Abstract
The transcription factor AhR modulates immunity at multiple levels. Here we report phagocytes exposed to apoptotic cells exhibited rapid activation of AhR, which drove production of interleukin 10. Activation of AhR was dependent on interactions between apoptotic-cell DNA and the pattern-recognition receptor TLR9 that was required for prevention of immune responses to DNA and histones in vivo. Moreover, disease progression in murine systemic lupus erythematosus (SLE) correlated with strength of the AhR signal, and disease course could be altered by modulation of AhR activity. Deletion of AhR in the myeloid lineage caused systemic autoimmunity in mice and an increased AhR transcriptional signature correlated with disease in patients with SLE. Thus, AhR activity induced by apoptotic cell phagocytes maintains peripheral tolerance.
Phagocytic removal of apoptotic cells (efferocytosis) initiates a series of immunoregulatory events including the expression of indoleamine 2,3 dioxygenase (IDO), interleukin 10 (IL-10) and transforming growth factor β (TGF-β) in myeloid cells and the recruitment of regulatory T cells1, 2, 3. However, when these regulatory processes are disrupted, apoptotic cells can induce significant inflammation that may overcome tolerogenic mechanisms1, 4. Defects in apoptotic cell recognition and clearance mechanisms or downstream tolerogenic pathways cause systemic autoimmunity in mice, generally with characteristics of systemic lupus erythematous (SLE). Similarly, genetic and experimental evidence suggest altered apoptotic cell clearance is a primary factor driving disease in SLE 5, 6, 7, 8.
The aryl hydrocarbon receptor (AhR) is a receptor and transcription factor important in xenobiotic metabolism9 and serves a key function in immunity. Upon activation, AhR is released from a chaperone complex that anchors it in the cytoplasm9, 10, 11, translocates to the nucleus and drives transcriptional activity10. In immune cells, AhR has a dominant impact on phenotype controlling the expression of cytokines, including IL-10, type I interferons, IL-12, IL-17 and TGF-β 10, 12, 13, 14, 15, 16, 17, 18.
Here we present genetic and pharmacologic evidence that DNA exposed by apoptotic cell death drove TLR9-dependent activation of AhR and downstream immune suppression and tolerance. Myeloid lineage AhR-deficient mice developed progressive pathology and autoimmunity reminiscent of SLE and an AhR transcriptional signature was associated with human SLE. These observations identify a previously unknown role of AhR in self-tolerance to apoptotic cells.
Results
Activation of AhR by apoptotic cells drives IL-10 production
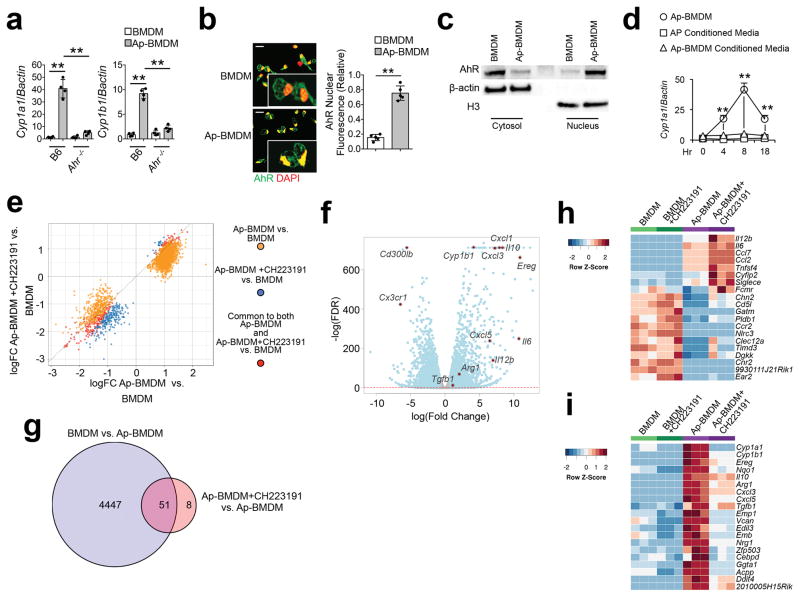
We examined the function of AhR in macrophages in an in vitro model of efferocytosis using bone marrow-derived macrophages (BMDM) or bone marrow-derived dendritic cells (BMDC) co-cultured with apoptotic thymocytes. Because cytochrome P4501A1 (Cyp1A1) and P450B1 (Cyp1B1) are strongly induced by AhR9, 10, 11, we used Cyp1a1 and Cyp1b1 mRNA as markers of AhR transcriptional activity. BMDM and BMDC co-cultured with apoptotic cells (hereafter defined as Ap-BMDM or Ap-BMDC) induced Cyp1a1 and Cyp1b1 mRNA by 8 hours of culture (Fig. 1a), which was abrogated in Ahr−/− BMDM (Fig. 1a). Immunofluorescence and Western blot analysis of BMDM before exposure to apoptotic cells showed cytoplasmic AhR localization, while addition of apoptotic cells induced a significant increase in nuclear AhR (Fig. 1b,c). We observed a similar pattern of AhR-dependent Cyp1a1 and Cyp1b1 mRNA expression in Ap-BMDCs (Supplementary Fig. 1a), indicating apoptotic cells induce AhR activity in efferocytic BMDC and BMDM.
Figure 1. Apoptotic cells activate AhR in resident macrophages driving regulatory polarization.
(a) BMDM of the indicated genotype were co-cultured with B6 apoptotic thymocytes for 8h and indicated mRNA were measured by sqPCR. Data are normalized to expression of β-actin. (b) Nuclear translocation of AhR determined by immunofluorescence 2h after co-culture described in a. Scale bar=20μm. (c) Immunoblot analysis of nuclear and cytoplasmic extracts 2h after co-culture as in a. β-actin and histone H3 were used as loading controls for cytoplasmic and nuclear extracts respectively. (d) B6 BMDM were cultured with apoptotic cells as described in a (Ap-BMDM) or cultured in conditioned media from apoptotic thymocyte cultures (Ap Conditioned Media), or from 8h Mϕ/apoptotic cell cultures (Ap-BMDM Conditioned Media) and Cyp1a1 mRNA induction was measured by sqPCR normalized against β-actin. (e) Quadrant plot of DARs identified from ATAC-seq analysis of BMDM versus Ap-BMDM +/− AhR inhibitor. (f) Volcano plot for differential expression based on transcriptome analysis of BMDM versus Ap-BMDM. Red dotted line marks FDR < 0.05. (g) Venn diagram showing significantly differentially expressed genes (FDR < 5%, logFC > ± 0.75) for the comparisons indicated. (h and i) Heat maps showing comparisons of up-regulated h or down-regulated i genes in Ap-BMDM +/− CH223191. For a n=4 and for b and d n=5 biologically independent samples per group +/− standard deviation and **P≤ 0.01 as determined by two sided Student’s t-test. Western blot in c is representative for 3 biologically independent samples and for ATAC- data is representative of 30,000 macrophages per experimental condition. All experiments were repeated three times with similar results.
Apoptotic cell-conditioned media (Fig. 1d) or apoptotic cell transwell cultures (Supplementary Fig. 1b) did not induce Cyp1a1 mRNA in BMDM, indicating AhR activation by apoptotic cells required cell-cell contact. Moreover, conditioned media from Ap-BMDM co-cultures did not induce Cyp1a1 mRNA in BMDM (Fig. 1d) and inhibition of protein synthesis with cycloheximide did not impact Cyp1a1 mRNA expression (Supplementary Fig. 1c), indicating apoptotic cells activated AhR through direct mechanism(s). Neither live nor necrotic cells induced AhR, and the ability to induce AhR in cells undergoing efferocytosis was acquired 3h post-induction of apoptosis (Supplementary Fig. 1d,e). Treatment of apoptotic cells with the pan-caspase inhibitor z-fad abrogated phagocytosis and Cyp1a1 mRNA induction in Ap-BMDMs (Supplementary Fig. 1f,g). Likewise, treatment of apoptotic cells with annexin V for 30 minutes prior to co-culture to mask phosphatidylserine (PS), or addition of cytochalasin D to Ap-BMDM co-cultures to inhibit phagocytosis prevented efferocytosis (Supplementary Fig. 1f) and significantly reduced Cyp1a1 mRNA induction in Ap-BMDM compared to control AP-BMDM cultures (Supplementary Fig. 1g). Thus, apoptosis and dead cell efferocytosis were required for AhR activation.
AhR modulates gene expression directly and indirectly, through epigenetic mechanisms10, 19. ATAC-seq analysis20 of BMDM showed efferocytosis drove large-scale changes in active chromatin, with 2736 differentially accessible regions (DARs) detected in Ap-BMDM compared to resting BMDM (Fig. 1e). In contrast, addition of the AhR inhibitor CH22319121 to Ap-BMDM co-cultures caused a modest change in active chromatin compared to Ap-BMDM without CH223191 addition, with only 230 unique DARs and 247 shared DARs in CH223191-treated Ap-BMDM compared to Ap-BMDM without AhR inhibition (Fig. 1e); however, changes were generally concordant between Ap-BMDM with or without Ahr inhibition (Fig. 1e), suggesting AhR had a weak impact on chromatin accessibility in Ap-BMDMs.
RNA-seq analysis of Ap-BMDM purified by FACS after 8 hour co-culture with apoptotic thymocytes showed altered expression of 4498 transcripts (FDR<0.05, logFC> +/− 0.75) in Ap-BMDM compared to resting BMDM (Fig. 1f). Among these, the most significant changes in expression occurred in genes with immune regulatory function (e.g. Il10) and linked to AhR responsiveness (e.g. Cyp1b1 and Ereg, encoding epiregulin) (Fig. 1f). Addition of the AhR inhibitor CH223191 altered the expression of 59 genes in Ap-BMDM compared to resting BMDM (Fig. 1g), but did not affect basal mRNA expression. Ingenuity pathway analysis showed AhR inhibition shifted the mRNA profile of Ap-BMDMs from one associated with homeostasis pathways to one associated with inflammation and cellular recruitment (Supplementary Fig. 2). Accordingly, transcript heat maps indicated the down regulation of mRNA associated with immune regulatory polarization (e.g. Il10, Arg1) and increased expression of transcripts associated with inflammation, including Il12p40, Il6 and Tnfs4 (OX40L) in Ahr-inhibited Ap-BMDMs (Fig. 1h,i). These data suggest AhR drives efferocytosis-dependent regulatory polarization in BMDMs.
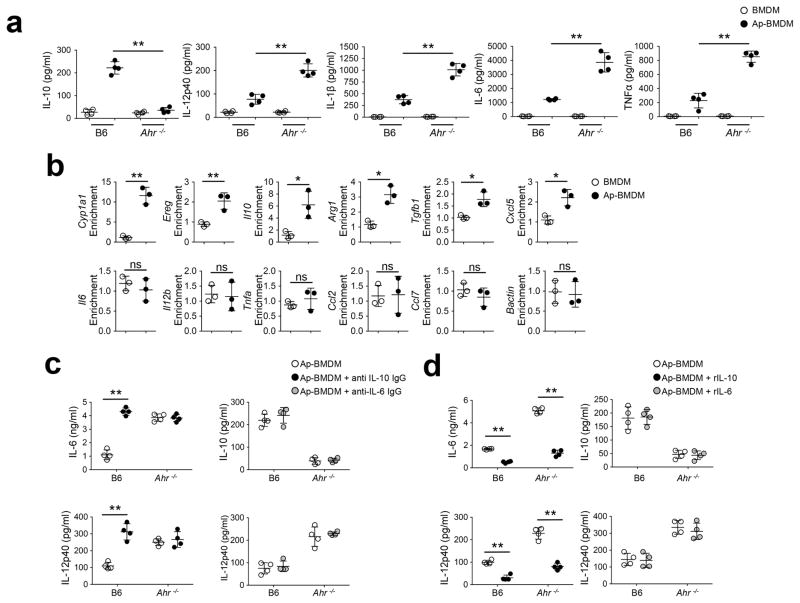
Elisa assays showed Ahr−/− Ap-BMDMs had significantly reduced IL-10 protein expression compared to wild-type Ap-BMDMs, but had increased expression of the proinflammatory cytokines IL-1β, IL-6, IL-12p40 and TNF (Fig. 2a). Because AhR modulates the activity of both the Il10 and Il6 promoters in myeloid cells19, 22, we used chromatin immune-precipitation (ChIP) to examine the binding to AhR responsive elements (AhRE) in Ap-BMDM. AhR association with the AhRE was enriched 13-fold in the Cyp1a1 promoter and 7 fold in the Il10 promoter in Ap-BMDMs compared to resting BMDMs (Fig. 2b). We did not detect AhR association with AhREs in promoters regulating proinflammatory cytokine expression in either Ap-BMDMs or resting BMDMs (Fig. 2b). Because IL-10 plays a key role in limiting inflammation and is induced rapidly in vivo in response to efferocytosis2, we tested whether AhR modulated Ap-BMDM polarization by inducing IL-10. Antibody neutralization of IL-10 increased IL-6 and IL-12p40 protein in wild-type Ap-BMDM cultures (Fig. 2c). IL-10 neutralization had no effect on Ahr−/− Ap-BMDMs (Fig. 2c), suggesting a lack of IL-10 production in Ahr−/− BMDMs post-efferocytosis, while addition of rIL-10 inhibited production of IL-6 or IL-12p40 in Ahr−/− Ap-BMDMs (Fig. 2d), indicating supplementation with IL-10 can restore regulatory responses. In contrast, manipulation of IL-6 activity by addition of neutralizing antibodies or recombinant IL-6 had no effect on the production of IL-10 or IL-12p40 in Ap-BMDM (Fig. 2c,d). Thus, apoptotic cells engulfment drives rapid induction of AhR-dependent IL-10 transcription in BMDMs, which has a key role in BMDM polarization following efferocytosis.
Figure 2. Apoptotic cell-activated AhR drives macrophage polarization and IL-10 production.
(a) BMDM of the indicated genotype were cultured with apoptotic cells as in Fig 1a and ELISA measurements of culture supernatants by ELISA was done after 12h. (b) BMDM were co-cultured with apoptotic cells for 3h and ChIP analysis was done to measure AhR interaction with AhREs in promoters for the indicated. Values represent fold-enrichment for AhREs over input controls. (c, d) ELISA measurements of Ap-BMDM co-culture supernatants done in the presence of neutralizing antibody against IL-10 or IL-6 c, or recombinant IL-10 or IL-6 d as indicated. For a, c, and d n=4 and for b n=3 biologically independent samples per group. Bars are mean value for groups +/− the standard deviation. *=Pvl≤ 0.05, **=P≤ 0.01, ns= not significant as determined by two-sided Student’s t-test. All experiments were repeated four times with similar results. r=recombinant.
Apoptotic cells activate AhR through a TLR9-dependent mechanism
Because AhR responds to diverse ligands generated downstream of IDO-dependent tryptophan metabolism, including kynurenines13, 14, we measured Ido1 expression and activity (i.e. kyurenine production) in Ap-BMDM. We did not detect any changes in Ido1 mRNA (Supplementary Fig. 3a) or increased kynurenine production in Ap-BMDM compared to BMDM (Supplementary Fig. 3b). Moreover, we could observe Cyp1a1 mRNA induction by sqPCR analysis in Ap-BMDMs treated with the IDO inhibitor 1MT (Supplementary Fig. 3c) or in Ido1−/− Ap-BMDMs (Supplementary Fig. 3c), suggesting that activation of AhR in Ap-BMDM is independent of IDO.
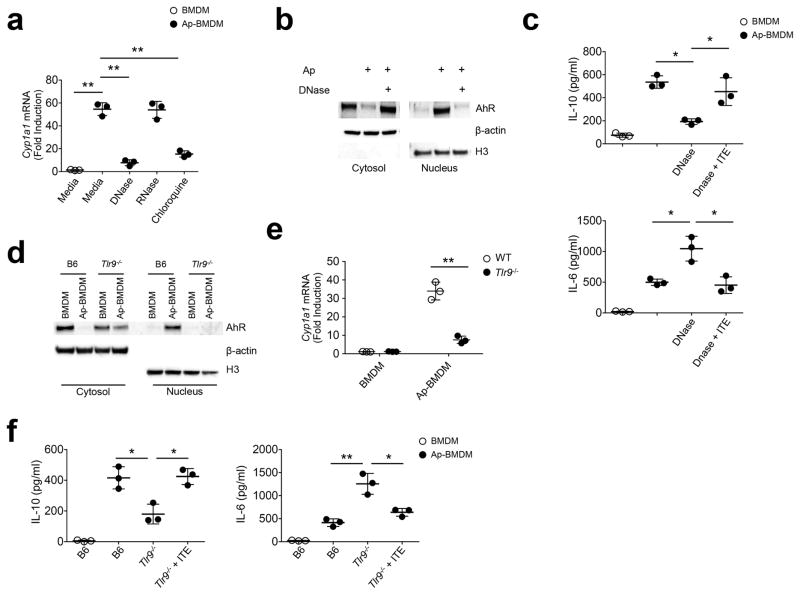
Because DNA exposed on apoptotic blebs was shown to induce expression of IL-10 in splenic B cells23, we examined if DNA from apoptotic cells could activate AhR in BMDMs. Treatment of apoptotic cells with DNAse23 prior to BMDM co-culture abrogated AhR induction (Fig. 3a) and nuclear accumulation (Fig. 3b) in Ap-BMDM and reduced induction of IL-10 compared to control Ap-BMDM co-cultures (Fig. 3c). The latter effect was overcome by the addition of the AhR agonist ITE to DNAse-treated Ap-BMDM co-cultures (Fig. 3c). Treatment of apoptotic cells with RNAse did not impact Cyp1a1 mRNA induction in Ap-BMDMs (Fig. 3a), while addition of chloroquine to Ap-BMDM co-cultures reduced Cyp1a1 expression in Ap-BMDMs (Fig. 3a), indicating lysosome acidification was required for AhR activation. Because TLR9 is the main DNA sensor in the lysosome, we tested its requirement for AhR induction in Ap-BMDMs. Tlr9−/− BMDMs did not show nuclear accumulation of AhR after efferocytosis (Fig. 3d) and both Tlr9−/− Ap-BMDMs and Ap-BMDM treated with the TLR9 antagonist IRS 86924 had significantly reduced induction of Cyp1a1 mRNA compared to wild-type, untreated Ap-BMDM cultures (Fig. 3e and Supplementary Fig. 3d). Inhibition of TLR7 with the oligonucleotide antagonist IRS 66125 did not impact Cyp1a1 mRNA expression (Supplementary Fig. 3d), suggesting RNA in apoptotic cells does not drive AhR activity in Ap-BMDMs. Furthermore, Tlr9−/− Ap-BMDMs showed reduced expression of IL-10 and increased expression of proinflammatory cytokines (Fig. 3f), which could be rescued by addition of the AhR agonist ITE to Ap-BMDM co-cultures (Fig. 3f). These data indicate that apoptotic cell-driven Ahr activation and subsequent IL-10 production requires TLR9 recognition of DNA from apoptotic cells.
Figure 3. Apoptotic cells activate AhR by a TLR9-dependent mechanism.
(a) Apoptotic cells were treated with DNAse (50 μg/ml) or RNAse (50 μg/ml) prior to culture with BMDM +/− 1μM chloroquine. After 8h Cyp1a1 mRNA was measured by sqPCR. (b) Immunoblot analysis of nuclear and cytoplasmic extracts 2h after Ap-BMDMco-culture +/− DNAse treated apoptotic cells. (c) BMDM were co-cultured with apoptotic cells for 8h (Ap-BMDM) with the indicated conditions alone or in combination with ITE at 1 μM. Culture supernatants were then measured for IL-6 and IL-10 by ELISA. (d) BMDM and Ap-BMDM of the indicated genotype were analyzed by immunoblot analysis of nuclear and cytoplasmic extracts 2h after co-culture as described in b. (e) Cyp1a1 mRNA was measured by sqPCR in BMDM and Ap-BMDM cultures of the indicated genotype. (f) Culture supernatants were measured for the indicated cytokines by ELISA in BMDM and Ap-BMDM after 8h co-culture +/− 1 μM ITE. For all graphs data n=3 biologically independent samples per group and bars in a, c, e, and f are the mean +/− standard deviation. *=P≤ 0.05, **=Pl≤ 0.01, ns= not significant as determined by two-sided Student’s t-test. All experiments were repeated three times with similar results.
AhR is required for efferocytosis-driven immunosuppression
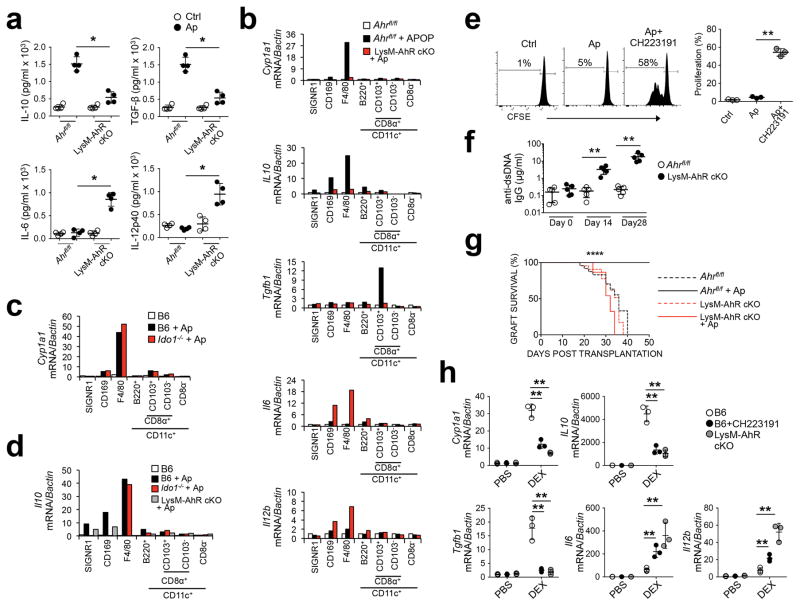
Because intra-venous administration of syngenic apoptotic cells induces expression of IL-10 and TGF-β in splenic macrophages and DC in mice2, 3, we next tested whether AhR suppressed responses to apoptotic cells in vivo. We challenged B6.AhRfl/f LysM-Cre+/− mice, which have a myeloid lineage-specific deletion of AhR (hereafter LysM-AhR cKO mice) intravenously (i.v.) with B6 thymocytes made apoptotic by irradiation and assessed cytokine production in whole spleen lysates 4 hours after administration. While we detected a significant increase in the expression of IL-10 and TGF-β protein in splenic lysates from littermate control mice (i.e. Ahrfl/fl without LysM-CRE) injected with apoptotic thymocytes (Fig. 4a), there was a significant increase in IL-6 and IL-12p40 protein, and no induction of IL-10 and TGF-β in LysM-AhR cKO mice (Fig. 4a).
Figure 4. Myeloid AhR is required for apoptotic cell-driven tolerance in vivo.
(a) Whole spleen lysate from mice 4 hours after apoptotic cell challenge (107 i/v) were measured for the indicated cytokines. (b, c, and d) Splenic macrophages and DC were sorted by FACS from mice of the indicated genotype 4 hours after apoptotic cell injection i.v. based on the markers indicated, and message for the indicated mRNA species was determined by sqPCR. (e) B6 mice +/− CH2231191 administration received CFSE-labeled Thy1.1+CD4+ OTII T cells i/v followed by challenge with OVA+ apoptotic cells. 3d later spleens were collected and CFSE intensity was determined by flow cytometry. (f) Mice were injected with apoptotic cells and serum was collected at the indicated time points and total IgG reactive against dsDNA was determined by ELISA. (g) Female mice received 107 male B6 apoptotic thymocytes i.v. and 7 days later received skin allografts from male B6 mice. N=10 mice/group. ****=P<0.0001 determined by log-rank test. (h) Control mice +/− CH223191 and B6.Ahr−/− mice received dexamethasone i.p. (0.2 mg per mouse). 24h later thymus was harvested and processed for quantification of mRNA by sqPCR. For a n=4, e and h n=3 biologically independent samples per group and bars are the mean +/− standard deviation. *=P≤ 0.05, =Pval≤ 0.01 as determined by two-sided Student’s t-test. For b, c, and d bars represent pooled samples from 4 mice/group. All experiments were repeated three times with similar results.
Next, we sorted SignR1+ marginal-zone macrophages, CD169+ metallophillic macrophages, and F4/80+ red pulp macrophages and CD11c+ DC populations sub-fractionated on the basis of CD8α, CD103 and B220 (Supplementary Fig. 4a) 1, 2, 3 4 hours after i.v. injection of apoptotic cells and quantified the transcripts for Cyp1a1, Il10, Tgfb1, Il6 and Il12b. We found robust induction of Cyp1a1 mRNA (>18-fold) and Il10 mRNA in wild-type, but not in LysM-AhR cKO F4/80+CD11c− red pulp macrophages (Fig 4b). Likewise, Tgfb1 mRNA was elevated in wild-type CD11c+CD8α+CD103+ DCs, but was not induced in LysM-AhR cKO CD11c+CD8α+CD103+ DCs (Fig. 4b). Similarly, LysM-AhR cKO F4/80+CD11c− red pulp macrophages and CD169+F4/80− metallophillic macrophages showed an increase in Il6 and Il12b mRNA expression compared to littermate controls after apoptotic cell administration (Fig. 4b). Cyp1a1 and Il10 mRNA were upregulated in Ido1−/− F4/80+CD11c− red pulp macrophages similar to wild-type following apoptotic cell i.v. injection (Fig. 4c,d), in agreement with lack of IDO expression after efferocytosis in these cells1. These data indicate AhR attenuates pro-inflammatory cytokine induction following i.v. apoptotic cell administration in mice through a pathway independent of IDO.
IL-10 production by macrophages and dendritic cells suppresses autoimmune T cell reactivity in EAE26, and lack of IL-10 induction in splenic macrophages is associated with expansion of pro-inflammatory T cells reactive to apoptotic cell antigens1, 27. As such, we tested whether AhR was required to suppress effector T cell responses to apoptotic antigens. We adoptively transferred 2×106 CFSE-labeled transgenic OTII T cells i.v. into B6 mice pre-treated with the Ahr antagonist CH223191 and challenged recipient mice with ovalbumin-expressing apoptotic thymocytes (Ova-Ap cells) from Act-mOVA mice, which express membrane-bound Ova driven by the chicken Actb promoter2. We detected no proliferation of CFSE-labeled OTII T cells in B6 mice receiving Ova-Ap cells +OTII T cells in comparison to B6 mice that received OTII T cells only (Fig. 4e). In contrast 55% of the CSFE-labeled OTII T cells had divided in B6 mice receiving Ova-Ap cells and the AhR inhibitor CH223191 (Fig. 4e), suggesting AhR was required to suppress effector T cell responses to antigens derived from apoptotic cells.
Chronic administration of apoptotic cells i.v. does not elicit autoimmunity in wild-type mice27, 28. However, deletion of splenic macrophages with clodronate liposomes triggers the development of autoantibodies to DNA and chromatin in response to apoptotic cell i.v. injection27. To ask whether AhR controlled autoimmunity induction upon chronic exposure to apoptotic cells, we i.v. injected syngeneic apoptotic cells (induced by irradiation) weekly for 4 weeks in LysM-AhR cKO mice. While littermate control mice did not show changes in serum anti-dsDNA IgG compared to baseline, LysM-AhR cKO mice showed a 60-fold increase in dsDNA IgG antibodies compared to baseline (Fig. 4f). Systemic administration of apoptotic cells induces allograft tolerance3. As such, we challenged female LysM-AhR cKO or littermate control mice i.v. with apoptotic thymocytes from male B6 mice (B6-Ap cells), followed by transplantation of skin grafts from male B6 mice 7 days later. Littermate control recipient mice that did not receive B6-Ap cells rejected male skin grafts with a mean survival time of 36d (Fig. 4g). While administration of B6-Ap cells i.v. resulted in 100% graft survival for 50d in Ahrfl/flLysM-CRE−/− littermate mice (Fig. 4g) 2, 3, administration of B6-Ap cells in LysM-AhR cKO mice resulted in mean graft survival time of 32d (Fig. 4g), indicating myelopid AhR function was required for apoptotic cell-induced allograft tolerance.
Administration of dexamethasone induces significant intrathymic thymocyte death followed by clearance by thymic-resident phagocytes29, 30. We used this model to test the impact of AhR on immune responses to cell death in B6 and LysM-AhR cKO mice. 24h after dexametasone administration we observed a 20-fold reduction in overall thymus cellularity for B6 mice with or without CH223191 administration and LysM-AhR cKO mice (Supplementary Fig. 5a) and a significant induction of Cyp1a1, Il10 and Tgfb1 mRNA in the thymus homogenate in B6 mice (Fig. 4h). In contrast, induction of these mRNAs was decreased in LysM-AhR cKO mice or B6 mice treated with CH223191, along with increased Il6 and Il12b expression (Fig 4h). These results suggest AhR is required as part of a general tissue response to apoptotic cells.
Manipulation of AhR modulates autoimmunity in lupus
Because resident phagocytes play a key role in limiting autoimmune disease27, we tested whether modulation of AhR impacted the development of autoimmunity. B6.Fcgr2b−/− (hereafter R2B) and MRLlpr mice, are prone to develop autoimmunity reminiscent of SLE. FcγRIIB is an IgG Fc receptor that limits B cell, macrophage, and DC activation. Deletion of the gene for FcγRIIB (Fcgr2b) causes fulminant autoimmunity with death by renal failure in B6 mice31. SqPCR analysis showed elevated Cyp1a1 mRNA in splenic F4/80+ macrophages and CD11c+CD8α+ and CD11c+CD8α− DCs in R2B and MRLlpr mice, compared to basal expression in age and sex matched B6 and MRL-MpJ control macrophages and DCs (Fig. 5a, Supplementary Fig. 5b). In particular, CD8α+ DC and F4/80+ macrophages in R2B and MRLlpr mice showed a large increase in Cyp1a1 mRNA over B6 and MRL-MpJ controls respectively (Fig. 5a and Supplementary Fig. 5b). Metabolites of IDO activity or derived from the gut microbiome are endogenous activators of AhR13, 14, 22, 32, and expression of IDO1 protein is increased in the spleen of R2B and MRLlpr mice compared to B6 and MRL-MpJ mice1, 2. Ido1 mRNA was primarily expressed in CD11c+B220+ splenic pDC in R2B and MRLlpr mice (Supplementary Fig. 5c,d). However, there was no increased Cyp1a1 mRNA in R2B and MRLlpr pDC compared to B6 and MRL-MpJ controls (Fig. 5a and Supplementary Fig. 5b). Moreover, treatment of R2B mice with broad-spectrum antibiotics to reduce intestinal microbiota did not effect Cyp1a1 mRNA in CD8α+ DC and F4/80+ macrophages (Supplementary Fig. 5d), suggesting AhR activity is independent of microbial metabolites in this context.
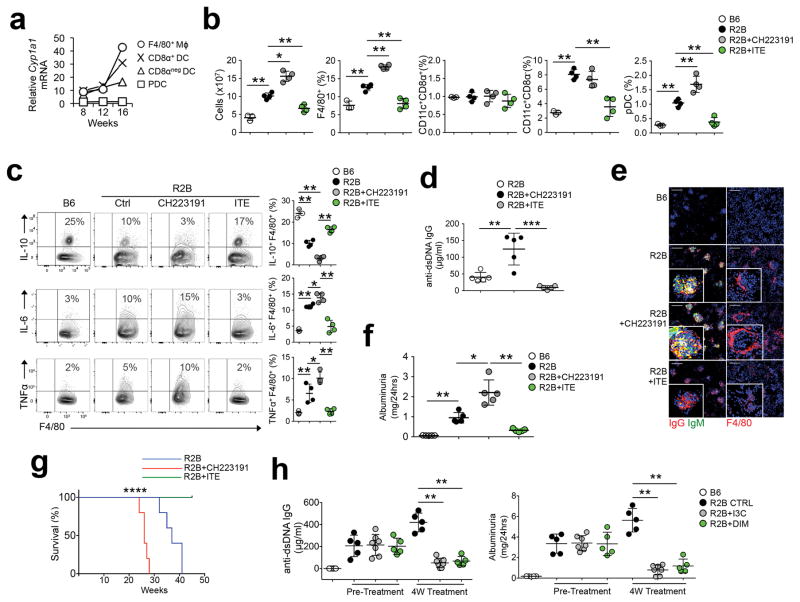
Figure 5. AhR limits disease severity in lupus prone mice.
(a) Splenic cells were FACS sorted from B6 or R2B mice at indicated ages via F4/80, CD11c, CD8α, and B220. Cyp1a1 mRNA was measured by sqPCR on pooled samples. Graph represents the ratio of Cyp1a1 expression of R2B over B6 controls normalized against β-actin. n=5 mice/group (b) Flow cytometry analysis of splenocytes from 20w-old R2B mice treated with CH223191 or ITE for 8w. (c) F4/80+ macrophages MACS-enriched from spleen treated as described in b were restimulated in vitro for 5 hours with PMA/ionomycin and intracellular FACS analysis for cytokines was done. (d) Sera were collected from R2B mice treated as described in b and IgG reactivity against dsDNA was measured by ELISA. (e) Kidney cryosections from mice in b were stained for IgG, IgM, and F4/80. Images are representative of 5-mice per group. Scale bar=100μm (f) Urine was collected from mice treated as in (b) and albumin concentration was determined. (g) Survival curve of female R2B mice treated as indicated. N= 10 mice/group. (h) 7 month old R2B mice were treated with I3C (25 mg/kg i/p, 2x weekly) or DIM (25 mg/kg i/p, 2x weekly) for 1 month and serum anti-dsDNA IgG and albuminuria was measured. For b, c, d, and f n=5 and for h n=6 biologically independent samples per group and bars are the mean +/− standard deviation. *=P≤ 0.05, **=P≤ 0.01, **=P≤ 0.001 as determined by two-sided Student’s t-test. For g ****=Pval<0.0001 determined by log rank test. All experiments were repeated 3 times with similar results.
We next tested the impact of AhR on autoimmune pathology in 24 weeks old R2B mice. Administration of the AhR antagonist CH223191 significantly increased splenomegaly in R2B mice, while the AhR agonist ITE reduced the total number of splenocytes by 40% relative to untreated R2B controls (Fig. 5b). AhR inhibition significantly increased the percentage of F4/80+ macrophages and pDC in R2B mice compared to the untreated R2B control group, while the percentage of CD8α+ and CD8α− DC did not change compared to untreated R2B controls (Fig. 5b). In contrast, ITE significantly reduced the percentages of F4/80+ macrophages, CD8α− DC and B220+ pDC relative to untreated R2B mice (Fig. 5b). Moreover, CH223191 increased the percentage of activated CD24lo splenic B cells in R2B mice compared to untreated R2B controls, while ITE reduced the percentage of CD24lo B cells and CD44hiCD4+ and CD44hiCD8+ T cells (Supplementary Fig. 6a,b). Compared to B6 mice, the percentage of IL-10+ F4/80+ spleen macrophages was reduced in age-matched R2B mice, while percentages of IL-6+ and TNF+ macrophages were increased (Fig. 5c); CH223191 treatment further increased IL-6+ and TNF+ macrophage frequency, while ITE significantly increased the percentage of IL-10+ macrophages (Fig. 5c). 24 week-old female R2B mice had higher serum anti-nuclear IgG (ANA) compared to B6 mice (Supplementary Fig. 6c). CH223191 administration further increased serum ANA, dsDNA and histone antibodies (Supplementary Fig. 6c,d), renal IgG and IgM deposition and periglomerular macrophage accrual (Fig. 5e) and exaggerated kidney pathology (sFig 6e) compared to untreated R2B mice, while ITE had the opposite effects (Fig. 5e and Supplementary Fig. 6c,d,e). Accordingly, CH223191 significantly increased albuminuria in R2B mice, while ITE significantly reduced it compared to R2B controls (Fig. 5f). Finally, CH223191-treated R2B mice showed 100% death by 28 weeks (Fig. 5g), while ITE-treated mice showed 100% survival at 45 weeks (Fig. 5g) compared to 100% mortality at 41 weeks in untreated control R2B mice. To test if AhR activation by other means had a protective effect, we treated 30 week-old R2B mice with established disease with the AhR pro-agonist indole-3-carbinol (I3C)33, 34 or its downstream metabolite 3,3-diindolylmethane (DIM)35, 36. A four-week course of either I3C or DIM significantly reduced serum dsDNA IgG and abrogated proteinuria (Fig. 5h). These results indicate that manipulation of AhR activity can prevent disease development and reverse established autoimmune pathology.
LysM-AhR cKO mice develop systemic autoimmunity with age
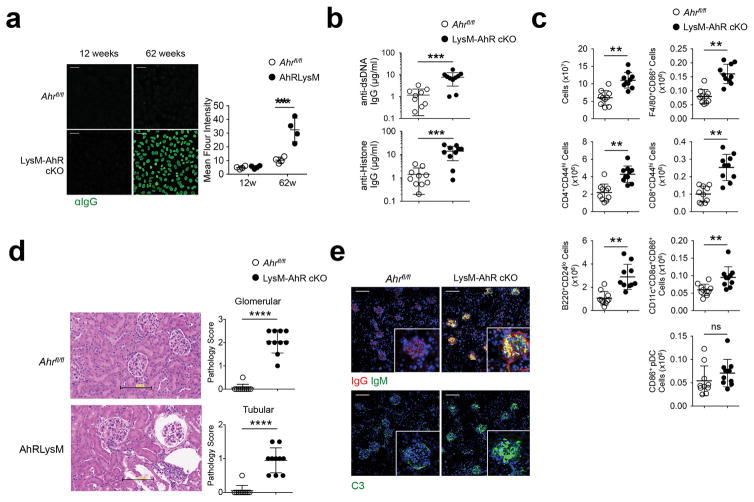
As AhR is known to limit inflammatory pathology13, 14, 15, 18, 32, next we tested whether homeostatic AhR activity was required to limit spontaneous autoimmunity. 12 weeks-old LysM-AhR cKO female mice showed no evidence of autoimmunity; however, aged (62 weeks) LysM-AhR cKO female mice showed prominent homogenous ANA staining patterns (Fig. 6a) and increased dsDNA and histone IgG antibodies compared to age and sex-matched littermate controls (Fig. 6b), suggesting loss of AhR drives autoimmunity. Aged LysM-AhR cKO females had a 2-fold increase in splenocyte numbers compared to age-matched littermates (Fig. 6c), with increased numbers of CD24lo B cells, CD44hiCD4+ and CD44hiCD8+ T cells, F4/80+ macrophages and CD8α+DC (Fig. 6c) suggesting chronic stimulation; however, there was no expansion of CD86+ pDC numbers compared to littermate controls (Fig. 6c). Aged LysM-AhR cKO female mice exhibited increased renal pathology (Fig. 6d) and immunofluorescence analysis of kidney showed increased IgG, IgM and complement deposition compared to littermate controls (Fig 6e), although there was no evidence of proteinuria. These data indicate AhR activity in myeloid cells was required for peripheral tolerance and prevention of spontaneous autoimmunity.
Figure 6. LysM-AhR cKO mice spontaneously develop systemic autoimmunity.
(a) Serum samples from mice were examined for anti nuclear IgG antibody (ANA) reactivity by immunofluorescence staining. Scale bar=50μm. (b) ELISA for serum autoreactivity to dsDNA and histone in 62w-old mice. (c) Flow cytometry analysis of spleens from 62w-old female mice of the indicated genotype. (d) Kidneys were collected and 5μm paraffin sections were stained with H&E and pathologic alteration of the glomeruli and tubules was scored in a blinded manner. Scale bar=100μm. (e) Mouse kidney cryosections were stained for IgG, IgM, and complement C3. All images are representative of 5 mice/group. Graph data points represent individual mice and bars are the mean value/group. Scale bar=100μm. For a n=5 and for b and c n=10 individual mice per group and bars are the mean +/− standard deviation. **=P≤ 0.01, ***=P≤ 0.001, ***=P≤ 0.0001 as determined by two-sided Student’s t-test. All experiments were repeated twice with similar results.
Human SLE is associated with an AhR signature
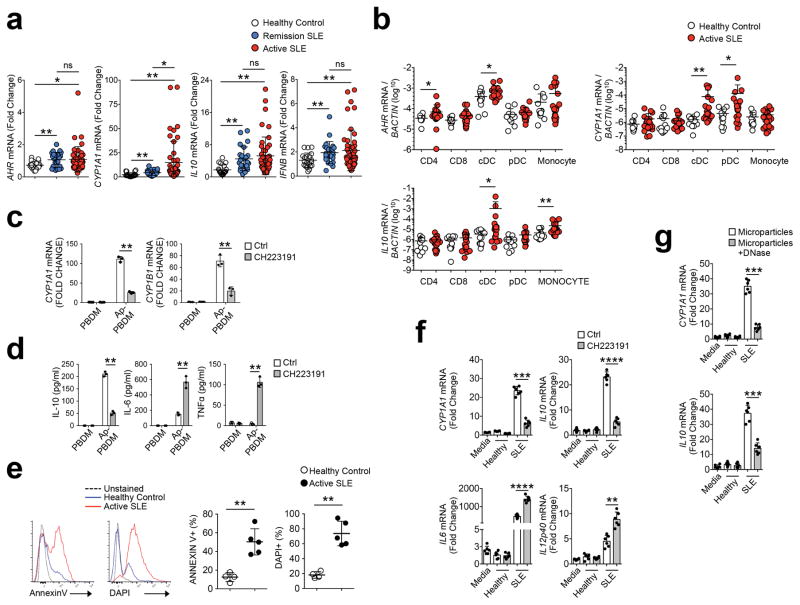
Next we asked whether AhR had a similar function in humans. We used PBMC samples from SLE patients with active disease or in remission and compared them with healthy donor controls (Supplementary Table 1,2,3). sqPCR indicated a significant increase in IFNB1 mRNA expression in SLE compared to healthy controls, although there was no differences between the two SLE groups (Fig. 7a). SLE patients had an increased AhR transcriptional signature (i.e. AHR, CYP1A1 and IL10) compared to healthy controls (Fig 7a); however, only CYP1A1 was higher in active disease versus remission (Fig 7a). We then used qPCR to test AhR activation in CD4+ T cells, CD8+ T cells, HLA-DR+CD14+ monocytes, HLA-DR+CD14−CD11c+CD123− conventional DCs (cDCs), and HLA-DR+CD14−CD11c−CD123+CD303+ pDC sorted from freshly collected blood samples of patients with active SLE or healthy controls (Supplementary Fig. 4b, Supplementary Table 4,5). While we did not detect macrophages in circulation (data not shown), AHR mRNA expression was 10-fold higher in CD11c+ cDC and CD14+ monocytes compared to T cells and CD123+CD303+ pDC in general (Fig. 7b). CD11c+cDC from SLE patients had significantly increased AhR transcriptional signatures (AHR, CYP1A1, IL10) compared to healthy donor controls, while CD123+CD303+ pDC had significant increases in CYP1A1 mRNA, but not AhR or IL10 compared to healthy donor controls (Fig. 7b).
Figure 7. AhR activity is increased in human SLE.
(a) PBMCs were collected: active disease (n=48), remission (n=24), healthy controls (n=20). Expression of indicated mRNA species was measured by sqPCR. (b) mRNA expression from enriched cell populations sorted from PBMCs were measured by sqPCR. For healthy control n=12, for SLE n=20. (c) macrophages derived from PBMCs (PBDM, from healthy donors) were co-cultured with apoptotic Jurkat cells (induced with staurosporine) for 8h in the presence of CH223191. Measurement of mRNA for CYP1A1 and CYP1B1 was done by sqPCR normalized against β-actin. N=3 biologic replicates per group. (d) Cytokine measurements of culture supernatants described in c were done by ELISA. (e) Purified microparticles were stained with Annexin V or DAPI and assessed by flow cytometry for phosphatidylserine exposure and exposed DNA content respectively. N= 5 patient or control samples. (f) Microparticles from SLE patients with active disease or healthy controls were added to cultures of healthy donor PBDM at a previously described concentration37 +/− CH223191 (5 μM). After 8h mRNA and culture supernatant was collected to measure the indicated mRNA. N=6 patient or control samples. (g) Microparticles purified from SLE patients or controls in e were treated with DNAse prior to culture with PBDM for 8h. RNA was collected for measure of the indicated mRNA normalized against BACTIN. N= 5 patient or control samples. For all panels bars are mean +/− standard deviation. *=Pval≤ 0.05, **=P≤ 0.01, ***=P≤ 0.001, ****=P≤ 0.0001 significance determined by the Wilcoxon rank-sum test (a and b) or Student’s t-test (c to g). Experiments were repeated three times with different donors with similar results.
To test if efferocytosis modulated Ahr activation in human myeloid cells we cultured PBMC-derived macrophages (hereafter PBDM) with apoptotic Jurkat T cells generated by irradiation or by 6 hour culture with staurosporine (hereafter Ap-PBDM). We observed increased expression of CYP1A1 mRNA by qPCR and IL-10 protein by ELISA in Ap-PBDM following 8h of culture, compared to control PBDM cultures without apoptotic Jurkat T cells, regardless of the mode of apoptosis induction (Fig. 7c and Supplementary Fig. 7a). AhR inhibition by addition of CH223191 to the co-culture media or treatment of apoptotic Jurkat T cells with DNAse prior to co-culture abrogated CYP1A1 mRNA and IL-10 production and increased IL-6 and TNF in Ap-PBDM compared to untreated Ap-PBDM (Fig. 7c,d and Supplementary Fig. 7b), indicating apoptotic cells activate the AhR pathway in human PBDM in a DNA-dependent manner.
To identify potential AhR-activating factors in SLE patients we used HPLC to analyze the corresponding plasma from SLE and healthy donors (Supplementary Table 1,2,3). The AhR agonists I3C, DIM and indole were not detected, while indole-3-propionic acid (IPA) was detected in all samples with a lower concentration in SLE patients relative to healthy controls (Supplementary Fig. 7c). Healthy controls and remission SLE patients exhibited similar concentrations of the IDO-tryptophan metabolite kynurenine (Kyn), while active SLE patients showed a slight, but non-significant, increase in serum Kyn (Supplementary Fig. 7c), indicating that the amounts of AhR agonists in the plasma did not correlate with the AhR signatures observed in SLE and healthy samples and suggesting other factors may contribute to AhR activity.
As circulating micro-particles with some characteristics of apoptotic cells can be found in the plasma of SLE patients37, we tested whether these microparticles may function as AhR agonists in SLE patients. Microparticles were present at similar concentration in plasma from SLE and healthy controls (Supplementary Fig. 7d), had a diameter of 100–400 nm and morphology typical for extracellular vesicles, as indicated by electron microscopy (Supplementary Fig. 7e). In both SLE and healthy donors these micro-particles were positive for CD9, CD63 and HLA-ABC, although we detected intra-group variability in the expression of these markers in both the SLE group and healthy donor controls, while HLA-DR was not detected (Supplementary Fig. 7f). As previously described37, microparticles from both SLE patients and healthy controls expressed the endothelial marker CD31 (Supplementary Fig. 7g), indicating endothelial origin, with microparticles in some patients expressing CD66b (Supplementary Fig. 7g), a marker usually assigned to neutrophils. However, microparticles from the plasma of SLE patients showed increased expression of phosphatidylserine compared to those from healthy plasma37 (Fig. 7e) and uniquely exhibited DNA on the membrane (Fig. 7e), as indicated by DAPI staining.
When incubated with PBDM, microparticles from SLE plasma induced CYP1A1 and IL10 mRNA expression in the PBDM, while microparticles from healthy plasma did not (Fig. 7f). Treatment with the AhR blocker CH223191 reduced IL-10 production and increased IL6 expression 2.5-fold in PBDM (Fig. 7f and Supplementary Fig. 7h). In addition, treatment of micro-particles from SLE plasma with DNase abrogated induction of CYP1A1 and IL10 mRNA in PBDM (Fig. 7g), indicating that DNA in microparticles induced AhR activation in PBDM. Thus, the data show that apoptotic cells activate AhR in human macrophages, and this is required for IL10 induction and restriction of proinflammatory cytokine production after apoptotic cell phagocytosis. Moreover, the data demonstrated microparticles with characteristics apoptotic cells (i.e. phosphatidylserine and DNA exposure) also induce DNA-dependent IL-10 expression in macrophages that correlates with increased AhR transcriptional signatures observed in SLE patients.
Discussion
Tolerance to apoptotic cells is required to maintain physiologic homeostasis and prevent autoimmunity. In this study we found phagocytosis of apoptotic cells triggered AhR activation in macrophages and DCs, which in turn restricted inflammatory responses to apoptotic cells by inducing the immune-regulatory cytokine IL-10. Moreover, we were able to extend observations in animal models to humans demonstrating that AhR is required for apoptotic cell-driven tolerogenic macrophage polarization. Thus, the data presented here identifies a previously unknown functional requirement of AhR in suppressing inflammatory immunity towards apoptotic cells, and attenuating systemic autoimmune disease pathogenesis.
AhR can bind to motifs in both the Il10 and Il6 promoter; however, in the context of efferocytosis AhR appears to interact directly with regulatory elements in the Il10 locus and does not bind to inflammatory cytokine promoters. While AhR interacts with multiple transcription factors and acts as a bridge between 5′ enhancers and downstream promoter elements38, the lack of large scale changes in either the extent of open chromatin or the transcriptome in macrophages suggests AhR has a specific effect on regulatory cytokine production.
We found that activation of AhR in macrophages was dependent on phagocytosis and the presence of DNA and was dependent on TLR9. Deletion of TLR9 was shown to accelerate the onset of disease pathology and mortality in MRLlpr mice39. Based on our observations, it is plausible that loss or reduction of AhR activation in Tlr9−/− mice might contribute to accelerated disease, suggesting that under certain circumstances innate sensors may be requisite for immune regulation. DNA exposed to the extracellular environment is rapidly degraded by extracellular DNAse in circulation, however in SLE patients DNA is protected by autoantibodies that mask it and likely aid in uptake by phagocytes40. Similarly, DNA exposed in apoptotic cells is rapidly opsonized by natural IgM and complement41, promoting phagocytosis and the anti-inflammatory effects of apoptotic cells. Thus we speculate that protection of the DNA and opsonized uptake is required for functional effects observed for apoptotic cells or microparticles in vivo.
Microparticles play a key role on cellular communication, facilitating inter-cellular transfer of material and information. Our results show microparticles from SLE patients activate AhR in human PBMC-derived macrophages, in a manner dependent on DNA. This suggests that apoptotic microparticles may regulate immunity systemically, dampening inflammation in chronic inflammatory diseases such as SLE. While our analysis was limited to microparticles in the circulation, it is probable that a similar process might occur in tissues. This would predict that tissue-resident phagocytes would function locally to promote tolerance to dying cells in situ.
Our data indicate that AhR is a key molecular regulator of inflammation in in the pathways involved in disposal of apoptotic cells in vivo. In mice, apoptotic cells induced inflammatory cytokine mRNA in LysM-AhR cKO macrophages and lead to autoimmune reactivity. Moreover, we detected an increased AhR-related transcriptional signature in patients with SLE, indicating that this anti-inflammatory pathway might be upregulated during autoimmunity to restrict pathology. Cyp1a1 mRNA was not elevated in pDC from R2B mice compared to matched B6 controls despite significant Ido1 expression; however, CYP1A1 mRNA was significantly increased in pDC from SLE samples compared to healthy donors. Mouse and human AhR binds to some of its ligands with different affinity42, 43. Thus it is possible that production of IDO and the gut microbiome metabolites may impact AhR activity differently in mouse and humans pDC. In particular, human AhR interacts with the bacterially-produced tryptophan derivative indirubin with a 10-fold higher affinity42, 43. Thus it is possible that human pDC may be more sensitive to the effects of IDO and gut-microbiome derived ligands resulting in the higher CYP1A1 expression observed in SLE versus healthy pDC. Nevertheless, our data indicate that phagocytosis of apoptotic cells directly induces AhR activity by a mechanism independent of IDO in both mice and human macrophages.
In conclusion, this report identifies an AhR-dependent mechanism of immunologic tolerance to self and provides new insights into the molecular control of peripheral tolerance, as well as provides new druggable targets for therapeutic intervention in systemic autoimmune disease.
Online Methods
Mice
C57BL/6J (B6), B6.Ahr−/−, B6.LysMCre, B6.Ido1−/−, B6.Act-mOVA-II (Act-mOVA), B6.OTII+Thy1.1+, B6.Fcgr2b−/−, B6.Ahrflox/flox, MRLlpr, and MRL-MpJ mice were bred and maintained in specific pathogen free conditions at the Princess Margaret Cancer Center animal facilities. Female mice between 8–12 weeks of age were used unless otherwise noted. All animal procedures were followed as per the University Health Network Institutional Animal Care and Use Committee guidelines. For animal and human studies there was no randomization of animal allocation or blinding during analysis and all samples were included (i.e. none were excluded).
SLE samples
SLE patients, satisfying 4 or more of the revised 1997 American College of Rheumatology classification criteria for SLE1, were recruited from the University of Toronto Lupus Clinic under full, informed consent according to UHN institutional guidelines. Blood and clinical data were obtained enabling calculation of disease activity using the SLE disease activity index (SLEDAI-2K) 2. Active patients (defined as a clinical SLEDAI-2K > 0, ie. not including the complement and anti-dsDNA Ab components of the SLEDAI-2K) had SLEDAI-2K scores from 4 to 32 (mean 13.5, Supplementary Table 1), and quiescent patients had SLEDAI scores from 0 to 4 (derived from low serum complements and/or high anti-dsDNA Ab levels alone, mean 1.8, Supplementary table 2). Control blood samples were obtained from healthy donors with no family history of SLE (Supplementary Table 3). In one set of experiments we collected fresh blood from active SLE patients (as defined above Supplementary Table 4) or healthy controls (Supplementary Table 5). PBMCs were isolated by Percoll gradient (GE Healthcare Life Sciences) and stained for flow cytometry sorting as described elsewhere in Methods. The study was approved by the Research Ethics Board of UHN with participants providing informed consent. Total RNA was isolated from blood archived in PAXgene tubes, collected at the time of a regular follow-up visit, utilizing the PAXgene Blood RNA Kit (Qiagen) with modifications to improve RNA yield and quality; addition of RNAse inhibitor, an off-column DNAse I digestion and final ethanol precipitation.
Culture of Mϕ and DC
Resident Mϕ were obtained from 6–8 week old female mice by peritoneal lavage with 3ml ice cold PBS and were cultured in RPMI 1640 + 10% fetal bovine serum (FBS), 100U/ml penicillin and streptomycin (ThermoFisher).
To generate mouse bone marrow derived Mϕ and DC, femurs and tibias were harvested from 6–8 week old female mice. Bone marrow was flushed from the cavity and single cell suspensions were generated and cultured in DMEM supplemented with 40 ng/ml MCSF for Mϕ or 20ng/ml GM-CSF for DC, (Peprotech) 10% fetal bovine serum (FBS), and 100U/ml penicillin and streptomycin. After 7d greater than 90% of the cells in the MCSF cultures were F4/80+CD11b+ Mϕ as determined by cytometry analysis. GM-CSF cultures generate a mixed population of Mϕ and DC3. Thus to isolate DC we collected 7d non-adherent cells fractions and FACS sorted the CD11C+CD11bintermediateMHCIIhigh cells representing the DC population3.
To generate human Mϕ, PMBCs from healthy donors (obtained from the Princess Margaret Cancer Center blood donor center) were cultured in the monocyte attachment media and incubated for 2h at 370C. Non-adherent cells were removed with extensive washes with PBS and adherent cells were cultured in M2-Mϕ generation medium as per the manufacturers instructions (PromoCell). After 10d the differentiated Mϕ were characterized by flow cytometry and used in experiments.
Generation of apoptotic cells and co-culture
Thymus from 4–6w old mice were collected and single cell suspensions were exposed to 40 Gy of radiation in a Gammacell® irradiator followed by 6h incubation at 370C in RPMI 1640 + 1% BSA (Sigma). For jurkat T cell (clone E6-1, ATCC) apoptosis, cells were treated with 5μM staurosporine (Sigma) at 370C in RPMI 1640 + 1% BSA for 6h or were exposed to 100 Gy of radiation in a Gammacell® Irradiator followed by culture as described above. In both cases cells were extensively washed with PBS prior to use. Flow cytometry analysis revealed >85% thymocytes were apoptotic and less than 1% were necrotic as determined by annexin V and propidium iodine staining. For some experiments a transwell co-culture system (Costar) was used to prevent direct contact of Mϕ with apoptotic cells. In some experiments, Mϕs were pre-treated with the AhR antagonist CH223191 (Sigma-Aldrich) dissolved in DMSO at 5μM for 2h.
To remove nucleic acids from apoptotic blebs prior to co-culture they were treated as previously described 4. Briefly, apoptotic cells were treated for 1h with DNAse (Roche) at 50 μg/ml dissolved in a storage buffer (20mM Tris-HCl, 1mM MgCl2, pH 7.5) or RNAse (Sigma) at 50 μg/ml dissolved in water. After incubation cells were washed extensively with ice-cold PBS+1%BSA to remove any residual enzyme..
In some experiments Mϕ/apoptotic cell co-cultures were supplemented with anti-IL-10 IgG at 1 μg/ml (Clone JES5-2A5, eBioscience), recombinant IL-10 at 100 ng/ml (R&D Systems), anti-IL-6 at 1 μg/ml (Clone MP5-20F3, eBioscience), and recombinant IL-6 at 1 μg/ml (R&D Systems). To test requirement of the IDO pathway for Cyp1a1 induction, MΦ/apoptotic cell cultures were pretreated with IDO1 inhibitor 1-methyl-D-tryptophan at 200 μM (Sigma) and measurements of kynurenine and tryptophan ratio were done using HPLC as previously described 5.
sqPCR
RNA from sorted cells was purified using RNeasy mini kit (Qiagen) and 250ng of RNA was reverse-transcribed using a random hexamer cDNA reverse transcription kit (Clontech). For sqPCR, 1μl of cDNA was amplified using primers for genes tested. Detailed information about the primers used is shown in Supplementary Table 6, 7. Demographic information for clinical samples used for RNA analysis of healthy human controls and patients with lupus is shown in Supplementary Table 1,2,3. iQ SYBR Green Supermix (Bio-Rad) was used for sqPCR and amplification was recorded on an iQ5 real-time PCR detection system (Bio-Rad). A typical 2-step real time PCR protocol per manufacturers instructions was performed (39 cycles of 10s at 950C and 30s at 600C followed by thermal dissociation step for the SYBR green detection). Results were analyzed with the accompanying software according to manufacturers instructions.
RNA sequencing analysis
RNA samples were quantified by qubit (Life Technologies) and an Agilent Bioananlyzer assessed the RNA quality. All samples had RIN above 8. Libraries were prepared using TruSeq Stranded mRNA kit (Illumina). Two hundred ng of total RNA were purified for polyA tail containing mRNA molecules using poly-T oligo attached magnetic beads, following purification RNA was fragmented. The cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using RNAse H and DNA Polymerase I. A single “A” base was added and adapter ligated followed by purification and enrichment with PCR to create cDNA libraries. Final cDNA libraries were size validated using an Agilent Bioanalyzer and concentration validated by qPCR. All libraries were normalized to 10nM and pooled together. Pooled libraries were further diluted to 2nM and denatured with 0.2N NaOH. 1.7pM of pool libraries were loaded onto a Nextseq cartridge for cluster generation and sequenced Pair-end 75 cycles V2 using Illumina Nextseq500 to achieve a minimum of ~35 million reads per sample.
A total of 1,325,194,544 sequencing reads were obtained in the four conditions, with each condition sequenced as a biological triplicate. After de-multiplexing and initial quality control, all sequencing reads were aligned against the mouse genome reference sequence GRCm38 with STAR v2.5.1a 6. We used the inherited 5′ trimming method of STAR as well as the inherited read counting. The entire command was as follows: STAR --outFilterMismatchNoverLmax 0.05 --outFilterType BySJout --outSAMstrandField intronMotif --outSAMattributes NH HI AS nM NM MD XS --outSAMmapqUnique 60 --quantMode GeneCounts --outFilterIntronMotifs RemoveNoncanonical --outFilterMultimapNmax 1 --clip5pNbases 13 --sjdbGTFfile $GRCm38.ensembl85.gtf. Read counts were measured for gene annotations downloaded from Ensembl Genes V85 7. This resulted in a total of 990,489,880 (74.74%) of overall reads mapping as pairs to annotated genes. Next, inter-sample normalization for read counts was applied with edgeR 8 resulting in counts per million (CPM). After filtering for lowly expressed genes (logCPM > 0 across all three replicates of at least one condition), normalized read counts were used for a differential gene expression analysis again using edgeR. We corrected for multiple testing, reporting the false discovery rate (FDR). All sequencing data was deposited at the European Nucleotide Archive (ENA), accession number PRJEB19473.
ATAC-seg (Assay for Transposase-Accessible Chromatin with high-throughput sequencing)
To determine the chromatin accessibility, ATAC-seq was performed 9. Briefly, 30,000 live Mϕ treated as indicated were washed with PBS and lysed for 5 minutes on ice (10mM Tris-HCl, pH7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630). After isolating crude nuclei, samples were treated for 30 minutes at 37°C with Tn5 transposase, which binds to the open chromatin of the cell, and the DNA was purified by MinElute PCR Purification Kit. Transposed DNA fragments were amplified using specific adapters followed by purification with MinElute PCR Purification Kit. Fragments from 240–360pb were selected in a PippinHT system. The quality of the library and its DNA concentration were assessed by Bioanalyzer instruments and ultimately submitted for sequencing using Illumina HiSeq 2500 sequencer, V4 chemistry. Bowtie2 was used to align fastq files to mm10, and samtools was used to remove duplicates, sort and index BAM files. ENSEMBL annotations for transcription start sites and FANTOM5 annotations for enhancers were used to identify windows of interest (TSS +/− 250bp) for read counting and bedtools multibamcov was used to retrieve read counts per sample per window. Limma-trend was used to identify significant Differentially Accessible Regions (DARs) at FDR < 0.1 for all pairwise comparisons between conditions.
Ingenuity® Pathway analysis (IPA)
Pathway analysis was performed using QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). We determined enrichments of DE genes between Ctrl vs. Apo and Ant_Apo vs. Apo (logFC ≥ ± 0.75, FDR ≤ 0.05) in IPA canonical pathways as well as in IPA diseases and functions.
Chromatin immunoprecipitation (ChIP) and sqPCR
Mϕ were fixed with 1% formaldehyde for 10 minutes, washed with PBS/BSA and then with PBS. Fixed cells were lysed in lysis buffer (1% SDS, 10mM EDTA, 50mM Tris-HCl pH8.1), incubated on ice for 10 minutes and sonicated using a diagenode bioruptor. Cells were sonicated for 30 cycles, 30 sec ON/30 sec OFF to generate fragments between 200–600bp. Lysates were clarified by centrifugation at 16,000 g at 4°C for 15 minutes and the supernatants were transferred to new tubes. 5μg of anti-AhR antibody (Enzo life sciences, cat# BML-SA210-0100) was prebound for 6h with Dynabeads A and G (ThermoFisher) and washed 3X with PBS/BSA. Ninety percent of the cross-linked chromatin were incubated overnight along with antibody-dynabeads complexes, whereas 10% were stored at 4°C as an input. Thereafter, samples were washed and decrosslinked overnight with a decrosslinking buffer (1% SDS and 0.1M NaHCO3) at 65°C. DNA fragments were purified using MinElute PCR purification kit (Qiagen), and were analyzed by sqPCR using SYBR green real time PCR (Bio-Rad). The ChIP primers have been previously described elsewhere or designed based on a DNA regions containing xenobiotic response element (XRE); 5′-GCGTG-3′, and a promoter region near transcription start site 10, 11, 12. Detailed information about the primers used is shown in Supplementary Table 8.
Western Blot
For nuclear extraction, samples were prepared using the NE-PER nuclear cytoplasmic extraction kit (Pierce) according to the manufacturer’s instruction. For Western blot proteins were separated on 4–12% SDS-PAGE gel. Following electrophoresis, proteins were transferred to a PVDF membrane and subsequently blocked for 1h (5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20) and then incubated with anti-AhR antibody (Enzo Life Sciences) overnight at 4oC. Blots were then washed 3 times in Tris-buffered saline containing 0.05% Tween20 and incubated for 1h with secondary antibody at room temperature 5, 13. All blots were stripped and reprobed with anti-β-actin (clone AC-15, Sigma) and anti-H3 (clone D1H2, Cell Signalling Technology) to ensure equal protein loading and purity of cytoplasmic and nuclear extracts. The protein bands were detected using an enhanced chemiluminescence kit (Bio-Rad).
In vivo treatments
AhR antagonist (CH223191, Sigma) and AhR agonists (ITE, Tocris Bioscience; I3C or DIM, Cayman Chemical) were dissolved in DMSO and diluted in 1X PBS prior to in vivo injections. Mice received CH223191 at 100μg per mouse (100μl) i/p two days prior to injections of apoptotic cells. To assess the therapeutic benefit of the AhR pathway in lupus prone mice, B6.R2b−/− or MRLlpr/lpr mice were treated mice with CH223191 and ITE at 100μg per mouse (100μl) i/p starting at eight weeks of age for 1–4 months. In one set of experiments 7 month-old female B6.R2b−/− mice with established proteinuria and high-titer anti-dsDNA IgG received DIM or IC3 at 25 mg/kg i/p two times a week for one month.
To examine the impact of microbiota on activation of the AhR pathway, B6.R2b−/− mice were treated with an antibiotic cocktail (1g/l ampicillin, 1g/l metronidazole, 1g/l neomycin, and 0.5g/l vancomycin, all from Sigma) given in their drinking water, which was replaced everyday for two weeks. For in vivo induction of apoptotic cells, mice were treated with a single injection of 0.2 mg of Dexamethasone (Sigma) per mouse (100μl) i/p. Mice injected with 1X PBS were used as controls.
Measurement of cytokines
Cytokine protein levels in culture supernatants and spleen tissues were measured by ELISA (eBioscience) as previously described 13, 14. For tissue cytokine measurements mice were challenged i/v with 2×107 apoptotic thymocytes by tail vein injections. 18h later spleens were collected and snap-frozen in liquid nitrogen. The spleen was weighed and homogenized with a pestle and mortar containing 50 mg sterile sand (Sigma) and 100 μl of PBS + protease inhibitor mixture (Sigma) per 100 mg of spleen. The mixture was centrifuged at 400 g and supernatant was used for measurement of cytokines.
Measurement of autoantibodies
To measure serum autoantibody levels, 50 μl of whole blood was collected from the tail vein, and the serum was separated using blood collection micro tubes (Sarstedt) according to the manufacturer’s directions. 96-well flat-bottom Immulon II plates (Dynatech) precoated with BSA (overnight at 4°C) were coated with 50μg/ml calf thymus dsDNA (Sigma) diluted in PBS overnight at 4°C to evaluate anti-DNA titers in serum. To assess anti-histone antibody titers plates were precoated with histone (calf thymus, Cayman Chemicals) overnight at 4°C. Plates were washed 3x (PBS, 0.1% Tween 20 (Acros Organics) followed by blocking (PBS + 3% BSA) for a minimum of 2 h at room temperature. Serum samples from mice challenged with apoptotic cells were prepared by diluting serum at 1:200 and assayed for auto-antigen reactivity against coated dsDNA and histone for 2h at room temperature. B6.Fcgr2b−/− mice treated with CH223191 and ITE were bled monthly and the serum samples were serially diluted with starting dilution of 1:500 to measure anti-DNA and anti-histone titers (all IgG subclasses and IgM). Plates were washed 5x and incubated with goat HRP-anti-mouse IgG, IgG1, IgG2a, IgG2b, IgG3 and IgM detection antibodies (Bethyl laboratories) to detect serum autoreactivity against dsDNA and histone for 1h at room temperature. After 5x washes, plates were developed by addition of TMB (KPL) and the absorbance values were read at 450/570 nm. Mouse reference serum (Bethyl Laboratories; RS10-101) was used to quantify absolute concentrations of antibodies in the serum.
HPLC analysis
For serum measurements of AhR agonists, 300μl acetonitrile was added to 100μl serum and immediately vortexed to precipitate protein. The mixture was then centrifuged for 10 min at max speed. The acetonitrile was evaporated from the supernatant using an Eppendorf vacufuge concentrator set at 30°C. After evaporation, the pellet was reconstituted in 85μl mobile phase A (5mM ammonium phosphate+0.05% acetic acid), vortexed, transferred to a centrifugal filter unit (PVDF 0.22 μm), and centrifuged for 10 min at 8000 x g. Lastly, 25μl of the supernatant was injected for HPLC separation. The supernatant was separated on a C18 3 x 50 x 4.6 mm column (Shimadzu), using mobile phase A and mobile phase B of 5mM ammonium phosphate in acetonitrile (ACN)+0.05% acetic acid. The supernatants were separated using a gradient elution as follows: 0–3 min 2.5% mobile phase B, 3–4 min 2.5%-12.5% mobile phase B, 4–8.5 min 12.5% mobile phase B, 8.5–21 min 85% mobile phase B at a flow rate of 1ml/min. Kynurenine was detected with a Shimadzu SPD-M20A UV detector at 360nm, tryptophan and 1-MT were detected at 280nm. Tryptophan, 1-MT, indole-3-carbinol (I3C), indole, indole-3-propionic acid (IPA), and DIM were detected using a Shimadzu RF-20A xs fluorescent detector at an excitation wavelength of 280nm and emission wavelength of 350nm. A mix of AhR agonists and the internal standard was prepared for the reference standard of retention times using kynurenine, tryptophan, I3C, IPA, indole, and DIM (all Sigma) at a final concentration of 2μM each, along with the internal standard, D-1-methyl-tryptophan [1-MT] (Sigma) at 5μM final concentration. All samples were measured in duplicate. Retention times were as follows: Kynurenine: 2.29 min; Tryptophan: 4.79 min; 1-methyl-trytophan (loading standard): 7.97 min; Indole-3-carbinol: 9.71 min; Indole-3-propionic acid: 14.11 min; Indole: 15.70 min; DIM: 19.20 min.
Immunofluorescence
To examine the nuclear translocation of AhR, Mϕ/apoptotic cell co-culture experiments were carried out in chamber slides (ThermoFisher). Mϕ were extensively washed 1X with PBS and slides were immediately fixed in −20°C methanol for 3 minutes. 1% non-fat milk (Sigma) was used for blocking followed by staining anti-AhR antibody (Enzo Life Sciences). Slides were washed 3X with PBS followed by secondary antibody AF546 conjugated goat anti-rabbit IgG antibody (Life Technologies) for 1h.. For staining anti-nuclear antibodies ANA test was performed on fixed Hep-2 slides (MBL). Serum samples were serially diluted in 1% non-fat milk (Sigma) and incubated on slides for 2h at room temperature. Slides were washed 3X with PBS followed by detection of bound serum antibodies using AF568 conjugated goat anti-mouse IgG antibody (Life Technologies) for 1h.
Immunofluorescence examinations of kidney cryosections were performed as described previously 13, 15. For kidney staining the organs were harvested and frozen in Tissue-Tek OCT compound (Sakura) on dry ice and stored at −80°C. 5μm cryosections were immediately fixed in −20°C methanol for 10 min. The sections were blocked with PBS containing 1% nonfat milk (Sigma) for 1–2h at room temperature and stained with AF568 conjugated goat anti-mouse IgG antibody (Life Technologies), IgM-FITC (Clone RMM-1, Biolegend), complement C3-FITC (MP Biomedical, LLC), αF4/80 (clone BM8, Biolegend) in blocking buffer for 1h. Slides were washed 3x with PBS and sections were mounted with Prolong Gold anti-fade with DAPI (Invitrogen). Fluorescent images were captured using a Zeiss 780 upright confocal microscope equipped with 405-, 458-, 488-, 514-, 543-, and 633-nm lasers.
Pathology and Immunohistochemistry
To assess extent of tissue pathology kidneys were fixed by immersion in 10% formalin solution for 48h and embedded in paraffin. Fixed kidney sections were stained with hematoxylin and eosin and Periodic acid-Schiff reagent (to examine basement membrane). Stained kidney sections were scanned on a Hamamatsu Nano-zoomer to capture whole kidney images and analyzed using NDP Nanozoomer software. Glomerular and tubular pathology was scored as previously described 13. To examine kidney function urine samples were collected over 24h and urinary albumin was measured by mouse albumin ELISA (Bethyl Labs).
Cell Sorting
Splenic phagocyte cell populations (Mϕ and DC) were sorted to assess cellular source of AhR activation and cytokine responses to apoptotic cells. Mice received 2X107 apoptotic cells i/v and 8h later harvested spleens were injected with 100U/ml of Collagenase IV (Sigma) in 2mL RPMI (supplemented with 10% FBS) and further incubated for 30 min at 37°C in 5 mL RPMI containing 400 U/mL of Collagenase IV (Sigma). From the digest single-cell suspensions were generated and incubated with αSignR1 (clone ERTR9), αCD169 (clone MOMA-1) (both from Serotec), α-CD8α (clone 53–6.7), α-CD11c (clone N418), α-F4/80 (clone BM8), and αCD103 (clone 2e7) (all from Biolegend).
For sorts from human samples, PBMC fractions were divided into 2 groups: for monocytic cell enrichment the PBMCs were stained with APC-labelled lineage markers- (αCD56/clone NCAM16.2, αCD19/clone HIB19, and αCD3/clone UCHT1) to remove T cells, B cells, and NK cells and markers for DC, pDC, and monocytes- αCD11c (clone B-Ly6), αCD123 (clone 7G3), αCD11b (clone ICRF44), αHLA-DR (clone G46-6), αBDCA-2/CD303 (clone V24-785), and αCD14 (clone 61D3) (all from BD Biosciences). For T cell enrichment the PBMC fraction was stained with αCD4 (clone RPA-T4), αCD8 (clone SK1), and αCD3 (clone SK7) (all from BD Biosciences). All cells were sorted on a Dako Cytomation MoFlo cell sorter into tubes containing RNA protect reagent (Qiagen).
Flow Cytometry
Single cell suspensions from spleens and kidneys were prepared. At least 106 were stained with fluorophore conjugated αF4/80 (clone BM8, Biolegend), αCD11c (clone N418, Biolegend), αCD8α (clone 53-6.7, Biolegend), αCD4 (clone GK1.5, Biolegend), αCD86 (clone GL1, BD Biosciences), αMHCII (clone M5/114.15.2, eBiosciences), αCD44 (clone IM7, BD Biosciences), αCD24 (clone M1/69, Biolegend), αB220 (clone RA3-6B2, eBiosciences), αCD23 (clone B3B4, BD Biosciences), αCD21/CD35 (clone 7G6, BD Biosciences), αCD103 (clone 2E7, Biolegend). For intracellular cytokine staining single cell suspensions of spleens from B6.R2b−/− or MRLlpr mice were incubated with Brefeldin A solution and Monensin (eBioscience) for 2h with shaking at 370C. Cells were fixed and permeabilized using an intracellular staining kit (eBioscience) and stained with fluorophore-conjugated αIL-10 (clone JES5-16E3, eBioscience), αIL-6 (clone MP5-20F3, BD Biosciences), α-TNFα (clone MP6-XT22, BD Biosciences), αF4/80 (clone BM8, Biolegend). For flow cytometry analysis, ≥105 (106 in the case of adoptive OTII transfer) events were collected on a LSR II flow cytometer, and all results were analyzed with FlowJo software (TreeStar).
To examine effector T cell responses to apoptotic cell associated antigens, 2X106 CD45.2+CD4+ OTII T cells labelled with 5-(and 6)- carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled (Invitrogen) were adoptively transferred i/v into CD45.1+ B6 mice. The next day mice were challenged with 107 membrane-bound ova expressing apoptotic thymocytes from Act-mOVA mice i/v. After three days spleens were harvested and assessed for proliferation of adoptively transferred OTII T cells by staining with anti anti CD45.2 (clone 104, eBioscience) and anti-CD4 (BD Pharmingen) followed by flow cytometry analysis.
Skin Transplantation
Female recipient mice were injected with 107 male apoptotic cells i/v. One week later, tail skin from donor male mice (~1 cm2) was placed onto the left thoracic flank region of female mice as previously described16. After 7–10d, bandages were removed and allografts were assessed for signs of rejection for 50d. Grafts were scored as rejected when ulceration and/or necrosis was evident in >80% of the graft as previously reported16.
Microparticle analysis
Plasma from active lupus patients or healthy controls was centrifuged at 500xg for 10 min at 40C. Supernatants were collected and centrifuged at 20,800xg for 10 min at 40C. The pellet was washed 2x with PBS containing 1% BSA.
Isolated MP corresponding to 2 mL of original plasma from healthy controls and patients with active SLE were bound to 4-μm aldehyde/sulfate latex beads (Molecular Probes, Paisley, United Kingdom), coupled with mouse anti-human CD9 (Clone HI9a; BioLegend), under gentle rotating agitation at room temperature overnight, as previously described17. The MP-bead complexes were then stained with phycoerythrin-conjugated antibodies specific for CD9 (Clone M-L13; BD Biosciences), CD63 (Clone H5C6; eBioscience, Inc.), HLA-ABC (Clone W6/32) and HLA-DR (Clone L243), CD31 (clone WM59), CD45 (clone 2D1), CD66b (clone G10F5), CD19 (clone HIB19), CD3 (clone UCHT1), annexin V (Cat.640920), all from BioLegend, DAPI (Thermo) and were assessed using flow cytometry and the corresponding isotype-matched antibodies (BioLegend).
For electron microscopy analysis MP preparations from plasma samples from healthy controls and patients with active SLE were subjected to EM with negative ion capture. In brief, an aliquot of 3 μL from each samples were added to a grid with a glow discharged carbon coated supporting film for 3 minutes. The excess solution was soaked off by a filter paper and the grid was rinsed by adding 5 μL distilled water for 10 seconds, and water was then soaked off by a filter paper. The grid was stained with 5 μL 1% uranyl acetate in water for 7 seconds. Excess stain was soaked off by a filter and the grid was air-dried. The samples were examined in a Hitachi HT 7700 (Hitachi, Tokyo, Japan) electron microscope at 80 kV and digital images were taken by a Veleta camera (Olympus, Münster, Germany).
Image and statistical analysis
For all in vitro studies, using Cyp1a1 or Il10 expression after apoptotic cell exposure the power of the comparison was 100% using n=3 samples per condition. Likewise, using cytokine data for splenic IL-10 protein production after apoptotic cell injection the power for the comparison was 100% using n=5/group. Thus these numbers were used for experiments unless otherwise noted. For survival studies the power of the comparison was 100% using n=10 mice/group. Image analysis was done using National Institutes of Health Image-J software unless otherwise indicated. Means, standard deviations, unpaired Student’s t-test, Wilcoxon rank-sum test, and one-way ANOVA were used to analyze the data using Prism© V6 software (GraphPad Software Inc). When comparing two groups, a p value of ≤ 0.05 was considered to be significant. Survival data for animals or skin allografts were analyzed with Kaplan-Meier survival plots followed by the log-rank test.
Supplementary Material
Acknowledgments
The authors would like to thank F. Barrat (Weill Cornell College of Medicine) for providing the TLR inhibitor and control oligonucleotides, M. Shlomchik and A. Marinov (University of Pittsburgh School of Medicine) and Z. Rahman (Pennsylvania State University School of Medicine) for assistance with analysis of SLE-prone mice, and N. Winegarden and the Princess Margaret Genomics Centre for assistance with sequence analysis. We also thank K. Hultenby (Department of Pathology, Karolinska Hospital Huddinge) for performing EM, J. C. Zuniga-Pflucker (University of Toronto) for advice during development of the research report, M. Butler (University of Toronto/Princess Margaret Cancer Centre) for assistance with healthy PBMC acquisition, and P. Ohashi and D. Brooks (University of Toronto/Princess Margaret Cancer Centre) for reading and critique of the manuscript. This work was supported by NIH grants AI105500, AR067763, CA190449, grants from the Medicine by Design/Canada First Research Excellence Fund (TLM), and the Swedish Medical Research Council and the Karolinska Institutet (SG).
Footnotes
Author Contributions
TLM designed and supervised the research. RS, KH, RD, DW, SL, ME, and SG executed the biochemical, cell biological and in vitro experiments. RS, BR, HL, KC, and MJH performed the animal experiments. AK and AT analyzed the RNA sequencing results. RS, AC, TDSM, and DDC performed the ATAC-seq experiments and analysis. MM scored renal pathology. KM, YB, ZT, JW, MM, SC, SG, and DM contributed reagents, human samples, and discussions. RS, KH, AK, MJH, AC, and TLM prepared figures and RS and TLM wrote the paper.
Competing interest statement
I declare that the authors have no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Data availability statement
The data supporting findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Ravishankar B, et al. Tolerance to apoptotic cells is regulated by indoleamine 2,3-dioxygenase. Proc Natl Acad Sci U S A. 2012;109:3909–3914. doi: 10.1073/pnas.1117736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravishankar B, et al. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc Natl Acad Sci U S A. 2015;112:10774–10779. doi: 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravishankar B, et al. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1320924111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravishankar B, et al. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc Natl Acad Sci U S A. 2014;111:4215–4220. doi: 10.1073/pnas.1320924111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–180. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGaha TL, Karlsson MC. Apoptotic cell responses in the splenic marginal zone: a paradigm for immunologic reactions to apoptotic antigens with implications for autoimmunity. Immunol Rev. 2016;269:26–43. doi: 10.1111/imr.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott RS, et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411:207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- 8.Hanayama R, et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 9.Hankinson O. The aryl hydrocarbon receptor complex. Annual review of pharmacology and toxicology. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 10.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 11.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nature reviews Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandhi R, et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada T, et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol. 2016;17:687–694. doi: 10.1038/ni.3422. [DOI] [PubMed] [Google Scholar]
- 15.Quintana FJ, et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010;107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mezrich JD, et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 18.Bessede A, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura A, et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen NT, et al. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miles K, et al. A tolerogenic role for Toll-like receptor 9 is revealed by B-cell interaction with DNA complexes expressed on apoptotic cells. Proc Natl Acad Sci U S A. 2012;109:887–892. doi: 10.1073/pnas.1109173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guiducci C, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrat FJ, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legge KL, et al. Coupling of peripheral tolerance to endogenous interleukin 10 promotes effective modulation of myelin-activated T cells and ameliorates experimental allergic encephalomyelitis. J Exp Med. 2000;191:2039–2052. doi: 10.1084/jem.191.12.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGaha TL, Chen Y, Ravishankar B, van Rooijen N, Karlsson MC. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood. 2011;117:5403–5412. doi: 10.1182/blood-2010-11-320028. [DOI] [PubMed] [Google Scholar]
- 28.Sharma MD, et al. The PTEN pathway in Tregs is a critical driver of the suppressive tumor microenvironment. Sci Adv. 2015;1:e1500845. doi: 10.1126/sciadv.1500845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 30.Cohen JJ. Glucocorticoid-induced apoptosis in the thymus. Semin Immunol. 1992;4:363–369. [PubMed] [Google Scholar]
- 31.McGaha TL, Sorrentino B, Ravetch JV. Restoration of tolerance in lupus by targeted inhibitory receptor expression. Science. 2005;307:590–593. doi: 10.1126/science.1105160. [DOI] [PubMed] [Google Scholar]
- 32.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjeldanes LF, Kim JY, Grose KR, Bartholomew JC, Bradfield CA. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci U S A. 1991;88:9543–9547. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiering C, et al. Feedback control of AHR signalling regulates intestinal immunity. Nature. 2017;542:242–245. doi: 10.1038/nature21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradlow HL, Zeligs MA. Diindolylmethane (DIM) spontaneously forms from indole-3-carbinol (I3C) during cell culture experiments. In Vivo. 2010;24:387–391. [PubMed] [Google Scholar]
- 36.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieker J, et al. Circulating Apoptotic Microparticles in Systemic Lupus Erythematosus Patients Drive the Activation of Dendritic Cell Subsets and Prime Neutrophils for NETosis. Arthritis Rheumatol. 2016;68:462–472. doi: 10.1002/art.39417. [DOI] [PubMed] [Google Scholar]
- 38.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Sisirak V, et al. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gronwall C, Vas J, Silverman GJ. Protective Roles of Natural IgM Antibodies. Front Immunol. 2012;3:66. doi: 10.3389/fimmu.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaveny CA, Perdew GH. Transgenic Humanized AHR Mouse Reveals Differences between Human and Mouse AHR Ligand Selectivity. Mol Cell Pharmacol. 2009;1:119–123. doi: 10.4255/mcpharmacol.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flaveny CA, Murray IA, Chiaro CR, Perdew GH. Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol Pharmacol. 2009;75:1412–1420. doi: 10.1124/mol.109.054825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 1.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 2.Gladman DD, Ibanez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29:288–291. [PubMed] [Google Scholar]
- 3.Helft J, et al. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Miles K, et al. A tolerogenic role for Toll-like receptor 9 is revealed by B-cell interaction with DNA complexes expressed on apoptotic cells. Proc Natl Acad Sci U S A. 2012;109:887–892. doi: 10.1073/pnas.1109173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinde R, et al. B Cell-Intrinsic IDO1 Regulates Humoral Immunity to T Cell-Independent Antigens. J Immunol. 2015;195:2374–2382. doi: 10.4049/jimmunol.1402854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates A, et al. Ensembl 2016. Nucleic acids research. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacsi SG, Reisz-Porszasz S, Hankinson O. Orientation of the heterodimeric aryl hydrocarbon (dioxin) receptor complex on its asymmetric DNA recognition sequence. Molecular pharmacology. 1995;47:432–438. [PubMed] [Google Scholar]
- 11.Mascanfroni ID, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nature medicine. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura A, et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravishankar B, et al. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc Natl Acad Sci U S A. 2015;112:10774–10779. doi: 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGaha TL, Chen Y, Ravishankar B, van Rooijen N, Karlsson MC. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood. 2011;117:5403–5412. doi: 10.1182/blood-2010-11-320028. [DOI] [PubMed] [Google Scholar]
- 15.Sharma MD, et al. The PTEN pathway in Tregs is a critical driver of the suppressive tumor microenvironment. Sci Adv. 2015;1:e1500845. doi: 10.1126/sciadv.1500845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravishankar B, et al. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1320924111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qazi KR, Gehrmann U, Domange Jordo E, Karlsson MC, Gabrielsson S. Antigen-loaded exosomes alone induce Th1-type memory through a B-cell-dependent mechanism. Blood. 2009;113:2673–2683. doi: 10.1182/blood-2008-04-153536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.