Abstract
Chromatid catenation is actively monitored in human cells, with progression from G2 to mitosis being inhibited when chromatids are insufficiently decatenated. Mitotic delay was quantified in normal and checkpoint-deficient human cells during treatment with ICRF-193, a topoisomerase II catalytic inhibitor that prevents chromatid decatenation without producing topoisomerase-associated DNA strand breaks. Ataxia telangiectasia (A-T) cells, defective in DNA damage checkpoints, showed normal mitotic delay when treated with ICRF-193. The mitotic delay in response to ICRF-193 was ablated in human fibroblasts expressing an ataxia telangiectasia mutated- and rad3-related (ATR) kinase-inactive ATR allele (ATRki). BRCA1-mutant HCC1937 cells also displayed a defect in ICRF-193-induced mitotic delay, which was corrected by expression of wild-type BRCA1. Phosphorylations of hCds1 or Chk1 and inhibition of Cdk1 kinase activity, which are elements of checkpoints associated with DNA damage or replication, did not occur during ICRF-193-induced mitotic delay. Over-expression of cyclin B1 containing a dominant nuclear localization signal, and inhibition of Crm1-mediated nuclear export, reversed ICRF-193-induced mitotic delay. In combination, these results imply that ATR and BRCA1 enforce the decatenation G2 checkpoint, which may act to exclude cyclin B1/Cdk1 complexes from the nucleus. Moreover, induction of ATRki produced a 10-fold increase in chromosomal aberrations, further emphasizing the vital role for ATR in genetic stability.
Human somatic cells replicate and segregate their genome with remarkable precision. This regulation depends in part on the activity of checkpoint surveillance systems that monitor the cell division cycle and impose delays when conditions are inopportune or inappropriate (1–3). Attenuation or ablation of checkpoint response by means of gene mutation distinguishes cell cycle delays that are the result of checkpoint systems from passive metabolic disturbances (3).
Cells delay mitosis under conditions of cellular stress such as incompletely replicated DNA and DNA damage (4). Mutations in RCC1 and ATM reduce mitotic delay in cells with unreplicated and damaged DNA, respectively, and confirm the checkpoint nature of the cycle delay responses (5–7). The topoisomerase II inhibitor, ICRF-193, which does not damage DNA, also induces mitotic delay when added to cells in G2 (8, 9). It was proposed that cells monitor the status of intertwined daughter chromatids after DNA replication and actively delay mitosis until chromatids are sufficiently decatenated by topoisomerase II (8). Sustained chromatid catenation as induced by ICRF-193, and other topoisomerase II inhibitors may stress chromosomes and contribute to the development of aneuploidy or polyploidy through aberrant mitosis.
The onset of mitosis is controlled by the activity and location of the cyclin B1/Cdk1 complex (10). During interphase, the protein kinases Wee1 and Myt1 maintain the complex in an inactive state by phosphorylating Thr-14 and Tyr-15 on Cdk1 (10). Cdk1 becomes activated when Thr-161 is phosphorylated by the Cdk7/cyclin H/mat1 complex and the Cdc25C phosphatase removes the inhibitory phosphates (10, 11). The G2 checkpoint response to DNA damage requires the activity of protein kinases that inhibit cyclin B1/Cdk1 kinase activity. Cells from patients with the familial cancer syndrome ataxia telangiectasia (A-T) display significantly reduced cell cycle delays in G1, S, and G2 after DNA damage (7, 12–14). The gene that is mutated in A-T (ATM) is a member of the phosphatidylinositol 3-kinase (PI3-kinase) superfamily, which also includes FRAP, DNA-PK, and (ATM- and rad3-related) ATR. Protein kinase activity associated with these gene products seems to mediate some of their biological activities, and one arm of the DNA damage G2 checkpoint relies on ATM-dependent inhibition of Cdk1 kinase activity (14). Overexpression of a kinase-inactive ATR (ATRki) allele has been associated with hypersensitivity to agents that induce DNA damage and replication blocks, and cell lines expressing ATRki lack a functional DNA damage G2 checkpoint (15, 16). Furthermore, ATR kinase activity is sensitive to inhibition by caffeine, which interferes with the G2 DNA damage checkpoint (17). Together, these results suggest that ATR also performs crucial functions during G2 checkpoint activation in human cells. ATR has been shown to phosphorylate and interact with BRCA1 in response to DNA damage and replication blocks, suggesting that it is a key regulator of BRCA1 function (18–20). Mouse embryo fibroblasts (MEFs) expressing BRCA1 protein containing an exon 11 deletion displayed an ablated G2 checkpoint response to ionizing radiation (IR; ref. 21). Moreover, expression of wild-type (wt) but not mutant BRCA1 in null cells restored IR-induced mitotic delay (22). These observations provide strong evidence that BRCA1 also participates in the DNA damage G2 checkpoint.
The effector kinase hChk1 may contribute to the DNA damage G2 checkpoint by phosphorylating Ser-216 of Cdc25C in a 14-3-3 binding site (23, 24), thereby preventing Cdc25C from activating cyclin B1/Cdk1. hChk1 is required to prevent mitotic entry in the presence of DNA damage and replication blocks in mouse embryos and mouse embryo fibroblasts (25, 26). Overexpression of fission yeast Chk1 in A-T cells partially restored G2 checkpoint function (27), and UCN-01, an agent that abrogates the G2 checkpoint, is a potent inhibitor of hChk1 (28, 29). ATR also has been shown to regulate hChk1 phosphorylation after treatment with ultraviolet radiation [wavelength band C 245 nm (UVC)] and hydroxyurea (HU; refs. 26, 30, and 31).
Cellular compartmentalization of cyclin B1/Cdk1 complexes also plays a role in checkpoint control of mitosis. During interphase, cyclin B1 is prevented from accumulating in the nucleus and is retained in the cytoplasm (32). Normal progression from G2 to mitosis seems to be initiated by phosphorylation of cyclin B1, which enhances nuclear import and inhibits nuclear export by interfering with binding to the nuclear export protein Crm1 (33–37). Overproduction of Myt 1 kinase in human cells perturbed cell cycle progression by sequestering cyclin B1/Cdk1 complexes in the cytoplasm (38). Expression of cyclin B1 containing a dominant nuclear localization sequence reversed IR-induced mitotic delay in HeLa cells (39), and expression of an export-resistant cyclin B1 in HeLa cells attenuated mitotic delay induced by the topoisomerase II inhibitor, etoposide (33). These observations suggest that the immediate G2 checkpoint response to DNA damage is enforced by at least two mechanisms, one that sustains inhibition of Cdk1 kinase activity and another that maintains nuclear exclusion of cyclin B1/Cdk1 complexes.
DNA topoisomerase II is required by eukaryotic cells to separate intertwined catenated daughter chromatids produced by DNA replication (40). ICRF-193 inhibits topoisomerase II by holding the enzyme in the form of a closed clamp that cannot form covalent complexes with DNA nor pass DNA strands, and thus is not associated with DNA strand breaks (8, 41, 42). As human cells respond to ICRF-193 with a severe mitotic delay (8, 9), it was hypothesized that cells monitor the status of chromatid decatenation after DNA replication and actively delay mitosis until chromatids are sufficiently decatenated (8). In this report, evidence is provided for the existence of a decatenation checkpoint in human cells that requires BRCA1 and relies on ATR-dependent signaling to maintain nuclear exclusion of cyclin B1/Cdk1 complexes.
Materials and Methods
Cell Culture.
Lymphoblastoid cell lines were obtained from the Coriell Institute for Medical Research (Camden, NJ). Normal lines included GM01815, GM02254A, GM03714A, and GM03657A. Cell lines from patients with A-T included GM09582, GM02782B, GM03332C, and GM03189D. Lymphoblasts were maintained in RPMI medium 1640 containing 15% FBS and 2 mM l-glutamine. The generation of SV40-immortalized GM847 human fibroblasts expressing an inducible ATRki allele has been described (15, 43). GM847 fibroblasts were grown in DMEM containing 10% FBS, 2 mM l-glutamine, and 0.4 mg/ml of G418-sulfate. Induction of ATRki was accomplished by incubating cells with 1 μg/ml of doxycycline for 48 h. Log-phase GM847 cells were prepared for metaphase spreads as described (44). The breast cancer line HCC1937, containing green fluorescent protein (GFP) fused to wt BRCA1, has been described (45). Diploid human fibroblasts expressing the catalytic subunit of telomerase (NHF1hTERT) and HeLa cells were maintained in MEM medium containing 10% FBS and 2 mM l-glutamine. All cell lines were grown at 37°C in a humidified atmosphere of 5% CO2.
Assay of Mitotic Delay.
Mitotic delay was enumerated by quantification of mitotic index in replicate cultures of treated and control cells (8, 14, 44). For treatment with IR (γ-rays), cells were irradiated in growth medium with a 137Cs source at a dose-rate of 0.9 Gy/min (Gammacell 40). Sham-treated controls were subjected to the same movements into and out of incubators as irradiated cells. For treatment with ICRF-193, cells were incubated in 0.5% DMSO alone or including ICRF-193 (0.1–6 μM) added directly to culture medium. For experiments testing inhibition of the nuclear export protein, Crm1, leptomycin B (LMB) was added at a final concentration of 6 ng/ml immediately preceding treatments (46). Overexpression of wt cyclin B1 or cyclin B1 containing a dominant nuclear localization sequence was accomplished in NHF1hTERT cells by using adenovirus to transduce the alleles (39), at a multiplicity of infection of 125–250. Twelve hours after infection, cells were harvested for assessment of protein expression or incubated with ICRF-193 in the presence of colcemid for an additional 6 h. Then cells were fixed with methanol/acetic acid (3:1), and cell nuclei were stained with DAPI (4′-6-diamidino-2-phenylindole, dihydrochloride). Interphase and mitotic cells were counted by fluorescence microscopy. Cdk1 kinase activity was quantified as histone H1 kinase activity in cyclin B1 immunoprecipitates as described (14). Incorporation of 32P from [γ-32P]ATP into histone H1 protein was quantified by using a Molecular Dynamics STORM 840 imaging system.
Western Immunoblots.
Cds1 phosphorylation was detected as described (47). For determination of Chk1 Ser-345 phosphorylation, cells were lysed with kinase lysis buffer (14) and subjected to SDS/PAGE. Immunoblots were performed with Abs specific for phosphorylated Ser-345 of Chk1 (1:1000, Cell Signaling Technology, Beverly, MA, no. 2341) or full-length Chk1 (1:2000; Santa Cruz Biotechnology; FL-476). Ser-317 phosphorylation was determined as described (30). ATR and cyclin B1 proteins were detected by using anti-ATR Ab (1:500; Oncogene, San Diego; no. PC128) and anti-cyclin B1 Ab (1:2000; Upstate Biotechnology, Lake Placid, NY; no. 05-158).
Localization of Cyclin B1 by Immunofluorescence Microscopy.
Lymphoblasts were sedimented onto glass microscope slides, fixed in 100% cold methanol for 10 min, and allowed to dry. Fixed cells were blocked with 10% FBS in PBS for 20 min followed by incubation with mAb to cyclin B1 (1:50; Santa Cruz Biotechnology, GSN1). Bound cyclin B1 was detected with a Cy3-conjugated sheep anti-mouse IgG (1:100; Sigma). After the final wash, DNA in the cell nuclei was stained with DAPI (4′-6-diamidino-2-phenylindole, dihydrochloride). Cells were examined with a Zeiss fluorescence microscope. Fluorescent images were captured with a charge-coupled device (CCD) camera by using separate red and blue filters. Images were reproduced by using Adobe Systems (Mountain View, CA) photoshop software.
Results
ICRF-193 Induces an ATM-Independent Mitotic Delay.
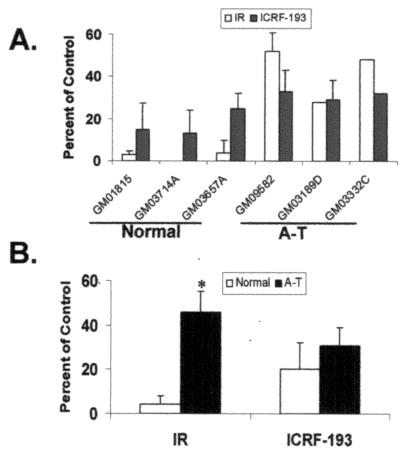
Treatment with 1 Gy of γ-rays induced severe mitotic delay in normal lymphoblastoid lines, whereas A-T lymphoblasts displayed the characteristic attenuation of G2 checkpoint response to DNA damage (Fig. 1 A and B). ICRF-193 also induced mitotic delay in human lymphoblastoid lines. Concentration response studies showed increasing inhibition of mitosis between 0.1 and 1 μM ICRF-193 with saturation of response at higher concentrations (results not shown). Mitotic inhibition was maximal 2 h after addition of the drug (data not shown), indicating cells were delayed in G2. When incubated with 2 μM ICRF-193, normal and A-T cells displayed 80 ± 10% and 69 ± 8% inhibition of mitosis, respectively (Fig. 1 A and B; P > 0.1). Mutations in ATM that attenuated the G2 checkpoint response to DNA damage had little effect on the mitotic delay induced by sustained chromatid catenation, indicating that the G2 arrest induced by ICRF-193 was ATM-independent.
Figure 1.
Sustained chromatid catenation induces mitotic delay in normal and A-T cells. (A) Normal and A-T lymphoblasts were sham-treated, irradiated with 1 Gy of γ-rays (IR), incubated in 2 μM ICRF-193, or incubated with DMSO solvent. Two hours after treatment or addition of drug, mitotic index was quantified as described in Materials and Methods. The results for IR and ICRF-193 are expressed as a percentage of the untreated control (mean + SD; n = 3–4 for all but GM03332C, where n = 2). (B) The mean values obtained in A were expressed as the average percentage of the untreated control for normal and A-T cells (mean + SD, n = 3). *, The mean inhibition of mitosis in A-T cells after IR was significantly less than in normal cells (Student's t test, P < 0.05).
ATR and BRCA1 Are Required for ICRF-193-Induced Mitotic Delay.
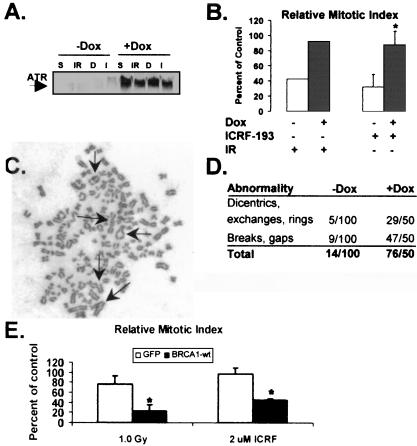
Caffeine has been shown to reverse the mitotic delay produced by ICRF-193 in rodent and human cells (8). The finding that both ATM and ATR kinases are inhibited by caffeine (17) in combination with the observation that A-T cells undergo mitotic delay in response to ICRF-193 suggested that ATR may be the relevant kinase which enforces the decatenation checkpoint. To test this hypothesis, SV40-immortalized human fibroblasts (GM847) expressing a doxycycline-inducible ATRki allele were examined for their ability to undergo mitotic delay in response to IR or ICRF-193. IR and ICRF-193 caused a reduction in mitotic index in uninduced GM847 cells. Induction of ATRki significantly reversed the DNA damage and decatenation responses (Fig. 2 A and B). As ATR seemed to be required for the decatenation checkpoint, it was hypothesized that cells overexpressing ATRki might enter mitosis with insufficiently decatenated chromosomes and ultimately display genomic instability. To test this hypothesis, metaphase preparations of uninduced and induced GM847 cells were analyzed for chromosomal aberrations. Although mitotic indices in GM847 cells were the same in the presence (+Dox) or absence (−Dox) of ATRki, 10 times more chromosomal aberrations were observed in the fibroblasts expressing ATRki (Fig. 2C). The chromosomal aberrations observed in the ATRki-expressing cells were diverse, consisting of chromatid breaks, gaps, rings, exchanges, and dicentrics (Fig. 2 C and D). Recent evidence implicates a functional interaction between ATR and BRCA1 (20). BRCA1 mutant HCC1937 cells expressing GFP alone or GFP-BRCA1wt were examined for their ability to undergo mitotic delay in response to IR or ICRF-193. HCC1937-GFP cells were defective in the response to IR and ICRF-193, and expression of wt BRCA1 restored IR- and ICRF-193-induced mitotic delay (Fig. 2E). These results suggest that the mitotic delay induced by ICRF-193 requires both functional ATR and BRCA1 and provide genetic evidence that the chromatid catenation-induced mitotic delay is an active checkpoint response and not the consequence of passive metabolic disturbance.
Figure 2.
ATR and BRCA1 are required for ICRF-193-induced mitotic delay. (A) Uninduced or induced GM847 cells were irradiated with 1.0 Gy of γ-rays (IR) or incubated with 2 μM ICRF-193 (I). Sham-treated and DMSO controls are denoted S and D, respectively. After 2 h, cells were harvested and Western immunoblot analysis was performed. (B) Uninduced (open bars) and induced GM847 (closed bars) cells were treated as described above. After 2 h, cells were assayed for mitosis. The results are expressed as the mean percentage of the untreated control (+ SD, for ICRF-193, n = 3; for IR, n = 2). *, The mean inhibition of mitosis in uninduced ICRF-193-treated cells was significantly different from that in induced ICRF-193-treated cells (Student's t test, P < 0.005). (C and D) Metaphase spreads of chromosomes from ATRki-induced GM847 cells. Arrows point to various chromosomal aberrations. (E) HCC1937 cells expressing GFP or BRCA1wt were irradiated with 1.0 Gy of γ-rays or incubated with 2 μM ICRF-193. After 2 h, cells were assayed for mitosis. The results are expressed as the mean percentage of the untreated control (+ SD, for ICRF-193, n = 3; for IR, n = 2). *, The mean inhibition of mitosis in GFP- expressing cells was significantly different from that in BRCA1wt-expressing cells (Student's t test, P < 0.005).
hCds1 and hChk1 Are Not Phosphorylated After Exposure to ICRF-193.
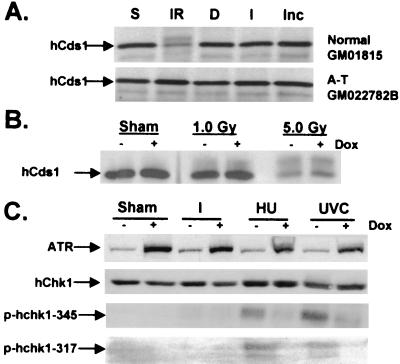
Next, the ability of ICRF-193 to induce the phosphorylation of the hChk1 and hCds1 effector checkpoint kinases was investigated. Consistent with recent reports, treatment of normal cells with IR caused an electrophoretic mobility shift in hCds1 indicative of phosphorylation, which was significantly reduced in irradiated A-T cells (refs. 47 and 48; Fig. 3A). Treatment with ICRF-193 failed to produce a detectable mobility shift of hCds1 in normal and A-T cells (Fig. 3A), supporting the observation that ICRF-193-induced mitotic delay was independent of ATM signaling and DNA damage (Fig. 1 A and B). It has been shown that ATR and ATM have similar substrates in vitro (49). To rule out the possibility that expression of ATRki also inhibited ATM kinase activity in vivo by interfering with endogenous substrates, GM847 cells expressing ATRki were tested for their ability to undergo IR-induced phosphorylation of hCds1. Treatment of GM847 cells with IR caused a mobility shift of hCds1 in both the presence and absence of ATRki (Fig. 3B), suggesting that induction of ATRki did not interfere with ATM-dependent signaling. Given that Chk1 is a substrate of ATR during checkpoint responses to UVC and HU (26, 30, 31), and ATR was required for ICRF-193-induced mitotic delay, we tested whether Chk1 was phosphorylated in cells treated with ICRF-193. As reported, treatment of GM847 cells with UVC or HU induced phosphorylation of hChk1 at Ser-345 and -317 (refs. 26 and 30; Fig. 3C). Induction of ATRki diminished phosphorylation of Ser-345 and -317, implying that these phosphorylation events were ATR-dependent (Fig. 3C). Incubation with ICRF-193 did not induce phosphorylation of either Ser-345 or -317 of hChk1 (Fig. 3C). Moreover, an electrophoretic mobility shift of Chk1 was observed in HeLa cells treated with UVC or HU but not ICRF-193 (data not shown). Thus, phosphorylations of Cds1 and Chk1 associated with DNA damage and replication blocks did not seem to be elements of decatenation checkpoint function.
Figure 3.
ICRF-193 does not induce phosphorylation of hCds1 or hChk1. (A) Normal and A-T lymphoblasts were sham-treated (S) or irradiated with 5 Gy of γ-rays (IR), incubated with DMSO solvent (D), incubated with 2 μM ICRF-193 (I), or retained untreated in the incubator (Inc). One hour after treatment or addition of drug, cells were harvested. (B) Uninduced or induced GM847 cells were harvested 2 h after sham treatment or irradiation with 1.0 or 5.0 Gy. Western immunoblot analysis was performed for expression of hCds1. (C) Uninduced or induced GM847 cells were harvested 2 h after sham treatment, irradiation with 50 J/m2 UVC, or incubation with 2 mM HU or 2 μM ICRF-193 (I). IP-Western or Western blot analysis was performed for expression of ATR, Chk1, and Ser-345- or -317-phosphorylated Chk1.
Nuclear Exclusion of Cyclin B1/Cdk1 Complexes Is Required for ICRF-193-Induced G2 Arrest.
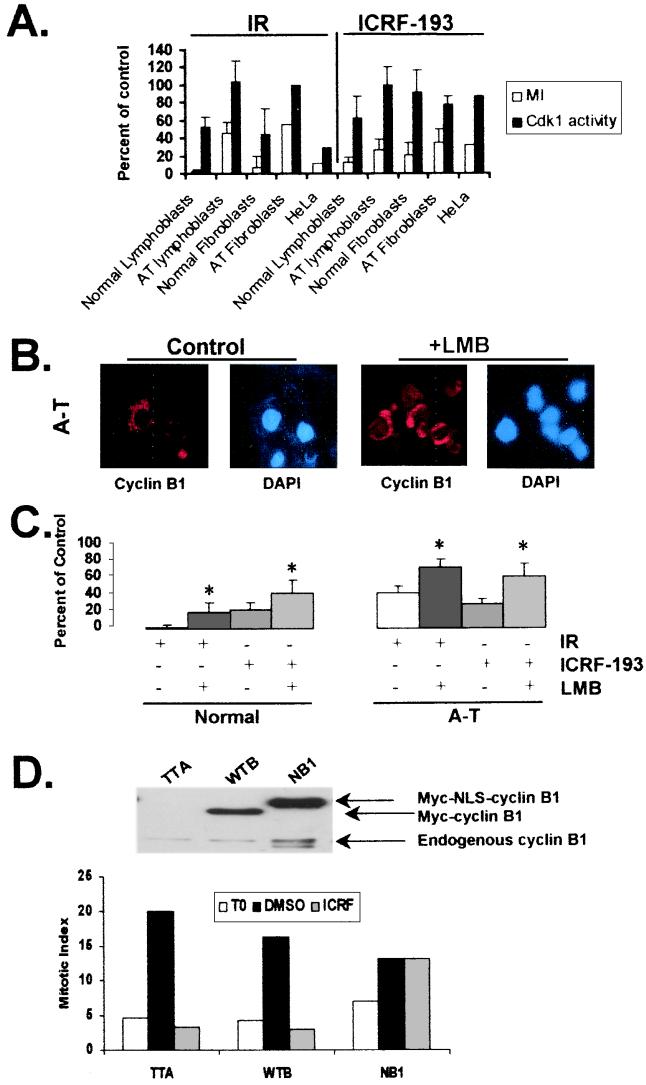
Cyclin B1-associated Cdk1 kinase activity decreases after cells are treated with DNA damaging agents, presumably reflecting ATM-dependent signaling to sustain inhibitory phosphorylation of Cdk1 (2, 14). To determine whether mitotic delay associated with sustained chromatid catenation also acts through regulation of Cdk1 activity, mitotic delay and cyclin B1-associated Cdk1 kinase activity were assessed in a variety of human cell types after irradiation with γ-rays or incubation with ICRF-193 (Fig. 4A). As expected, IR induced an ATM-dependent inhibition in Cdk1 activity and mitosis (Fig. 4A). The ICRF-193-induced checkpoint response occurred despite substantial Cdk1 activity. All cell types underwent a strong ICRF-193-induced inhibition of mitosis even though Cdk1 kinase activity was significantly inhibited only in normal lymphoblasts (Fig. 4A). As entry into mitosis depends on both the activity and location of cyclin B1/Cdk1 complexes, it was hypothesized that the decatenation checkpoint carries out mitotic delay in part by preventing nuclear accumulation of cyclin B1/Cdk1 complexes. To test this theory, ICRF-193-induced mitotic inhibition was assessed under two conditions that promote nuclear localization of cyclin B1/Cdk1 complexes. These included inhibition of nuclear export by LMB treatment and overexpression of cyclin B1 containing a dominant nuclear localization sequence. Treatment with LMB to inhibit the nuclear exporter Crm1 (33, 46) altered the staining pattern of cyclin B1 from mostly cytoplasmic to both nuclear and cytoplasmic in normal (data not shown) and A-T lymphoblasts (Fig. 4B). Incubation of cells with LMB alone did not have an effect on chromosome condensation or entry into mitosis. The mitotic index (% mitotic cells) in cultures that had been treated with LMB for 2 h was similar to that of controls (data not shown). In contrast, LMB attenuated the mitotic delay observed in IR- and ICRF-193-treated cells (Fig. 4C). Because LMB is expected to inhibit the nuclear export of many cellular proteins, we next examined the individual contribution made by cyclin B1 localization to the ICRF-193-induced mitotic delay (Fig. 4D). To carry out these experiments, cells were infected with recombinant adenoviruses encoding the tetracycline transactivator (TTA) alone or together with virus-encoding wt cyclin B1 (WTB) or a form of cyclin B1 that is constitutively nuclear (NB1; ref. 39). As seen in Fig. 4D, ICRF-193 treatment reduced the number of mitotic cells from 20% to 3.4% in cells expressing TTA and from 16% to 2.9% in cells expressing WTB. In contrast, attenuation of the ICRF-193-induced mitotic delay was observed in cells expressing NB1. The mitotic indices in DMSO- and ICRF-193-treated NB1 cells were 13.2% and 13.1%, respectively. Taken together, these results suggest that the decatenation checkpoint relies, in part, on the nuclear exclusion of cyclin B1/Cdk1 complexes.
Figure 4.
Nuclear cyclin B1 abrogates ICRF-193-induced mitotic delay. (A) Human cell lines (normal and A-T lymphoblasts and fibroblasts, HeLa) were irradiated or incubated with ICRF-193 or DMSO. Two hours after treatment or addition of drug, cells were harvested and assayed for mitotic index and Cdk1 kinase activity. The results are expressed as a percentage of the untreated control. There was not a significant correlation between the reduction in mitotic index and Cdk1 kinase activity after incubation with ICRF-193 (R2 = 0.03, P > 0.5). (B) Normal and A-T lymphoblastoid cell lines were treated with and without 6 ng/ml of LMB for 2 h, after which cells were fixed onto glass slides and immunofluorescence microscopy was performed. Representative images are shown for GM03189D (A-T). (C) Normal (GM03714A, GM01815, and GM03657A) and A-T lymphoblasts (GM09582, GM03189D, and GM03332C) were pretreated with and without 6 ng/ml of LMB and then were sham-treated, irradiated with 1 Gy of γ-rays, or incubated with 2 μM ICRF-193 or DMSO. Two hours after treatment or addition of drug, mitotic index was quantified. The results are expressed as the average percentage of control mitotic index in normal or A-T cells. (mean + SD, n = 3 for IR and n = 5 for ICRF-193). *, The mean inhibition of mitosis in LMB-treated cells was significantly different from the mean inhibition of mitosis in IR- or ICRF-193-treated normal and A-T cells (Student's t test, P < 0.05). (D) Log-phase NHF1hTERT cells were infected with adenovirus containing tetracycline transactivator (TTA) alone or coinfected with TTA and wt cyclin B1 (WTB) or cyclin B1 containing a dominant nuclear localization sequence (NB1). Twelve hours after infection, cells were harvested for cyclin B1 Western blot analysis, fixed for mitotic index determination, or incubated with DMSO or 2 μM ICRF-193 in the presence of 100 ng/ml of colcemid. After 6 h, cells were fixed and mitotic index was determined. The results are expressed as the percentage of mitotic cells at time of addition of DMSO or ICRF-193 (T0) and after 6 h of incubation in colcemid and ICRF-193 or DMSO. The results depicted were reproduced in two additional experiments.
Discussion
The findings presented here suggest that the status of chromatid decatenation is monitored and communicated to the machinery of the cell cycle through a G2 checkpoint system. The mitotic delay response to ICRF-193 does not seem to be a DNA damage checkpoint as it was independent of ATM and did not involve phosphorylations of hCds1 or hChk1, which are induced by DNA damage. Cell cycle checkpoints depend on active gene expression and are defined by loss of function mutations that alleviate cell cycle arrest (3). The observations that expression of an ATRki allele ablated the mitotic delay induced by ICRF-193 and wt BRCA1 restored ICRF-193-induced G2 arrest in a BRCA1 mutant cell line provide genetic evidence to support the existence of a decatenation checkpoint. The results also suggest that ATR and BRCA1 function to regulate the decatenation checkpoint in addition to the DNA damage and replication checkpoints. Dependency checkpoints are control mechanisms that enforce the completion of early events before the initiation of late events in the cell cycle (4). As decatenation is essential for accurate separation of chromatids at anaphase, the G2 arrest induced by ICRF-93 seems to represent a dependency checkpoint response.
ATR has recently been shown to phosphorylate Chk1 after treatment with UVC and HU, and it has been suggested that an ATR–Chk1 signaling pathway regulates the replication and DNA damage checkpoints (26, 30, 31). In vitro, ATR phosphorylates hChk1 on two serine residues (Ser-317 and -345), and mutation of these residues severely impairs the ability of Chk1 to be activated in vivo during a checkpoint response (30). Given that phosphorylation of Chk1 at Ser-317 and -345 was not observed after ICRF-193 treatment, it is unlikely that the decatenation checkpoint signals to hChk1. However, it is possible that ATR phosphorylates Chk1 on Ser-317 and/or -345 at too low a level to be detected by our Abs or to cause a shift in the electrophoretic mobility of Chk1 on SDS gels.
Studies of the cell cycle characteristics of blastocytes from ATR- and Chk1-null embryos demonstrated essential roles for ATR and Chk1 as null cells entered mitosis with severely fragmented chromosomes resulting in apoptosis (27, 28, 50). The similarity between this phenomenon and mitotic catastrophe in cells defective for the DNA damage G2 checkpoint suggests that ATR and hChk1 are involved in regulating the transition between DNA replication and mitosis. Our results are consistent with a role for ATR in checkpoint control mechanisms that enforce the completion of dependent events in the S and G2 phases of the cell cycle. Expression of ATRki increased the frequency of chromosomal aberrations in proliferating cells by 10-fold, implying that loss of ATR function allows cells to enter mitosis inappropriately, for example, when DNA is incompletely replicated or chromatids insufficiently decatenated. Entry into mitosis with catenated chromatids may cause stress on the chromosomes during anaphase and result in chromatid breakage or nondisjunction. Recently, BRCA1 was reported to exist in a multimeric complex with several proteins that participate in DNA repair processes and cell cycle control (51). The finding that BRCA1 was required for IR- and ICRF-193-induced mitotic delay supports the idea that BRCA1 plays a central role in checkpoint responses by facilitating interactions between checkpoint transducers and effectors (22, 51).
ICRF-193-induced mitotic delay was abrogated when cyclin B1/Cdk1 complexes were allowed to accumulate in the nucleus. Inhibition of Crm1-mediated nuclear export significantly reversed mitotic delay induced by sustained chromatid catenation, implying that nuclear export of cyclin B1/Cdk1 complexes is important for decatenation checkpoint function. The mechanism that prevents nuclear accumulation of cyclin B1/Cdk1 complexes seems to be independent of ATM signaling, as nuclear cyclin B1 reversed ICRF-193-induced mitotic delay in normal and A-T cells. Other components of the checkpoint may sequester cyclin B1/Cdk1 complexes in the cytoplasm or block import. The decatenation G2 checkpoint may prevent nuclear accumulation of cyclin B1/Cdk1 complexes by modulating protein kinases that phosphorylate cyclin B1.
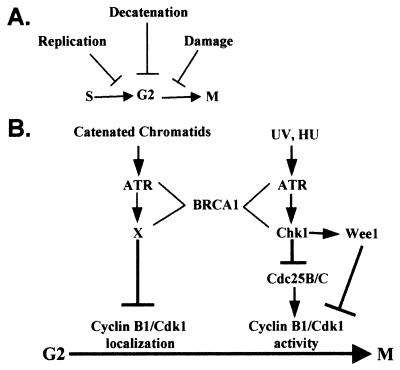
Cellular checkpoints that regulate the onset of mitosis include the damage checkpoint, which responds to DNA double-strand breaks, the replication checkpoint, which responds to inhibition of DNA synthesis, and the decatenation checkpoint, which responds to sustained catenation of daughter chromatids (Fig. 5A). Fig. 5B depicts a model for ATR-dependent G2 checkpoints. Depending on what signals activate ATR (UVC, HU, or catenated chromatids), different effectors mediate mitotic delay. In the case of UVC and HU, ATR phosphorylates Chk1, which then acts to inhibit Cdk1 activity possibly through Wee1 or Cdc25B/C directly (23, 52, 53). In the presence of sustained chromatid catenation, ATR signals to an as yet unidentified downstream effector molecule (X) that acts to prevent the nuclear accumulation of cyclin B1/Cdk1 complexes. BRCA1 acts as a scaffolding protein that facilitates the interaction between ATR and its substrates. Future studies should focus on the mechanisms whereby the decatenation checkpoint sustains nuclear exclusion of cyclin B1/Cdk1 complexes, and whether inactivation of decatenation checkpoint function contributes to genetic instability.
Figure 5.
(A) Mitotic entry checkpoints. (B) Model for ATR-dependent G2 checkpoint function.
Acknowledgments
We thank Dr. Dennis Simpson for the generation of NHF1hTERT cells and Dr. Randal Tibbetts for technical advice; Drs. Thomas Petes, Marila Cordeiro-Stone, and Randal Tibbetts for critical reading of the manuscript; Dr. Minoru Yoshida (University of Tokyo) for providing LMB; Dr. Robert Abraham (Duke University) for the GM847 cells; Dr. Ralph Scully (Dana–Farber Cancer Institute) for the HCC1937 cells expressing wt BRCA1; Dr. Robert Weinberg (Whitehead Institute for Biomedical Research) for the hTERT construct; and Drs. David Morgan (University of California at San Francisco) and Amy Mauser (University of North Carolina at Chapel Hill) for the cyclin B1 adenoviruses. This work was supported in part by grants from the American Heart Association, Heartland Affiliate (to P.R.G.), the Ulster Cancer Foundation (to C.S.D.), and from the National Institutes of Health (to W.K.K. and H.P.W.). H.P.W. is an Investigator of the Howard Hughes Medical Institute. P.B.D. was supported by a National Institute on Environmental Health Sciences Environmental Pathology training grant.
Abbreviations
- A-T
ataxia telangiectasia
- ATM
A-T mutated
- ATR
ATM- and rad3-related
- GFP
green fluorescent protein
- IR
ionizing radiation
- LMB
leptomycin B
- ATRki
kinase-inactive ATR
- IR
ionizing radiation
- UVC
ultraviolet radiation, wavelength band C
- HU
hydroxyurea
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann W K, Paules R S. FASEB J. 1996;10:238–247. doi: 10.1096/fasebj.10.2.8641557. [DOI] [PubMed] [Google Scholar]
- 3.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Sekiguchi M, Hayashida H, Kuma K, Miyata T, Fukushige S, Murotsu T. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- 6.Anderson H, Roberge M. Cell Growth Differ. 1996;7:83–90. [PubMed] [Google Scholar]
- 7.Zampetti-Bosseler F, Scott D. Int J Radiat Biol Relat Stud Phys Chem Med. 1981;39:547–558. doi: 10.1080/09553008114550651. [DOI] [PubMed] [Google Scholar]
- 8.Downes C S, Clarke D J, Mullinger A M, Gimenez-Abian J F, Creighton A M, Johnson R T. Nature (London) 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann W K, Kies P E. Mutat Res. 1998;400:153–167. doi: 10.1016/s0027-5107(98)00041-4. [DOI] [PubMed] [Google Scholar]
- 10.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 11.Morgan D O. Nature (London) 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 12.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 13.Canman C E, Wolff A C, Chen C Y, Fornace A J J, Kastan M B. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 14.Paules R S, Levedakou E N, Wilson S J, Innes C L, Rhodes N, Tlsty T D, Galloway D A, Donehower L A, Tainsky M A, Kaufmann W K. Cancer Res. 1995;55:1763–1773. [PubMed] [Google Scholar]
- 15.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright J H, Munar E, Jameson D R, Andreassen P R, Margolis R L, Seger R, Krebs E G. Proc Natl Acad Sci USA. 1999;96:11335–11340. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkaria J N, Busby E C, Tibbetts R S, Roos P, Taya Y, Karnitz L M, Abraham R T. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 18.Chen J. Cancer Res. 2000;60:5037–5039. [PubMed] [Google Scholar]
- 19.Gatei M, Zhou B, Hobson K, Scott S, Young D, Khanna K. J Biol Chem. 2001;276:17276–17280. doi: 10.1074/jbc.M011681200. [DOI] [PubMed] [Google Scholar]
- 20.Tibbetts R S, Cortez D, Brumbaugh K M, Scully R, Livingston D, Elledge S J, Abraham R T. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Weaver Z, Linke S P, Li C, Gotay J, Wang X W, Harris C C, Ried T, Deng C X. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 22.Xu B, Kim S t, Kastan M B. Mol Cell Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 25.Takai H, Tominaga K, Motoyama N, Minamishima Y A, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, Guntuku S, Cui X S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P, Gatei M, O'Connell M J, Khanna K K, Bugg S J, Hogg A, Scott S P, Hobson K, Lavin M F. Oncogene. 1999;18:249–256. doi: 10.1038/sj.onc.1202257. [DOI] [PubMed] [Google Scholar]
- 28.Graves P R, Yu L, Schwarz J K, Gales J, Sausville E A, O'Connor P M, Piwnica-Worms H. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 29.Busby E C, Leistritz D F, Abraham R T, Karnitz L M, Sarkaria J N. Cancer Res. 2000;60:2108–2112. [PubMed] [Google Scholar]
- 30.Zhao H, Piwnica-Worms H. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z, Kumagai A, Wang S X, Dunphy W G. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pines J. Results Probl Cell Differ. 1998;22:57–78. doi: 10.1007/978-3-540-69686-5_3. [DOI] [PubMed] [Google Scholar]
- 33.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Meyer A N, Donoghue D J. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagting A, Jackman M, Simpson K, Pines J. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 37.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Rothblum-Oviatt C, Ryan C E, Piwnica-Worms H. Mol Cell Biol. 1999;19:5113–5123. doi: 10.1128/mcb.19.7.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin P, Hardy S, Morgan D O. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm C. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 41.Ishida R, Miki T, Narita T, Yui R, Sato, Utsumi K R, Tanabe K, Andoh T. Cancer Res. 1991;51:4909–4916. [PubMed] [Google Scholar]
- 42.Roca J, Ishida R, Berger J M, Andoh T, Wang J C. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Cliby W A, Shieh S Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaufmann W K, Schwartz J L, Hurt J C, Byrd L L, Galloway D A, Levedakou E, Paules R S. Cell Growth Differ. 1997;8:1105–1114. [PubMed] [Google Scholar]
- 45.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston D M. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 46.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 47.Brown A L, Lee C H, Schwarz J K, Mitiku N, Piwnica-Worms H, Chung J H. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge S J. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. . (First Published September 5, 2000; 10.1073/pnas.190030497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S T, Lim D S, Canman C E, Kastan M B. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 50.Brown E J, Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 52.Raleigh J M, O'Connell M J. J Cell Sci. 2000;113:1727–1736. doi: 10.1242/jcs.113.10.1727. [DOI] [PubMed] [Google Scholar]
- 53.Lee J, Kumagai A, Dunphy W G. Mol Biol Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]