Chem. 1997;272:33283–33289. doi: 10.1074/jbc.272.52.33283. [DOI] [PubMed] [Google Scholar]
- 14.Michels A A, Kanon B, Bensaude O, Kampinga H H. J Biol Chem. 1999;274:36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- 15.Nollen E A, Kabakov A E, Brunsting J F, Kanon B, Hohfeld J, Kampinga H H. J Biol Chem. 2001;276:4677–4682. doi: 10.1074/jbc.M009745200. [DOI] [PubMed] [Google Scholar]
- 16.Nollen E A, Brunsting J F, Song J, Kampinga H H, Morimoto R I. Mol Cell Biol. 2000;20:1083–1088. doi: 10.1128/mcb.20.3.1083-1088.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroder H, Langer T, Hartl F U, Bukau B. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michels A A, Nguyen V T, Konings A W, Kampinga H H, Bensaude O. Eur J Biochem. 1995;234:382–389. doi: 10.1111/j.1432-1033.1995.382_b.x. [DOI] [PubMed] [Google Scholar]
- 19.Nollen E A, Brunsting J F, Roelofsen H, Weber L A, Kampinga H H. Mol Cell Biol. 1999;19:2069–2079. doi: 10.1128/mcb.19.3.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginisty H, Sicard H, Roger B, Bouvet P. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 21.Zeiner M, Gebauer M, Gehring U. EMBO J. 1997;16:5483–5490. doi: 10.1093/emboj/16.18.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch W J, Feramisco J R. J Biol Chem. 1984;259:4501–4513. [PubMed] [Google Scholar]
- 23.Welch W J, Suhan J P. J Cell Biol. 1986;103:2035–2052. doi: 10.1083/jcb.103.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelham H R B. EMBO J. 1984;3:3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerber D A, Souquere-Besse S, Puvion F, Dubois M F, Bensaude O, Cochet C. J Biol Chem. 2000;275:23919–23926. doi: 10.1074/jbc.M002697200. [DOI] [PubMed] [Google Scholar]
- 26.Olson M O, Dundr M, Szebeni A. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Mata R, Bebok Z, Sorscher E J, Sztul E S. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston J A, Ward C L, Kopito R R. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidair C A, Huang R N, Doxsey S J. Int J Hyperthermia. 1996;12:681–695. doi: 10.3109/02656739609027676. [DOI] [PubMed] [Google Scholar]
- 30.Wigley W C, Fabunmi R P, Lee M G, Marino C R, Muallem S, DeMartino G N, Thomas P J. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]