Abstract
The antimicrobial peptides (AMP) produced by intestinal epithelial cells (IEC) play crucial roles in the regulation of intestinal homeostasis by controlling microbiota. Gut microbiota has been shown to promote IEC expression of RegIIIγ and certain defensins. However, the mechanisms involved are still not completely understood. In this report, we found that IEC expression of RegIIIγ and β-defensins 1, 3, and 4 was lower in G protein-coupled receptor (GPR)43−/−mice compared to that of wild-type (WT) mice. Oral feeding with short chain fatty acids (SCFA) promoted IEC production of RegIIIγ and defensins in mice. Furthermore, SCFA induced RegIIIγ and β-defensins in intestinal epithelial enteroids generated from WT but not GPR43−/−mice. Mechanistically, SCFA activated mTOR and STAT3 in IEC, and knockdown of mTOR and STAT3 impaired SCFA induction of AMP production. Our studies thus demonstrated that microbiota metabolites SCFA promoted IEC RegIIIγ and β-defensins in a GPR43-dependent manner. The data thereby provides a novel pathway by which microbiota regulates IEC expression of AMP and intestinal homeostasis.
Keywords: SCFA, GPR43, IEC, RegIIIγ, Defensins
Introduction
In spite of being home to a diverse community of large numbers of indigenous microorganisms, host intestines live in harmony with microbiota under homeostatic conditions. Among multiple mechanisms involved in the regulation of host responses to microbiota in maintaining intestinal homeostasis, antimicrobial peptides (AMP) that are produced by intestinal epithelial cells (IEC) and Paneth cells serve as the first line of defense against microbiota and occasionally pathogens1,2. AMP regulate host responses to microbiota and are involved in the maintenance of intestinal homeostasis. Several distinct families of AMP have been identified including defensins, cathelicidins, and C-type lectins (such as the regenerating islet-derived protein (REG) family). AMP function to kill or inactivate microorganisms rapidly through different mechanisms1,2. The spectrum of antimicrobial activity varies for each particular antimicrobial peptide. In general, defensins affect both Gram-positive and Gram-negative bacteria and, in some cases, fungi, viruses, and protozoa3. RegIIIγ, on the other hand, is selective for Gram-positive bacteria4. Mutually, gut microbiota may also differentially regulate AMP production. For example, although expression of α-defensins is independent of gut microbiota, the expression of RegIIIγ is virtually absent in germ-free (GF) mice and increased upon re-colonization with gut microbiota4,5.
How microbiota promotes the expression of AMP remains unclear. In mice deficient in MyD88, an adaptor molecule common to most TLRs, IEC expression of RegIIIγ and RegIIIβ is decreased compared to that in wild-type (WT) mice, suggesting that IEC can directly sense bacteria through TLRs and promote the expression of RegIIIγ and RegIIIβ6,7. In addition, innate lymphoid cell (ILC) production of IL-22 has also been shown to promote IEC expression of RegIIIγ mRNA. Notably, microbiota drive IL-22 production in ILCs, as demonstrated by the low levels of IL-22 produced by ILCs from GF mice8. Thus, gut microbiota can regulate AMP expression, at least partially, through interaction with IEC TLR or stimulating ILC IL-22 production.
Accumulating evidence indicates that the host immune system can sense gut bacterial metabolites in addition to TLR ligands. Recognition of these small molecules greatly impacts the host immune response9,10. Among gut microbiota metabolites, short-chain fatty acids (SCFA), which are solely metabolized from indigestible carbohydrates by intestinal bacteria11, have been shown to have immense impact on the host12. Intact production of SCFA is associated with a reduced risk of different diseases, including IBD. As the most abundant SCFA, acetate, propionate, and butyrate, are present in the colonic lumen of humans at high concentrations, ranging from 50 to 150 mM11,13. SCFA can function through the activation of mammalian G protein-coupled receptors (GPCR)14. GPR41 and GPR43 are two major host receptors for most SCFA15. SCFA can also inhibit histone deacetylase activity (HDAC) to downregulate macrophage pro-inflammatory cytokine production and activate peroxisome proliferator-activated receptor γ to stimulate human colonic epithelial cell expression of angiopoietin-like protein 416,17. In addition, SCFA promote intestinal Treg cell development which is probably also mediated by their HDAC inhibitor activity but independent of GPR43, although it is still debatable as conflicting data have been published10,18,19.
SCFA butyrate has been demonstrated to induce LL-37/hCAP18, a member of cathelicidin family, in various human colon cell lines20. However, the underlying molecular mechanisms by which SCFA regulate IEC AMP production are still not completely understood. It has been reported that MAP kinases respond differently to butyrate stimulation21. The MEK-ERK pathway regulates butyrate-induced LL-37 expression, whereas p38/MAP kinase exhibits little effect on AMP production21,22. C-Jun N-terminal kinase (JNK) also positively regulates cathelicidin production in tissue epithelial cells23. The current study was undertaken to investigate the mechanisms of AMP production induced by SCFA. We report here that SCFA promoted RegIIIγ and β-defensin expression in IEC through GPR43, which is mediated by mTOR and STAT3.
Results
1. Expression of RegIIIγ and β-defensins in IEC is impaired in GPR43−/−mice
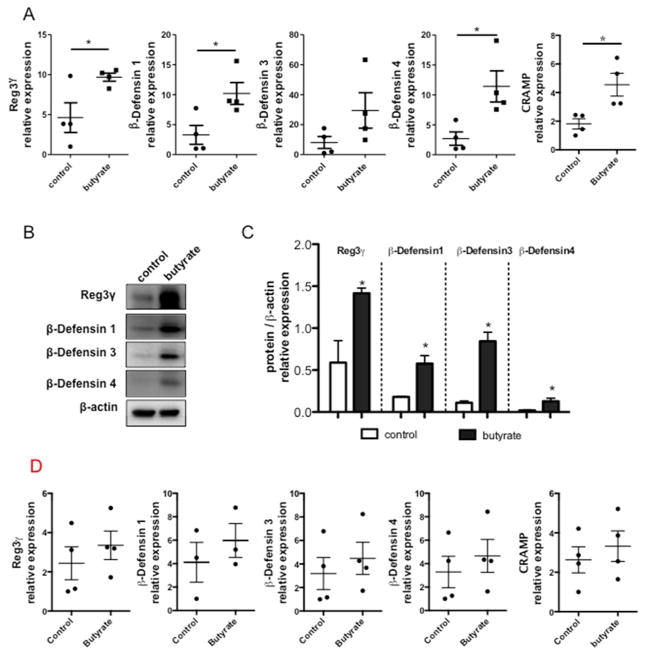
It has been previously reported that microbiota regulate IEC AMP expression differentially4,5. As microbiota-derived SCFA can modulate IEC activity through GPR43, we investigated whether GPR43 regulates AMP expression in IEC. We measured the expression of RegIIIγ and β-defensins 1, 3, 4 in C57BL/6 (WT) and GPR43−/−mice at both RNA and protein levels. Quantitative real-time PCR (qRT-PCR) revealed that the transcription of RegIIIγ and β-defensin 1, 3, 4 mRNA was compromised in GPR43−/−IEC compared to the WT control (Fig. 1A). Consistent with gene expression studies, protein levels of RegIIIγ and β-defensin 1, 3, 4 in GPR43−/−mice were significantly lower compared to WT mice (Fig. 1B and 1C). Collectively, these data indicated that SCFA recognition by GPR43 promotes IEC production of RegIIIγ and β-defensins.
Figure 1. Decreased production of RegIIIγ and β-defensins in IEC of GPR43−/−mice.
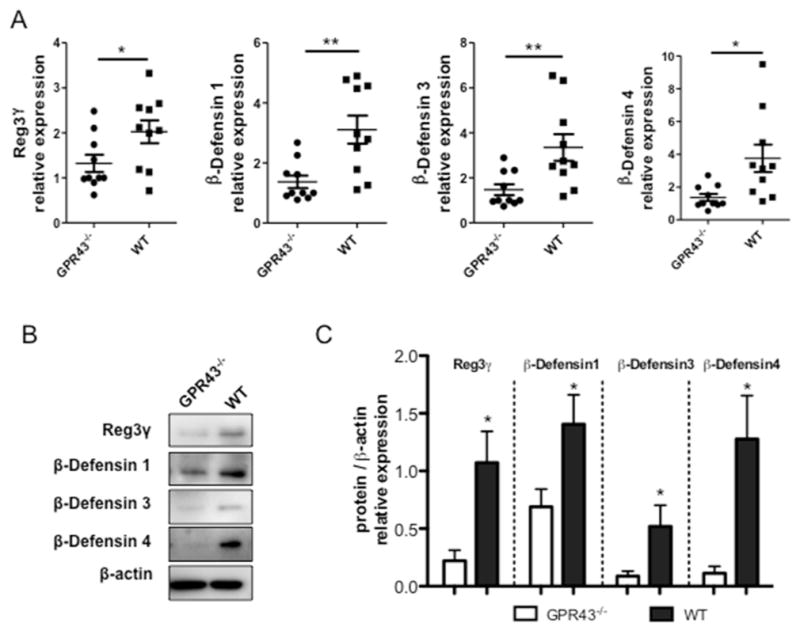
IEC from small intestine were isolated from WT and GPR43−/−mice respectively. (A) IEC expression of RegIIIγ and β-defensins 1, 3, 4 in GPR43−/−and WT mice determined by qRT-PCR and normalized against gapdh. Pooled data from 2 independent experiments. n=5 mice/group. (B) Protein levels of RegIIIγ and β-defensins were determined by Western blot. Data are reflective of 2 independent experiments. n=4 mice/group. (C) Protein/β-actin relative expression of RegIIIγ and β-defensins were compared between GPR43−/−and WT mice. Data were combined from 2 independent experiments. *p<0.05; **p<0.01.
2. Feeding SCFA promotes IEC expression of RegIIIγ and β-defensins in WT but not GPR43−/−mice
As the most potent agonists for GPR43, SCFA play critical roles in intestinal barrier function24,25. Butyrate, in particular, has been demonstrated to induce cathelicidin AMP and β-defensin 1 in human lung epithelial cells23. We thus investigated whether SCFA can elicit a similar effect on IEC, and whether GPR43 mediates such effects. Because gut microbiota produces high levels of SCFA in the intestines which may confound the data interpretation, we first treated mice with broad-spectrum antibiotics for 10 days to eliminate gut microbiota as well as their metabolites SCFA, followed by 21 days of butyrate and antibiotics administration in drinking water. IEC were then isolated and assessed for AMP expression. We found decreased expression of RegIIIγ, but no significant difference of β-defensins in mice receiving antibiotics compared to the control mice (Fig. S1). As expected, in WT mice, feeding butyrate enhanced the expression of RegIIIγ and β-defensin 1, 3, 4 compared to controls in IEC (Fig. 2A). Increased protein levels of RegIIIγ and β-defensins were also detected by Western blot (Fig. 2B and 2C). Feeding butyrate also increased expression of cathelicidin-related antimicrobial peptide (CRAMP) (Fig. 2A). Additionally, mice fed with acetate also slightly increased expression of RegIIIγ and β-defensins in IEC compared to control (Fig. S2). However, feeding butyrate did not affect the expression of RegIIIγ and β-defensin 1, 3, 4 in IEC of GPR43−/−mice (Fig. 2D). Together, these data suggested that SCFA serve as potent inducers of AMP production by IEC, which is mediated by GPR43.
Figure 2. Butyrate feeding promotes IEC expression of RegIIIγ and β-defensins in vivo.
WT and GPR43−/−C57BL/6 mice were treated with antibiotics in drinking water for 10 days, and then fed with or without 300 mM butyrate in drinking water for 21 days. (A) IEC expression of RegIIIγ and β-defensins 1, 3, 4, and CRAMP in WT mice were determined by qRT-PCR and normalized against gapdh. Data are reflective of 2 independent experiments. n=4 mice/group. (B) Protein levels of RegIIIγ and β-defensins determined by Western blot in WT mice. Data are reflective of 2 independent experiments. (C) Protein/β-actin relative expression of RegIIIγ and β-defensins were compared between control and butyrate-treated WT mice. Data were combined from 2 independent experiments. *p<0.05. (D) IEC expression of RegIIIγ and β-defensins 1, 3, 4 in GPR43−/−mice were determined by qRT-PCR and normalized against gapdh. Data are reflective of 2 independent experiments. n=4 mice/group.
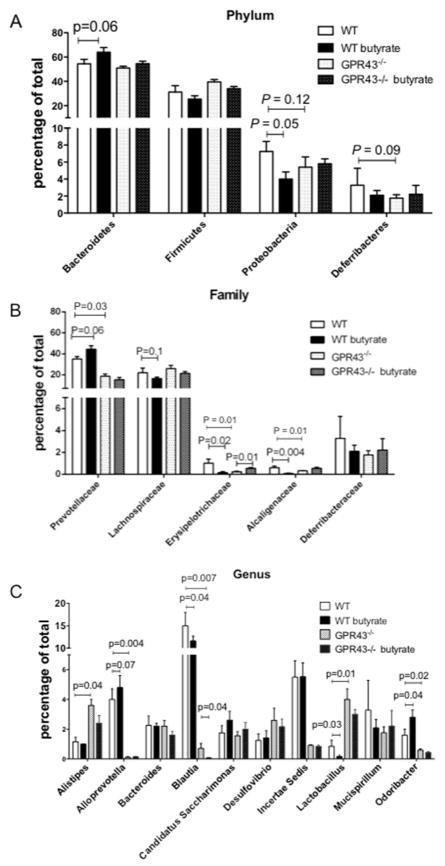
We then investigated whether GPR43 and feeding butyrate affect gut microbiota, which could potentially mediate butyrate-induction of AMP production. We fed WT and GPR43−/−mice with 300 mM butyrate for 21 days, and gut microbiota assessed by 16s rRNA pyrosequencing of fecal samples. In GPR43−/−mice, the abundance of families of Prevotellaceae, Erysipelotrichaceae, and Alcaligenaceae was decreased compared to WT mice (Fig. 3B). At genus level, Alistipes and Lactobacillus were increased while Alloprevotella, Blautia, and Odoribacter were decreased (Fig. 3C), indicating that GPR43 signaling regulated gut microbiota composition. Feeding WT mice butyrate resulted in deviation of the microbial population toward an increased proportion of Bacteroidetes phylum (54.7% to 64.2%, including Prevotellaceae, p=0.06) but decreased proportion of Proteobacteria phylum (7.3% to 4.0%, including Helicobacteraceae and Alcaligenaceae, p=0.05) and Firmicutes phylum (31.4% to 25.5%, including Lachnospiraceae and Erysipelotrichaceae) (Fig. 3A). However, in GPR43−/−mice, the increased proportion of Bacteroidetes phylum by feeding butyrate was decreased (from 51% to 55%) compared with WT mice. Furthermore, butyrate did not affect proportion of Proteobacteria phylum in GPR43−/−mice. We also found that butyrate feeding increased the abundance of Prevotellaceae family but decreased the abundance of Alcaligenaceae family in WT mice but not in GPR43−/−mice (Fig. 3B). At genus level, butyrate feeding increased abundance of Alloprevotella and Odoribacter but decreased abundance of Blautia and Lactobacillus in WT mice but not in GPR43−/−mice (Fig. 3C). However, in contrast to the mice treated with antibiotics (Fig. 2), feeding butyrate without depletion of gut bacteria did not increase IEC expression of RegIIIγ and β-defensins in both WT and GPR43−/−mice (Fig. S3), possibly due to the high levels of pre-existing SCFA produced by gut microbiota in the intestines.
Figure 3. Butyrate feeding changes gut microbiota.
WT and GPR43−/−C57BL/6 mice were fed with 300 mM butyrate for 21 days. Gut microbiota prior and after feeding butyrate was determined by 16s rRNA sequencing. Bar charts for fecal bacterial composition of phylum level (A), family level (B), and genus level (C). N=4–5 mice/group.
3. SCFA promote expression of RegIIIγ and β-defensins in both mouse and human IEC in vitro in a restricted time window
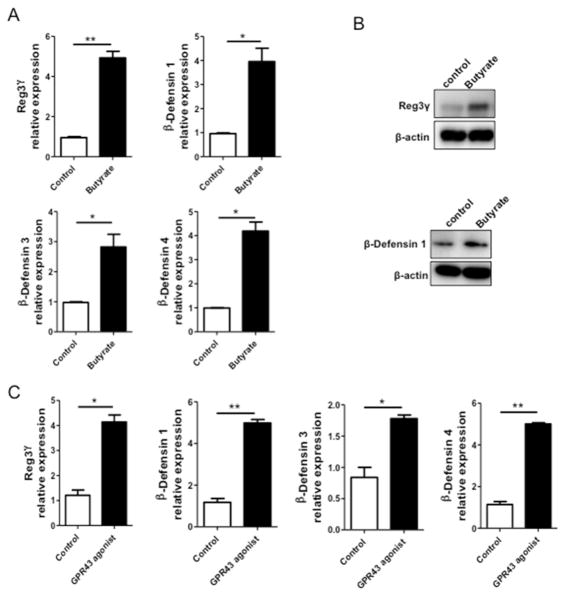
To determine whether SCFA directly promote the IEC expression of AMP in vitro, we utilized intestinal epithelial cell lines and examined RegIIIγ and β-defensins expression after butyrate stimulation. We first used mouse small intestinal epithelial (MSIE) cells, a conditionally immortalized epithelial cell line that retains properties of primary IEC26. We treated MSIE cells with butyrate for 48 h and measured RegIIIγ and β-defensins at both gene expression and protein levels. Treatment with butyrate significantly induced the expression of RegIIIγ, β-defensin 1, 3, 4 (Figs. 4A and B). We then determined the kinetics of the butyrate-induced IEC expression of RegIIIγ and β-defensins. We treated the MSIE cells with butyrate and measured RegIIIγ and β-defensins with respect to time over the course of 72 h. Butyrate treatment did not induce expression of RegIIIγ and β-defensins in early time points of 1 – 12 hrs. Their expression was slightly increased at 24 h, but did not reach significance. Consistently, the expression of RegIIIγ and β-defensins was significantly increased at 48 h, but returned to levels similar to those in controls by 72 h (Fig. S4), indicating that butyrate stimulation occurs within a narrow time window. Treatment with GPR43 agonist elicited similar effect on RegIIIγ and β-defensin 1, 3, 4 expression (Fig. 4C), indicating that GPR43 may mediate the butyrate-induced expression of RegIIIγ and β-defensins in IEC.
Figure 4. Butyrate and GPR43 agonist induce expression of RegIIIγ and β-defensins in mouse IEC.
MSIE cells were treated with 0.5 mM butyrate for 48 h. (A) The expression of RegIIIγ and β-defensins mRNA determined by qRT-PCR and normalized against gapdh. (B) Protein levels of RegIIIγ and β-defensin 1 determined by Western blot. (C) MSIE cells were treated with 5 μM GPR43 agonist. The expression of RegIIIγ and β-defensins were determined by qRT-PCR at 48 h. *p<0.05; **p<0.01. Data are reflective of 3 independent experiments.
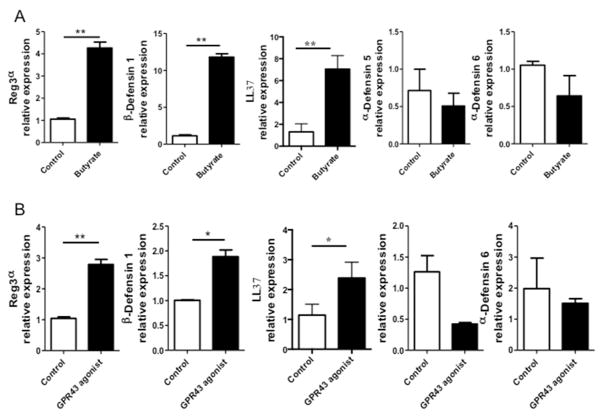
To investigate if SCFA also regulate expression of AMP in human IEC, a human colonic epithelial cell line (HT-29) was stimulated with 0.5 mM butyrate for 48 h. Consistently, mRNA expression levels of RegIIIα, the human ortholog of mouse RegIIIγ, β-defensin 1, and LL-37 were enhanced in butyrate-treated HT-29 cells (Fig. 5A). However, butyrate did not affect the expression of α-defensin 5 and α-defensin 6 in HT-29 cells (Fig. 5A). We also demonstrated a similar effect of acetate and propionate on HT-29 cells. While propionate induced mRNA expression of both RegIIIα and β-defensin 1, the expression of α-defensins remained unaffected. Acetate induced β-defensin 1 but not RegIIIα and α-defensins (Fig. S5). Moreover, GPR43 agonist promoted HT-29 cell expression of RegIIIα, β-defensin 1, and LL-37 but not α-defensin 5 and α-defensin 6 (Fig. 5B).
Figure 5. Butyrate and GPR43 agonist induce expression of RegIIIγ and β-defensins in human IEC.
HT-29 cells were treated with 0.5 mM butyrate for 48 h. (A) The expression of α-defensins, β-defensins, RegIIIα, and LL37 were determined by qRT-PCR and normalized against gapdh. (B) HT-29 cells were treated with 5 μM GPR43 agonist 48 h, and the expression of α-defensin 6, β-defensin 1, RegIIIα, and LL37 was determined by qRT-PCR. *p<0.05; **p<0.01. Data are reflective of 3 independent experiments.
4. SCFA induce expression of RegIIIγ and β-defensins in WT but not GRP43−/−small intestinal epithelial enteroids
We next sought to determine if IEC expression of GPR43 directly mediates SCFA-induced AMP production. We established an ex-vivo 3-D enteroid culture to recapitulate the comprehensive intestinal microenvironment. Enteroids were generated from WT and GRP43−/−mice respectively, and treated with butyrate or PBS control for 48 h. Butyrate treatment did not affect the viability of enteroids as well as the expression of MKI67 (gene for proliferation) and Lgr5 (gene for stemness) (Fig. S6). As shown in Fig 6, butyrate treatment induced the expression of RegIIIγ and β-defensin 1 in WT enteroids, whereas the effect was abrogated in GPR43−/−organoids. This data confirmed that GPR43 mediates IEC production of RegIIIγ and β-defensins induced by butyrate.
Figure 6. Butyrate induces expression of RegIIIγ and β-defensins in WT but not GPR43−/−intestinal epithelial enteroids.
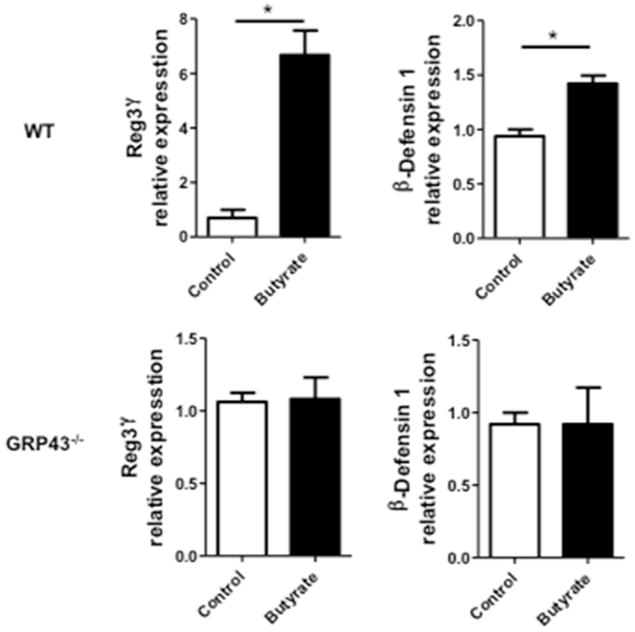
Intestinal epithelial enteroids were generated from either WT or GPR43−/−mice, and treated with 0.5 mM butyrate. The expression of RegIIIγ and β-defensins were determined by qRT-PCR at 48 h and normalized against gapdh. *p<0.05. Data are reflective of 2 independent experiments.
5. mTOR mediates SCFA induction of RegIIIγ and β-defensins in IEC
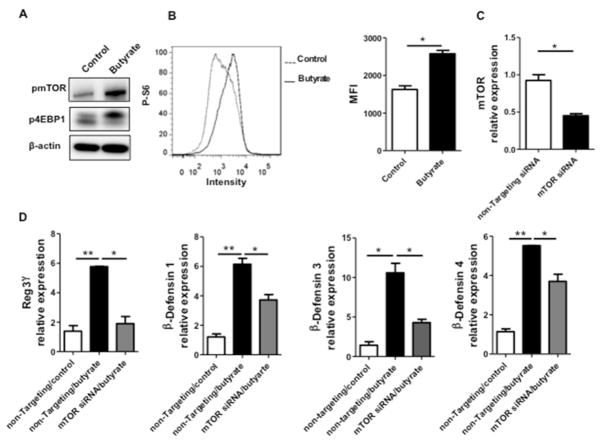
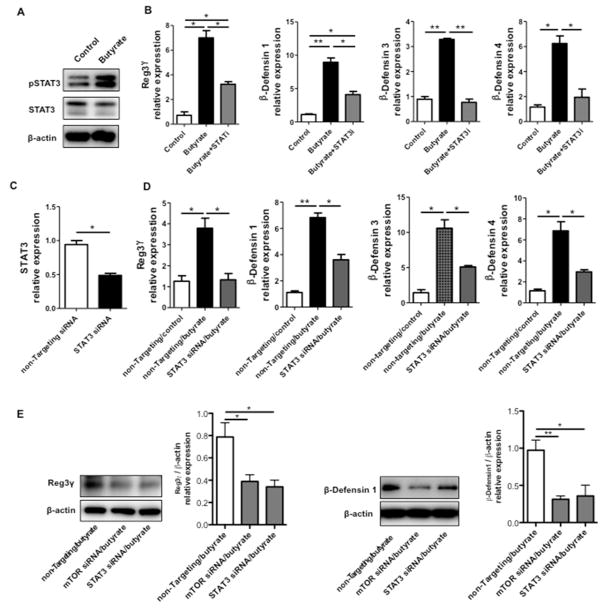
The mammalian target of rapamycin (mTOR), an evolutionarily conserved integrative serine-threonine kinase, is a crucial regulator of physiology of various cells in response to environmental cues and nutrients27. mTOR functions through phosphorylating the eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and p70 ribosomal S6 kinase 1 (S6K1). SCFA have been shown to activate mTOR in T cells and dendritic cells (DC)13,28. We thus investigated whether the mTOR pathway is involved in SCFA-induced RegIIIγ and β-defensins production in IEC. We first treated MSIE cells with butyrate and measured mTOR activation by detecting both phosphorylated-mTOR (p-mTOR) and phosphorylated-4EBP1 (p-4EBP1). As shown in Fig. 7A, butyrate promoted the expression of p-mTOR and p-4EBP1. Consistently, we detected activation of S6K1 by flow cytometry in butyrate-treated MSIE cells (Fig. 7B). Moreover, butyrate activation of mTOR signaling was via GPR43, as evidenced by the activation of mTOR downstream effector S6K1 by a GPR43 agonist (Fig. S7). To determine that butyrate-induced mTOR activation contributes to IEC production of RegIIIγ and β-defensins, we used mTOR siRNA to specifically knockdown mTOR. MSIE cells were transfected with mTOR siRNA or control non-targeting siRNA, with a transfection efficiency around 50% (Fig. 7C). The cells were then treated with butyrate. Knockdown of mTOR impaired butyrate-induced expression of RegIIIγ and β-defensins at the levels of both mRNA (Fig. 7D) and proteins (Fig. 8E). Collectively, these data demonstrated that butyrate activates the mTOR pathway to positively regulate RegIIIγ and β-defensins production in IEC.
Figure 7. mTOR regulates butyrate induction of RegIIIγ and β-defensins in IEC.
MSIE cells were treated with 0.5 mM butyrate for 1 h. (A) Phosphorylation of mTOR and 4E-BP1 were determined by Western blot, with β-actin as a loading control. (B) The expression of phosphorylated S6K1 was determined by flow cytometry at 48 h post butyrate treatment. Combined median fluorescence intensity (MFI) was presented. (C and D) MSIE cells were transfected with mTOR siRNA or control non-targeting siRNA, followed by 0.5 mM butyrate for 48 h. (C) siRNA knockdown efficiency was confirmed by RT-PCR at 40 h post-transfection. (D) The expression of RegIIIγ and β-defensins were determined by qRT-PCR and normalized against gapdh 48 h post-treatment. *p<0.05; **p<0.01. Data are reflective of 3 independent experiments.
Figure 8. STAT3 regulates butyrate induction of RegIIIγ and β-defensin in IEC.
MSIE cells were treated with 0.5 mM butyrate for 1 h. (A) Phosphorylation of STAT3 was determined by Western blot, with total STAT3 and β-actin as loading controls. (B) MSIE cells were treated with 0.5 mM butyrate in the presence or absence of 5 μM STAT3 inhibitor HJC0152. The expression of RegIIIγ and β-defensins was determined by qRT-PCR at 48 h and normalized against gapdh. (C and D) MSIE cells were transfected with STAT3 siRNA or control non-targeting siRNA and then treated with 0.5 mM butyrate. (C) siRNA knockdown efficiency was confirmed by RT-PCR at 40 h post-transfection. (D) The expression of RegIIIγ and β-defensins were determined by qRT-PCR 48 h post-transfection and normalized against gapdh. (E) Protein levels of RegIIIγ and β-defensin were determined by Western blot and combined protein/β-actin relative expression was presented. *p<0.05; **p<0.01. Data are reflective of 3 independent experiments.
6. STAT3 regulates butyrate promotion of RegIIIγ and β-defensins production in IEC
It has been shown that signal transducers and activator of transcription 3 (STAT3) is required for RegIIIγ production in lung epithelium29. Likewise, it is also necessary for IL-22-induced RegIIIγ and β-defensins production in human IEC and alveolar epithelium30–32. We postulated that STAT3 regulates SCFA induction of AMP production in IEC. Administration of butyrate led to activation of STAT3 in MSIE cells, evidenced by enhanced phosphorylation of STAT3 (Fig. 8A). Blockade of STAT3 using a specific STAT3 inhibitor HJC015233 greatly attenuated butyrate-induced RegIIIγ and β-defensins mRNA expression (Fig. 8B). Secondly, we knocked down STAT3 in MSIE cells with specific siRNA. The obtained transfection efficiency was about 50% (Fig. 8C). In accordance with the data of the STAT3 inhibitor, the knockdown of STAT3 resulted in decreased butyrate-induced IEC expression of RegIIIγ and β-defensins at the levels of both mRNA and proteins (Figs. 8D and E). Together, these data indicated that STAT3 is critical for the butyrate-induced AMP production in IEC.
Discussion
The interplay between gut commensal bacteria and the host provides beneficial effects on host metabolism as well as immune regulation34–36. In this report, we have demonstrated that gut microbiota metabolites SCFA promoted IEC expression of RegIIIγ and β-defensins 1, 3, and 4 through GPR43, essential for the maintenance of intestinal homeostasis.
Gut microbiota has been shown to be important for the production of certain AMP in the intestines, in that RegIIIγ and certain β-defensins are virtually absent in GF mice4,5. Mice lacking TLR ligands also displayed impaired production of RegIIIγ and RegIIIβ in IEC compared to WT mice, suggesting that microbiota can directly affect IEC AMP production through interaction with TLRs6,7. However, substantial amounts of AMP have been detected in MyD88−/−mice housed under SPF conditions4–7. This indicates the presence of multiple sensors and mechanisms for AMP regulation by microbiota, in addition to direct TLR-TLR ligand interactions. In the current report, we investigated whether SCFA regulated the IEC production of RegIII and the defensin family. Indeed, extraneous butyrate administration promoted IEC production of RegIIIγ, β-defensin 1, 3, and 4, and CRAMP (Fig. 2). This was further supported by in vitro studies using human and murine intestinal cell lines (Fig. 4 and 5). Interestingly, a previous report showed that SCFA also promoted β-defensin expression in porcine IEC by using porcine IPEC-J2 intestinal epithelial cells37, indicating that SCFA can function across different species to promote IEC expression of AMP.
It has been shown that SCFA bind cell-surface receptors such as GPR41, GPR43, and GPR109α13,38. SCFA-GPR43 interaction has been reported in regulating intestinal inflammatory responses39, in that GPR43−/−mice demonstrated exacerbated colitis after dextran sodium sulfate (DSS) insult40. This further indicates that SCFA affects the intestine via the GPR43 receptor. More recently, it was discovered that low fiber diets induced and perpetuated intestinal inflammation, whereas high fiber diets protected against colitis41. The beneficial effect of high fiber intake derives from gut microbiota metabolic products, and subsequent SCFA binding to GPR43 and GPR109a in the intestines. Further studies revealed that SCFA activated the NLRP3 inflammasome through GPCR signaling, which conferred resistance to colitis41. Our data demonstrated that production of RegIIIγ and β-defensins in IEC was impaired in GPR43−/−mice (Fig. 1). Additionally, administration of a GPR43 agonist enhanced AMP production in both murine and human IEC, indicating that GPR43 is involved in AMP production. Furthermore, butyrate promoted expression of RegIIIγ and β-defensins in WT but not in GPR43−/−intestinal epithelial enteroids (Fig. 6). SCFA, especially butyrate, have been shown as HDAC inhibitors, and butyrate-induced HDAC3 inhibition regulates IEC production of retinoic acid, which can potentially contribute to maintenance of intestinal homeostasis42. Our data do not rule out the involvement of HDAC-inhibition in SCFA induction of AMP in IEC. It is very likely that SCFA regulate IEC function via multiple mechanisms.
mTOR is a key regulator in various physiological aspects43. Among different cell types, SCFA have been shown to activate mTOR in T cells and DCs27,44. We showed that SCFA activated the mTOR pathway in IEC (Fig. 7A). Knockdown of mTOR attenuated SCFA-induced AMP production, indicating that activation of mTOR facilitated RegIIIγ and β-defensins production in IEC. Interestingly, butyrate also activated STAT3, which has been implicated in the regulation of immune responses of both innate and adaptive immune cells in the intestines45. The blockade of STAT3 signaling compromised RegIIIγ and β-defensins production in SCFA-treated IEC, suggesting that STAT3 is indispensable in SCFA induction of AMP production in IEC (Fig. 8). As activation of STAT3 impacts epithelial cell viability, it is very likely that SCFA activation of STAT3 supports intestinal organoid stemness proliferation, which will affect the AMP secretion by an indirect mechanism as well. Therefore, SCFA activation of mTOR and STAT3 could collaboratively promote AMP expression in IEC.
RegIIIγ and β-defensins secreted by IEC are known for limiting bacteria and manipulating species composition, especially Gram-positive species. Our findings that SCFA induce IEC RegIIIγ and β-defensins production in IEC suggest a possible autoregulation of microbiota. In light of this, drugs targeting the intrinsic pathways of SCFA-mediated AMP production might serve as promising candidates for clinical applications by promoting host barrier defense and correcting dysbiosis, and restoring homeostatic balance. Nevertheless, the modification of AMP requires carefully monitoring, and dysregulated AMP production can have negative impacts on intestinal homeostasis. Indeed, it was discovered that excessive lipocalin-2 and calprotectin production induced by IL-22 can, in turn, suppress commensal bacteria, resulting in pathogen colonization46. In order to preserve intestinal homeostasis, critical elements of host immunity and microbial colonization need to counterbalance each other. To that end, we present evidence that metabolites from commensal microbiota are capable of inducing host innate immunity to regulate the intestinal environment.
Moreover, we demonstrated that GPR43 signaling regulated gut microbiota and feeding butyrate resulted in alterations in the composition of the microbiota (Fig. 3). Among the bacteria affected by GPR43 signaling, the abundance of Prevotellaceae was decreased in GPR43−/−mice compared to WT mice. Feeding WT mice but not GPR43−/−mice butyrate increased the abundance of Prevotellaceae. A recent report by Jiminez et al demonstrated more potent effects of butyrate on gut microbiota by rectal administration of 140 mM butyrate, especially when mice developed colitis upon infection with Citrobacter rodentium47. The difference of the data between Jiminez et al and our current study is likely due to the different routes of butyrate administration. While butyrate by rectal administration would mostly go to the colon, due to potential absorption, only a small portion of oral administered butyrate would reach the intestinal lumen to affect gut microbiota. Even so, in consistent with our results, their study also demonstrated an increased proportion of Bacteroidetes phylum but decreased Firmicutes phylum after rectal administration of butyrate47. While our data demonstrated that butyrate promotes IEC AMP production, which can in turn affects microbiota composition, direct antimicrobial effects of butyrate have also been reported48, both of which can contribute to butyrate-induced microbiota alterations. Whether these changes directly or indirectly affect butyrate regulation of IEC expression of AMP cannot be stated at this point; however, we showed that SCFA-producing phyla were altered by feeding butyrate. While the Bacteroidetes phylum, which produces mainly acetate and propionate, was increased, members of the Firmicutes phylum, which preferentially produce butyrate49, were decreased. Thus, we are still not clear, at this point, whether and at what levels the butyrate-mediated alternations of gut microbiota contribute to butyrate induction of IEC AMP expression.
Materials and Methods
Mice
C57BL/6 mice were purchased from the Jackson Laboratory and housed in the Animal Resource Center at UTMB. GPR43−/−(Ffar2tm1Lex) mice were obtained from Bristol-Myers Squibb and maintained in the Animal Resource Center at UTMB. All experiments were reviewed and approved by the Institutional Animal Care and Use Committees of UTMB.
Reagents
OPTI-MEM were purchased from Life Technologies. GPR43 agonist (phenylacetamide agonist #44, Amgen Inc)50 was kindly provided by Dr. Warren N. D’Souza, Amgen Inc. STAT3 inhibitor HJC0152 was kindly provided by Dr. Jia Zhou, University of Texas Medical Branch. McCoy’s 5A and Lipofectamine RNAiMAX Transfection Reagent were purchased from Thermo Fisher Scientific. Sodium butyrate (MKBZ0227V), sodium propionate (SLBL9015V), and sodium acetate (SLBH4840V) were purchased from Sigma-Aldrich. Predesigned siRNA directed against mouse mTOR (SI01005683), STAT3 (SI01435287), and negative control siRNA (1027310) were purchased from Qiagen. Antibodies against phosphor-STAT3 (Tyr705), phosphor-mTOR (S2448), phosphor-4E-BP1 (T70), phosphor-p44/42 MAPK (Thr202/Tyr204), phosphor-NF-κB p65 (Ser536), phosphor-S6k1 Ribosomal protein (S235/236) (D57.2.2E), β-actin, and HRP-conjugated anti-rabbit secondary Ab were purchased from Cell Signaling Technology.
SCFA treatment
Mice were given drinking water containing 1g/l metronidazole (Sigma-Aldrich), 0.5g/l vancomycin (Hospira), 1g/l ampicillin (Sigma-Aldrich), and 1g/l kanamycin (Fisher Scientific) for 10 days, followed by SCFA feeding. Butyrate or acetate were added to drinking water with a final concentration of 300 mM. Mice were sacrificed 21 days later for subsequent studies.
Isolation of IEC
After cleaning, small intestines were sliced into small pieces (<0.5 cm), and incubated with 5mM EDTA in HBSS buffer containing 5% FBS at 37°C for 40 min. Cells were collected by passaging supernatant through a 100-μM cell strainer (BD Falcon). After washing with PBS, IEC were separated from a 20%/40% Percoll interface (Amersham Pharmacia Biotech).
Epithelial cell culture
MSIE cells26 were maintained in RPMI 1640 medium with 5 U/ml murine IFNγ, ITS (5 μg/ml insulin, 5 μg/ml transferrin, and 5 ng/ml selenous acid), 100 U/ml penicillin/streptomycin, and 5% FBS at 33°C and 5% CO2. After reaching 80% confluence, cells were starved at 37°C for 16 h before subsequent experiments. HT-29 cells were cultured in complete McCoy's 5A Medium with 10% FBS at 37°C and 5% CO2.
siRNA transfection
siRNA transfection in MSIE cells was performed by using Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer’s instructions. Briefly, 3×105 MSIE cells were incubated with 50 nM siRNA and 10 μl LipofectamineRNAiMAX Transfection reagent in 1ml OPTI-MEM medium for 6 h at 33°C, followed by normal growth medium for 40 h at 33°C. MSIE cells were then starved for 6 h at 33°C before treatment. Transfection efficiency was determined at 40 h post transfection. The expression of RegIIIγ and β-defensins was determined at 48 h after administration with SCFA.
Flow cytometry
MSIE cells were treated with SCFA for 48 h then stained for Live/Dead dye and phospho-S6k1 (S235/236). Cells were sampled on a LSRII Fortessaflow cytometer (BD Biosciences), and data analyzed by using FlowJo7.6 software (Tree Star).
Western blot
Total protein of cells was extracted and the concentration determined by a BCA Protein Assay kit. Four μg of protein was separated electrophoretically by NuPAGEBis-Tris mini gels (Life Technologies) and probed with phospho-mTOR, phospho-4EBP1, phospho-STAT3, phosphor-p44/42 MAPK, or phosphor-NF-κB p65 Abs overnight, followed by 1h HRP-conjugated anti-rabbit secondary Ab. Membranes were stripped and re-probed for actin as loading control. Protein/β-actin relative expression was calculated using ImageJ (10.2) software.
Quantitative Real-time PCR
Total RNA was extracted with TRIzol (Life Technologies, Carlsbad, CA) and quantified for cDNA synthesis. Quantitative real-time PCR reactions were performed by using SYBR Green Gene Expression Assays (Bio-Rad, Hercules, CA, USA). The primers were listed in supplementary Table 1. All data were normalized to GAPDH mRNA expression.
Enteroid culture
The intestines were cut longitudinally, minced, and treated with 2 mM EDTA. The tissues were then treated with PBS containing 43.3 mM sucrose and 54.9 mM sorbitol, and filtered through a 70-μm cell strainer. Supernatant was collected and centrifuged. The resulting pellet containing detached crypts was re-suspended in Matrigel (BD Bioscience) with 0.5μg/ml rEGF (R&D Systems2028-EG), 1μg/ml rNoggin (R&D Systems1967-NG/CF), 5μg/ml rR-spondin (R&D Systems 3474-RS), and 1μg/ml rWnt3a (R&D Systems 35036-WN/CF). 50μl Matrigel with 500 crypts was plated, and advanced DMEM/F12 media (Invitrogen 12634-010) with 1x N2 supplement (R&D Systems AR009), and 1x B27 (Invitrogen 12587-010). Five days later, the enteroids were treated with 0.5 mM butyrate.
16S rRNA pyrosequencing analysis
Fecal bacterial DNA was isolated using a MoBio PowerFecal kit (MoBio, USA). The isolated DNA was amplified using universal V3-V4 16S rRNA V3-V4 region primers. Sequencing was performed with an Illumina MiSeq instrument. The raw sequencing reads were trimmed and filtered based on initial quality assessment using FASTQC. The reads were trimmed to 350 bases and filtered to exclude reads with low quality, two or more unknown characters, sequencing adapters. To identify the presence of known bacteria, the subsequences were analyzed using CLC Genomics Workbench 8.5 Microbial Genomics Module. Reference based OTU picking was performed using the SILVA SSU v119 97% database. Sequences present in more than one copy but not clustered to the database were then placed into de-novo OTUs (97% similarity) and aligned against the database with 80% similarity threshold using MUSCLE alignment.
Statistical analysis
Student’s t test in Prism 5.0 (Graphpad) was used to determine levels of significance for comparisons between samples. Where appropriate, mean ± SEM is represented on graphs. *p< 0.05; **p < 0.01.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK098370, DK105585, and DK112436, and John Sealy Memorial Endowment Fund (to YC). We appreciate Dr. Linsey Yeager of The University of Texas Medical Branch for proofreading the manuscript.
Footnotes
Author Contributions: Y.Z. and F.C. performed experiments, analyzed data and wrote the manuscript. W.W., M.M, A.J.B., S.Y., Y.X., and X.H. performed part of experiments, analyzed data and reviewed the manuscript. G.G., Y.X., and Y.F. performed and analyzed 16s rRNA pyrosequencing. T.D.E., W.D., Q.Z., and Z.L. provided the reagents and revised the manuscript. Y.C. conceived the project, designed the experiments, and wrote the manuscript.
Conflict of Interest: No authors have conflicting financial interests.
References
- 1.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nature reviews. Microbiology. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nature immunology. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature immunology. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 6.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. The Journal of experimental medicine. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nature immunology. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghikia A, et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity. 2015;43:817–829. doi: 10.1016/j.immuni.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Smith PM, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macia L, et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunological reviews. 2012;245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune network. 2014;14:277–288. doi: 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. American journal of physiology. Cell physiology. 2014;307:C979–985. doi: 10.1152/ajpcell.00228.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AJ, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. The Journal of biological chemistry. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 16.Alex S, et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Molecular and cellular biology. 2013;33:1303–1316. doi: 10.1128/MCB.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 20.Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun. 2002;70:953–963. doi: 10.1128/iai.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schauber J, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauber J, et al. Histone-deacetylase inhibitors induce the cathelicidin LL-37 in gastrointestinal cells. Mol Immunol. 2004;41:847–854. doi: 10.1016/j.molimm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Steinmann J, Halldorsson S, Agerberth B, Gudmundsson GH. Phenylbutyrate induces antimicrobial peptide expression. Antimicrob Agents Chemother. 2009;53:5127–5133. doi: 10.1128/AAC.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly CJ, et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut. 1994;35:S35–38. doi: 10.1136/gut.35.1_suppl.s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehead RH, Robinson PS. Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. American journal of physiology. Gastrointestinal and liver physiology. 2009;296:G455–460. doi: 10.1152/ajpgi.90381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurav A, et al. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. The Biochemical journal. 2015;469:267–278. doi: 10.1042/BJ20150242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi SM, et al. Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. The Journal of experimental medicine. 2013;210:551–561. doi: 10.1084/jem.20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li A, et al. IL-22 Up-Regulates beta-Defensin-2 Expression in Human Alveolar Epithelium via STAT3 but Not NF-kappaB Signaling Pathway. Inflammation. 2015;38:1191–1200. doi: 10.1007/s10753-014-0083-z. [DOI] [PubMed] [Google Scholar]
- 31.Murano T, et al. Hes1 promotes the IL-22-mediated antimicrobial response by enhancing STAT3-dependent transcription in human intestinal epithelial cells. Biochemical and biophysical research communications. 2014;443:840–846. doi: 10.1016/j.bbrc.2013.12.061. [DOI] [PubMed] [Google Scholar]
- 32.Sovran B, et al. IL-22-STAT3 pathway plays a key role in the maintenance of ileal homeostasis in mice lacking secreted mucus barrier. Inflammatory bowel diseases. 2015;21:531–542. doi: 10.1097/MIB.0000000000000319. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, et al. Discovery of -Alkylamino Tethered Niclosamide Derivatives as Potent and Orally Bioavailable Anticancer Agents. ACS medicinal chemistry letters. 2013;4:180–185. doi: 10.1021/ml3003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 35.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature reviews. Microbiology. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng X, et al. Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PLoS One. 2013;8:e72922. doi: 10.1371/journal.pone.0072922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Current opinion in pharmacology. 2013;13:869–874. doi: 10.1016/j.coph.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 39.McKenzie CI, Mackay CR, Macia L. GPR43 - A Prototypic Metabolite Sensor Linking Metabolic and Inflammatory Diseases. Trends in endocrinology and metabolism: TEM. 2015;26:511–512. doi: 10.1016/j.tem.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macia L, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nature communications. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 42.Schilderink R, et al. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. American journal of physiology. Gastrointestinal and liver physiology. 2016;310:G1138–1146. doi: 10.1152/ajpgi.00411.2015. [DOI] [PubMed] [Google Scholar]
- 43.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haidinger M, et al. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. Journal of immunology. 2010;185:3919–3931. doi: 10.4049/jimmunol.1000296. [DOI] [PubMed] [Google Scholar]
- 45.Ernst M, Thiem S, Nguyen PM, Eissmann M, Putoczki TL. Epithelial gp130/Stat3 functions: an intestinal signaling node in health and disease. Seminars in immunology. 2014;26:29–37. doi: 10.1016/j.smim.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Behnsen J, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiminez JA, Uwiera TC, Abbott DW, Uwiera RRE, Inglis GD. Butyrate Supplementation at High Concentrations Alters Enteric Bacterial Communities and Reduces Intestinal Inflammation in Mice Infected with Citrobacter rodentium. mSphere. 2017;2 doi: 10.1128/mSphere.00243-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Namkung H, Yu H, Gong J, Leeson S. Antimicrobial activity of butyrate glycerides toward Salmonella Typhimurium and Clostridium perfringens. Poult Sci. 2011;90:2217–2222. doi: 10.3382/ps.2011-01498. [DOI] [PubMed] [Google Scholar]
- 49.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, et al. The first synthetic agonists of FFA2: Discovery and SAR of phenylacetamides as allosteric modulators. Bioorganic & medicinal chemistry letters. 2010;20:493–498. doi: 10.1016/j.bmcl.2009.11.112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.