Abstract
Nuclear pore complexes (NPCs) are channels connecting the nucleus with the cytoplasm. We report that loss of the tissue-specific NPC component Nup210 causes a severe deficit of naïve CD4+ T cells. Nup210-deficient CD4+ T lymphocytes develop normally but fail to survive in the periphery. The decreased survival results from both an impaired ability to transmit tonic TCR signals and increased levels of Fas, which sensitize Nup210−/− naïve CD4+ T cells to Fas-mediated cell death. Mechanistically, Nup210 regulates these processes by modulating the expression of Caveolin-2 (Cav2) and cJun at the nuclear periphery. While the TCR-dependent and CD4+ T cell-specific upregulation of Cav2 is critical for proximal TCR signaling, cJun expression is required for STAT3-dependent repression of Fas. Our results uncover an unexpected role for Nup210 as a cell-intrinsic regulator of TCR signaling and T cell homeostasis; and expose NPCs as key players in the adaptive immune system.
INTRODUCTION
T lymphocytes are integral players in the adaptive immune response. After development in the thymus, mature T cells recirculate between the blood, lymph and secondary lymphoid organs, where they scan antigen-presenting cells (APCs) for their cognate antigen1. The maintenance of the circulating naïve T cell population is the result of a balance between thymic output, survival, and homeostatic proliferation1, 2. Naïve T cell homeostasis is essential for maintaining the functional TCR repertoire necessary for ensuring immunity against foreign antigens while avoiding self-reactivity3, 4.
Nuclear pore complexes (NPCs) are aqueous channels that span the nuclear envelope5. Traditionally known as regulators of nucleocytoplasmic transport, it has become evident that they also play multiple transport-independent functions including the regulation of gene expression and chromatin organization6. NPCs are built from 32 different proteins known as nucleoporins5. While the structure of the NPC is conserved in all cells, the expression of several nucleoporins varies among different cell types and tissues, and mutations in various nucleoporins result in tissue-specific diseases7. This indicates that NPCs can be specialized to perform cell type-specific functions7. Supporting this idea, we recently identified that the tissue-specific nucleoporin Nup2108, is a critical regulator of skeletal muscle physiology9, 10. While Nup210 expression is absent in myoblasts, its incorporation into the NPCs of differentiating myotubes is both required and sufficient for myogenesis and myofiber maturation9, 10.
Here we identified that Nup210 deletion in mice specifically reduces the number of circulating naïve CD4+ T lymphocytes. We discovered that Nup210-deficient CD4+ T cells have reduced tonic TCR signaling, which compromises their survival in the periphery, and fail to properly activate in response to TCR ligation. We found that Nup210 mediates proximal TCR signaling by modulating the induction of the lipid raft protein Caveolin-2 (Cav2) following TCR activation. The findings that the Cav2 gene is present at NPCs and that its efficient activation requires Nup210, support the emerging idea that NPCs act as scaffolds for the regulation of inducible genes10, 11, 12. We also identified that Nup210 is critical for the proper expression of cJun, which together with STAT3, prevents the expression of the Fas death receptor. Our findings reveal a cell intrinsic role for Nup210 in the regulation of CD4+ T cell homeostasis, and establish tissue-specific NPCs as key modulators of TCR signaling.
RESULTS
Nup210−/− mice show reduced numbers of CD4+ T lymphocytes
While analyzing Nup210 mRNA levels in mouse adult tissues we found that this nucleoporin shows high expression levels in immune organs, including spleen, lymph nodes, and bone marrow (Fig. 1a). Analysis of immune cell subsets revealed that T and B lymphocytes express higher levels of Nup210 than eosinophils, macrophages, monocytes and neutrophils (Supplementary Fig. 1a). These results are consistent with publicly available ImmGen data13. To investigate the function of Nup210 in the immune system we generated a constitutive Nup210 knockout mouse line (Nup210−/−) by deletion of exon 2, which completely abolishes expression of the protein (Supplementary Fig. 1b, c). Nup210−/− mice are viable, fertile, have normal weight, and display no apparent phenotype. Consistent with our previous findings9, Nup210−/− cells showed correct localization of nucleoporins (Fig. 1b; Supplementary Fig. 1d), no nuclear envelope alterations or NPC clustering (Fig. 1c), and normal nucleocytoplasmic transport (Fig. 1d; Supplementary Fig. 1e, f), indicating no major defects in NPC assembly or function.
Fig. 1. Nup210−/− mice have normal NPC structure and functionality.
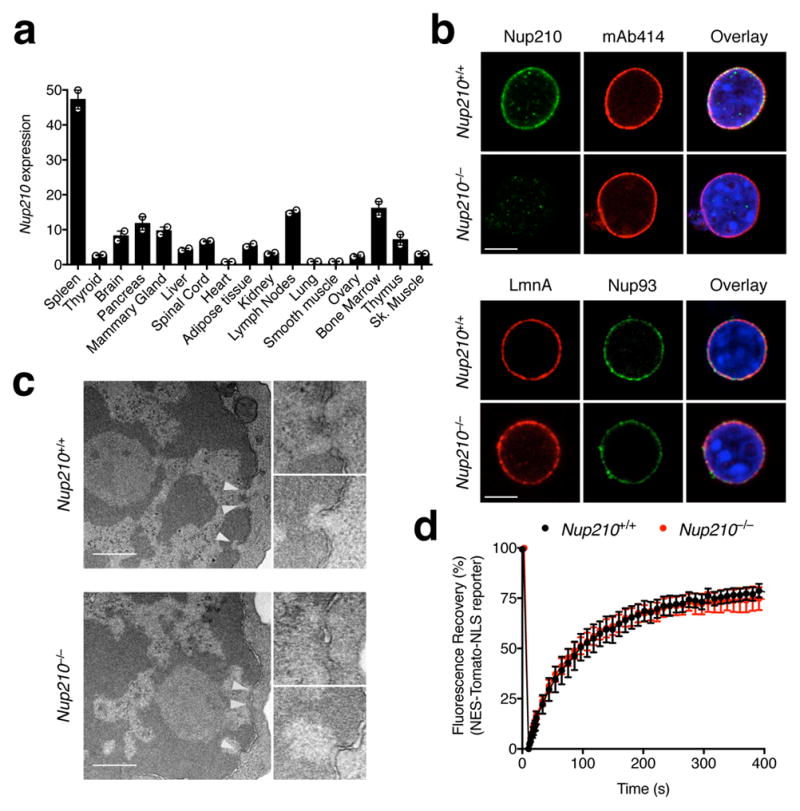
(a) qPCR analysis showing Nup210 mRNA levels in mouse tissues. Nup210 expression was normalized to β-actin, Gapdh, and Hprt. (b) Confocal microscopy showing nuclei isolated from Nup210+/+ or Nup210−/− primary hepatocytes stained with the indicated antibodies and co-stained with Hoechst. mAb414: reacts with several FXFG repeat-containing nucleoporins, including Nup62, Nup153, Nup214, and Nup358/RanBP2. LmnA: Lamin A. Scale bar: 5 μm. (c) Immunoelectron micrographs of flow cytometrically-purified Nup210+/+ and Nup210−/− naïve CD4+ T cells. White arrowheads indicate NPCs. Images to the right show magnified NPCs. Scale bar: 0.5 μm. (d) Fluorescence recovery after photobleaching (FRAP) showing nuclear import of the NES-Tomato-NLS reporter in primary hepatocyte cells derived from Nup210+/+ and Nup210−/− mice, as described9. The percentage of fluorescence recovery from photobleaching of the nucleus is shown. See Supplementary Figures 1 e and f. (a) Mean ± s.e.m, n = 3 mice per group, two independent experiments pooled; (b) representative of two independent experiments; (c) representative of two biological samples (male and female) for each genotype; samples were prepared pooling cells from n = 3 or 4 mice per group; (d) mean ± s.e.m, n = 7–9 cells per group, representative of two independent experiments.
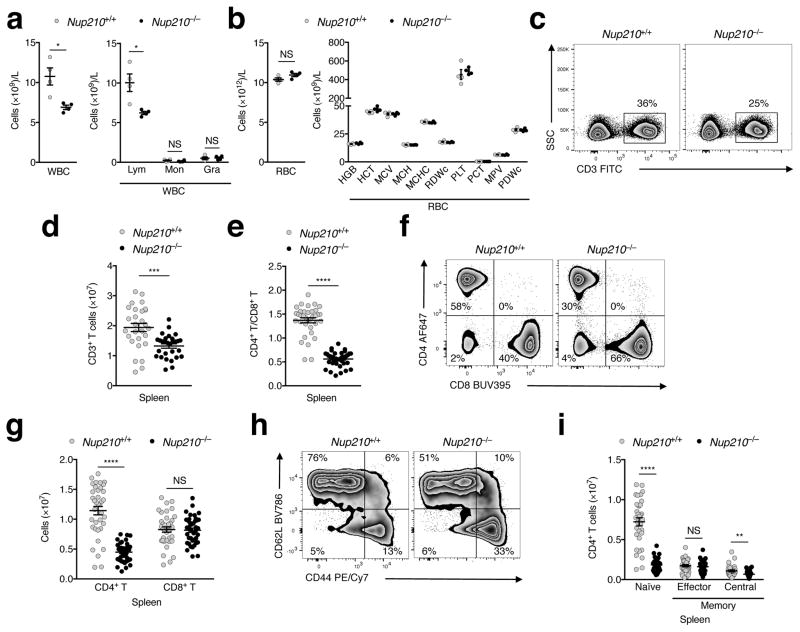
Analysis of blood and bone marrow in Nup210−/− mice showed a significant decrease in the number of white blood cells (WBCs) specific to lymphocytes, with no detectable alterations in erythrocytes or myeloid cells (Fig. 2a, b; Supplementary Fig. 2a). Detailed characterization of the lymphocyte populations in spleen revealed no changes in B cells or spleen cellularity (Supplementary Fig. 2b, c), but decreased proportion and number of CD3+ T lymphocytes (Fig. 2c, d). Further analysis of the CD3+ population showed an abnormal CD4+/CD8+ T cell ratio (1.4 ± 0.05 in Nup210+/+ vs. 0.6 ± 0.02 in Nup210−/−; Fig. 2e) that results from a strong reduction (~60%) in the number of CD4+ T lymphocytes (Fig. 2f, g). Importantly, we found no alterations in the number of CD8+ T cells, although their proportion within the CD3+ population was increased due to the reduced number of CD4+ T cells (Fig. 2f, g). Within the CD4+ T cell population, we observed a decreased percentage of naïve cells and an increased percentage of the effector and central memory subsets (Fig. 2h). This abnormal distribution results from a striking decrease (>75%) in the number of naïve CD4+ T cells (Fig. 2i). A similar decrease in naïve CD4+ T cells was observed in blood (Supplementary Fig. 2d–f). Numbers of TCRγδ T cells, natural killer (NK) cells, NKT cells, and regulatory T cells were not altered in Nup210−/− mice (Supplementary Fig. 2g–j). These findings indicate that Nup210 is critical for the maintenance of the naïve CD4+ T cell population.
Fig. 2. Nup210 is required for naïve CD4+ T cell homeostasis.
Analysis of White Blood Cell (WBC) (a) and Red Blood Cell (RBC) (b) concentration in blood of Nup210+/+ and Nup210−/− mice. Lym: lymphocytes; Mon: monocytes; Gra: granulocytes; HGB: Hemoglobin; HCT: Hematocrit; MCV: Mean Cell Volume; MCH: Mean Corpuscular Hemoglobin; MCHC: Mean Corpuscular Hemoglobin Concentration; RDWc: Red Cell Distribution Width; PLT: Platelet; Pct: Platelet hematocrit; MPV: Mean Platelet Volume; PDWc: Platelet Distribution Width. (c–i) Flow cytometric analysis of Nup210+/+ and Nup210−/− spleens. (c) Gating strategy used to define splenic CD3+ T cells. (d) Splenic CD3+ T cell numbers. (e) Splenic CD4+/CD8+ T cell ratios. (f) Gating strategy used to define splenic CD4+ and CD8+ T cell populations. (g) Splenic CD4+ and CD8+ T cell numbers. (h) Gating strategy used to identify naïve (CD62LhiCD44lo), effector memory (CD62LloCD44hi) and central memory (CD62LhiCD44hi) splenic CD4+ T cells. (i) Splenic naïve and memory CD4+ T cell numbers. (a,b,d,e,g,i) mean ± s.e.m; each symbol represents an individual mouse; mice per group: (a,b) n = 4, (d) n = 28 or 29, (e) n = 35 or 39, (g) n = 37 or 41, and (i) n = 33 or 36. Data are representative of (a,b) two, (c) eight, (f) eleven, and (h) ten independent experiments, or are pooled from (d) eight, (e,g) eleven, and (i) ten independent experiments. NS, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test).
Nup210−/− mice have normal T cell development and circulation
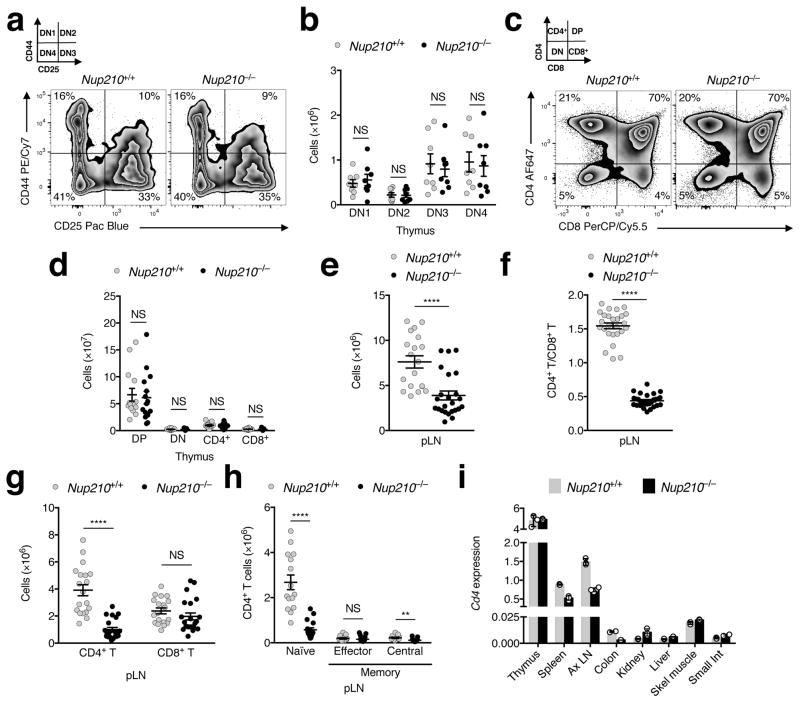
To address whether the deficit in peripheral naïve CD4+ T lymphocytes is caused by abnormal T cell development, we analyzed T cell precursors in the thymus. We found no differences in the number or proportion of double negative (DN1–DN4) or double positive (DP) thymocyte populations between Nup210+/+ and Nup210−/− mice (Fig. 3a–d; Supplementary Fig. 3a, b). Furthermore, we observed equal numbers of single positive CD4+ and CD8+ T cells (Supplementary Fig. 3c) and confirmed that Nup210−/− mice produce equal numbers of naïve CD4+ T cells as Nup210+/+ mice (Supplementary Fig. 3d). These findings indicate that T cells develop normally in Nup210−/− mice, and suggest that the decrease in the naïve CD4+ T cells is caused by abnormalities in the periphery.
Fig. 3. Nup210−/− mice have normal T cell development and circulation.
(a–d) Flow cytometric analysis of thymic populations of Nup210+/+ and Nup210−/− mice. (a) Gating strategy used to subdivide double negative (DN) populations into DN1 (CD44+CD25−), DN2 (CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44−CD25−). (b) Quantification of thymocyte populations from (a). (c) Gating strategy used to define single positive CD4+ and CD8+, double positive (DP), and DN thymocyte populations. (d) Quantification of thymocyte populations from (c). (e–h) Flow cytometric analysis of Nup210+/+ and Nup210−/− mice peripheral lymph nodes (pLN: axillary, brachial and inguinal). (e) Total cell number. (f) CD4+/CD8+ T cell ratios. (g) CD4+ and CD8+ T cell number. (h) Number of naïve/memory CD4+ T cells. (i) CD4 mRNA levels in different tissues from Nup210+/+ and Nup210−/− mice were analyzed by qPCR. CD4 expression was normalized to Gapdh and Hprt. (b,d–h) mean ± s.e.m, each symbol represents (b,d–h) an individual mouse, (i) mean ± s.d, each symbol represents a technical replicate; mice per group: (b) n = 8, (d) n = 14 or 15, (e) n = 19 or 24, (f) n = 26 or 28, (g) n = 19 or 22, (h) n = 15 or 17; (i) n = 3 technical replicates of one biological sample from each genotype, each prepared pooling cells from n = 2 mice per group. Data are representative of (a,i) two, (c) four independent experiments, or are pooled from (b) two, (d,h) four, (e,g) five, (f) nine independent experiments. NS, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test).
Once they exit the thymus, naïve T cells continuously recirculate between blood, lymph, and secondary lymphoid organs. The reduced number of CD4+ T cells in spleens and blood of Nup210−/− mice could result from abnormal retention in lymph nodes, as shown for mice lacking the kinase TBK114, or from the aberrant migration to non-lymphoid organs, as observed in the transcription factor KLF2−/− mouse15. Analysis of peripheral and mesenteric lymph nodes showed the same decrease in naïve CD4+ T cells and no changes in the CD8+ T cell population (Fig. 3e–h; Supplementary Fig. 3e–g), indicating that Nup210 deficiency does not cause T cell retention in secondary lymphoid organs. To determine whether naïve Nup210−/− CD4+ T cells abnormally localize to non-lymphoid organs, RNA was isolated from tissues of Nup210+/+ and Nup210−/− mice and the distribution of CD4+ T cells was analyzed by measuring Cd4 mRNA levels15. Consistent with reduced numbers of CD4+ T cells in spleen and lymph nodes we observed decreased CD4 expression in these tissues but no compensatory increases in any organ analyzed (Fig. 3i). These results indicate that Nup210 depletion does not cause naïve CD4+ T cells to be abnormally retained in lymph nodes or mislocalized to non-lymphoid tissues.
Nup210 has a cell-intrinsic role in naïve CD4+ T cell survival
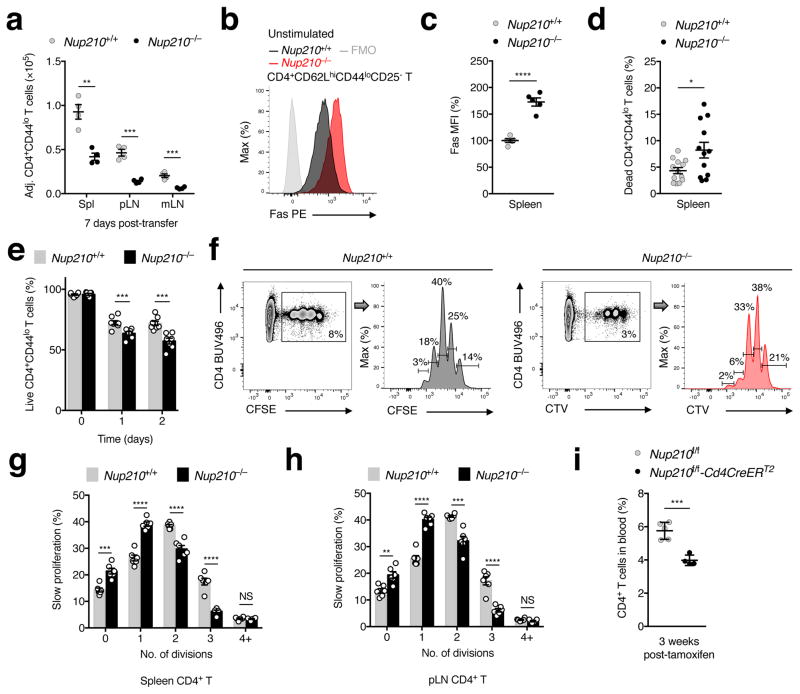
The normal development and circulation of CD4+ T cells in Nup210−/− mice suggests that the phenotype observed could result from survival defects in the periphery. To investigate this, T cells from Nup210+/+ and Nup210−/− mice were isolated and labeled with the fluorescent tracking dyes CFSE and CTV, mixed at a 1:1 ratio and transferred into lymphoreplete recipient mice. Consistent with an intrinsic survival defect, the number of Nup210−/− naïve CD4+ T cells recovered from spleens and lymph nodes seven days after co-transfer was significantly lower than control cells (Fig. 4a; Supplementary Fig. 4a). Moreover, naïve CD4+ T cells isolated from Nup210−/− mice exhibited increased expression of the pro-apoptotic factor Fas, increased staining with a cell death marker, and died faster than their wild-type counterparts when cultured ex vivo (Fig. 4b–e; Supplementary Fig. 4b). Additionally, in a lymphopenia-induced proliferation (LIP) model where a mixture of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− T cells were adoptively transferred into sublethally irradiated wild-type hosts, Nup210−/− T cells underwent less homeostatic proliferation, suggesting a reduced ability to sense survival signals (Fig. 4f–h).
Fig. 4. Nup210 is required for peripheral naïve CD4+ T lymphocyte survival.
(a) Flow cytometry showing the number of naïve CD4+ (CD4+CD44lo) T cells recovered from spleens (Spl), peripheral (pLN) and mesenteric lymph nodes (mLN) 7 days after co-transfer of a 1:1 mixture of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− T cells into lymphoreplete Nup210+/+ hosts. Values were normalized to the ratio at time of transfer. (b,c) Flow cytometry showing Fas expression (b) and mean fluorescence intensity (MFI) (c) in resting naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells (FMO, fluorescence minus one). MFI is shown relative to that of Nup210+/+ mice. (d) Flow cytometry showing the percentage of dead naïve CD4+ (CD4+CD44lo) splenic T cells determined using a viability dye. (e) Flow cytometry showing the percentage of live naïve CD4+ (CD4+CD44lo) T cells over time in ex vivo cultured splenocytes. (f) Flow cytometry showing cell division of naïve CD4+ (CD4+CD44lo) T cells recovered from spleens and pLN 8 days after co-transfer of a 1:1 mixture of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− T cells into sublethally irradiated Nup210+/+ hosts. Cell proliferation was determined by dilution of the CFSE and CTV labels. The percentage of slow proliferating cells in each peak of division is indicated. (g,h) The percentage of cells from (g) that have divided 1–4+ times in spleens (g) and pLN (h) is shown. (i) Flow cytometry showing the percentage of circulating CD4+ T cells in Nup210f/f-Cd4CreERT2 mice 3 weeks after CD4+ T cell-specific Nup210 depletion by tamoxifen treatment. (a,c–e,g–i) mean ± s.e.m; each symbol represents an individual mouse; mice per group: (a) n = 4, (c) n = 5, (d) n = 12 or 13, (e) n = 7 or 8, (g,h) n = 6, (i) n = 4 or 5. Data are representative of (a,i) two, (b,c,f–h) three independent experiments, or are pooled from (d) five, (e) two independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test).
To further confirm that Nup210 has a cell-intrinsic function in CD4+ T cell survival we crossed mice in which exon 2 of Nup210 is flanked by loxP sites (Nup210f/f) with Cd4CreERT2 mice, to specifically delete Nup210 in CD4-expressing cells16. Consistent with our previous data, we observed a decrease in the CD4+ T cell population within 3 weeks of tamoxifen-induced Nup210 ablation (Fig. 4i). These findings indicate that Nup210 expression is necessary for the survival and maintenance of peripheral naïve CD4+ T cells.
Nup210 is required for efficient TCR signaling
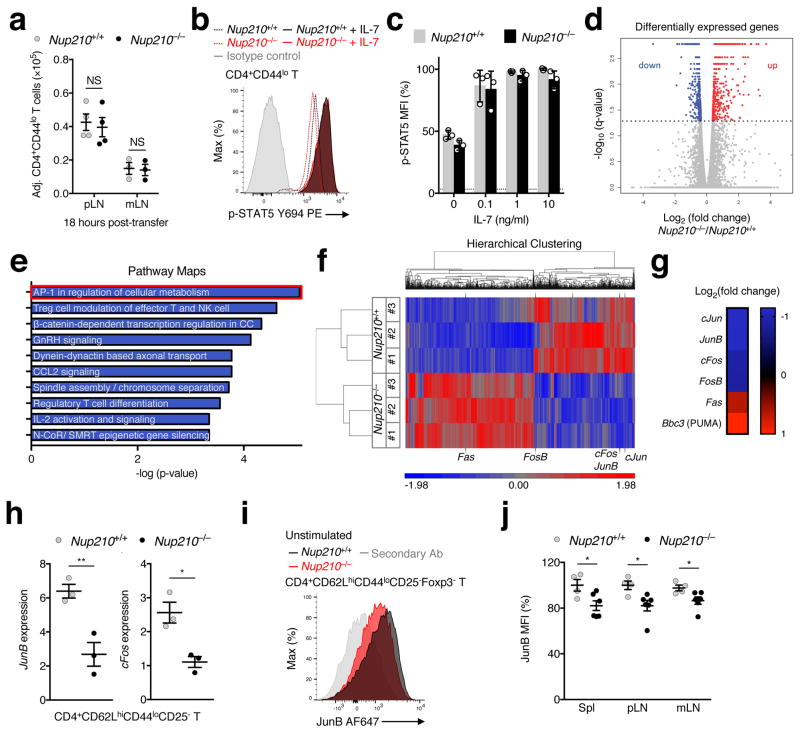
Naïve T cell homeostasis is largely maintained by two signals: the survival cytokine IL-7, and interactions of the T cell receptor (TCR) with self-peptides loaded on major histocompatibility complexes (self-pMHC), also known as tonic TCR signaling1, 2. These homeostatic signals are received locally in secondary lymphoid organs. Therefore the ability of naïve T cells to enter peripheral lymph nodes, known as homing, is essential for their survival1. We observed that Nup210−/− naïve CD4+ T cells had a slight downregulation in the expression of CD62L (Supplementary Fig. 5a), a homing receptor required for this process17. To determine if Nup210−/− naïve CD4+ T cells were able to migrate into peripheral lymph nodes, we adoptively transferred a mixture of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− T cells into wild-type recipient mice. Analysis of peripheral and mesenteric lymph nodes eighteen hours post-transfer showed equal recovery of Nup210+/+ and Nup210−/− cells (Fig. 5a; Supplementary Fig. 5b), indicating that naïve CD4+ T cells lacking Nup210 can efficiently home, and further confirming their ability to circulate normally.
Fig. 5. Nup210−/− naïve CD4+ T cells show features of altered TCR signaling.
(a) Flow cytometry showing the number of naïve CD4+ (CD4+CD44lo) T cells recovered from peripheral (pLN) and mesenteric lymph nodes (mLN) 18 hours after co-transfer of a 1:1 mixture of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− T cells into lymphoreplete Nup210+/+ hosts. Values were normalized to the ratio at time of transfer. (b) Flow cytometric analysis of STAT5 (pY694) expression in unstimulated (dashed lines) or IL-7-treated (solid lines) CD4+CD44lo T cells. (c) Mean fluorescence intensity (MFI) from (b) is shown relative to that of Nup210+/+ mice. (d–g) RNA-seq analysis of flow cytometric-purified Nup210+/+ and Nup210−/− naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells. (d) Scatter plot depicting the differentially expressed genes (DEG) in Nup210−/− cells. Genes were organized by log2 (fold change) and -log10 (q-value). Genes below the cutoff (dashed line: q-value of 0.05) are shown in gray. Downregulated genes are shown in blue, and upregulated genes are shown in red. (e) The most significantly altered pathways from (d) ranked by p-value are shown. The AP-1 pathway is highlighted in red. (f) Hierarchical clustering of DEG (q-value < 0.05). Each sample (#1–3) is a pool of 4 mice. AP-1 pathway members are depicted. Red and blue colors show upregulated and downregulated genes, respectively. (g) Heatmap illustrating the log2 (fold change) values of selected genes in Nup210−/− relative to Nup210+/+ animals. (h) qPCR analysis of JunB and cFos expression in naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells normalized to Hprt. (i,j) Flow cytometric analysis of JunB expression (i) and MFI (j) in unstimulated CD4+CD62LhiCD44loCD25−Foxp3− T cells. Secondary Ab: fluorescently labeled secondary antibody stained sample. MFI values are shown as a percentage of Nup210+/+ cells. (a,h,j) mean ± s.e.m. (c) mean ± s.d; each symbol represents (a,j) an individual mouse or (c) a technical replicate or (h) a biological sample; (a) n = 3 or 4 host mice, (c) n = 3 technical replicates of one biological sample from each genotype, each prepared pooling cells from n = 2 or 3 mice per group, (h) n = 3 biological samples from each genotype, each prepared pooling cells from n = 4 mice per group, (j) n = 4 or 5 mice per group. Data are representative of (a–c,j) two, (d–g) three independent experiments, or are pooled from (h,i) three independent experiments. (d–g) false discovery rate (FDR) adjusted P-values were calculated using the Benjamini-Hochberg correction for multiple testing with an allowed false discovery rate of 0.05. NS, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.01 (two-tailed unpaired Student’s t-test).
These findings suggest that the survival defects of Nup210−/− naïve CD4+ T cells might result from alterations in sensing survival signals in secondary lymphoid organs. Within lymph nodes, T cells encounter the survival cytokine IL-7, which upon binding to the IL-7 receptor (IL-7R) triggers STAT5 phosphorylation and promotes cell survival18. To determine whether Nup210 depletion affects IL-7 signaling, Nup210+/+ and Nup210−/− naïve CD4+ T cells were co-cultured in varying concentrations of IL-7 and STAT5 phosphorylation was measured. We observed no difference in the dose-dependent increase in p-STAT5 levels between Nup210+/+ and Nup210−/− cells (Fig. 5b, c), indicating that Nup210−/− naïve CD4+ T cells can sense IL-7 efficiently.
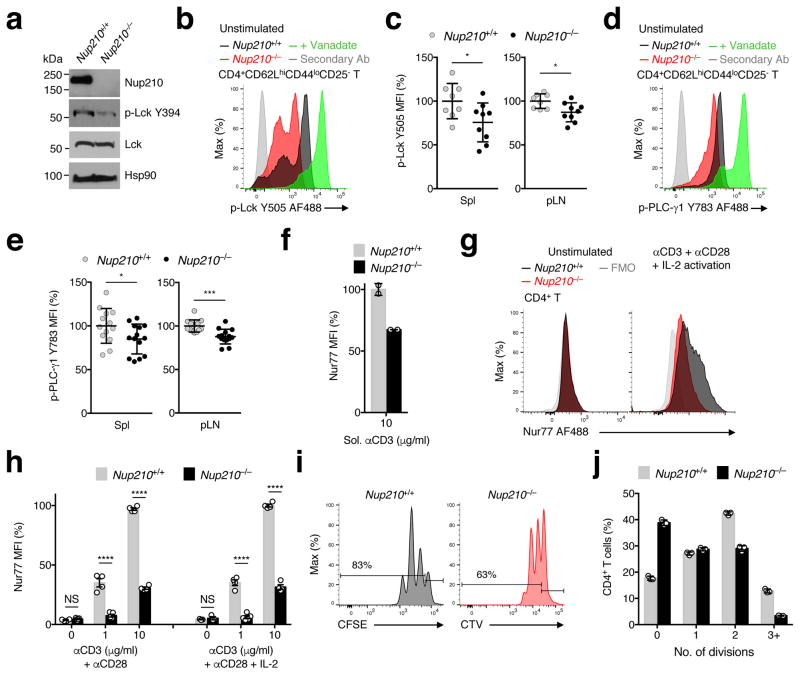
The second survival signal received in lymph nodes results from TCR engagement with self-pMHC displayed on APCs. These interactions result in low levels of TCR signaling (tonic signaling), which are essential for naïve T cell survival19, 20, 21, 22. TCR stimulation initiates a signaling cascade that results in activation of the transcription factor NFAT and the up-regulation of AP-1 family members Jun and Fos23. Therefore, expression levels of AP-1 factors have long been used as a readout for TCR signaling24. Our whole-transcriptome sequencing (RNA-seq) analysis of unstimulated Nup210+/+ and Nup210−/− naïve CD4+ T cells (>99% purity; Supplementary Fig. 5c) showed significantly reduced levels of members of the AP-1 family, being this pathway the most significantly altered in the dataset (Fig. 5d–g; Supplementary Table 1). Our analysis also showed significant alterations in the NFAT pathway (data not shown) and increased levels of apoptotic markers Fas and PUMA in Nup210−/− naïve CD4+ T cells (Fig. 5g), consistent with their decreased survival. The reduced levels of AP-1 factors JunB and cFos in unstimulated Nup210−/− naïve CD4+ T lymphocytes were confirmed by real-time PCR and flow cytometric analysis (Fig. 5h–j). These findings suggest that Nup210−/− naïve CD4+ T cells have reduced basal TCR signaling. To confirm this, we isolated unstimulated Nup210+/+ and Nup210−/− naïve CD4+ T cells and determined the phosphorylation levels of downstream TCR effectors, including Lck; Zap70,; and PLC-γ125. Because Lck is catalytically active when phosphorylated at the activating Tyr394 residue alone, or in combination with the inhibitory Tyr505 residue26, we analyzed the levels of both Lck modifications. As shown in Figure 6a–e and Supplementary Fig. 6a, b, all these factors showed reduced phosphorylation levels in Nup210−/− naïve CD4+ T cells at steady state, indicating that Nup210 depletion disrupts TCR signaling events. To further confirm that Nup210-deficient cells have defective basal TCR signaling naïve CD4+ T cells were stimulated with soluble anti-CD3 monoclonal antibody (mAb), which has previously been used to mimic in vivo tonic signaling27. We found that induction of the early TCR responsive gene Nur7728 was strongly impaired in Nup210−/− CD4+ T cells (Fig. 6f). Altogether, these observations indicate that Nup210 is required to sustain basal/tonic TCR signaling of naïve CD4+ T cells to promote survival.
Fig. 6. Nup210 is required for TCR signaling.
(a) Immunoblot analysis of p-Lck (Y394) and total Lck protein levels in Nup210+/+ and Nup210−/− naïve CD4+ T cells. Hsp90 used as loading control. (b,c) Flow cytometric analysis of p-Lck (Y505) expression (b) and mean fluorescence intensity (MFI) (c) in resting naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells from Nup210+/+ and Nup210−/− mice. Activation with sodium peroxyvanadate used as positive control. Secondary Ab: fluorescently labeled secondary antibody stained sample. MFI is shown relative to that of Nup210+/+ mice. (d,e) Flow cytometric analysis of p-PLC-γ1 (Y783) expression (d) and MFI (e) in resting naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells from Nup210+/+ and Nup210−/− mice. (f) Flow cytometric analysis of Nur77 expression in Nup210+/+ or Nup210−/− CD4+CD44lo T cells stimulated for 16 hours with soluble (sol.) anti-CD3 mAb. Values are displayed relative to sol. anti-CD3-stimulated Nup210+/+ cells. (g) Flow cytometric analysis of Nur77 expression in unstimulated (left) or TCR-stimulated (right) Nup210+/+ and Nup210−/− CD4+CD44lo T cells. TCR stimulation was performed with plate-bound anti-CD3 and anti-CD28 mAbs, and soluble IL-2 for 14 hours. FMO, fluorescence minus one. (h) Flow cytometric analysis of Nur77 levels in CD4+ T cells stimulated as in (g) or without IL-2 supplementation. Values shown are relative to control treated cells. (i) Flow cytometric analysis of CFSE and CTV dilutions 48 hours after stimulation of CFSE-labeled Nup210+/+ and CTV-labeled Nup210−/− CD4+CD44lo T cells with plate-bound anti-CD3/ anti-CD28 mAbs and soluble IL-2. The percentage of cells that have undergone proliferation is indicated. (j) The percentage of cells from (i) that underwent 1–3 divisions was quantified. (c,e,h) mean ± s.e.m., each symbol represents an individual mouse, (f,j) mean ± s.d.; each symbol represents a technical replicate; (a) biological samples prepared pooling cells from n = 4 or 8 mice; mice per group: (c) n = 8 or 9, (e) n = 13 or 14, (h) n = 4 or 5; (f) n = 2 technical replicates of one biological sample from each genotype, each prepared pooling cells from 4 mice; (j) n = 3 technical replicates of one biological sample from each genotype, each prepared pooling cells from 5 or 10 mice per group. Data are representative of (a,f,i,j) two, (b,d,g,h) three independent experiments, or are pooled from (c,e) three independent experiments. NS, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test).
Nup210 is required for CD4+ T lymphocyte activation
The discovery that Nup210 is required for proper tonic TCR signaling in naïve CD4+ T cells raised the question of whether this nucleoporin is also important for TCR-dependent activation. To investigate this, naïve populations of Nup210+/+ and Nup210−/− CD4+ T cells were cultured in the presence of immobilized CD3 mAb plus costimulatory CD28 mAb, in the presence or absence of exogenous IL-2, and Nur77 protein levels were measured. In contrast to control cells, Nup210−/− cells failed to increase Nur77 following TCR ligation regardless of IL-2 stimulation (Fig. 6g, h). As previously reported, IL-2 addition did not alter Nur77 induction29, 30. We also detected a reduced frequency of Nup210−/− naïve CD4+ T cells that upregulated the early activation marker CD69 relative to control cells (Supplementary Fig. 6c, d) and decreased cell division in response to TCR engagement (Fig. 6i, j). Altogether, these findings indicate that the TCR signaling defect caused by Nup210 deficiency cannot be rescued by optimal activation conditions, and demonstrate that Nup210 is critical for proper TCR signaling during CD4+ T cell activation and proliferation.
Nup210 regulates early TCR signaling by promoting Caveolin-2 expression
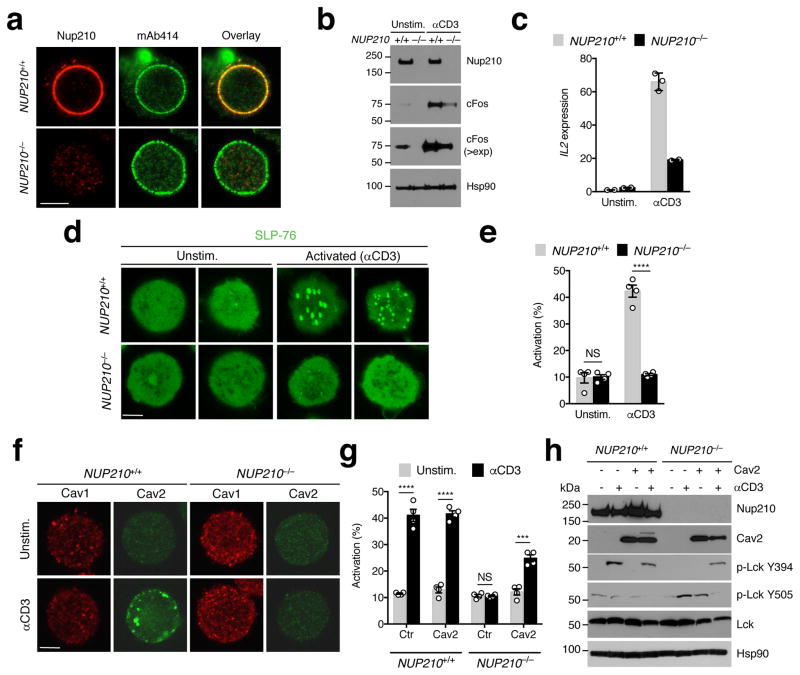
To dissect the molecular mechanism of Nup210 regulation TCR signaling and survival we knocked it out in human Jurkat J14 T cells expressing the TCR adaptor protein SLP-76 tagged with EYFP31 using CRISPR/Cas9. Single guide RNA oligonucleotides targeting exon 2 resulted in full depletion of Nup210 in these cells (Fig. 7a, b). Analogous to the primary Nup210−/− naïve CD4+ T cells, Nup210-depleted J14 cells showed lower levels of cFos in unstimulated conditions (Fig. 7b), and reduced activation in response to CD3-induced TCR stimulation (Fig. 7b, c; Supplementary Fig. 7a). Because TCR ligation leads to the rapid formation of SLP-76-containing TCR microclusters at the plasma membrane31, 32, clustering of SLP76-EYFP in these cells has been used as a reporter for proximal TCR activation31. Although it is unknown if clustering occurs during tonic TCR signaling in vivo, we found that stimulation with soluble anti-CD3 antibodies, which mimics tonic TCR signaling in vitro27, leads to SLP-76 clustering in J14 T cells (Fig. 7d). Nup210 depletion in these cells completely abolished SLP-76 cluster formation in response to TCR activation (Fig. 7d, e), confirming that Nup210 is critical for early TCR signaling and indicating a conserved role for Nup210 in TCR activation in humans.
Fig. 7. Nup210 is required for the TCR-dependent induction of Cav2 expression.
(a) Immunofluorescence analysis of Nup210 and mAb414 in NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. (b) Immunoblot analysis of Nup210 and cFos in resting and soluble anti-CD3-activated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. Hsp90 was used as loading control (>exp: higher exposure). (c) qPCR analysis of Il2 mRNA levels in resting or soluble anti-CD3-activated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. Il2 expression was normalized to Hprt1. (d) Immunofluorescence analysis of SLP-76 clustering on the surface of unstimulated or anti-CD3-activated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. (e) The percentage of cells with SLP-76 foci from (d) was quantified. (f) Immunofluorescence analysis of Cav1 and Cav2 in unstimulated or anti-CD3-activated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. (g) The percentage of activated (SLP-76 cluster-positive cells) NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells expressing control (empty vector) or Cav2 was quantified after stimulation with soluble anti-CD3. (h) Immunoblot analysis of Nup210, Cav2, p-Lck Y394, p-Lck Y505, and total Lck in unstimulated or anti-CD3-activated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells expressing control or Cav2. Hsp90 was used as loading control. (a,b,d,f) representative of three independent experiments; (c) mean ± s.d., each symbol represents a technical replicate n = 3, representative of three independent experiments; (e,g) mean ± s.e.m, pooled from four independent experiments, n = 4 biological samples, n ≥ 200 cells per group; (h) representative of two independent experiments. NS, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test). Scale bar: 5 μm.
The formation of clusters/signaling assemblies induced by TCR ligation is mediated by actin cytoskeletal rearrangements32, and the lipid raft protein Caveolin-1 (Cav1) has been shown to be required for this process33. The caveolin protein family includes two additional members, Cav2 and Cav3. While Cav3 is selectively expressed in muscle tissues34, Cav1 and Cav2 are more ubiquitous and can form hetero-oligomeric complexes35. Immunofluorescence analysis of Cav1 and Cav2 expression in J14 SLP-76-EYFP cells showed that both proteins were expressed at low levels, but only Cav2 was upregulated in response to TCR stimulation with soluble anti-CD3 antibodies (Fig. 7f). Cav2 upregulation was not observed in Nup210-depleted cells, indicating a critical role for Nup210 in its induction (Fig. 7f). Because Cav1 was found to be required for TCR signaling in CD8+ but not CD4+ T cells36, and J14 cells express the CD4 but not the CD8 coreceptor37, these findings raise the possibility that Cav2 is needed for TCR signaling specifically in CD4+ T cells. This would explain the CD4+ T cell-restricted phenotype of Nup210−/− mice. To confirm that Nup210 modulates proximal TCR signaling through Cav2 we performed rescue experiments. For this, the ability of wild-type and Nup210-depleted J14 T cells transduced with control or Cav2-expressing lentiviruses to initiate TCR signaling was determined by analyzing SLP-76 clustering and Lck phosphorylation. Cav2 overexpression did not induce spontaneous activation or increased the activation of wild-type cells, but was sufficient to partially reestablish SLP-76 clustering upon TCR activation in Nup210-depleted cells (Fig. 7g; Supplementary Fig. 7b). Ectopic Cav2 expression also restored Lck phosphorylation at the activating Tyr394 residue and promoted the dephosphorylation of the inhibitory Tyr505 (Fig. 7h). These results confirm that Nup210 mediates proximal TCR signaling by promoting Cav2 expression. Interestingly, Cav2 was not able to rescue the TCR-induced increase in the expression of downstream genes (Supplementary Fig. 7c), indicating that Nup210 regulates TCR-induced gene expression through another mechanism.
The Cav2 gene localizes to the nuclear periphery
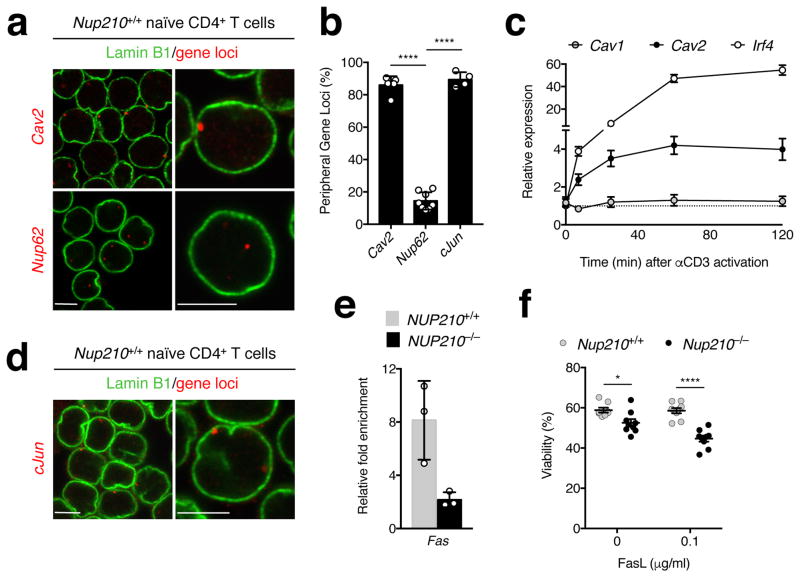
Our previous work identified that Nup210 regulates muscle physiology by modulating gene expression at the nuclear periphery10. To determine the intranuclear localization of the Cav2 gene we performed fluorescence in situ hybridization (FISH) in primary naïve CD4+ T cells. We found that the Cav2 gene, but not the Nup62 gene whose expression is not regulated by Nup210, localizes to the nuclear periphery (Fig. 8a, b). Recent work suggests that NPCs can act as scaffolds for regulation of inducible poised genes10, 11, 12. Interestingly, analysis of histone modifications from published data38, 39 shows that the Cav2 promoter has active (H3K4me3) and repressive (H3K27me3) marks (Supplementary Fig. 8b). The presence of bivalent histone marks in a promoter is believed to maintain genes in a silent/low expression state while keeping them poised for rapid activation or stable silencing40. Notably, the Cav1 gene, which is positioned next to the Cav2 gene, only shows the repressive H3K27me3 mark (Supplementary Fig. 8b), indicating that localizing at the nuclear periphery is not sufficient to maintain a poised state. To determine if the Cav2 gene at NPCs is poised for rapid activation in response to TCR stimulation we determined its kinetics of expression by analyzing the mRNA levels of Cav1 and Cav2 in primary naïve CD4+ T cells activated ex vivo. While no changes in Cav1 expression were observed under these conditions, we found that Cav2 expression was rapidly upregulated, with a kinetics similar to the early TCR-responsive gene Irf4 (Fig. 8c). Upregulation of Cav2 was not observed in CD8+ T lymphocytes (Supplementary Fig. 8c), further supporting the idea that Cav2 function might be restricted to CD4+ T cells. Our results are consistent with earlier observations that Cav1 and Cav2 genes are independently regulated at the transcriptional level35, and indicate that in CD4+ T cells, the Cav2 gene is set to be rapidly activated at the nuclear periphery in response to TCR stimulation in a Nup210-dependent manner.
Fig. 8. Cav2 and cJun genes localize to the nuclear periphery and Nup210 is required for the repression of the proapoptoticreceptor Fas.
(a) DNA-FISH analysis of Cav2 and Nup62 in Nup210+/+ naïve CD4+ T cells. Gene loci are shown in red and the nuclear periphery marker Lamin B1 in green. (b) The percentage of nuclei from (a) that show one or both loci associated with the nuclear periphery was quantified from 3D nuclear reconstructions (see Supplementary Fig. 8a). (c) qPCR analysis of Cav1, Cav2, and Irf4 mRNA levels in Nup210+/+ naïve CD4+ T cells unstimulated or activated with soluble anti-CD3. Expression was normalized to Hprt. (d) DNA-FISH of cJun in Nup210+/+ naïve CD4+ T cells. Gene loci (red) and Lamin B1 (green). (e) Chromatin immunoprecipitation (ChIP) analysis of STAT3 binding to the Fas promoter in unstimulated NUP210+/+ or NUP210−/− J14 SLP-76-EYFP cells. Binding of STAT3 was quantified by qPCR. (f) Flow cytometric viability analysis of Nup210+/+ or Nup210−/− naïve CD4+ T cells co-cultured with splenocytes from TCRbeta- TCRdelta- mice and soluble FasL. Viability was determined using annexin V and propidium iodide staining. (a,d) representative of three independent experiments; (b) mean ± s.d. pooled from three independent experiments, n ≥ 200 cells quantified from n = 4, 5 or 8 individual fields, symbols show individual field quantification; (c) mean ± s.e.m, n = 5 mice per time point, representative of one experiment; (e) mean ± s.d., each symbol represents a technical replicate, n = 3, representative of two independent experiments, (f) mean ± s.e.m, each symbol represents an individual mouse, n = 9 mice per group, data pooled from two independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 (two-tailed unpaired Student’s t-test). Scale bar: 5 μm.
Nup210 is required for the repression of the proapoptotic receptor Fas
Our RNA-seq analysis identified that AP-1 was the most significantly altered pathway in Nup210-deficient naïve CD4+ T cells (Fig. 5e–j). FISH analyses in naïve CD4+ T lymphocytes showed that the cJun gene also localizes to NPCs (Fig. 8b, d), suggesting that Nup210 might regulate its activity at the nuclear periphery. Previous studies found that cJun works with STAT3 to repress the expression of the cell death receptor Fas41. Because the association of STAT3 with the Fas promoter has depends on cJun41, and this AP-1 factor is strongly downregulated in Nup210-deficient cells, our findings suggest that in the absence of Nup210, recruitment of STAT3 to the Fas promoter might be impaired, resulting in de-repression of this gene. To test this, we analyzed the levels of STAT3 at the Fas gene promoter in wild-type and Nup210-depleted J14 cells using chromatin immunoprecipitation (ChIP). We found that NUP210−/− cells exhibited a significant decrease in the recruitment of STAT3 to the Fas promoter (Fig. 8e), even though the levels and localization of STAT3 were not affected (Supplementary Fig. 8d, e). These findings explain the higher levels of FasR in naïve CD4+ T lymphocytes of Nup210−/− mice (Fig. 4b, c; Fig. 5f, g). To test if increased Fas levels render these cells more susceptible to cell death, we incubated Nup210+/+ and Nup210−/− naïve CD4+ T cells with Fas ligand (FasL), and measured cell survival. We found that Nup210−/− cells showed increased cell death (Fig. 8f). The same was observed using wild-type and Nup210-depleted J14 cells (Supplementary Fig. 8f). Altogether, our findings indicate that the death of peripheral naïve CD4+ T cells in Nup210−/− mice results from a combination of their inability to sense survival tonic TCR signals and their increased sensitivity to Fas-mediated cell death (Supplementary Fig. 8g).
DISCUSSION
Accumulating evidence indicates the existence of specialized NPCs7, however the physiological functions of these structures remain largely unknown. Here, we discovered that deletion of the cell type-specific nucleoporin Nup210 in mice results in a dramatic reduction of circulating naïve CD4+ T cells. While the development and migration of these cells occurs normally, the survival of peripheral naïve CD4+ T lymphocytes in Nup210−/− animals is compromised. The increased death of CD4+ T cells results from impaired tonic TCR signaling, which prevents the proper transmission of the survival signals provided by the TCR-self peptide/MHC interactions, and from increased levels of Fas, which sensitizes naïve CD4+ T lymphocytes to cell death. We found that Nup210 regulates these processes by modulating the expression of Cav2 and cJun genes present at NPCs. While TCR stimulation-dependent upregulation of Cav2 is critical for proximal TCR signaling, cJun expression is required for STAT3-dependent repression of Fas receptor. Our results uncover an unexpected role for Nup210 as a cell-intrinsic regulator of TCR signaling and T cell homeostasis; and expose NPCs as key players in the adaptive immune system.
Connections between NPCs and the immune system have recently emerged42, 43, 44, 45. Mice heterozygous for the nucleoporins Nup96 and Sec13 show reduced levels of MHC class I and class II in APCs43, 44. Decreased MHC expression in Nup96+/− mice indirectly affects T cell expansion and the response to vesicular stomatitis virus infections43. Our findings that Nup210-deficient naïve CD4+ T cells show decreased survival and activation resulting from their inability to properly transmit TCR signals uncovered that NPCs also regulate T lymphocyte activity in a cell-intrinsic manner. Interestingly, the SLP-76 adaptor protein was recently found to interact with the NPC-associated protein RanGAP1 during T cell activation. This association was found to be required for the nuclear accumulation of NFAT and NF-κB42. This suggested that TCR activation can modulate the transport function of NPCs. Reciprocally, our findings indicate that NPCs modulate proximal and distal TCR signaling, allowing us to propose the existence of a positive regulatory loop between NPCs and TCRs that ensures the proper transmission of tonic and activation TCR signals.
Several reports have shown a critical role for Cav1 in the initiation and downstream TCR signaling cascade33, 36, 46. Cav1 was previously shown to be important for TCR signaling in CD8+, but not CD4+ T cells36. Here we report that TCR activation in naïve CD4+ T cells, but not in CD8+, results in the specific upregulation of Cav2. This selective, TCR activation-induced increase in Cav2 expression is required for proximal TCR signaling and is dependent on the expression of Nup210. These findings raise the possibility that Cav1 and Cav2 have specialized roles selectively regulating TCR signaling in different T lymphocyte subsets. In this context, the CD4+-specific upregulation of Cav2 and its dependence on Nup210 could explain why depletion of this nucleoporin affects only CD4+ T cell survival.
In muscle, Nup210 regulates genes that localize to NPCs by recruiting the transcription factor MEF2C10. The finding that the Cav2 and cJun genes localize to the nuclear periphery indicates that the role of Nup210 in regulating NPC-associated genes is conserved in T cells. The identification that the nuclear periphery-associated Cav2 gene is rapidly induced in response to TCR activation supports the idea that nuclear pores act as scaffolds for the regulation of inducible/poised genes10, 11, 12, and suggests that NPCs also act as a hub for the regulation of TCR signaling-related genes. What transcription factors regulate the activity of TCR signaling genes at the nuclear periphery is currently unknown, but it is worth noting that NFATc1, a central transcription factor of TCR signaling, was recently found to interact with Nup210 in T cells47. How Nup210, with its small NPC/nuclear-facing domain48, regulates gene expression is unclear. Most likely, and similar to its yeast counterpart Pom152, Nup210 interacts with other nuclear envelope49 components and other soluble factors to form a transcriptional regulatory complex at the nuclear periphery. It is also important to consider that Nup210 could be working outside NPCs. Lapetina et al. recently described novel non-NPC structures at the nuclear envelope of Saccharomyces cerevisiae that contain several nucleoporins and are involved in chromatin regulation50.
ON-LINE METHODS
Mice
Nup210flox/flox (Nup210f/f) mice were generated by nuclear injection of homologous DNA (The Center for Mouse Genome Modification (CMGM); University of Connecticut Health Center). To generate Nup210−/− mice, Nup210f/f mice were then crossed with HprtCre mice (Stock No. 004302, The Jackson Laboratory). A marker-assisted speed congenic breeding strategy was used to 99.9% backcross the mice to the C57BL/6J strain. To generate Nup210f/f-Cd4CreERT2 mice, Nup210f/f mice were crossed with Cd4CreERT2 mice. Cd4CreERT2 and TCRbeta- TCRdelta- (B6.129P2-Tcrbtm1Mom Tcrdtm1Mom/J) mice were from The Jackson Laboratory (Stock No. 022356 and 002122). All animals were bred in specific-pathogen-free facilities at the Sanford Burnham Prebys Medical Discovery Institute (SBP). All experiments were approved by the Institutional Animal Care and Use Committee of SBP and were performed in accordance with institutional guidelines and regulations. Male and female mice were used at 6–10 weeks of age.
In vivo tamoxifen treatment
Nup210f/f-Cd4CreERT2 mice were administered 3 mg of tamoxifen (Sigma) suspended in corn oil intraperitoneally once daily for four days. Cre-induced Nup210 deletion was verified by PCR genotyping and immunofluorescence.
Cell culture
Mouse naïve CD4+ T cells (1.0 x 106/ml) were cultured in RPMI-1640 medium containing 2.05 mM L-Glutamine and supplemented with 10% heat-inactivated FBS, 10 mM HEPES, 1% non-essential amino acids, 1 mM sodium pyruvate, penicillin-streptomycin, and 55 μM 2-mercaptoethanol. The J14 SLP-76-EYFP cell line was a kind gift of Dr. Lawrence E. Samelson (Laboratory of Cellular and Molecular Biology, NCI, NIH). J14 cells were maintained in RPMI-1640 medium containing 2.05 mM L-Glutamine and supplemented with 10% FBS, 2 mM GlutaMAX, and penicillin-streptomycin.
Viral transduction
For CRISPR/Cas9 gene editing51 J14 SLP-76-EYFP cells transduced with tetracycline-inducible Cas9 lentivirus, followed by selection with 1.25 μg/ml Puromycin (Gibco). Cas9-expressing cells were then transfected with lentiviruses expressing scrambled or Nup210 CRISPR gRNAs52, and selected with 10 μg/ml Blasticidin (Gibco). Cas9 expression was induced with 2 μg/ml Doxycycline (Clontech) for three days. Doxycycline was removed from the medium and cells were single cell cloned. Clones depleted of Nup210 were identified by immunofluorescence. Two or more clones for each cell line were used for functional studies. For the Cav2 rescue studies, scramble or Nup210 knockout CRISPR cells were transduced with lentiviruses expressing human Cav2 (NM_001233.4) or empty vector and selected with 0.5 mg/ml Hygromycin (Gibco). Lentiviral vectors were produced by VectorBuilder.
Ex vivo and in vitro T cell activation
Naïve CD4+ T cells were prepared from spleens and peripheral lymph nodes by negative selection. Cells were incubated with CD16/32 (93) and with biotin-labeled antibodies against B220 (RA3-6B2), CD11b (M1/70), CD11c (N418), CD19 (MB19-1), CD24 (M1/69), CD8 (53–6.7), CD25 (PC61), and CD44 (IM7), followed by immunomagnetic isolation using EasySepTM Mouse Streptavidin RapidSpheresTM Isolation Kit (STEMCELL Technologies). Efficacy of enrichment was analyzed by flow cytometry and the same number of viable, CD4+CD44loCD25− T cells were used per sample. Naïve CD8+ T cells were negatively isolated using the same methodology, except that a biotin-labeled CD4 antibody (RM4-5) was used instead of CD8. For TCR activation experiments, 0.2 x 106 cells were cultured on 96-well plates at 1.0 x 106/ml in T cell medium either with soluble or plate-bound anti-CD3 (1, 2 or 10 μg/ml; 145-2C11, LEAFTM purified, BioLegend) alone or in combination with soluble anti-CD28 (5 μg/ml; 37.51, LEAFTM purified, BioLegend), and recombinant murine IL-2 (20 ng/ml, PeproTech) for 14–16 hours at 37°C (for Nur77 analysis) or 48–72 hours (for proliferation analysis). For p-STAT5 activation, 0.2 x 106 cells were cultured at 1.0 x 106/ml in T cell medium supplemented with IL-7 (0.1 to 10 ng/ml; PeproTech) for 16 hours at 37°C. For J14 SLP-76-EYFP TCR activation, cells (1.0 x 106/ml) were incubated in complete medium with or without 2% FBS and with 2 μg/ml soluble anti-CD3 antibody (OKT-3, BioXcell).
Fas-mediated apoptosis
Naïve CD4+ T cells (1.0 x 106/ml) were co-cultured for 16–24 hours with splenocytes (4.0 x 106/ml) from TCRbeta- TCRdelta- mice in medium supplemented with 0.1 μg/ml soluble Fas ligand (Enzo Life Sciences, Inc.), followed by flow cytometric analysis of cell viability using Annexin V (1/20 dilution in Annexin V binding buffer; BioLegend) or Propidium Iodide (10 μg/ml; Sigma). J14 SLP-76-EYFP cells were cultured for 16–24 hours in medium supplemented with 1% FBS and 0.01 or 0.1 μg/ml soluble Fas ligand (FasL).
Proliferation
T cells (10–20 x 106/ml) were labeled with 5 μM CFSE or CTV (Life Technologies) for 10 minutes at 37°C, washed with cold DMEM medium supplemented with 0.5% FBS and 10 mM HEPES, and cultured as described. Labeling efficiency was confirmed by flow cytometry and was >99%. CFSE and CTV dilutions were analyzed 48 or 72 hours later for in vitro experiments, and 7 or 8 days later for in vivo experiments.
Adoptive transfer
T cells were isolated by negative enrichment, labeled with CFSE or CTV, and co-injected (normalized to 2 x 106 CD4+CD44lo cells of each) intravenously into wild-type mice. Spleen and lymph nodes were harvested 18 hours or 7 days later. For homeostatic proliferation, enriched T cells were labeled and co-injected (1 x 106 T cells of each) into wild-type hosts sub-lethally irradiated using 600 Rad. CFSE and CTV dilutions were analyzed in spleens and peripheral lymph nodes 8 days later.
Blood composition analysis
Blood was retro-orbitally collected in EDTA-coated tubes and analyzed by Vetscan HMII. For flow cytometric analysis, retro-orbital blood collection was performed using heparinized capillary tubes directly into 5 ml of red blood cell (RBC) lysis buffer (0.16 M Ammonium Chloride, 0.17 M Tris-HCl, pH 7.20). RBC lysis was performed for 1 hour at room temperature (RT). Samples were washed twice in HBSS containing 1% FBS, 0.5% HEPES, and penicillin-streptomycin and stained as indicated in the Flow Cytometry section.
Flow Cytometry
Staining and washes were performed in 96-well plates. Unless otherwise stated, all incubations were performed in Staining Buffer (HBSS containing 1.2% FBS) for 30 minutes at 4°C. Primary antibodies were used at a final dilution of 1:100; except anti-Nur77, which was used at 1 μg per sample, and anti-p-STAT5, which was used at 0.06 μg per sample. Anti-Rabbit secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 647 (Life Technologies) were used at 1:1,000. For cell surface staining, freshly isolated cells (2 x 106 cells per sample) were stained with LIVE/DEAD® Fixable Aqua Dead Cell Stain Kit (Life Technologies) or with Zombie NIRTM Fixable Viability Kit (BioLegend) diluted at 1:500 in PBS at RT for 10 minutes, followed by incubation with fluorochrome-conjugated primary antibodies. For CCR7 surface staining, incubation was performed for 30 minutes at 37°C. Cells were either immediately analyzed by flow cytometry or fixed in 4% formaldehyde in PBS for later analysis. For intracellular staining of p-Lck, p-PLC-γ1, p-Zap70, and p-STAT5, after viability and surface staining, resting or IL-7 treated cells (0.2 x 106 cells per sample) were fixed in 1.85% formaldehyde for 7 minutes at 37°C and post-fixed in 90% methanol for 30 minutes at 4°C. Cells were then stained with antibodies against p-Lck, p-PLC-γ1, p-Zap70, p-STAT5, or matching isotype control. A fluorescently conjugated secondary antibody was subsequently used. For intracellular staining of Nur77, JunB, and Foxp3, after viability and surface staining, resting or TCR-stimulated cells (0.2 x 106 cells per sample) were fixed in BD Cytofix/Cytoperm (BD Biosciences) followed by post-fixation using the Foxp3/Transcription Factor Fixation/Permeabilization solution (eBioscience). Intracellular staining was performed with Nur77, JunB or Foxp3 antibodies diluted in Permeabilization Buffer (eBioscience). For JunB, a fluorescently conjugated secondary antibody diluted in Permeabilization Buffer was subsequently used. Flow cytometry data was acquired on a BD LSRFortessaTM (BD Biosciences), or on a BD LSRFortessaTM X-20 (BD Biosciences) using the BD FACSDIVATM Software (BD Biosciences); or on a ZE5TM Cell Analyzer (Bio-Rad) using the EverestTM software (Bio-Rad). Flow cytometry data were analyzed using FlowJo software v10.0.8r1 (Tree Star, Inc.).
Unless otherwise stated, all antibodies were purchased from BioLegend. Antibodies used: CD115 (AFS98); CD11b (M1/70); CD11c (N418); CD19 (MB19-1); CD24 (M1/69); CD16/32 Fc Block (93); CD197/CCR7 (4B12, eBioscience); CD25 (PC61); CD25 (3C7); CD3ε (145-2C11); CD3 (17A2, BD Biosciences); CD4 (GK1.5); CD4 (RM4-5); CD44 (IM7); CD45 (30-F11, eBioscience); CD45R/B220 (RA3-6B2); CD49b (DX5); CD62L (MEL-14); CD69 (H1.2F3, eBioscience); CD8a (53-6.7); cFos (9F6, Cell Signaling Technology); CD95 (Fas; Jo2, BD Biosciences); CD127 (A7R34); CD127 (SB/199, BD Biosciences); F4/80 (BM8); Foxp3 (FJK-16s, eBioscience); JunB (C37F9, Cell Signaling Technology); Ly-6C (AL-21, BD Biosciences); Ly-6G (1A8); NK1.1 (PK136); Nur77 (12.14, eBioscience); Siglec-F (E50-2440, BD Biosciences); Phospho-Lck (Tyr505) (Cell Signaling Technology); TCR β chain (H57-597); TCR γ/δ (GL3); Phospho-PLC-γ1 (Tyr783) (D6M9S, Cell Signaling Technology); Phospho-Zap70 (Tyr319)/Syk (Tyr352) (65E4, Cell Signaling Technology); and p-STAT5 (pY694) (47, BD Phosflow™; mouse IgG1 platelet control γ1 used as isotype control).
Fluorescence Activated Cell Sorting (FACS)
Single cell suspensions were prepared from spleens and peripheral lymph nodes or bone marrow, and stained for flow cytometry as indicated in the flow cytometry section. Before sorting, cells were resuspended in FACS buffer (PBS containing 1% FBS). Live CD4+CD25−CD62LhiCD44lo T cells were sorted at 4°C on a BD FACSAria I or II Cell Sorter (BD Biosciences) using a 70 μm nozzle into PBS supplemented with 2% FBS. Purity was verified after each sort and was >99%. RiboLock RNase Inhibitor (10 U per sample; Thermo Fisher Scientific) was used for staining neutrophils (CD45+Ly-6G+CD11b+F4/80−), macrophages (CD45+F4/80hiCD11b+CD115−), monocytes (CD45+CD115+Ly-6G−Ly6C+CD11bmed), eosinophils (CD45+Siglec-F+), CD4+ T cells (CD45+CD3+CD4+CD8−), CD8+ T cells (CD45+CD3+CD8+CD4−), and B cells (CD45+B220+CD3−) were sorted at 4°C on a BD FACSAria II Cell Sorter (BD Biosciences) using an 85 μm nozzle into Lysis Buffer (PureLinkTM RNA Micro Kit, Thermo Fisher Scientific). Bone marrow (BM) cells were flushed from tibia and femur using PBS containing 1.2% FBS and 5 mM EDTA.
RNA sequencing
A total of three biological replicates of FACS-sorted cells were used, each consisting of cells pooled from 4 mice. RNA extraction was performed using the RNeasy Plus Micro Kit (QIAGEN) and RNA integrity was confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies). PolyA RNA was isolated using the NEBNext® Poly(A) mRNA Magnetic Isolation Module and barcoded libraries were made using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich MA). Libraries were pooled and single end sequenced (1X75) on the Illumina NextSeq 500 using the High output V2 kit (Illumina Inc., San Diego CA). Read data was processed in BaseSpace (basespace.illumina.com). Reads were aligned to Mus musculus genome (mm10) using the BaseSpace RNA-Seq Alignment app v1.1 app and Tophat2 aligner (http://tophat.cbcb.umd.edu/) with default settings. Differential transcript expression was determined using the Cufflinks Cuffdiff package (http://cufflinks.cbcb.umd.edu/). Pathway analysis was performed using MetaCoreTM software (Thomson Reuters) and Ingenuity® Pathway Analysis (IPA®, QIAGEN). Hierarchical clustering of differentially expressed genes (q-value < 0.05) in three Nup210+/+ and three Nup210−/− naïve CD4+ T cell samples was performed on standardized data (gene expression rescaled for each sample to mean value of 0 and standard deviation of 1) in Partek Genomic Suite 6.6. Clustering was performed with average linkage algorithm and Euclidian distance as dissimilarity measure.
qPCR
Total RNA extraction was performed from primary cells or homogenized tissues (QIAGEN TissueLyser LT, 5 cycles of 1 minute at 50 Hz) using the PureLink RNA Mini kit (Thermo Fisher Scientific), and up to 1 μg was used to synthesize cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN). For the characterization of Nup210 expression in different tissues, commercial RNA (pooled from tissues of three 10-week old female or male mice; Zyagen) was used. qPCR was carried out using SYBR Green (BioRad or ThermoFisher Scientific) or TaqMan gene expression master mix (ThermoFisher Scientific). qPCR data were collected in a ABI 7900HT Real-Time PCR System (ThermoFisher Scientific) or in a CFX384 Real-Time PCR Detection System (Bio-Rad). The following primers were used to detect mouse transcripts. CD4: 5′-CCAACAGCGCCAGGCA-3′ and 5′-GCACTGGCAGGTCTTCTTCT-3′; Hprt53: 5′-TCATTATGCCGAGGATTTGGA-3′ and 5′-CAGAGGGCCACAATGTGATG-3′; GAPDH: 5′-CTTTGTCAAGCTCATTTCCTGG-3′ and 5′-TCTTGCTCAGTGTCCTTGC-3′ ; JunB: 5′-AGGCAGCTACTTTTCGGGTC-3′ and 5′-TTGCTGTTGGGGACGATCAA-3′; cFos: 5′-TTTCAACGCCGACTACGAGG-3′ and 5′-GCGCAAAAGTCCTGTGTGTT-3′ ; Cav1: 5′-ACGTAGACTCCGAGGGACA-3′ and 5′-GCGCGTCATACACTTGCTTC-3′ ; Cav2: 5′-TCAACTCTCATCTCAAGCTAGGC-3′ and 5′-AGGCAAGACCATTAGGCAGG-3′ ; and Irf4: 5′-GCTCATCACAGCTCATGTGGA-3′ and 5′-AACTCGTAGCCCCTCAGGAA-3′. The following primers were used to detect human transcripts. Egr2: 5′-ACGTCGGTGACCATCTTTCC-3′ and 5′-TTGATCATGCCATCTCCGGC-3′ ; cFos: 5′-GGAGAATCCGAAGGGAAAGGA-3′ and 5′-AGTTGGTCTGTCTCCGCTTG-3′ ; Hprt1: 5′-CCTGGCGTCGTGATTAGTGA-3′ and 5′-CGAGCAAGACGTTCAGTCCT-3′ ; Nfatc1: 5′-CAAGCCGAATTCTCTGGTGGT-3′ and 5′-ATGGCGTTACCGTTGGCG-3′ ; Il-2: 5′-CTGGAACTAAAGGGATCTGAAACA-3′ and 5′-AGTGTTGAGATGATGCTTTGACA-3′ ; Nur77: 5′-TACGAAACTTGGGGGAGTGC-3′ and 5′-CTGCACCCTACCCGGC-3′; and Cd69: 5′-GATGCCACCAGTCCCCATTT-3′ and 5′-TTGGCCCACTGATAAGGCAAT-3′. In addition, the following TaqMan® Gene Expression Assays (ThermoFisher Scientific) were used: Nup210 (Mm00497713_m1), β-actin (Mm00607939_s1), Gapdh (Mm99999915_g1), and Hprt (Mm00446968_m1).
Immunofluorescence and confocal microscopy
Primary liver cells fixed in 4 methanol-free % PFA for 5 minutes were blocked and stained in IF buffer (1x PBS, 10 mg/ml BSA, 0.02% SDS, 0.1% Triton X-100). Primary and secondary antibodies was incubated for 1 hour at RT or overnight at 4°C with IF buffer washes between and after incubations. Cells were labeled with Hoechst 33342 (Life Technologies) for 5 minutes before mounting using VECTASHIELD (Vector Laboratories). Antibodies used: mAb414 (Covance or BioLegend); anti-Nup210 (Bethyl Laboratories, Inc.); anti-Pom121 (ThermoFisher Scientific); anti-Nup88 (22, BD Transduction Laboratories); anti-Nup107 (a kind gift from Martin Hetzer9, The Salk Institute, La Jolla CA, USA); anti-Nup98 (39A3, Cell Signaling Technology); anti-Nup93 (F2, Santa Cruz BioTechnology); anti-Lamin A (Sigma Aldrich); anti-Lamin B1 (Abcam); anti-Cav1 (Cell Signaling Technology), and anti-Cav2 (65, BD Biosciences). Images were taken using a Leica SP8 confocal microscope and analyzed using the Leica Application Suite X software v3.1.5.16308, ImageJ v2.0.0-rc-54/1.51h (NIH), and Adobe Photoshop CS5.1 v12.1 x64.
Fluorescence recovery after photobleaching (FRAP)
Primary hepatocytes were isolated as described previously54. Cells were transfected with a plasmid expressing NES-Tomato-NLS9 using Lipofectamine 3000 (Life Technologies). 48 hours post-transfection, cells were imaged on a Leica SP8 confocal microscope. Nuclear tomato signal was photobleached with maximum laser power for 3 seconds. Recovery was recorded for 10 minutes and transport rates were analyzed using the LAS X software.
DNA fluorescence in situ hybridization (FISH)
Immunomagnetic isolated cells were spotted onto poly-L-lysine-coated coverslips and left to settle for 15–30 minutes at RT. Cells were then processed for immune-DNA FISH as described previously10, with the following modifications. Digoxigenin (DIG)-labeled probes were prepared by labeling 1 μg of BAC DNA using DIG-Nick Translation Mix (Sigma-Aldrich) and cells were subjected to IF with an anti-DIG antibody (Sigma-Aldrich). The following BAC probes from BACPAC resources, CHORI were used: Cav2 BAC (RP23-448I4), cJun BAC (RP24-282B18), and Nup62 BAC (RP23-403D23).
Electron microscopy
FACS-purified naïve CD4+ T cells were fixed in 2.5% glutaraldehyde with 2% paraformaldehyde in 0.15 M cacodylate buffer containing 2 mM calcium chloride, pH 7.4 for 18 hours at 4˚C. Cells were then embedded in 2% low melting point agarose (Sigma). The pellet was fixed in 1% osmium tetroxide/1.5% potassium ferrocyanide in buffer at 4˚C in the dark and then en bloc stained with 2% uranyl acetate at 4˚C in the dark, followed by a graded dehydration in acetone (50%, 70%, 90%, 100%, 100%). Samples were then rapidly infiltrated in Spurr’s resin using a Ted Pella PELCO BioWave microwave processing unit, embedded in flat bottom tube, and cured at 60˚C overnight. 70 nm ultrathin sections were then cut on a Leica UC7 ultramicrotome and cells were examined on a Zeiss Libra 120kV PLUS Energy Filtered Transmission Electron Microscope at nominal magnifications of 8,000x and 16,000x.
Immunoblotting
Immunoblotting was performed as described previously10, with the following modifications. Protein extracts were obtained using RIPA lysis buffer (tissues) or Tris-NP40 buffer (50mM Tris-HCL, 150mM NaCl, 0.5% NP-40, 1mM EDTA, 1mM MgCl2, 1mM DTT) (J14 cells) containing protease and phosphatase inhibitors (Pierce Halt Protease and Phosphatase Inhibitor Cocktail, ThermoFisher Scientific) and 1 mM PMSF (Sigma). Tissues were lysed using a QIAGEN TissueLyser LT (5 cycles of 30 seconds at 50 Hz). Protein concentration was determined using the Pierce BCA reagent (ThermoFisher Scientific). LDS Sample Buffer premixed with NuPAGE® Sample Reducing Agent (Life Technologies) was added, and samples were incubated for 10 minutes at 70°C. For western blot analysis, 100 μg of protein were resolved by SDS-PAGE on NuPAGE® Novex 3–8% Tris-Acetate or Bolt 4–12% Bis-Tris Plus protein gels (Life Technologies), and blotted to nitrocellulose membranes using an iBlot2. The following antibodies were used: anti-CD3ε (CD3-12, Cell Signaling Technology); anti-Nup210 (Bethyl Laboratories, Inc.; targets the C-terminal domain of Nup210, between residues 1837 and 1887); anti-Hsp90 (341320, R&D Systems); anti-cFos (9F6, Cell Signaling Technology); anti-Cav2 (65, BD Biosciences); anti-JunB (C37F9, Cell Signaling Technology); anti-Phospho-Lck (Tyr505) (Cell Signaling Technology); anti-Phospho-Lck (Tyr394) (755103, R&D Systems); anti-Lck (3A5, Santa Cruz BioTechnology); anti-human Lck (LCK-01, BioLegend); and anti-STAT3 (124H6, Cell Signaling Technology).
Chromatin Immunoprecipitation (ChIP)
Cells were diluted to 1 x 106 cells/ml and incubated for 2 hours at 37°C in complete medium without FBS, in the presence (activated) or absence (control) of 2 μg/ml of anti-human CD3 antibody (OKT-3, BioXcell). ChIP assays were performed using the Zymo-Spin CHIP (Zymo Research) Kit with the following modifications. Cells were fixed in 1% formaldehyde (Sigma) for 10 min at RT followed by 5 min quenching with 125 mM glycine. Cells were then scraped, washed 3 times with ice-cold PBS, and centrifuged at 1,000 x G for 10 min. Cell pellets containing 1 x 107 cells were resuspended in 500 μl lysis buffer with Protease Inhibitors and 1 mM PMSF. Chromatin shearing was performed with a Misonix S4000 Sonicator until DNA bands were between 300–500 bp. The chromatin solution was clarified by centrifugation and an aliquot of sheared chromatin was put aside for preparation of input sample. The remaining chromatin was immunoprecipitated using 10 μg of STAT3 (124H6, Cell Signaling Technology) or the mouse control IgG (Millipore) and Chip blocked Protein A/G magnetic beads (Millipore). Protein complexes were eluted and cross-links were reversed following the manufacturer’s protocol. Immunoprecipitated DNA fragments were purified using spin columns and analyzed by qPCR using primers (5′-GAAGCCTTTAGAAAGGGCAGGA-3′, 5′-GGGAGGGCTCCATTGATTCAG-3′) designed to amplify a 189 bp fragment of the Fas core promoter previously described41.
Data analysis
GraphPad Prism software v7.0a (GraphPad Software, Inc.) was used to prepare graphs and to perform statistical analysis. The scatter plot depicting the differentially expressed genes in Nup210−/− naïve CD4+ T cells was prepared using R. ChIP-seq data for H3K4me3 and H3K27me3 in mouse naïve CD4+ T cells was accessed using the Cistrome Data Browser55 and visualized using the UCSC Genome Browser56.
Statistical analysis
Two-tailed unpaired Student’s t test was used to compare outcomes (GraphPad Prism) and resulting P-values are indicated. For the RNA-seq samples, false discovery rate (FDR) adjusted P-values were calculated using the Benjamini-Hochberg correction for multiple testing with an allowed false discovery rate of 0.05 (Cufflinks).
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
The RNA-seq datasets generated during the current study are available in the NCBI biorepository at https://www.ncbi.nlm.nih.gov/bioproject/438343. The remaining data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
List of the differentially expressed genes in Nup210+/+ and Nup210−/− naïve CD4+ T cells. Flow cytometric-purified Nup210+/+ and Nup210−/− naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells were analyzed by RNA-seq. Data were collected from three independent experiments. A total of 3 biological replicates from each genotype, each prepared pooling cells from n = 4 mice per group, were sequenced. Differential transcript expression was determined using the Cufflinks Cuffdiff package (http://cufflinks.cbcb.umd.edu/). False discovery rate (FDR) adjusted P-values (Q-values) were calculated using the Benjamini-Hochberg correction for multiple testing with an allowed false discovery rate of 0.05. Further details can be found in the Methods section.
Acknowledgments
Financial support was provided by: We thank Dr. Lawrence E. Samelson (National Institutes of Health) for kindly providing the SLP-76-EYFP cell line. J.B. was supported by the American Heart Association Award #15POST22600000 and the SBP Fishman Fund Fellowship. M.A.D. was supported by the Pew Biomedical Science Scholar Award and by a Research Scholar Grant, RSG-17-148-01-CCG, from the American Cancer Society. This work was also supported by the National Institutes of Health (Awards #RO1AR065083 and #RO1AR065083-S1). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by the NCI Cancer Center grant P30 CA030199, which supports the animal, flow cytometry, genomics and bioinformatics cores at the SBP La Jolla campus. The EM work was supported by the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from NIH-NCI CCSG: P30 014195, NINDS Neuroscience Core Grant and the Waitt Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
J.B. designed experimental approach, performed experiments, analyzed data and co-wrote the manuscript; S.S. performed experiments, analyzed data, and provided critical input; M. R. performed experiments, analyzed data, and provided critical input; F.C. assisted with adoptive transfer experiments and provided critical input; R.T. assisted with viral work and provided critical input; L.M.B. provided reagents and critical expertise; M.A.D. designed experimental approach, analyzed data, provided oversight and critical expertise, and co-wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
References
- 1.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 2.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Viret C, Wong FS, Janeway CA. Designing and maintaining the mature TCR repertoire: the continuum of self-peptide:self-MHC complex recognition. Immunity. 1999:559–568. doi: 10.1016/s1074-7613(00)80055-2. [DOI] [PubMed] [Google Scholar]
- 4.Blattman JN, et al. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 6.Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 8.Olsson M, Scheele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292:359–370. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 9.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raices M, et al. Nuclear Pores Regulate Muscle Development and Maintenance by Assembling a Localized Mef2C Complex. Dev Cell. 2017;41:540–554e547. doi: 10.1016/j.devcel.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascual-Garcia P, et al. Metazoan Nuclear Pores Provide a Scaffold for Poised Genes and Mediate Induced Enhancer-Promoter Contacts. Mol Cell. 2017;66:63–76e66. doi: 10.1016/j.molcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Urso A, Brickner JH. Epigenetic transcriptional memory. Curr Genet. 2016 doi: 10.1007/s00294-016-0661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heng TS, Painter MW Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun. 2015;6:6074. doi: 10.1038/ncomms7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 16.Aghajani K, Keerthivasan S, Yu Y, Gounari F. Generation of CD4CreER(T(2)) transgenic mice to study development of peripheral CD4-T-cells. Genesis. 2012;50:908–913. doi: 10.1002/dvg.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradley LM, Watson SR, Swain SL. Entry of naive CD4 T cells into peripheral lymph nodes requires L-selectin. J Exp Med. 1994;180:2401–2406. doi: 10.1084/jem.180.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. 2012;24:209–217. doi: 10.1016/j.smim.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labrecque N, et al. How much TCR does a T cell need? Immunity. 2001;15:71–82. doi: 10.1016/s1074-7613(01)00170-4. [DOI] [PubMed] [Google Scholar]
- 20.Martin B, Becourt C, Bienvenu B, Lucas B. Self-recognition is crucial for maintaining the peripheral CD4+ T-cell pool in a nonlymphopenic environment. Blood. 2006;108:270–277. doi: 10.1182/blood-2006-01-0017. [DOI] [PubMed] [Google Scholar]
- 21.Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells. J Exp Med. 1997:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seddon B, Zamoyska R. TCR Signals Mediated by Src Family Kinases Are Essential for the Survival of Naive T Cells. The Journal of Immunology. 2002;169:2997–3005. doi: 10.4049/jimmunol.169.6.2997. [DOI] [PubMed] [Google Scholar]
- 23.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 24.Foletta VC, Segal DH, Cohen DR. Transcriptional regulation in the immune system: all roads lead to AP-1. J Leukoc Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 25.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 26.Nika K, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32:766–777. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho JH, et al. CD45-mediated control of TCR tuning in naive and memory CD8+ T cells. Nat Commun. 2016;7:13373. doi: 10.1038/ncomms13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu ZG, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367:281–284. doi: 10.1038/367281a0. [DOI] [PubMed] [Google Scholar]
- 29.Au-Yeung BB, et al. A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc Natl Acad Sci U S A. 2014;111:E3679–3688. doi: 10.1073/pnas.1413726111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunnell SC, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. The Journal of Cell Biology. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fooksman DR, et al. Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schonle A, et al. Caveolin-1 regulates TCR signal strength and regulatory T-cell differentiation into alloreactive T cells. Blood. 2016;127:1930–1939. doi: 10.1182/blood-2015-09-672428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song KS, et al. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 35.Scherer PE, et al. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 36.Tomassian T, et al. Caveolin-1 orchestrates TCR synaptic polarity, signal specificity, and function in CD8 T cells. J Immunol. 2011;187:2993–3002. doi: 10.4049/jimmunol.1101447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandberg Y, et al. Human T-cell lines with well-defined T-cell receptor gene rearrangements as controls for the BIOMED-2 multiplex polymerase chain reaction tubes. Leukemia. 2007;21:230–237. doi: 10.1038/sj.leu.2404486. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov VN, et al. Cooperation between STAT3 and c-jun suppresses Fas transcription. Mol Cell. 2001;7:517–528. doi: 10.1016/s1097-2765(01)00199-x. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, et al. The Immune Adaptor SLP-76 Binds to SUMO-RANGAP1 at Nuclear Pore Complex Filaments to Regulate Nuclear Import of Transcription Factors in T Cells. Mol Cell. 2015;59:840–849. doi: 10.1016/j.molcel.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faria AM, et al. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity. 2006;24:295–304. doi: 10.1016/j.immuni.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Moreira TG, et al. Sec13 Regulates Expression of Specific Immune Factors Involved in Inflammation In Vivo. Sci Rep. 2015;5:17655. doi: 10.1038/srep17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu Y, et al. Nuclear Pore Permeabilization Is a Convergent Signaling Event in Effector-Triggered Immunity. Cell. 2016;166:1526–1538e1511. doi: 10.1016/j.cell.2016.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohnuma K, et al. Caveolin-1 triggers T-cell activation via CD26 in association with CARMA1. J Biol Chem. 2007;282:10117–10131. doi: 10.1074/jbc.M609157200. [DOI] [PubMed] [Google Scholar]
- 47.Gabriel CH, et al. Identification of Novel Nuclear Factor of Activated T Cell (NFAT)-associated Proteins in T Cells. J Biol Chem. 2016;291:24172–24187. doi: 10.1074/jbc.M116.739326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–1502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell. 2011;22:1375–1388. doi: 10.1091/mbc.E10-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lapetina DL, Ptak C, Roesner UK, Wozniak RW. Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J Cell Biol. 2017;216:3145–3159. doi: 10.1083/jcb.201609049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Severgnini M, et al. A rapid two-step method for isolation of functional primary mouse hepatocytes: cell characterization and asialoglycoprotein receptor based assay development. Cytotechnology. 2012;64:187–195. doi: 10.1007/s10616-011-9407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mei S, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017;45:D658–D662. doi: 10.1093/nar/gkw983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the differentially expressed genes in Nup210+/+ and Nup210−/− naïve CD4+ T cells. Flow cytometric-purified Nup210+/+ and Nup210−/− naïve CD4+ (CD4+CD62LhiCD44loCD25−) T cells were analyzed by RNA-seq. Data were collected from three independent experiments. A total of 3 biological replicates from each genotype, each prepared pooling cells from n = 4 mice per group, were sequenced. Differential transcript expression was determined using the Cufflinks Cuffdiff package (http://cufflinks.cbcb.umd.edu/). False discovery rate (FDR) adjusted P-values (Q-values) were calculated using the Benjamini-Hochberg correction for multiple testing with an allowed false discovery rate of 0.05. Further details can be found in the Methods section.
Data Availability Statement
The RNA-seq datasets generated during the current study are available in the NCBI biorepository at https://www.ncbi.nlm.nih.gov/bioproject/438343. The remaining data that support the findings of this study are available from the corresponding author upon request.