Abstract
Ubiquitin chains regulate distinct signaling events through cooperative interactions with effector proteins and deubiquitinases. Measuring the strength of these interactions is often challenging; either large amounts of material are required or one of the binding partners must be labeled for detection. We sought to develop a label-free method for measuring binding of ubiquitin chains to the proteasome-associated deubiquitinase UCH37 and its binding partner RPN13. The method we describe here is based on a fluorescence polarization competition (FPcomp) assay in which fluorescent monoubiquitin is competed off the UCH37•RPN13 complex by the addition of an unlabeled ubiquitin chains. We show that the UCH37•RPN13 complex displays higher affinity toward chains with more than two ubiquitin subunits. Removing the ubiquitin-binding PRU domain of RPN13 does not change affinities. These results suggest UCH37•RPN13 acts to selectively recruit proteins modified with long chains (>2 subunits) to the proteasome for degradation. We also demonstrate that the FPcomp assay is suitable for high-throughput screening, which is important considering both UCH37 and RPN13 are potential targets for cancer therapy.
Introduction
Protein ubiquitination is a post-translational modification process that orchestrates almost every cellular pathway, including protein degradation, DNA repair, and cell communication [1, 2]. Ubiquitin (Ub) is covalently attached to substrate proteins through a series of enzymatic reactions and removed by a class of isopeptidases referred to as deubiquitinases or DUBs. Similar to glycosylation, Ub can modify itself during multiple rounds of conjugation in the form of Ub chains [2]. There are several ways in which chain formation can occur owing to the eight amino groups: Met1, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63. Specific chains are recognized by effector proteins (i.e., proteins containing Ub-binding domains) and DUBs, resulting in precise control over signaling pathways [3,4]. Understanding how the information embedded in different chains is translated into different biological outcomes is key to defining the ‘Ub code’.
Measuring interactions between Ub chains and their binding partners is, however, rather challenging. Common methods for characterizing these interactions include isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and fluorescence spectroscopy [5–8]. In the context of Ub chains and their interactions with Ub-binding proteins (UBPs), each of these methods has their drawbacks. ITC requires large amounts of material, which is often difficult to achieve with chains longer than two subunits. SPR has the issue of protein immobilization, which can adversely affect native interactions and requires immobilization tags on either the Ub chain or UBP. Similarly, fluorescence-based approaches necessitate the introduction of fluorophore tags, which can also incur non-native binding properties.
To circumvent these issues, we sought to devise a new method for measuring interactions between Ub chains and UBPs. The method described herein is based on a fluorescence polarization competition (FPcomp) assay [9]. FPcomp starts with a high-polarization state in which fluorophore-labeled Ub is bound to a UBP. Titrating in unlabeled Ub then leads to a decrease in polarization as the labeled Ub is forced to dissociate. The resulting titration curve yields an inhibitory constant, which can be treated as an apparent dissociation constant (Ki). Performing the same experiments with unlabeled Ub chains instead of mono-Ub then furnishes a range of Kis whose differences can be correlated with chain length. While this method still utilizes a fluorophore-tagged Ub derivative, the Ub chains themselves are unlabeled. Thus, the FPcomp assay retains the sensitivity of fluorescence measurements and allows comparisons in binding interactions between label-free chains.
To explore the utility of our FPcomp assay, we chose the Ub C-terminal hydrolase UCH37 also known as UCHL5 [10]. UCH37 is a cysteine-dependent DUB involved in regulating several fundamental cellular processes such as cell cycle progression, genome stability, cell migration, and TGF-β signaling [11–14]. Most of these cellular functions have been ascribed to the ability of UCH37 to interact with two distinct multi-subunit complexes: the 26S proteasome and the INO80 chromatin remodeling complex [15]. The UBP RPN13/ADRM1 recruits UCH37 to the proteasome and INO80G/NFRKB interacts with UCH37 in the INO80 complex [15–19]. RPN13 and INO80G have opposing effects on UCH37; RPN13 activates UCH37 toward model mono-Ub substrates and INO80G acts as an inhibitor.
Structural studies have revealed Ub binding sites in both UCH37 and RPN13. UCH37 engages Ub through a large surface exposed region (~2300 Å2) of the catalytic UCH domain [20, 21]. By binding the C-terminal UCH37-like domain (ULD) of UCH37, the C-terminal DEUBAD domain of RPN13 enhances the interaction between Ub and the UCH domain. RPN13 also contains an N-terminal pleckstrin-like receptor for Ub (PRU) domain that preferentially binds Lys48 di-Ub over mono-Ub [22]. Based on the presence of multiple Ub-binding domains, we speculated that the UCH37•RPN13 complex could exhibit high affinity toward Ub chains. Using the FPcomp assay we discovered that a catalytically dead variant of the UCH37•RPN13 complex (UCH37C88A•RPN13) does indeed favor Ub chains over mono-Ub; chains with more than two subunits are preferred. What is surprising, however, is that in the absence of the PRU domain of RPN13, the UCH37C88A•RPN13DEUBAD complex still exhibits a strong preference for longer Ub chains. Indeed, the binding affinities we measured by FPcomp are quite similar for both UCH37-containing complexes. Lastly, we show that the FPcomp assay can be applied in a high-throughput manner for the purpose of identifying inhibitors. This is important considering both UCH37 and RPN13 have emerged as cancer targets [23–25].
Materials and Methods
Cloning, protein expression and purification
Site directed mutagenesis was performed to generate catalytically inactive UCH37 (UCH37 C88A) and the Ub cysteine variants Ub K6C, Ub K11C, Ub K29C, Ub K48C and Ub K63C. The gene encoding UCH37 C88A was cloned into pVP16 vector with 8×His MBP tag and TEV protease cleavable site. 6×His-tagged RPN13 was expressed from a pGXT4 vector. UCH37 C88A and RPN13 were co-purified using amylose resin and Ni2+ affinity column chromatography, followed by size exclusion chromatography with a HiLoad 26/60 Superdex 200 column prep grade column (GE Healthcare). Ub variants were expressed and purified as previously described [26].
Synthesis and purification of Ub chains
Lys48-linked ubiquitin chains were prepared using the E1 activating enzyme UBA1 and the E2 conjugating enzyme UBE2R1 in buffer containing 40 mM Tris-HCl pH 8, 50 mM NaCl, 10 mM ATP, 5 mM MgCl2 and 0.6 mM DTT. Ub monomer, dimer, trimer and tetramer were separated by size exclusion chromatography using a HiLoad 26/60 Superdex 75 prep grade column (GE Healthcare).
Synthesis of fluorescein-labeled Ub
Ub was labeled with 5-iodoacetamido-fluorescein (5-IAF). The iodoacetamido group of 5-IAF allows it to react with free sulfhydryl groups to form stable thioether bonds. Labeling reactions were performed by dissolving a Ub cysteine variant (0.2 mM) and 5-IAF (1 mM) in a buffer containing 50 mM HEPES pH 7.4, 25 mM KCl, 5 mM MgCl2, 1mM TCEP and 10% DMSO. The reaction mixture was then incubated at 37°C and monitored by MALDI-TOF. After the reaction reached completion, unreacted 5-IAF was quenched by adding an aqueous solution of 10 mM DTT. The resulting mixture was buffer exchanged into H2O. The crude Ub-fluorescein conjugate was purified using a combination of a desalting spin column to remove free 5-IAF and reverse-phase HPLC.
Concentration determination
The concentration of Ub-fluorescein was determined by measuring fluorescein absorbance at 494 nm using an extinction coefficient (ε) of 68,000 M−1 cm−1. For unlabeled mono-Ub and Ub chains, BCA assays were performed. In each case, Ub standards were prepared by dissolving known amounts of lyophilized Ub into 50 mM Tris-HCl pH 7.5.
Fluorescence polarization binding assay
Complexes containing catalytically inactive UCH37 (UCH37C88A•RPN13 and UCH37C88A•RPN13DEUBAD) were serial diluted using the FP buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 0.01% Brij-35) into a 384-well, black nonbinding polystyrene plate (Corning). A fixed concentration of Ub-fluorescein (50 nM) was then added into each well. Fluorescence polarization (FP) was measured after a 1 h incubation at 25 °C using plate reader (Biotek Snergy 2) with polarized filters and optical modules for fluorescein (λex = 480 nm, λem = 535 nm). FP values (mP) were calculated from raw parallel and perpendicular fluorescence intensities. Graphpad Prism 6.0 was used to fit the data into eq. 1 and the equilibrium dissociation constant Kd was calculated as previously described [27]. In this equation, total FP is dependent on the fluorescent molar emissivity ratio (Q), which corresponds to the ratio of bound to unbound Ub-fluorescein. [Ub*]tot = [Ub-fluorescein]tot
| (eq.1) |
where
Fluorescence polarization-based competition assay and data analysis
FP assays require a large molecular weight difference between an unbound fluorescent protein and a bound protein. This criterion is satisfied with mono-Ub and UCH37C88A•RPN13, but not when Ub chains are the binding partner. To then use FP to measure the binding of Ub chains to UCH37C88A•RPN13 and UCH37C88A•RPN13DEUBAD, we turned to a FP competition (FPcomp) assay in which the binding ability of Ub-fluorescein is still measured but in the presence of an unlabeled competitor. A decrease in FP occurs because unlabeled mono-Ub and Ub chains compete with Ub-fluorescein for binding UCH37C88A•RPN13 and UCH37C88A•RPN13DEUBAD. To setup the assay, either mono-Ub (800 μM) or a Lys48-linked Ub chain of a specified length (400 μM) is first serial diluted into a 384-well plate. A fixed concentration of UCH37C88A•RPN13 (15 μM) or UCH37C88A•RPN13DEUBAD (20 μM) along with Ub-fluorescein (50 nM) was then added. The resulting mixture was equilibrated for 1 h at 25 °C prior to measuring FP. The data was fit to eq. 2 using Graphpad Prism 6.0 as described previously [9, 28]. The Kd2 is nominally an apparent dissociation constant (Kd,App) for the individual competitors. Kd1 is the dissociation constant for Ub-fluorescein binding. [Ub*]tot = [Ub-fluorescein]tot; [Ub]tot = [unlabeled Ub]tot.
| (eq.2) |
where
a = Kd1 + Kd2 + [Ub*]tot + [Ub]tot − [DUB]tot
b = Kd1([Ub]tot − [DUB]tot) + Kd2([Ub*]tot − [DUB]tot) + Kd1Kd2
c = −Kd1Kd2[DUB)tot
Results and Discussion
We aimed to develop a method with low material consumption that allows measurement of binding strengths between Ub chains and ubiquitin binding proteins (UBPs). These efforts led to a fluorescence polarization competition (FPcomp) assay in which unlabeled ubiquitin chains compete with fluorophore-labeled mono-Ub (Ub-fluorescein) for binding UBPs (Figure 1). As our model system, we focused on the proteasome-associated DUB UCH37 and its partner protein RPN13, which has an N-terminal PRU domain capable of binding Ub with low affinity. The UCH37•RPN13 subcomplex of the proteasome is thought to process substrates tagged with Ub chains and is a potential cancer target.
Figure 1.
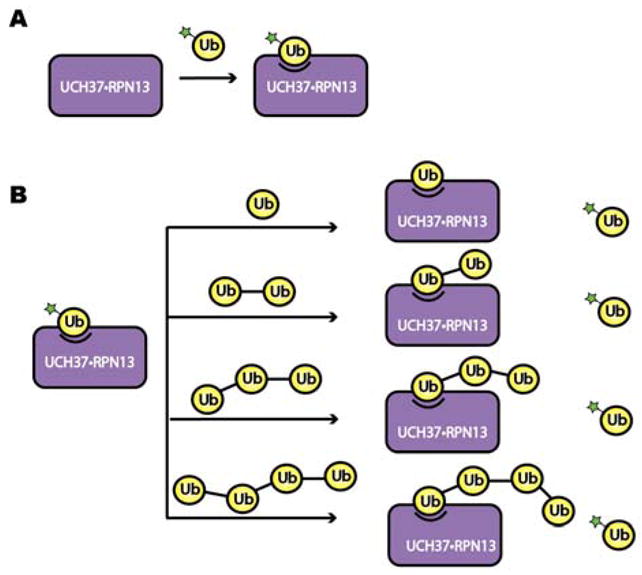
Schematic of the fluorescence polarization-based competition (FPcomp) assay. (A) The binding affinity of Ub-fluorescein and UCH37C88A•RPN13 was measured through a direct binding assay. (B) The binding affinity of unlabeled mono-Ub, Lys48 linked Ub2, Ub3, and Ub4 was then measured by a competitive binding assay, in which the unlabeled Ub derivative competes for binding with the fluorescein-labeled Ub.
Tracer design and assay optimization
To minimize the amount of protein and tracer required for a strong FP signal, it is crucial for the tracer to bind with relatively high affinity to its target. Previous studies have shown that the UCH37•RPN13DEUBAD complex binds mono-Ub with a dissociation constant (Kd) in the low μM range. Thus, we surmised that a fluorescent mono-Ub derivative could be used as a tracer in competition binding assays. To test this, several Ub cysteine variants (K6C, K11C, K29C, K48C, and K63C) were labeled with fluorescein and purified. Varying amounts of the catalytically inactive UCH37C88A•RPN13 complex were then titrated into solutions containing each fluorescein-labeled Ub derivative. The data show that fluorescein-labeled Ub K48C (Ub K48C-Fl) binds UCH37C88A•RPN13 significantly tighter than the other variants (Figure 2), with a change in FP signal between the unbound and bound states of 121 ± 2 mP units. Using 50 nM of the tracer, we obtained Kds of 2.4±0.4 and 3.4±0.7 μM for binding UCH37C88A•RPN13 (Figure 3A and Table 1) and UCH37C88A•RPN13DEUBAD (Figure 3B and Table 2), respectively. These values are nearly identical to those obtained using orthogonal methods. For example, using ITC, Ub-Gly-Ser-Thr binds UCH37•RPN13DEUBAD with a Kd of 4.5 μM, and stopped-flow fluorescence measurements have shown that Ub-Lys-Gly-TAMRA binds UCH37•RPN13DEUBAD with a Kd of 2.3 μM [19].
Figure 2.
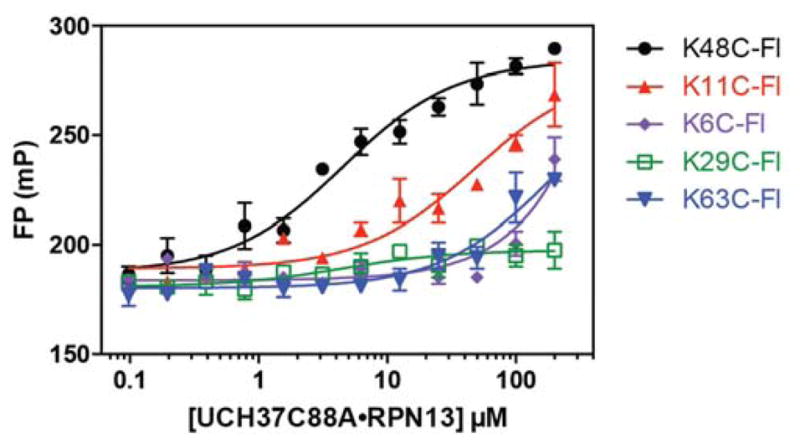
Direct binding of UCH37C88A•RPN13 to five different fluorescein-labeled mono-Ub derivatives (Ub K6C-Fl, Ub K11C-Fl, Ub K29C-Fl, Ub K48C-Fl and Ub K63C-Fl). Data were fit to eq. 1 using nonlinear regression analysis, while holding the total fluorescent mono-Ub concentration constant and maintaining Q=0.8. Data reflect mean ± SEM of two independent experiments.
Figure 3.
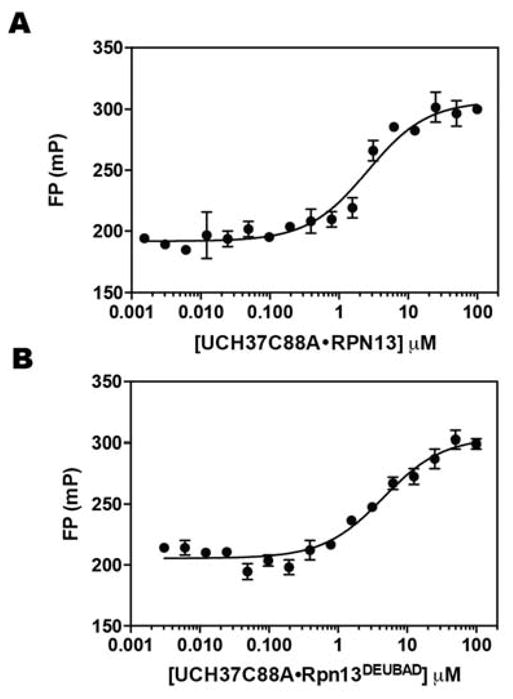
Direct binding of Ub K48C-Fl to UCH37C88A•RPN13 (A) and UCH37C88A•RPN13DEUBAD (B). Data were fit to eq. 1 using nonlinear regression analysis, while holding the total fluorescent mono-Ub concentration constant and maintaining Q=1.1. Data reflect mean ± SEM of two independent experiments.
Table 1.
FP analysis of the interactions between UCH37C88A•RPN13 and mono-Ub/Lys48 Ub chains.
| Substrate | Ligand | Kd (μM) |
|---|---|---|
| UCH37C88A•RPN13 | Ub K48C-Fl | 2.4 ± 0.4 |
| Ub | 7.8 ± 2 | |
| K48 Ub2 | 1.0 ± 0.3 | |
| K48 Ub3 | 0.14 ± 0.08 | |
| K48 Ub4 | 0.074 ± 0. 04 |
Table 2.
FP analysis of the interactions between UCH37C88A•RPN13DEUBAD and mono-Ub/Lys48 Ub chains.
| Substrate | Ligand | Kd (μM) |
|---|---|---|
| UCH37C88A• RPN13DEUBAD | Ub K48C-Fl | 3.4 ± 0.7 |
| Ub | 2.5 ± 0.9 | |
| K48 Ub2 | 1.9 ± 0.6 | |
| K48 Ub3 | 0.30 ± 0.1 | |
| K48 Ub4 | 0.10 ± 0. 06 |
Competition FP assay
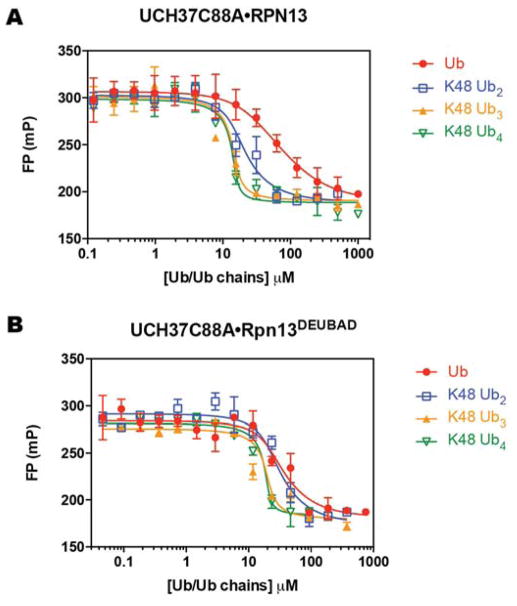
After establishing an FP assay that affords Kds close to those reported using different methods, we shifted our attention to a competition FP assay in which unlabeled Ub chains compete with the tracer for binding. Affinities of the unlabeled competitors were quantified as apparent dissociation constants (Ki), which we calculated from inhibition curves. Unlabeled mono-Ub competes with the Ub K48C-Fl tracer for binding UCH37C88A•RPN13 (Figure 4A) and UCH37C88A•RPN13DEUBAD (Figure 4B); Ki values of 7.8±0.2 and 2.5±0.9 μM were calculated for binding of mono-Ub to UCH37C88A•RPN13 (Table 1) and UCH37C88A•RPN13DEUBAD (Table 2), respectively. Unlabeled mono-Ub therefore binds the two complexes with similar affinity as Ub K48C-Fl, indicating that the fluorophore does not contribute to the binding energy, and more importantly, the FPcomp assay can inform on the binding constants for unlabeled Ub chains.
Figure 4.
Competitive displacement of Ub K48C-Fl from UCH37C88A•RPN13 using unlabeled mono-Ub, Lys48 Ub2, Ub3 and Ub4 (A). Competitive displacement of Ub K48C-Fl from UCH37C88A•RPN13DEUBAD using unlabeled monoUb, Lys48 Ub2, Ub3 and Ub4 (B). Data were fit to eq. 2 using nonlinear regression analysis, while holding the total fluorescent mono-Ub concentration constant. Data reflect mean ± SEM of two independent experiments.
We then explored longer chains as competitors. Titrating in unlabeled Lys48-linked di-Ub (Lys 48 Ub2) reveals a decrease in Ki with UCH37C88A•RPN13 (Figure 4A and Table 1), but not with UCH37C88A•RPN13DEUBAD (Figure 4B and Table 2). Using Lys48 Ub3, we observed a marked increase in affinity toward both complexes; with UCH37C88A•RPN13 the Ki decreases by nearly an order of magnitude to 0.14±0.08 μM relative to Lys48 Ub2, and with UCH37C88A•RPN13DEUBAD the Ki decreases by 6-fold to 0.3±0.1 μM. Addition of a fourth subunit further enhances binding affinity by 2- to 3-fold compared to Lys48 Ub4. These results indicate that increasing the valency of a Lys48 chain by increasing chain length leads to more avid binding. Interestingly, this holds true regardless of whether the Ub-binding PRU domain is present in RPN13, suggesting there is little cooperation between the PRU domain and UCH37. Thus, high avidity is likely to manifest from either multiple Ub binding sites on UCH37 or oligomerization of UCH37.
Assay quality assessment
With growing interest in targeting the UCH37•RPN13 subcomplex of the proteasome to prevent tumorigenesis [29], we wanted to assess the suitability of the FPcomp assay in a high-throughput format. To this end, we examined the FP signal of different concentrations of the Ub K48C-Fl tracer in the absence of UCH37-containing complexes. The mP value does not change between 3 and 200 nM of the tracer (Figure 5). Since the tracer concentration used in our standard assay format is within this range (50 nM), these results demonstrate that the FP signal should not be affected by fluctuations in concentrations between separate assays. We then performed a Z′ factor test, which measures the difference in signal between the free and bound tracer across the complete 384-well plate (Figure 6). Typically, a Z′ factor between zero and 0.5 indicates a small separation between the high and low values [30]. The ideal situation is for the Z′ factor to be greater than 0.5. We calculated a Z′ factor of 0.51, which indicates the assay is robust enough for high-throughput screening.
Figure 5.
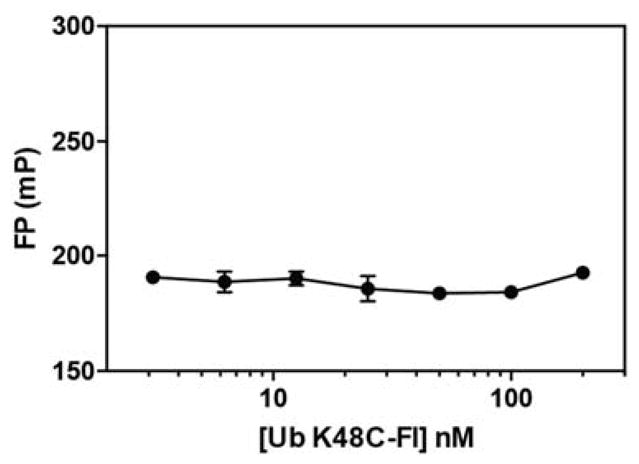
FP measurements for free Ub K48C-Fl tracer at different concentrations. Data reflect mean ± SEM of two independent experiments.
Figure 6.
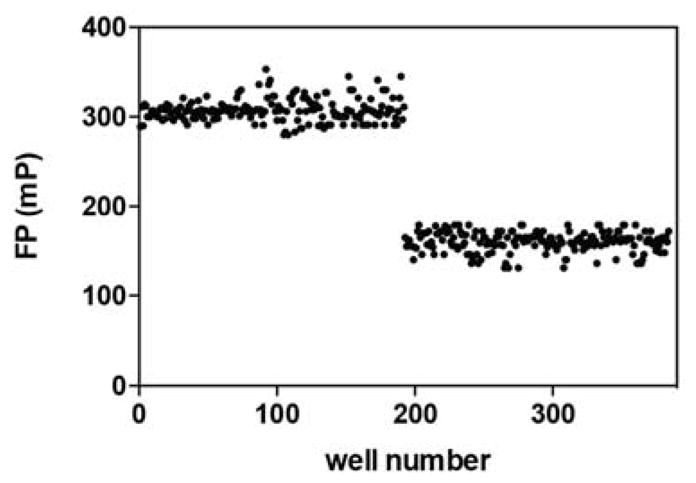
Z′ factor test. UCH37C88A•RPN13 and Ub K48C-Fl were added to wells 1–192 to obtain mP values for the bound species. Free Ub K48C-Fl tracer was added to wells 193–384 to obtain mP values for the free species.
Conclusion
Here, we report on a FPcomp assay for measuring the binding affinity between a deubiquitinase complex, UCH37•RPN13, and unlabeled mono-Ub or Ub chains. The FPcomp assay overcomes many of the limitations of conventional binding measurements; there is low sample consumption, high sensitivity, and fluorophore labeling is only required for the tracer, not the actual Ub derivative of interest. We also demonstrate that this assay can be used for high-throughput screening, which is not feasible with other label-free techniques, e.g., ITC. Using this FPcomp assay, we discovered that UCH37 has a higher affinity towards longer Ub chains. This is significant, as it suggests UCH37 in collaboration with RPN13 could play a role in selecting ubiquitinated substrates carrying long Ub chains for proteasomal degradation. Finally, since Ub chains are recognized and processed by many DUBs, we envision that the FPcomp assay can become a general method for evaluating the binding properties between these two partners.
Supplementary Material
Acknowledgments
This research was supported by a National Institutes of Health (NIH) grant (GM110543 to E.R.S.). J. D. was supported in part by a NIH Chemistry and Biology Interface Predoctoral Training Grant (GM008515).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. the Ubiquitin System. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18:579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 5.Toma A, Takahashi TS, Sato Y, Yamagata A, Goto-Ito S, Nakada S, Fukuto A, Horikoshi Y, Tashiro S, Fukai S. Structural basis for ubiquitin recognition by ubiquitin-binding zinc finger of FAAP20. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0120887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozlov G, Nguyen L, Lin T, De Crescenzo G, Park M, Gehring K. Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD. J Biol Chem. 2007;282:35787–35795. doi: 10.1074/jbc.M705655200. [DOI] [PubMed] [Google Scholar]
- 7.Tirat A, Schilb A, Riou V, Leder L, Gerhartz B, Zimmermann J, Worpenberg S, Eidhoff U, Freuler F, Stettler T, Mayr L, Ottl J, Leuenberger B, Filipuzzi I. Synthesis and characterization of fluorescent ubiquitin derivatives as highly sensitive substrates for the deubiquitinating enzymes UCH-L3 and USP-2. Anal Biochem. 2005;343:244–255. doi: 10.1016/j.ab.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roehrl MHA, Wang JY, Wagner G. A General Framework for Development and Data Analysis of Competitive High-Throughput Screens for Small-Molecule Inhibitors of Protein–Protein Interactions by Fluorescence Polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 10.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 11.Randles L, Anchoori RK, Roden RBS, Walters KJ. The proteasome ubiquitin receptor hrpn13 and its interacting deubiquitinating enzyme Uch37 are required for proper cell cycle progression. J Biol Chem. 2016;291:8773–8783. doi: 10.1074/jbc.M115.694588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Ten Dijke P, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-β signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- 13.Nan L, Jacko AM, Tan J, Wang D, Zhao J, Kass DJ, Ma H, Zhao Y. Ubiquitin carboxyl-terminal hydrolase-L5 promotes TGFβ-1 signaling by de-ubiquitinating and stabilizing Smad2/Smad3 in pulmonary fibrosis. Sci Rep. 2016;6:1–11. doi: 10.1038/srep33116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H, Xue R, Liu T, Huang X, Dong L, Wu H, Shen X. Ubiquitin C-terminal Hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta - Mol Cell Res. 2013;1833:559–572. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. Distinct Modes of Regulation of the Uch37 Deubiquitinating Enzyme in the Proteasome and in the Ino80 Chromatin-Remodeling Complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 18.Jiao L, Ouyang S, Shaw N, Song G, Feng Y, Niu F, Qiu W, Zhu H, Hung LW, Zuo X, Eleonora Shtykova V, Zhu P, Dong YH, Xu R, Liu ZJ. Mechanism of the Rpn13-induced activation of Uch37. Protein Cell. 2014;5:616–630. doi: 10.1007/s13238-014-0046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahtoe DD, vanDijk WJ, ElOualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 Activation and Inhibition by DEUBAD Domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishio K, Kim SW, Kawai K, Mizushima T, Yamane T, Hamazaki J, Murata S, Tanaka K, Morimoto Y. Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem Biophys Res Commun. 2009;390:855–860. doi: 10.1016/j.bbrc.2009.10.062. [DOI] [PubMed] [Google Scholar]
- 21.VanderLinden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP. Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase. Mol Cell. 2015;57:901–911. doi: 10.1016/j.molcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, Hamazaki J, Murata S, Burke TR, Jr, Walters KJ. Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat Commun. 2017;8:15540. doi: 10.1038/ncomms15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Chen YJ, Xu K, Wang YY, Shen XZ, Tu RQ. High expression of UCH37 is significantly associated with poor prognosis in human epithelial ovarian cancer. Tumor Biol. 2014;35:11427–11433. doi: 10.1007/s13277-014-2446-3. [DOI] [PubMed] [Google Scholar]
- 26.Trang VH, Valkevich EM, Minami S, Chen YC, Ge Y, Strieter ER. Nonenzymatic polymerization of ubiquitin: Single-step synthesis and isolation of discrete ubiquitin oligomers. Angew Chemie - Int Ed. 2012;51:13085–13088. doi: 10.1002/anie.201207171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Mayhood T, Lipari P, Wang Y, Durkin J, Syto R, Gesell J, McNemar C, Windsor W. Fluorescence polarization assay and inhibitor design for MDM2/p53 interaction. Anal Biochem. 2004;331:138–146. doi: 10.1016/j.ab.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Peterson KJ, Sadowsky JD, Scheef EA, Pal S, Kourentzi KD, Willson RC, Bresnick EH, Sheibani N, Gellman SH. A fluorescence polarization assay for identifying ligands that bind to vascular endothelial growth factor. Anal Biochem. 2008;378:8–14. doi: 10.1016/j.ab.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Wang X, Wang Z, Banerjee S, Yang J, Huang L, Dixon JE. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–212. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JH, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.